Open Access Article
Open Access ArticleA DNAzyme amplifier-based immunoassay for small molecule detection
Han
Pang
ab and
Qiang
Zhao
 *abc
*abc
aState Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, 100085, China. E-mail: qiangzhao@rcees.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing, 100049, China
cSchool of Environment, Hangzhou Institute for Advanced Study, UCAS, Hangzhou 310024, China
First published on 11th November 2025
Abstract
Detection of small molecules is of urgent demand for many applications, such as drug discovery, disease diagnosis, food safety, and environmental monitoring. Developing simple, selective, and sensitive methods to detect small molecules has attracted wide attention from researchers. In this work, we report a DNAzyme amplifier-based immunoassay for sensitive detection of small molecules. This strategy combines the benefits of the specific binding between antibodies and antigens with the superior signal amplification capability of DNAzymes. Antigen-labeled 10–23 DNAzyme binds with immobilized antibodies on a microplate, catalyzing the cleavage of numerous fluorescently labeled substrate molecules in the presence of Mg2+, thus generating amplified fluorescence signals. In the presence of small molecule targets, the targets compete with the antigen-labeled DNAzyme for binding with the immobilized antibodies on the microplate, resulting in a decrease in the fluorescence signal. We successfully detected small molecules, digoxin and folic acid, at picomolar levels through the proposed immunoassay. The immunoassay can specifically identify small molecules and enable their detection in complex sample matrices. This strategy provides a simple and sensitive approach to selectively detect small molecules, inspiring researchers to develop DNAzyme-based immunoassays with multiple functions.
1. Introduction
Small molecules (SMs) are compounds with a low molecular weight, including drugs, toxins, hormones, environmental pollutants, and so on. Detection of SMs is urgently required in various fields such as disease diagnosis, food safety, and environmental monitoring.1–4 Developing simple, sensitive, and selective methods to quantify SMs is one of the popular research areas.5,6 Given their high sensitivity, high selectivity, and ease of operation, immunoassays are widely utilized for SM detection. Immunoassays operate in two basic modes: competitive and non-competitive immunoassays. Competitive immunoassays are the most common and suitable methods for SM detection because SMs usually have only one binding site and cannot bind to two antibodies.7 Generally, enzymes or luminescent compounds serve as labels on antibodies or antigens to produce signals, enabling the detection of SMs. Enzyme labels, such as horseradish peroxidase (HRP), facilitate sensitive detection of SMs; however, they may suffer from complicated preparation, high cost, and susceptibility to activity reduction due to pH-, heat-, or chemical-induced denaturation.8 Fluorophores and quantum dots (QDs) mitigate these drawbacks but often produce weak signals, which diminish the sensitivity of these methods.5,8,9 Although efforts have been made to enhance the signal intensities, most proposed strategies tend to be complex, uneconomical, and labor-intensive.10–13Immunoassays based on binding a single target to trigger reactions with multiple turnovers can produce significant signal amplification.14 The amplification of signals allows the detection of targets at undetectable concentrations using a conventional immunoassay. Deoxyribozymes (DNAzymes) have attracted broad interest due to their potential as biosensors for various applications in clinical and environmental settings.15,16 These DNAzymes are a class of DNA strands with enzymatic properties that mediate multiple chemical reactions.17 Some can effectively catalyze the cleavage of their substrates in the presence of cofactors, rapidly generating numerous DNA fragments.16 Compared to conventional amplification techniques, such as rolling circle amplification or CRISPR-based systems, DNAzyme-based methods operate efficiently under isothermal conditions without requiring a protein enzyme.18 This intrinsic property eliminates equipment dependency and reaction complexity associated with thermocycling-dependent processes. DNAzymes show advantages in terms of high catalytic efficiency, high stability, ease of preparation, and low synthesis cost. DNAzymes have the potential to be ideal labels with multi-turnover reactivity for developing immunoassays with amplification signals.19,20 Sang et al. developed an immunomethod using DNAzymes to sensitively detect small molecules (e.g., chloramphenicol, 17β-estradiol, and aflatoxin M1).20 DNAzyme 8–17 and antibodies were conjugated to the surface of gold nanoparticles (AuNPs) to prepare a detection probe. However, the preparation of AuNP probes involves complex procedures that hinder immediate deployment. Additionally, steric effects from AuNPs impair antibody binding to immobilized antigens and restrict DNAzyme accessibility to substrates.20
