 Open Access Article
Open Access ArticleRecent advances and applications of capillary electrophoresis-sodium dodecyl sulfate for characterization and fragment identification of monoclonal antibodies and their derivatives
Yalan
Yang
ab,
Meng
Li
ab,
Gangling
Xu
ab,
Yongbo
Ni
ab,
Luyun
Guo
ab and
Chuanfei
Yu
*ab
aNational Institutes for Food and Drug Control, Key Laboratory of the Ministry of Health for Research on Quality and Standardization of Biotech, Beijing 102629, China. E-mail: yucf@nifdc.org.cn
bState Key Laboratory of Drug Regulatory Science, Beijing 102629, China
First published on 19th December 2025
Abstract
Capillary electrophoresis-sodium dodecyl sulfate (CE-SDS), a high-resolution and high-sensitivity analytical technique, is an essential tool for analyzing the critical quality attributes (CQAs) of monoclonal antibodies (mAbs) and their derivatives, including antibody–drug conjugates (ADCs) and bispecific antibodies (bsAbs). This study systematically reviews the applications of CE-SDS in analyzing the purity and fragments of mAbs, characterizing positional isomers of ADCs, and identifying mismatch impurities in bsAbs. Focusing on the core technical challenge that CE-SDS cannot be directly coupled with mass spectrometry (MS) for fragment structure identification, the study summarizes technical solutions based on indirect identification approaches and offline/online coupling strategies with Capillary Zone Electrophoresis-Mass Spectrometry (CZE-MS). In addition, from a regulatory science perspective, this study details the key considerations for method validation, establishment of quality standards, and preparation of regulatory submissions for CE-SDS. This study aims to provide a systematic reference for the development and quality control of related biopharmaceuticals, highlighting future development directions, including high-throughput analysis, coupling techniques, and degradation prediction.
1. Introduction
Recently, monoclonal antibody (mAb) therapies have advanced rapidly, demonstrating significant clinical efficacy in treating cancers, autoimmune disorders, and viral infections. To date, over 212 antibody-based drugs have received regulatory approval, providing therapeutic benefits to millions of patients.1 Compared to small-molecule drugs, mAbs are derived from cellular secretions and are characterized by high molecular weight, structural complexity, and heterogeneity in molecular size and composition. Molecular size heterogeneity is recognized as a critical quality attribute (CQA) of mAb products.2,3 Fragments represent one of the most common impurities in mAbs, with these size variants possibly arising from proteolytic degradation, incomplete assembly, or exposure to physical stresses during production, storage, and transport.4,5Fragment analysis remains a critical and challenging aspect of mAb characterization.6–9 Insufficient sensitivity and accuracy in monitoring fragment content can hinder process optimization and formulation screening and may also lead to incorrect assessments of product stability.10 Moreover, simply determining the relative abundance of fragments is insufficient; additional analysis is required to determine their properties, identify cleavage sites, and evaluate their potential effects on critical quality attributes. Without a comprehensive understanding of fragment characteristics, establishing scientifically sound quality standards becomes challenging.11 Therefore, developing a highly sensitive and robust method for fragment separation and identification is essential for process development, quality control, and regulatory submission of mAb products. However, in complex mAb samples, fragments often occur at low levels, and conventional size-exclusion chromatography (SEC) methods may lack sufficient sensitivity to effectively separate and detect these low-abundance fragments.
Capillary electrophoresis-sodium dodecyl sulfate (CE-SDS) is employed to determine the molecular size variations in mAb samples.12 The technique separates sample components by applying a high-voltage direct current field within the capillary filled with polyacrylamide gel. Under the influence of the electric field, the sample is introduced into the capillary, after which the voltage is increased to initiate separation. As different components pass sequentially through the detection window, signals are captured by the detector, ultimately generating an electrophoretic profile.13 Owing to the porous nature of the polyacrylamide gel, a molecular sieve is formed within the capillary, enabling the separation of sample components with slight differences in molecular weight. Moreover, during sample preparation, sodium dodecyl sulfate (SDS) is added, which acts as a denaturant to unfold proteins and confer numerous negative charges, thereby minimizing intrinsic charge differences and allowing migration and separation of the samples based on molecular weight under the applied electric field. CE-SDS analysis for mAb purity is performed under denaturing conditions and is generally divided into reduced and nonreduced modes. In reduced CE-SDS (RCE-SDS), reducing agents such as β-mercaptoethanol are added during sample preparation to open the disulfide bonds in mAb molecules. This mode is used to analyze the purity of light and heavy chains, alongside any cleavage fragments. Consequently, the electropherogram of a reduced mAb sample displays two distinct peaks corresponding to the light and heavy chains, while additional peaks are attributed to fragment-related species. In the non-reduced CE-SDS (NRCE-SDS) mode, the analysis primarily targets intact mAb molecules and fragments connected by disulfide bonds. During sample preparation, alkylating reagents such as iodoacetamide are added to protect free sulfhydryl groups. In NRCE-SDS, the electropherogram typically shows one dominant peak representing the intact mAb. Peaks appearing before the primary peak are identified as fragments, while those appearing after are identified as aggregates.14–16 Compared with SEC, the CE-SDS method improves separation efficiency owing to the high viscosity of the gel filling, which can effectively reduce diffusion. However, for high-molecular-weight impurities, co-elution may occur owing to incomplete separation, thereby affecting the accurate quantitation of aggregates. Therefore, in practical applications, CE-SDS is often combined with techniques such as SEC-HPLC to comprehensively evaluate molecular size variants in mAb samples.17 Additionally, while CE-SDS can determine the relative abundance of fragments, it does not provide structural identification. While mass spectrometry (MS) is a powerful tool for fragment identification, significant technical challenges remain when coupling CE-SDS directly with MS for integrated analysis. These challenges are primarily attributed to the incompatibility of SDS with MS, inhibiting the direct coupling of CE-SDS with MS for fragment peak identification.18 In addition, the rapid advancement of antibody therapeutics has contributed to the continuous emergence of novel antibody formats, including bispecific antibodies (bsAbs) with symmetric or asymmetric architectures, antibody–drug conjugates (ADCs), and nanobodies. These increasingly complex and structurally diverse modalities pose new challenges for the applicability and optimization of CE-SDS methods. This study aims to systematically summarize recent advances in CE-SDS for identifying fragment peaks in mAb products and characterizing novel antibody formats. It also provides insights into regulatory registration, thereby offering a systematic reference for the development and quality control of related biopharmaceuticals.
2. Application of capillary electrophoresis-sodium dodecyl sulfate in the critical quality attributes analysis of monoclonal antibodies and their derivatives
2.1 Analysis of monoclonal antibody purity and fragments
With its high resolution and robust performance,19 CE-SDS has become a routine method for determining the purity of mAb drugs and is widely applied in characterization studies, release testing, and stability testing of mAbs. The CE-SDS platform method has been included in the pharmacopoeias of numerous countries globally.15,20–22 The General Chapter 3127 of Part III of the Chinese Pharmacopoeia specifies the procedures for the optimized and validated CE-SDS platform method for determining mAb molecular size variants. Advances in electrophoretic techniques have led to the development of several novel CE-SDS methods for analyzing recombinant protein therapeutics, including mAb products and adeno-associated virus (AAV) capsid proteins.23 For example, CE-SDS methods incorporating laser-induced fluorescence (LIF) detectors provide significantly higher sensitivity for detecting mAb size variants than conventional UV detection.24 Microchip electrophoresis significantly reduces analysis duration while increasing throughput.25 Furthermore, optimizing separation parameters, including temperature, voltage,26 and gel buffer composition (incorporating sodium hexadecyl sulfate [SHS], which exhibits greater hydrophobicity than SDS)27–29—effectively eliminates spurious peaks in the CE-SDS profiles of specific mAbs. These methods serve as extensions and complements to the CE-SDS platform approach. Collectively, they also indicate that CE-SDS has become a well-established technique for the separation of size variants in mAb products.ADCs are constructed by linking a small-molecule toxin to a mAb using a chemical linker. The mAb specifically binds antigens overexpressed on the surface of tumor cells. Following binding, the antigen–antibody complex is internalized through endocytosis to form an endocytic vesicle, which subsequently fuses with lysosomes,30 where the toxin is released. Cleavable linkers release the payload under the acidic lysosomal environment or through protease-mediated cleavage, while non-cleavable linkers require complete antibody degradation.31–33 Payloads are primarily classified into three categories: (1) microtubule inhibitors (MMAE, MMAF, DM1, DM4), which disrupt tubulin polymerization, causing mitotic arrest and apoptosis; (2) DNA-damaging agents (calicheamicin, PBD dimers), which induce DNA double-strand breaks or crosslinking, thereby inhibiting DNA replication and transcription; and (3) topoisomerase I inhibitors (DXd, SN-38), which stabilize topoisomerase I–DNA complexes and convert transient single-strand breaks into irreversible damage. Additionally, some payload is released into the extracellular environment and internalized by neighboring cells, a process known as the bystander effect.34,35 ADCs combine the target specificity of mAbs with the cytotoxic potency of small-molecule drugs, leading to significant clinical benefits.36 Several conjugation methods—including lysine, interchain cysteine, and glycan conjugations37–39—pose distinct challenges for purity analysis using CE-SDS.
Currently, lysine and cysteine conjugations are the primary conjugation methods used in approved ADCs. Lysine conjugation, a non-specific approach (e.g., Kadcyla, ELAHERE), links the small-molecule payload to surface-exposed lysine residues on the antibody without significantly altering its overall structure. These ADCs exhibit a CE-SDS profile closely resembling that of the unconjugated (naked) antibody.40,41 In contrast, cysteine-conjugated ADCs are generated by reducing the four interchain disulfide bonds of the antibody to expose free thiol groups used for linker–payload attachment. The number of thiols available for conjugation is determined by the degree of disulfide bond reduction, leading to variation in the number of attached small molecules. This variation leads to distinct CE-SDS peak profiles corresponding to ADCs with different drug-to-antibody ratios (DARs). Fig. 1 illustrates representative profiles of clinically approved cysteine-conjugated ADCs, which typically exhibit DAR values ranging from 2 to 8. For DAR2 and DAR4 ADCs (e.g., polatuzumab vedotin, brentuximab vedotin, and loncastuximab tesirine), the NR CE-SDS profile exhibits six major peaks: LC, HC, HL, HH, HHL, and the intact antibody structure (HHLL).40 For DAR8 ADCs (e.g., trastuzumab deruxtecan and sacituzumab govitecan), complete reduction of interchain disulfide bonds results in nearly identical nonreduced and reduced CE-SDS profiles,42 both exhibiting two dominant peaks (LC and HC). However, for sacituzumab govitecan under non-reducing conditions, the relative peak areas of HL and HH are approximately twice those observed under reducing conditions, indicating that ≈50% of these components exist as disulfide-linked dimers. The CE-SDS profile of glycan-conjugated ADCs closely resembles that of the unconjugated (naked) antibody.
A nanobody is a heavy-chain antibody (HCAb) without light chains. This antibody comprises two constant regions (CH2 and CH3): a hinge region and a heavy-chain variable region (VHH).43,44 Nanobodies offer several advantages: their small size facilitates efficient penetration through tissue barriers to reach tumor sites; they exhibit low aggregation propensity and high thermal stability; they demonstrate strong antigen-binding affinity and can recognize epitopes inaccessible to conventional antibodies; and they show low immunogenicity.45,46 These properties make nanobodies particularly promising candidates for developing ADCs, bsAbs, and multispecific antibodies. However, challenges such as short half-life and renal retention persist and must be addressed through rational design strategies.47,48 Envafolimab, a single-domain antibody–Fc fusion protein targeting PD-L1, comprises a single-domain antibody component derived from a humanized camelid heavy-chain variable domain, which binds human PD-L1 with high affinity and specificity. By blocking the interaction between PD-L1 and its receptor PD-1, Envafolimab reverses PD-1/PD-L1-mediated suppression of the T-cell activation and proliferation pathway, thereby enhancing immune-mediated antitumor activity. Fusion of the VHH domain to the Fc region of human IgG extends the half-life and duration of action of the drug.49,50 Envafolimab forms a homodimer linked by interchain disulfide bonds. Reduced and NR CE-SDS analyses show a single major peak.
bsAbs are engineered antibodies that simultaneously bind two different antigens or distinct epitopes on the same antigen. Beyond their antigen-targeting function, bsAbs exert several biological effects, including recruiting T cells to eliminate target cells, simultaneously inhibiting two immune checkpoints or signaling pathways, binding distinct epitopes on a single antigen to enhance binding affinity and mitigate drug resistance mechanisms,51 and targeting a tumor-associated antigen and an immune checkpoint to enhance the antitumor response.52 Structurally, bsAbs are more complex than conventional mAbs.53 IgG-like bsAbs maintain the symmetric structure of traditional IgG molecules, with each arm binding a distinct antigen while retaining Fc-mediated effector functions and exhibiting a favorable serum half-life. Representative examples include talquetamab54 and mosunetuzumab. In contrast, blinatumomab,55 a bispecific T-cell engager (BiTE), is constructed in a single-chain variable fragment (scFv) format, comprising two scFvs connected by a peptide linker that forms two oppositely oriented antigen-binding sites. This compact and flexible structure facilitates immune synapse formation and T cell-mediated cytotoxicity against tumor cells. Tarlatamab represents an optimized BiTE format containing an Fc fragment fused to the molecule. This enhances stability and serum half-life. Glofitamab incorporates two CD20-binding domains and one CD3-binding domain; this 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding ratio improves specificity and B-cell targeting. The CE-SDS profile for Glofitamab is presented in Fig. 2. Cadonilimab56 and ivonescimab are generated by conjugating two scFvs to the C-terminus of the HCs, producing a tetravalent bispecific structure. This format resolves chain mispairing but presents challenges, including aggregation and increased fragment formation. Owing to the structural complexity of bispecific antibodies, CE-SDS analysis may require method optimization using a platform-based approach. Cheng et al.57 identified a cluster of noncovalently aggregated high molecular weight (HMW) species exhibiting concentration-dependent behavior. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) band recovery experiments demonstrate that these HMWs are reversible and form noncovalently during electrophoretic separation. In the study, these HMW peaks were eliminated by adding 0.3% SHS to the gel buffer, increasing the separation temperature, and decreasing the separation voltage.