The 10–23 DNAzyme is the most efficient DNAzyme known to date and has a broad range of target RNAs with its only requirement for substrates being a purine–pyrimidine dinucleotide.16,21,22 It exhibits tremendous activity against its target RNA with a kcat of ∼10 min−1 under high Mg2+ concentrations (10 mM or above).16,23 The 10–23 DNAzyme has been used in sensing applications of ions, small molecules, nucleic acids and proteins due to its small catalytic cores, high catalytic activity, and capability independent of the arm sequence.16,24–26 For example, Xiong et al. reported a DNAzyme-mediated genetically encoded sensor for ratiometric imaging of metal ions,24 and Cao et al. successfully used the 10–23 DNAzyme and its multicomponent nucleic acid enzymes to detect nucleic acids and proteins.25
Herein, we have reported a simple immunoassay based on the 10–23 DNAzyme as a signal amplifier for sensitive detection of SMs. The proposed strategy features straightforward preparation, avoids steric interference from solid-phase matrices, and enables rapid and sensitive detection of small molecules. It constructs an efficient signal transduction pathway converting immunorecognition to nucleic acid catalysis. In this immunoassay, the SM labeled 10–23 DNAzyme, serving as a detection probe, binds to antibodies immobilized on the wells of a microplate through the antibody–antigen interaction, achieving efficient signal transduction and signal amplification. The DNAzyme bound to the immobilized antibodies catalyzes the cleavage of substrate molecules modified with a quencher (BHQ1) at one end and a fluorophore (FAM) at the other end, generating amplified fluorescence signals. When the target SMs are added, they compete with the SM labeled DNAzyme for binding with the immobilized antibodies, causing a decrease in the amount of DNAzyme on the antibodies and fluorescence signals, realizing the detection of target SMs. Because one DNAzyme molecule can catalytically cleave numerous substrate molecules, this DNAzyme amplifier-based immunoassay exhibits significant signal amplification compared with the immunoassay based on a SM labeled DNA fluorescent probe. In this study, digoxin, a cardiac glycoside drug with a narrow treatment window,27 and folic acid, which plays a key role in nucleotide synthesis and methylation reactions,28 were employed as model SM targets to demonstrate proof-of-principle and generality verification for the concept of the proposed strategy. The DNAzyme amplifier-based immunoassay enabled the detection of the two targets at picomolar levels, displaying great selectivity, and successfully determined target SMs in a complex sample matrix. The proposed strategy offers a paradigm to develop a simple, sensitive, and selective approach for SM detection with high universality.
2. Experimental section
2.1. Materials and reagents
All the oligonucleotide sequences (listed in Table S1) were synthesized and purified by either Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China) or Takara Biotechnology (Dalian) Co., Ltd (Dalian, China), and stored at −20 °C. All materials and reagents used in this work are listed in the SI.2.2. Preparation of antibody-coated microplates
A Na2CO3 solution (0.1 M, pH 9.6) of 100 μL containing 1 μg mL−1 monoclonal antibodies for digoxin (anti-Dig) or 0.5 μg mL−1 monoclonal antibodies for folic acid (anti-FA) was added to the wells of a microplate, which was then incubated overnight at 4 °C. After washing the wells three times with 200 μL of washing buffer, 200 μL of blocking buffer was added to the wells, and then the microplate was shaken gently for 1 h at room temperature. Finally, the wells were washed with 300 μL of washing buffer to finish the preparation of the antibody-coated microplate.2.3. Competitive assay for digoxin or folic acid
For detecting digoxin or folic acid with the DNAzyme amplifier-based immunoassay, 100 μL of DNAzyme solutions containing various concentrations of digoxin or folic acid were added to the corresponding antibody-immobilized wells, which were incubated for 30 min at room temperature under gentle shaking. Then, 200 μL of binding buffer was used to wash the wells three times. After that, 100 μL of reaction buffer containing 50 nM substrate was added to each well. The microplate was placed in a plate reader to record the fluorescence changes over 1.5 h at 3-minute intervals.For the DNA fluorescent probe-based immunoassay, 100 μL of DNA probe solutions containing different concentrations of digoxin or folic acid were added to the corresponding antibody-immobilized wells. The microplate was incubated for 30 min at room temperature under gentle shaking and then washed with 200 μL of binding buffer three times. The fluorescence intensity of each well after adding 100 μL of reaction buffer was recorded with a plate reader.
3. Results and discussion
3.1. Principle of the DNAzyme amplifier-based immunoassay for SM detection
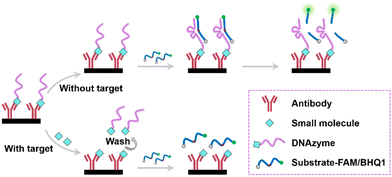
Scheme 1 shows the principle of the DNAzyme amplifier-based immunoassay for SM detection. High-affinity monoclonal antibodies are immobilized on the surface of wells, and the 10–23 DNAzyme is bound to the immobilized antibodies through the interaction between its SM label and the antibodies. The bound DNAzyme molecules catalyze the cleavage of substrate molecules labeled with a BHQ1 and a FAM molecule at their 5′-end and 3′-end, generating amplified fluorescence signals. In the presence of target SMs, the SMs in solution compete with DNAzyme molecules for binding with the immobilized antibodies. The efficiency of the competition is governed by the ratio of their dissociation constants, KD, SM/KD, DNAzyme, where KD, SM and KD, DNAzyme represent the affinities of the target SM and SM-labelled DNAzyme for the antibody, respectively. In the proposed strategy, the ligation sites of the DNAzyme differ from key epitope regions of small molecules recognized by antibodies. The use of a spacer of carbon chains can reduce steric hindrance. Thus, DNAzyme modified antigens and unlabeled antigens have similar affinity for antibodies, allowing for efficient competition with high sensitivity. As the concentration of SMs increases, the amount of DNAzyme bound to the antibodies decreases, resulting in reduced fluorescence signals. Sensitive detection of SMs is achieved by measuring the fluorescence changes with a plate reader.3.2. DNAzyme amplifier-based immunoassay for digoxin detection
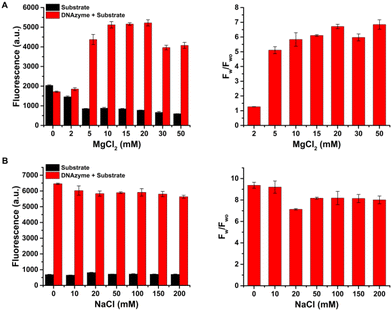
To demonstrate the principle of the proposed strategy, we first constructed a DNAzyme amplifier-based immunoassay to detect digoxin using the high-affinity monoclonal antibody of digoxin (anti-Dig) and digoxin-labeled 10–23 DNAzyme. Digoxin is a cardiac drug with a harmful effect at high concentrations and a narrow treatment window,27 and digoxin detection is important for drug analysis, clinical test, and food safety. Fig. 1A shows the feasibility of the proposed immunoassay. The DNAzyme bound with the immobilized antibodies and catalyzed the cleavage of substrate molecules to generate amplified fluorescence signals (black line), while the system without the DNAzyme maintained low fluorescence signals (blue line). The fluorescence signals reduced as 5 nM digoxin (named Dig) was added (red line).The concentration of cations in the reaction buffer has effects on the cleavage reaction mediated by the 10–23 DNAzyme. The concentration of MgCl2 affects the activity of the 10–23 DNAzyme and promotes hybridization between the DNAzyme and its substrate. The DNAzyme catalyzed the cleavage of substrate molecules in the presence of MgCl2, and the signal-to-background ratio (Fw/Fwo, Fw and Fwo are the fluorescence signals of systems with the DNAzyme and without the DNAzyme, respectively) reached a plateau at 20 mM MgCl2 (Fig. 2A). The presence of MgCl2 does not affect the integrity or stability of the pre-formed antigen–antibody complex. The concentration of NaCl has little effect on the cleavage reaction (Fig. 2B). A reaction buffer containing 10 mM Tris-HCl (pH 7.5) and 20 mM MgCl2 was used for DNAzyme cleavage.