1 binding ratio improves specificity and B-cell targeting. The CE-SDS profile for Glofitamab is presented in Fig. 2. Cadonilimab56 and ivonescimab are generated by conjugating two scFvs to the C-terminus of the HCs, producing a tetravalent bispecific structure. This format resolves chain mispairing but presents challenges, including aggregation and increased fragment formation. Owing to the structural complexity of bispecific antibodies, CE-SDS analysis may require method optimization using a platform-based approach. Cheng et al.57 identified a cluster of noncovalently aggregated high molecular weight (HMW) species exhibiting concentration-dependent behavior. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) band recovery experiments demonstrate that these HMWs are reversible and form noncovalently during electrophoretic separation. In the study, these HMW peaks were eliminated by adding 0.3% SHS to the gel buffer, increasing the separation temperature, and decreasing the separation voltage.
The complex structure of bispecific antibodies presents additional challenges, including rare fragment formation. CE-SDS is a powerful tool that resolves these species, enables their identification, and improves structural understanding. Cao et al.58 report a shoulder peak preceding the main peak in the NRCE-SDS electropherogram of an IgG-like bsAb. This shoulder primarily arises from cleavages at specific sites within the CH2 and CH1 domains, with the resulting fragments remaining linked by intrachain disulfide bonds. In reducing CE-SDS, this shoulder peak corresponds to two new fragment peaks but cannot be enriched by hydrophobic interaction chromatography (HIC). Furthermore, no complementary small-fragment peaks were observed in the NRCE-SDS profile, supporting the hypothesis that these fragments are generated through cleavage events, with the resulting pieces remaining connected via disulfide bonds. Reduced LC-MS and peptide mapping analyses reveal that the CH2 and CH1 domains contain cleavage sites at leucine 306 (L306), leucine 309 (L309), and leucine 182 (L182), respectively. Subsequent IgdE enzymatic digestion to generate Fc subunits, followed by nonreduced RP-LC-MS analysis, confirms the presence of these disulfide-linked fragments, evidenced by multiple Fc variants exhibiting a mass increase of ∼18 Da. The study shows that L306/L309 cleavage is a common phenomenon in IgG1 and IgG4 antibodies. The resulting fragments cannot be effectively resolved using size-exclusion chromatography (SEC); however, NRCE-SDS resolves these species and quantitatively monitors the extent of CH2 cleavage based on shoulder peak area. Lin et al.59 detected an aberrant peak near the LC in NRCE-SDS. The intensity of this peak strongly correlates with the abundance of the HHL peak. MS reveals that its mass is approximately 944 Da higher than that of the LC, suggesting that the species represents an LC-related sequence variant. This variant was enriched using denaturing SEC-HPLC and characterized through peptide mapping and LC-MS/MS. mRNA sequencing indicates that the variant sequence results from an aberrant splicing event, a transcriptional error that produces a C-terminal amino acid extension and increases the MW. Functional evaluations demonstrate that the variant retains its biological activity and thermostability. However, this non-native structure may present immunogenicity risks. The use of NRCE-SDS as a product-release method facilitates effective detection of the aberrant peak and monitoring of its level.
2.2 Analysis of the drug-to-antibody ratio and positional isomers of the antibody–drug conjugate
CE-SDS can reflect the coupling structure of ADCs and characterize positional isomers of cysteine-coupled ADCs. Van den Berg et al.60 demonstrated that NRCE-SDS effectively characterizes positional isomers of cysteine-conjugated ADCs. While HIC shows that the DAR remains constant with increasing reduction time, NRCE-SDS reveals shifts in the relative abundances of ADC species (LC, HC, HL, HH, HHL, and HHLL). Specifically, conjugation in the Fab region decreases (LC abundance decreases from 20.3% to 16.7%), while conjugation in the hinge region increases (HL increases from 9.1% to 19.2%, and HH decreases from 11.0% to 4.9%). These findings indicate that, with increasing reduction time, ADC positional isomers transition from a Fab- to a hinge-oriented configuration. To explain this transition, an “intramolecular thiol–disulfide exchange” mechanism was proposed: a reduced cysteine residue in the Fab domain attacks and cleaves a disulfide bond in the hinge region while simultaneously being re-oxidized to form a new disulfide bond. Consequently, NRCE-SDS enables ADC purity monitoring and provides insight into conjugation-site consistency, highlighting its application in process development and quality control.In a study of a novel anti-CD22 nanobody-drug conjugate (NDC), Ziaei et al.61 revealed significant differences in the NRCE-SDS profiles pre- and post-conjugation. The NDC was conjugated to DM1 via lysine residues using the SMCC linker, yielding a DAR of approximately 2.04. Following conjugation, the main peak in the NRCE-SDS profile of the NDC exhibits broadening and minor shoulder peaks. Owing to the fact that NDC is expressed in a prokaryotic system, glycosylation heterogeneity is excluded as a contributing factor. These findings are attributed to conjugated species with varying DARs, further suggesting that impurities from the small-molecule toxin may be responsible.
2.3 Mismatch and fragment analysis of bispecific antibodies
CE-SDS can separate fragments and also analyze mismatch impurities and homodimers in IgG-like bispecific antibodies.Cao et al.62 reported a novel mispair impurity in an asymmetric IgG-like bsAb with a Knob-into-Hole (KiH) architecture. This impurity, identified as a light–heavy–light chain (LHL) mispair, arises from the integration of the KiH format with a stabilizing disulfide bond in the CH3 domain and an engineered CH1–CL disulfide bond in one Fab arm. An interchain disulfide bond forms between Fc-engineered cysteine (C349) on the hole HC and CL-engineered cysteine (C121) on the lambda LC, producing the LHL mispair. Uncharacterized peaks observed in the HIC chromatogram of the heat-stressed IgG-like bsAb were hypothesized, based on intact mass analysis, to correspond to a heterotrimer comprising a kappa LC, an HC, and a lambda LC. This hypothesis was subsequently confirmed using various techniques, including disulfide bond mapping and IdeS digestion. CE-SDS effectively separates the LHL mispair peak and distinguishes it from the large hinge fragment, which lacks one Fab arm and exhibits a mass difference of approximately 3 kDa. Consequently, CE-SDS serves as a product-release method for monitoring the level of the LHL mispair and evaluating the clearance of process-related impurities in a bsAb product.