We optimized the concentration of the immobilized antibodies. Fig. 1B shows that higher fluorescence signals were obtained with increasing antibody concentration. Considering the detection sensitivity, 1 μg mL−1 anti-Dig was selected to modify the wells of a microplate. The concentration of DNAzyme affects the level of the amplified fluorescence signal and the progress of the competitive reaction. 4 nM DNAzyme was selected for the immunoassay (Fig. 1C). We also tested the concentration of the substrate to obtain a cost-effective and efficient protocol for constructing the proposed immunoassay. The substrate at 50 nM was used for the cleavage reaction (Fig. 1D).
To demonstrate that the strategy possesses signal amplification capability derived from the 10–23 DNAzyme, we made comparison experiments in which a DNA probe labeled with FAM and digoxin (Table S1) competes with SMs to bind with the immobilized antibodies (Fig. 3A). The strategy of the DNA fluorescent probe-based immunoassay allowed the detection of digoxin (Fig. 3B). We used the same concentration of anti-Dig (1 μg mL−1) for microplate coating to construct the DNA fluorescent probe-based immunoassay. When digoxin was absent, the DNA probe bound to the immobilized antibodies, and low fluorescence signals (about 300 a.u.) were observed. This initial fluorescence intensity is derived from the DNA fluorescent probe bound to the immobilized antibodies. Compared with the DNA fluorescent probe-based immunoassay, the fluorescence intensity of the proposed DNAzyme amplifier-based immunoassay was much higher (Fig. 1C), demonstrating the superior performance of DNAzyme-based assays in signal amplification because one DNAzyme molecule can catalyze the cleavage of many substrate molecules labeled with a BHQ1 molecule at one end and a FAM molecule at the other end.
We used the proposed DNAzyme amplifier-based immunoassay to detect digoxin under optimal conditions. As shown in Fig. 4, the fluorescence intensity gradually decreased with increasing digoxin concentration. The immunoassay allowed digoxin detection in the concentration range from 0.02 nM to 160 nM, with a detection limit of 0.03 nM. The proposed strategy showed better detection sensitivity than some previously reported methods (Table S2),29–32 and the detection limit is much lower than the digoxin concentration (0.7 to 0.9 ng mL−1) stated in the 2010 practice guideline of the Heart Failure Society of America (HFSA).33 Compared with the immunoassay using fluorescent DNA labeled with digoxin under the same reaction conditions (Fig. S1), the DNAzyme amplifier-based immunoassay shows a larger signal decrease caused by the target, a wider detection range and a lower detection limit, and the DNAzyme amplifier-based immunoassay achieved about 16-fold signal amplification (Fig. 4).
We confirmed that the immunoassay had good selectivity by testing several different small molecule samples, including L-ascorbic acid, β-estradiol, tetracycline, ampicillin, uric acid, L-asparagine, and L-tyrosine (Fig. 5). Only the samples containing digoxin showed significant fluorescence decreases. High selectivity benefits from the specific antigen–antibody interactions.
 | ||
| Fig. 5 The selectivity of the DNAzyme amplifier-based immunoassay for digoxin detection. The fluorescence intensities at 60 min were used. The concentration of small molecules was 5 nM. | ||
The proposed DNAzyme amplifier-based immunoassay enables sensitive detection of digoxin in a complex sample matrix. We successfully detected digoxin in 50-fold diluted human serum samples, and the digoxin concentration range is from 0.02 nM to 80 nM, with a detection limit of 0.09 nM (Fig. S2). We further achieved the detection of digoxin in 20-fold diluted urine samples with a LOD of 0.2 nM (Fig. S3). These results indicate the potential application of the DNAzyme amplifier-based immunoassay in SM analysis.