3. Strategy of capillary electrophoresis-sodium dodecyl sulfate in the identification of monoclonal antibodies
The identification of fragment peaks is critical for mAb quality control, particularly when peak levels increase significantly during long-term storage or under accelerated degradation conditions. Identifying and characterizing these peaks facilitates the understanding of potential cleavage sites and the propensity for heterogeneity in mAb molecules. This provides valuable guidance for optimizing manufacturing processes, buffer formulations, and storage conditions.3.1 Indirect identification strategy
Duhamel et al.63 and Fei et al.64 employed a similar workflow, as shown in Fig. 3, excluding the use of capillary electrophoresis–mass spectrometry (CE-MS), to identify fragment peaks in CE-SDS. First, to determine whether a fragment peak was associated with a disulfide bond, the mAb samples were partially reduced, generating standard fragments (LC, HL, HH, HHL). Subsequently, the disulfide-bonded fragments in the sample were identified by comparing their CE-SDS migration times with those of the standards. To determine whether glycosylation heterogeneity contributes, samples were deglycosylated using PNGase F under native/denaturing conditions, with non-glycosylated heavy chain (NGHC) standards prepared. Confirmation was achieved by comparing migration times. To localize cleavage regions, the size of uncharacterized fragments was estimated based on a linear regression between CE-SDS migration time and MW. Alternatively, the presence of disulfide bonds was assessed by comparing RCE-SDS and NRCE-SDS profiles. IdeS enzyme digestion was used to determine whether cleavage occurred in the Fab or Fc region. Finally, MS was conducted on the identified region to determine the exact cleavage site(s). Duhamel et al. induced fragment peak formation in an IgG1 mAb through a forced degradation strategy including thermal stress (incubation at 40 °C for 2 weeks), pH stress (treatment with pH 3.0 or pH 9.0 buffers), and oxidative stress (incubation with 0.1–1.0% H2O2 for 20 h). The degraded samples were analyzed using reduced and NRCE-SDS to screen for fragment peaks exhibiting >2% increases in peak area for subsequent identification. Cleavage sites were confirmed through peptide mapping-based LC-MS. The cleavage sites are shown in Fig. 4. Conversely, Fei et al. revealed naturally occurring fragments in process samples and characterized the fragments using MS techniques combining reversed-phase LC-MS (RPLC-MS) and size-exclusion LC-MS (SEC-MS). Duhamel et al. reported four core types of fragment peaks in thermally degraded IgG1 mAb samples. Fei et al. revealed impurity peaks in seven mAb products, providing traceability data to support process development and Biologics License Application (BLA) submissions.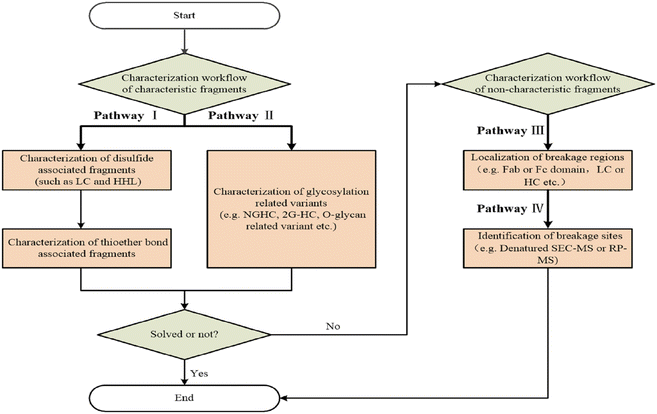 | ||
| Fig. 3 Workflow for identifying and characterizing monoclonal antibody fragments using CE-SDS. Reproduced from ref. 64 with permission from John Wiley and Sons, copyright 2024. Abbreviations: HC, heavy chain; LC, light chain; 2G-HC, two glycosylated heavy chains; HHL, heavy–heavy–light fragment; NGHC, non-glycosylated heavy chain; CE-SDS, capillary electrophoresis-sodium dodecyl sulfate; SEC-MS, size-exclusion chromatography-mass spectrometry; RP-MS, reversed-phase liquid chromatography-mass spectrometry. | ||
 | ||
| Fig. 4 Electropherogram showing the cleavage sites on an IgG1 antibody. The heat-stressed sample (red trace) is compared to the untreated control (black trace). Peaks corresponding to fragments generated at specific cleavage sites are labeled in the CE-SDS profile. Reproduced from ref. 63 with permission from Springer Nature, copyright 2019. Abbreviations: IS, internal standard; NG-IgG, non-glycosylated IgG; AU, absorbance units. | ||
3.2 Offline and online modes based on capillary zone electrophoresis–mass spectrometry
Capillary zone electrophoresis–mass spectrometry (CZE-MS) combines the high-resolution separation capability of capillary electrophoresis with the precise molecular identification capability of MS. Giorgetti et al.65 comprehensively characterized seven clinically approved mAb drugs using sheathless CZE-ESI-MS at the following three analytical levels: intact protein, IdeS-digested and reduced fragments, and peptides. At the intact protein level, primary glycoforms and several high-mass post-translational modifications (PTMs) are identified. At the IdeS-digested level, additional PTMs are detected through the formation of Fc and fragment antigen-binding (F(ab’)) fragments. At the peptide level, each PTM is precisely localized and quantitatively assessed. This study highlights the value of multi-level analysis for comprehensive mAb characterization. Xu et al.66 established a novel CZE-MS method by optimizing parameters, including sheath liquid composition and background electrolyte, to analyze cysteine-linked ADC fragments and determine the oligonucleotide-to-antibody ratio (OAR) of antibody–oligonucleotide conjugates (AOCs). Additionally, this method effectively separates fragment impurities, including half-antibodies conjugated with one or two small-molecule drug molecules, LCs conjugated with one or two small-molecule drug molecules, LCs with cleaved C-terminal cysteine residues, and HC clip variants. These impurities are often difficult to separate using traditional LC-MS methods owing to co-elution or signal suppression. AOC67 combines the targeted delivery capability of antibodies with the gene-specific precision of oligonucleotides, including small interfering RNA (siRNA) and antisense oligonucleotides (ASOs). Oligonucleotides are highly negatively charged, water-soluble, and typically >10 kDa in MW. Conventional hydrophobicity-based chromatographic methods, such as – HIC and reversed-phase HPLC (RP-HPLC), are generally ineffective for separating antibodies conjugated to different oligonucleotides. Conversely, the novel CZE-MS method enables characterization and relative quantification of samples with varying OARs, demonstrating higher separation efficiency than HIC-MS and RPLC-MS.Two primary strategies are currently used for detecting impurity peaks of CE-SDS-separated fragments using CZE-MS. The first is an offline approach. Liu et al.68 treated NIST mAb with H2S to induce accelerated degradation, generating various disulfide bond-related molecular fragments (e.g., HHL, HC, and LC). These degraded samples were analyzed concurrently using LabChip GXII microfluidic chip electrophoresis and CZE-MS. A linear calibration curve was constructed by plotting the log10(MW) against the electrophoretic mobilities (calculated as the inverse of migration times [1/migration time]) determined using GXII. Consequently, the MWs of uncharacterized impurity peaks separated using the LabChip GXII were accurately estimated by substituting their migration times into the calibration curve. This approach is applied in multiple forced degradation studies. For example, the method is used for identifying fragment peaks in rituximab resulting from cleavage at asparagine–proline bonds under alkaline conditions. Further, it is used to characterize NISTmAb fragments formed by alanine–alanine and phenylalanine–isoleucine cleavages on the LC following cathepsin-mediated degradation. Li et al.69 developed a complementary CE-MS workflow that facilitates the identification and characterization of a clipped variant in bevacizumab, resulting from cleavage of the Ser105–Ser106 peptide bond during long-term storage. Analysis of bevacizumab stored for 36 months reveals a previously uncharacterized shoulder peak to the left of the main peak in the NRCE-SDS electropherogram. An additional unidentified peak is observed in the reduced CE-SDS profile. Using the established linear relationship between migration time and the theoretical MW of reference peaks, the approximate MW of this shoulder peak observed in the NRCE-SDS was estimated at 138.4 kDa. Subsequently, intact-protein analysis using CZE-MS and imaged capillary isoelectric focusing-MS (iCIEF-MS) confirms the MW of the variant as 137.5 kDa, further revealing its acidic nature. Following sample reduction and IdeS enzymatic digestion, CZE-MS analysis reveals a cleavage site within the Fab region of the HC, resulting in the loss of N-terminal residues 1–105 owing to cleavage at the Ser105–Ser106 peptide bond. This integrated CE-MS workflow facilitates the discovery, estimation, precise characterization, and localization of the variant, with results corroborated across methods, thereby addressing the limitation of CE-SDS in directly identifying uncharacterized peaks.
The second strategy operates in an online mode. Römer et al.70 developed an integrated online CE-SDS–CZE–top-down MS (TDMS) platform for the comprehensive characterization of the intact LC of the NIST mAb. An eight-port nanoliter valve was employed to connect the CE-SDS and CZE 2-dimensional capillary systems, as depicted in Fig. 5. Following CE-SDS separation (first dimension, 1D, separated according to their molecular weight), peaks of interest were monitored in real time through UV detection. When the target peak migrated into the sample loop, the voltage in the 1D was halted, and the valve was switched to transfer the analyte to the second dimension (CZE). Water and cetyltrimethylammonium bromide (CTAB) were simultaneously injected into the CZE capillary. The addition of water reduces the concentration of SDS, while the cationic surfactant CTAB interacts with SDS to form electrically neutral complexes.
 | ||
| Fig. 5 Schematic illustration of the CE-SDS–CZE–TDMS system. An 8-port nanoliter valve was used to connect CE-SDS separation to an Orbitrap mass spectrometer. CE-SDS analysis (green section) represents the first separation dimension. Decomplexation zones containing water (red section) and CTAB (blue section) bridge the first and second separation dimensions via dedicated valve interfaces. The second-dimension separation is CZE-MS analysis (orange section). Reproduced from ref. 70 with permission from Elsevier, copyright 2021. Abbreviations: CTAB, Cetyltrimethylammonium bromide; CZE-TDMS, Capillary zone electrophoresis-top-down mass spectrometry. | ||
This interaction mitigates ionization suppression and promotes efficient protein desorption. Subsequently, the target protein underwent secondary separation according to charge/size in an acidic CZE buffer. Coupling to MS was accomplished using a dual-capillary nanoflow sheath liquid interface.71 During the analysis, the sheath liquid capillary provides a continuous flow to ensure stable electrospray ionization and eliminate non-target components. When the separation voltage is applied in the CZE dimension, the target analyte is mixed with the sheath liquid and subsequently introduced into the Orbitrap Fusion Lumos MS for detection. For MS2 characterization, the parameters of three fragmentation techniques—higher-energy collisional dissociation, electron transfer dissociation, and ultraviolet photodissociation—were optimized and applied in combination. By analyzing the fragment ions and comparing them with the antibody sequence database, the precise sites of fragmentation can be accurately identified. This analytical platform was employed to successfully identify low-abundance degradation fragments that accounted for only 1–1.5% of the total peak area in stressed mAb samples. In this study, an online TDMS platform was established to enable comprehensive characterization of unidentified CE-SDS peaks. This strategy was employed to reduce analytical complexity, minimize sample consumption, and ensure the unambiguous correlation of identified mAb fragments with CE-SDS profiles.
3.3 Method comparison and selection
All three fragment identification strategies are capable of accurately determining the molecular weight and cleavage site of the resulting fragments. However, the underlying principles of these three methods differ. The indirect identification approach initially determines the cleavage site of the fragment through sample pretreatment (e.g., partial reduction and enzymatic digestion) and subsequently verifies the molecular weight of the corresponding region using mass spectrometry. The offline CZE-MS method leverages the precise molecular weight obtained from CZE-MS to correlate with and confirm the peaks separated by CE-SDS. However, confirming the cleavage site still necessitates additional procedures, such as reduction and enzymatic digestion, to systematically narrow down the possibilities. In contrast, the online CE-SDS-CZE-MS method integrates CE-SDS and CZE-MS through an online SDS removal technique, thereby enabling a direct correlation between CE-SDS peaks and MS data. This platform not only provides molecular weight information but can also directly identify cleavage sites through fragment ion matching. Each of these strategies possesses distinct advantages and disadvantages and applicable scenarios. The optimal selection depends on the specific laboratory capabilities and requirements. A comprehensive comparison is presented in Table 1.| Identification method | Advantage | Limitation | Instrument cost | Application scenario |
|---|---|---|---|---|
| Indirect identification | Easy to implement in most laboratories | Multi-step process; time-consuming and dependent on sample preparation | Low | Early-stage discovery, process development, and routine quality control laboratories |
| Offline CZE-MS | Providing MW information with higher precision | No direct sequence data; dependent on sample preparation to identify cleavage sites; | Medium | Characterizing new fragments in stability studies |
| Online CZE-MS | Direct peak correlation; simultaneous acquisition of MW and cleavage sites; automated workflow | High instrument cost; challenging method development | High | Characterization of very low abundance impurities (<1%); samples with limited availability |
4. Regulatory considerations
4.1 The critical role of capillary electrophoresis-sodium dodecyl sulfate in biopharmaceutical development
CE-SDS is an essential analytical tool for monitoring size variants of monoclonal antibodies and is routinely included in the quality standards for release and stability testing. Non-reducing CE-SDS is typically employed to assess the relative contents of the main peak and fragment peaks. Beyond assessing the relative contents of light chains, heavy chains, and fragments, reducing CE-SDS analysis should also be employed to evaluate the content of non-glycosylated heavy chains. The establishment of quality standards for CE-SDS is generally based on statistical analyses across multiple batches, clinical experience, and structure–activity relationship studies. As a release method, CE-SDS requires comprehensive and systematic methodological validation; therefore, the corresponding validation data must be included in the regulatory submission materials.CE-SDS, a key analytical technique in the comprehensive characterization of antibodies, is discussed in the sections addressing characteristic identification and impurities. A comprehensive analysis of mAb size variants typically employs several orthogonal techniques, including CE-SDS, analytical ultracentrifugation (AUC), and SEC.27 SEC is employed to characterize fragments under natural conditions, while AUC serves as an orthogonal technique for fragment characterization. CE-SDS is employed to analyse fragments under denaturing and reducing conditions. In the characteristic identification and impurity section of the regulatory submission, the CE-SDS data are presented in detail. Furthermore, the detected fragments are subsequently characterized through release and dedicated characterization methods to identify their precise cleavage sites and origins.