3.3. DNAzyme amplifier-based immunoassay for folic acid detection
We further applied the immunoassay for folic acid (FA) detection to demonstrate its potential application as a general approach for SM analysis. Folic acid is a class of water-soluble vitamins that play an important role in human health and development.28 We used 0.5 μg mL−1 monoclonal antibodies for FA (anti-FA) and 1 nM FA-labeled DNAzyme to construct the immunoassay for FA detection (Fig. S4). The immunoassay could sensitively detect FA with a detection limit of 0.02 nM (Fig. 6), which is lower than the normal concentration in the human body (5–15 ng mL−1),34 and FA at concentrations ranging from 0.01 nM to 320 nM was detected (Fig. 6). The detection limit of FA is lower than that of some reported methods (Table S3),35–38 demonstrating its great capability in SM analysis. The immunoassay could selectively detect FA (Fig. S5), and successfully detected FA in 50-fold diluted human serum samples and 20-fold diluted urine samples with LODs of 0.04 nM and 0.2 nM, respectively (Fig. S6 and S7).As comparison, we also constructed a FA-labeled DNA fluorescent probe-based immunoassay. Under the same concentration of immobilized anti-FA (0.5 μg mL−1), the use of 1 nM FA labeled DNA probe conjugated with FAM (Table S1) resulted in the highest fluorescence signal value (about 100 a.u.) (Fig. S8), which was much lower than that produced by the DNAzyme amplifier-based immunoassay (Fig. S4B). We used the DNA probe-based immunoassay to detect folic acid (Fig. S9), which shows much lower sensitivity than the DNAzyme amplifier-based immunoassay (Fig. 6). These results indicate that the proposed DNAzyme amplifier-based immunoassay has high signal amplification capability and exhibits better detection sensitivity for SMs.
In our strategy, the intrinsic catalytic amplification capability of the DNAzyme significantly enhances assay sensitivity. It exhibits rapid response kinetics at room temperature, completing detection within one hour, which is suitable for timely diagnostics. Unlike conventional point-of-care testing (POCT) methods using CRISPR or isothermal amplification, DNAzyme-based sensors do not need multi-step operation and costly enzyme reagents, enabling “sample-to-result” one-step detection. The proposed immunoassay holds significant promise for integration with portable readout systems or compatibility with established POCT platforms. Besides the 10–23 DNAzyme, other DNAzyme sequences can also be used as labels of small molecules, and we can use the corresponding substrate modified with other fluorophores. In this way, multiple small molecule targets can be detected by measuring the fluorescence signals from different fluorophores with the DNAzyme based immunoassays.
4. Conclusions
In summary, a simple DNAzyme amplifier-based immunoassay was reported for detecting SMs. SMs in solution compete with the antigen-labeled DNAzyme for binding to immobilized antibodies, leading to changes in the amplified fluorescence signals generated by the cleavage of the substrate of the DNAzyme. The fluorescence signals decrease with increasing SM concentration. The proposed immunoassay has the advantages of simple design and no need for pre-amplification and complex operation. The strategy integrates the signal amplification from the DNAzyme and the high selectivity of the immunoassay. The modular design of the DNAzyme amplifier-based immunoassay provides a possibility for combining with a cascading amplification strategy to further improve assay sensitivity. In addition, this method is suitable for the development of portable devices since the catalytic cleavage reaction of the DNAzyme can be performed at room temperature. The immunoassay holds significant potential for achieving multiplex detection by rationally designing DNAzyme sequences and may provide a pathway for developing an easily manufacturable point-of-need sensor.Author contributions
Han Pang: conceptualization, data curation, investigation, writing – original draft. Qiang Zhao: conceptualization, funding acquisition, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information include materials and reagents, oligonucleotide sequences, and other experimental results. See DOI: https://doi.org/10.1039/d5an01013f.Acknowledgements
We are thankful for the support from the National Key R&D Program of China (2024YFA0918802) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0750100).References
- L. Li, Q. Zhang, Y. Wang and C. Xu, J. Clin. Lab. Anal., 2023, 37, e24865 CrossRef CAS PubMed
.