The detection data of CE-SDS constitute a critical component of mAb stability studies. Its profile must be monitored throughout long-term, accelerated, and stress stability studies to ensure consistent product quality. Furthermore, in comparability studies of biosimilar drugs and manufacturing process changes, data obtained from CE-SDS are also an important aspect for comparison.
4.2 Method validation
The validation of CE-SDS methodologies is conducted in accordance with the ICH Q2(R2) guideline,72 along with specific requirements from relevant pharmacopoeias. Key validation parameters—namely specificity, range, accuracy, precision, and robustness—collectively ensure that the method accurately reflects the content of size variants within mAb samples. For this category of purity-assessment method, the following aspects require particular attention and are areas that regulatory authorities will pay close attention to:① Units of validation parameters and specifications should be consistent: Quality specifications for CE-SDS are generally defined in terms of the relative percent abundances for the main peak, fragments, or the sum of heavy and light chains. Consequently, validation should not depend exclusively on samples exhibiting varying protein concentrations. Instead, the study should utilize samples exhibiting varying levels of purity. This approach ensures that the validation results accurately demonstrate the appropriateness of the method for the designated application.
② Establishment of the Limit of Quantitation (LOQ): The LOQ should not be merely estimated. Demonstration should be performed using a sample at an appropriate concentration, with validated accuracy and precision at that level. Critically, the LOQ must be expressed in units consistent with those employed in the corresponding quality specification.
③ Comprehensive coverage of controlled species: Validation should cover all components specified in the quality control strategy. For example, if the specification for non-reducing CE-SDS establishes limits for the main peak and total fragments, validation should encompass not only the main peak but also a representative, specific fragment peak.
4.3 How method development trends influence quality standards and regulatory expectations
The newly developed CE-SDS analytical method described above has profoundly influenced quality standards and regulatory expectations. The continuous optimization of the CE-SDS method—such as incorporating a more sensitive LIF detector and introducing novel denaturants—has significantly enhanced the sensitivity of the method, minimized operational errors, and enabled more accurate quantification of size variants in antibody samples. At the same time, this advancement enhances the understanding of the product among researchers and regulators, thereby fostering more scientifically grounded quality standards. It also facilitates more reliable monitoring of process consistency and batch-to-batch uniformity.Furthermore, as the biopharmaceutical industry continues to mature, regulatory expectations for fragment analysis have expanded beyond simple content control to encompass identification and origin assessment, aligning with the principles of Quality by Design (QbD). Emerging CE-SDS technology, particularly when integrated with mass spectrometry, represents a critical tool for addressing this shift. These techniques enable researchers and regulators to gain a comprehensive understanding of fragment characteristics, thereby facilitating the evaluation of their potential influence on product safety and efficacy. Such in-depth molecular-level understanding of critical quality attributes (CQAs) provides robust scientific evidence to support process validation, establish appropriate impurity control strategies, and ultimately secure regulatory approval.
5. Outlook
5.1 High-throughput capillary electrophoresis-sodium dodecyl sulfate detection platform
Most current CE-SDS detection instruments employ single-capillary systems. When processing many samples, this configuration considerably extends analysis time. Recent developments in CE-SDS technology are moving toward high-throughput designs, featuring instruments with 8 or 12 parallel capillaries capable of generating multiple results simultaneously.73 However, achieving such high-throughput analysis introduces substantial challenges in ensuring reproducibility across capillaries. In practical sample analysis, when discrepancies arise in results, it is often challenging to discern whether they stem from intrinsic sample variability or systematic deviations among capillaries. Future development should focus on establishing methods for real-time monitoring and correction of capillary performance.5.2 Sodium dodecyl sulfate removal technology
The method developed by Jennifer Römer et al. for eliminating the SDS interface and integrating CE-SDS with CZE-MS in an online configuration offers a novel approach toward fragment analysis. Undoubtedly, this currently represents the most advanced, sensitive, and streamlined technique for identifying monoclonal antibody fragments. However, it is equally evident that the technique entails substantial instrument costs and significant challenges in method development. Future research should focus on designing more efficient SDS-removal interfaces or identifying denaturants compatible with mass spectrometry. Achieving widespread implementation of online CE-SDS coupling with high-resolution mass spectrometry would constitute a transformative advancement in capillary electrophoresis.5.3 Prediction
Predictive analysis of monoclonal antibodies will constitute an important direction for future development. Harnessing the high separation efficiency of CE-SDS while integrating multidimensional analytical techniques such as SEC, HIC, and mass spectrometry remains essential. By accumulating comprehensive data on the various fragment forms of novel antibodies generated under diverse stress conditions, a predictive model of antibody degradation pathways can be established. This model will provide a robust scientific basis for molecular design, formulation screening, and process optimization.74 Ultimately, the objective is to build a knowledge framework capable of not only addressing quality issues but also anticipating and preventing them.6. Conclusions
CE-SDS has become a key analytical technology in biopharmaceutical development for evaluating the CQAs of antibody-based therapeutics, including purity. This review summarizes the application of CE-SDS in purity and fragment analysis of mAbs and their derivatives, characterization of ADC positional isomers, and identification of mismatch impurities in bispecific antibodies, highlighting its high resolution, high sensitivity, and excellent method stability. To address the fundamental technical challenge of CE-SDS incompatibility with mass spectrometry and the associated difficulty in directly identifying fragment peaks, this article emphasizes an indirect identification approach alongside offline and online integration strategies based on CZE-MS, thereby providing a clear technical framework for resolving fragment structure identification issues. Additionally, this review summarizes strategies for effectively incorporating CE-SDS technology into regulatory submissions, addressing key aspects such as quality standard establishment, stability studies, and method validation. Finally, based on the current industry landscape and emerging technological trends, it presents a forward-looking perspective on the future development of CE-SDS in antibody drug analysis, providing valuable insights for its broader application in process development and quality control.Author contributions
Yalan Yang: writing – original draft and methodology. Meng Li and Gangling Xu: writing – review & editing. Yongbo Ni and Luyun Guo: data curation. Chuanfei Yu: methodology and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
The raw data from this study are available in the supporting information (SI).Acknowledgements
This work was supported by the State Key Laboratory of Drug Regulatory Science Project (2025SKLDRS0318) and the National Drug Standards Enhancement Program (2025S14).References
- A. C. Chan, G. D. Martyn and P. J. Carter, Nat. Rev. Immunol., 2025, 10, 745–765 CrossRef PubMed.
- T. K. Ha, D. Kim, C. L. Kim, L. M. Grav and G. M. Lee, Biotechnol. Adv., 2022, 54, 107831 CrossRef CAS PubMed.
- Y. Xu, D. Wang, B. Mason, T. Rossomando, N. Li, D. Liu, J. K. Cheung, W. Xu, S. Raghava, A. Katiyar, C. Nowak, T. Xiang, D. D. Dong, J. Sun, A. Beck and H. Liu, mAbs, 2019, 11, 239–264 CrossRef CAS PubMed.
- F. Torkashvand and B. Vaziri, Iran. Biomed. J., 2017, 21, 131–141 CrossRef PubMed.
- Q. Luo, M. K. Joubert, R. Stevenson, R. R. Ketchem, L. O. Narhi and J. Wypych, J. Biol. Chem., 2011, 286, 25134–25144 CrossRef CAS PubMed.
- E. Tamizi and A. Jouyban, Eur. J. Pharm. Biopharm., 2016, 98, 26–46 CrossRef CAS PubMed.
- W. Li, B. Yang, D. Zhou, J. Xu, W. Li and W. C. Suen, J. Chromatogr. B:Anal. Technol. Biomed. Life Sci., 2017, 1048, 121–129 CrossRef CAS PubMed.
- W. H. Wang, J. Cheung-Lau, Y. Chen, M. Lewis and Q. M. Tang, mAbs, 2019, 11, 1233–1244 CrossRef CAS PubMed.
- J. Vlasak and R. Ionescu, mAbs, 2011, 3, 253–263 CrossRef PubMed.
- S. Tang, J. Tao and Y. Li, Antibody Ther., 2024, 7, 1–12 CrossRef CAS PubMed.
- Y. Atsumi, A. Yamada, Y. Kojima, Y. Yagi, K. Nishimura and K. Wakamatsu, J. Pharm. Sci., 2022, 111, 3243–3250 CrossRef CAS PubMed.
- R. R. Rustandi, M. W. Washabaugh and Y. Wang, Electrophoresis, 2008, 29, 3612–3620 CrossRef CAS PubMed.
- C. E. Sänger-van de Griend, Electrophoresis, 2019, 40, 2361–2374 CrossRef PubMed.
- Y. Leblanc, C. Ramon, N. Bihoreau and G. Chevreux, J. Chromatogr. B:Anal. Technol. Biomed. Life Sci., 2017, 1048, 130–139 CrossRef CAS PubMed.
- M. Li, C. Yu, W. Wang, G. Wu and L. Wang, Electrophoresis, 2021, 42, 1900–1913 CrossRef CAS PubMed.
- R. Bhimwal, R. R. Rustandi, A. Payne and M. Dawod, J. Chromatogr. A, 2022, 1682, 463453 CrossRef CAS PubMed.
- O.O Dada, R. Rao, N. Jones, N. Jaya and O. Salas-Solano, Comparison of SEC and CE-SDS methods for monitoring hinge fragmentation in IgG1 monoclonal antibodies, J. Pharm. Biomed. Anal., 2017, 145, 91–97, DOI:10.1016/j.jpba.2017.06.006 . Epub 2017 Jun 17. PMID: 28654781.
- H. A. Alhazmi and M. Albratty, Pharmaceuticals, 2023, 16, 291 CrossRef CAS PubMed.
- B. Nunnally, S. S. Park, K. Patel, M. Hong, X. Zhang, S.-X. Wang, B. Rener, A. Reed-Bogan, O. Salas-Solano, W. Lau, M. Girard, H. Carnegie, V. Garcia-Cañas, K. C. Cheng, M. Zeng, M. Ruesch, R. Frazier, C. Jochheim, K. Natarajan, K. Jessop, M. Saeed, F. Moffatt, S. Madren, S. Thiam and K. Altria, Chromatographia, 2006, 64, 359–368 CrossRef CAS.
- A. L. Esterman, A. Katiyar and G. Krishnamurthy, J. Pharm. Biomed. Anal., 2016, 128, 447–454 CrossRef CAS PubMed.
- R. Kumar, A. Guttman and A. S. Rathore, Electrophoresis, 2022, 43, 143–166 CrossRef CAS PubMed.
- X. Song, H. Tian and G. Zhang, J. Liq. Chromatogr. Relat. Technol., 2024, 47, 44–54 CrossRef CAS.
- Z. Zhang, J. Park, H. Barrett, S. Dooley, C. Davies and M. F. Verhagen, Hum. Gene Ther., 2021, 32, 628–637 CrossRef CAS PubMed.
- D. A. Michels, M. Parker and O. Salas-Solano, Electrophoresis, 2012, 33, 815–826 CrossRef CAS PubMed.
- H. Cai, Y. Song, J. Zhang, T. Shi, Y. Fu, R. Li, N. Mussa and Z. J. Li, J. Pharm. Biomed. Anal., 2016, 120, 46–56 CrossRef CAS PubMed.
- L. Zhang, M. Fei, Y. Tian, S. Li, X. Zhu, L. Wang, Y. Xu and M. H. Xie, J. Pharm. Biomed. Anal., 2020, 190, 113527 CrossRef CAS PubMed.
- Q. Guan, R. Knihtila, J. Atsma, R. Tulsan, S. Singh, S. Kar, J. Beckman, J. Ding and Z. J. Li, J. Chromatogr. B:Anal. Technol. Biomed. Life Sci., 2020, 1152, 122230 CrossRef CAS PubMed.
- J. Beckman, Y. Song, Y. Gu, S. Voronov, N. Chennamsetty, S. Krystek, N. Mussa and Z. J. Li, Anal. Chem., 2018, 90, 2542–2547 CrossRef CAS PubMed.
- H. Gao, S.-T. Wang, F. Hu, B.-B. Shen, M.-F. Sun, H. Wang, L. Li and W.-J. Fang, Pharm. Res., 2022, 39, 1959–1968 CrossRef CAS PubMed.
- J. V. Leyton, Expert Opin. Biol. Ther., 2023, 23, 1067–1076 CrossRef CAS PubMed.
- S. Balamkundu and C.-F. Liu, Biomedicines, 2023, 11, 3080 CrossRef CAS PubMed.