- C. Griesche and A. J. Baeumner, TrAC, Trends Anal. Chem., 2020, 128, 115906 CrossRef CAS
.
- S. Mitra, R. K. Saran, S. Srivastava and C. Rensing, Sci. Total Environ., 2024, 935, 173026 CrossRef CAS PubMed
.
- K. Nomiyama, R. Sato, F. Sato and A. Eguchi, Sci. Total Environ., 2024, 933, 173212 CrossRef CAS PubMed
.
- Z. Wang, Q. Zhou, A. Seth, S. Kolla, J. Luan, Q. Jiang, P. Rathi, P. Gupta, J. J. Morrissey, R. R. Naik and S. Singamaneni, Biosens. Bioelectron., 2022, 200, 113918 CrossRef CAS PubMed
.
- W. Wang, J. Wu, Z. Zhao, Q. Li, B. Huo, X. Sun, D. Han, M. Liu, L. C. Cai, Y. Peng, J. Bai and Z. Gao, ACS Appl. Mater. Interfaces, 2022, 14, 54914–54923 CrossRef CAS
.
- X. Cui, M. Jin, P. Du, G. Chen, C. Zhang, Y. Zhang, Y. Shao and J. Wang, Food Agric. Immunol., 2018, 29, 638–652 CrossRef CAS
.
- Y. Gao, Y. Zhou and R. Chandrawati, ACS Appl. Nano Mater., 2020, 3, 1–21 CrossRef CAS
.
- J. Wang, X. Huang, H. Liu, C. Dong and J. Ren, Anal. Chem., 2017, 89, 5230–5237 CrossRef CAS
.
- Z. Li, L. Luo, C. Chen, Q. Li, Q. Zhao, G. M. Mari, C. Xing, X. Xia, J. Shen, X. Yu, Z. Wang and Y. Pan, Food Chem., 2025, 480, 143928 CrossRef CAS
.
- X. Jiang, H. Wang, Y. Shen, N. Hu and W. Shi, Sens. Actuators, B, 2022, 350, 130891 CrossRef CAS
.
- T. Hang, C. Zhang, F. Pei, M. Yang, F. Wang, M. Xia, Q. Hao and W. Lei, ACS Sens., 2024, 9, 6779–6788 CrossRef CAS
.
- X. Liu, Z. Xu, Z. Han, L. Fan, S. Liu, H. Yang, Z. Chen, T. Sun and B. Ning, Talanta, 2021, 234, 122703 CrossRef CAS
.
- R. J. Lake, Z. Yang, J. Zhang and Y. Lu, Acc. Chem. Res., 2019, 52, 3275–3286 CrossRef CAS
.
- M. Liu, D. Chang and Y. Li, Acc. Chem. Res., 2017, 50, 2273–2283 CrossRef CAS
.
- E. M. McConnell, I. Cozma, Q. Mou, J. D. Brennan, Y. Lu and Y. Li, Chem. Soc. Rev., 2021, 50, 8954–8994 RSC
.
- I. Cozma, E. M. McConnell, J. D. Brennan and Y. Li, Biosens. Bioelectron., 2021, 177, 112972 CrossRef CAS PubMed
.
- C.-H. Lau, Q.-L. Liang, X. Chen, J. Li, Y. Shi, Y. Huang, Q. Xia, H. Zhu and G. Chen, Biosens. Bioelectron., 2025, 288, 117778 CrossRef CAS PubMed
.
- M. Wang, Z. Liu, C. Liu, W. He, D. Qin and M. You, Biosens. Bioelectron., 2024, 251, 116122 CrossRef CAS PubMed
.
- P. Sang, G. Lu, D. Yu, X. Song, Y. Guo, Y. Xie, W. Yao, H. Qian and Z. Hu, J. Agric. Food Chem., 2022, 70, 12681–12688 CrossRef CAS
.
- S. W. Santoro and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4262–4266 CrossRef CAS PubMed
.
- M. J. Cairns, A. King and L. Q. Sun, Nucleic Acids Res., 2003, 31, 2883–2889 CrossRef CAS PubMed
.
- S. W. Santoro and G. F. Joyce, Biochemistry, 1998, 37, 13330–13342 CrossRef CAS PubMed
.
- M. Xiong, Z. Yang, R. J. Lake, J. Li, S. Hong, H. Fan, X.-B. Zhang and Y. Lu, Angew. Chem., Int. Ed., 2020, 59, 1891–1896 CrossRef CAS PubMed
.
- Y. Cao, H. Zhang and X. C. Le, Anal. Chem., 2021, 93, 15712–15719 CrossRef CAS PubMed
.
- W. Li, F. Wang, Y. Chen, X. Weng and X. Zhou, Anal. Biochem., 2019, 581, 113350 CrossRef CAS
.
- J. Patocka, E. Nepovimova, W. Wu and K. Kuca, Environ. Toxicol. Pharmacol., 2020, 79, 103400 CrossRef CAS
.
- N. A. Wani, A. Hamid and J. Kaur, IUBMB Life, 2008, 60, 834–842 CrossRef
.
- Y.-T. Lin, J.-R. Liou, H.-H. Liang, Y.-H. Lin and Y.-L. Chen, Anal. Methods, 2024, 16, 8148–8156 RSC
.
- K. Li, P. Xiao, C. Wang, Z. Zhang, J. Feng, Y. Cai, Y. Xiang, X. Bai, G. Zuo and B. Han, Microchem. J., 2024, 200, 110452 CrossRef CAS
.
- H. Shi, X. Mao, X. Chen, Z. Wang, K. Wang and X. Zhu, Biosens. Bioelectron., 2017, 91, 136–142 CrossRef CAS PubMed
.
- M. Hansen-Bruhn, L. D. F. Nielsen and K. V. Gothelf, ACS Sens., 2018, 3, 1706–1711 CrossRef CAS PubMed
.
- J. Lindenfeld, N. M. Albert, J. P. Boehmer, S. P. Collins, J. A. Ezekowitz, M. M. Givertz, S. D. Katz, M. Klapholz, D. K. Moser, J. G. Rogers, R. C. Starling, W. G. Stevenson, W. H. Tang, J. R. Teerlink and M. N. Walsh, J. Card. Failure, 2010, 16, e1–194 CrossRef
.
- E. H. Reynolds, B. B. Gallagher, R. H. Mattson, M. Bowers and A. L. Johnson, Nature, 1972, 240, 155–157 CrossRef CAS PubMed
.
- Y. Li, Y. Jiang, Q. Zhang, Y. Zhao, L. Zhang, G. Song, K. Huang and Z. Yao, Talanta, 2020, 219, 121222 CrossRef CAS
.
- J. Chen, Q. Zhang, F. Xu and S. Li, Microchem. J., 2021, 170, 106673 CrossRef CAS
.
- M. Zhou, Z. Zhang, Q. Zheng, M. Yu, M. Si, S. Wu, Y. Zhang, S. Ding and D.-Y. Fu, Microchim. Acta, 2025, 192, 231 CrossRef CAS PubMed
.
- W. Li, Z. Xingzhuo, W. Yan, R. Wang, Z. Yang, Y. Hu, Y. Liu, Z. Jia and Y. Li, Talanta, 2023, 251, 123789 CrossRef CAS
.
| This journal is © The Royal Society of Chemistry 2026 |