- J. Z. Drago, S. Modi and S. Chandarlapaty, Nat. Rev. Clin. Oncol., 2021, 18, 327–344 CrossRef PubMed.
- C. H. Chau, P. S. Steeg and W. D. Figg, Lancet, 2019, 394, 793–804 CrossRef CAS PubMed.
- C. Dumontet, J. M. Reichert, P. D. Senter, J. M. Lambert and A. Beck, Nat. Rev. Drug Discovery, 2023, 22, 641–661 CrossRef CAS PubMed.
- S. Modi, W. Jacot, T. Yamashita, J. Sohn, M. Vidal, E. Tokunaga, J. Tsurutani, N. T. Ueno, A. Prat, Y. S. Chae, K. S. Lee, N. Niikura, Y. H. Park, B. Xu, X. Wang, M. Gil-Gil, W. Li, J.-Y. Pierga, S.-A. Im, H. C. Moore, H. S. Rugo, R. Yerushalmi, F. Zagouri, A. Gombos, S.-B. Kim, Q. Liu, T. Luo, C. Saura, P. Schmid, T. Sun, D. Gambhire, L. Yung, Y. Wang, J. Singh, P. Vitazka, G. Meinhardt, N. Harbeck, D. A. Cameron and DESTINY-Breast04 Trial Investigators, N. Engl. J. Med., 2022, 387, 9–20 CrossRef CAS PubMed.
- J. Cortés, S.-B. Kim, W.-P. Chung, S.-A. Im, Y. H. Park, R. Hegg, M. H. Kim, L.-M. Tseng, V. Petry, C.-F. Chung, H. Iwata, E. Hamilton, G. Curigliano, B. Xu, C.-S. Huang, J. H. Kim, J. W. Y. Chiu, J. L. Pedrini, C. Lee, Y. Liu, J. Cathcart, E. Bako, S. Verma and S. A. Hurvitz, N. Engl. J. Med., 2022, 386, 1143–1154 CrossRef PubMed.
- Q. Yang and Y. Liu, RSC Med. Chem., 2025, 16, 50–62 RSC.
- H. Xu, H. Zhang, W. Guo, X. Zhong, J. Sun, T. Zhang, Z. Wang and X. Ma, BMC Cancer, 2022, 22, 923 CrossRef CAS PubMed.
- P. Gogia, H. Ashraf, S. Bhasin and Y. Xu, Cancers, 2023, 15, 3886 CrossRef CAS PubMed.
- T. Chen, Y. Chen, C. Stella, C. D. Medley, J. A. Gruenhagen and K. Zhang, J. Chromatogr. B:Anal. Technol. Biomed. Life Sci., 2016, 1032, 39–50 CrossRef CAS PubMed.
- A. Wagh, H. Song, M. Zeng, L. Tao and T. K. Das, mAbs, 2018, 10, 222–243 CrossRef CAS PubMed.
- Sciex.com. Analytical characterization of the antibody drug conjugate Enhertu using multi-capillary electrophoresis. (2024, March 28), from https://sciex.com/tech-otes/biopharma/analytical-characterization-of-the-antibody-drug-conjugate-enher.
- C. Hamers-Casterman, T. Atarhouch, S. Muyldermans, G. Robinson, C. Hammers, E. B. Songa, N. Bendahman and R. Hammers, Nature, 1993, 363, 446–448 CrossRef CAS PubMed.
- S. Muyldermans, Annu. Rev. Biochem., 2013, 82, 775–797 CrossRef CAS PubMed.
- S. Sun, Z. Ding, X. Yang, X. Zhao, M. Zhao, L. Gao, Q. Chen, S. Xie, A. Liu, S. Yin, Z. Xu and X. Lu, Int. J. Nanomed., 2021, 16, 2337–2356 CrossRef PubMed.
- S. Kunz, M. Durandy, L. Seguin and C. C. Feral, Int. J. Mol. Sci., 2023, 24, 13229 CrossRef CAS PubMed.
- V. M. Medina Pérez, M. Baselga and A. J. Schuhmacher, Cancers, 2024, 16, 2681 CrossRef PubMed.
- P. Tamburini, D. V. Pedersen, D. Devore, J. Cone, R. Patel, T. Hunter, F. Sun, G. R. Andersen and J. Hunter, mAbs, 2024, 16, 2415060 CrossRef PubMed.
- E. Alexander and K. W. Leong, J. Nanobiotechnol., 2024, 22, 661 CrossRef PubMed.
- B. Li, X. Qin and L.-Z. Mi, Nanoscale, 2022, 14, 7110–7122 RSC.
- D. L. Niquille, K. M. Fitzgerald and N. Gera, mAbs, 2024, 16, 2310890 CrossRef PubMed.
- J. Zhang, J. Yi and P. Zhou, Antibody Ther., 2020, 3, 126–145 CrossRef CAS PubMed.
- A. F. Labrijn, M. L. Janmaat, J. M. Reichert and P. W. H. I. Parren, Nat. Rev. Drug Discovery, 2019, 18, 585–608 CrossRef CAS PubMed.
- J. Jeong, J. Park, G. Y. Mo, J. Shin and Y. Cho, J. Mol. Biol., 2024, 436, 168748 CrossRef CAS PubMed.
- M. Zhu, B. Wu, C. Brandl, J. Johnson, A. Wolf, A. Chow and S. Doshi, Clin. Pharmacokinet., 2016, 55, 1271–1288 CrossRef CAS PubMed.
- X. Pang, Z. Huang, T. Zhong, P. Zhang, Z. M. Wang, M. Xia and B. Li, mAbs, 2023, 15, 2180794 CrossRef PubMed.
- J. Cheng, Q. Lv, Y. Ji, C. Zhou, J. Guo, X. Li and J. Hu, J. Pharm. Biomed. Anal., 2025, 255, 116673 CrossRef CAS PubMed.
- M. Cao, Y. Jiao, C. Parthemore, S. Korman, J. Ma, A. Hunter, G. Kilby and X. Chen, mAbs, 2021, 13, 1981806 CrossRef PubMed.
- J. Lin, M. Xie, D. Liu, Z. Gao, X. Zhao, H. Ma, S. Ding, S. Mei Li, S. Li, Y. Liu, F. Zhou, H. Hu, T. Chen, H. Chen, M. Xie, B. Yang, J. Cheng, M. Ma, Y. Nan and D. Ju, Front. Chem., 2022, 10, 994472 CrossRef CAS PubMed.
- E. B. A. van den Berg, J. C. W. Hendriks, E. W. Elsinga, M. Eggink and E. H. C. Dirksen, Electrophoresis, 2023, 44, 62–71 CrossRef CAS PubMed.
- V. Ziaei, A. Ghassempour, F. Davami, B. Azarian, M. Behdani, H. Dabiri and M. Habibi-Anbouhi, Mol. Cell. Biochem., 2024, 479, 579–590 CrossRef CAS PubMed.
- M. Cao, C. Parthemore, Y. Jiao, S. Korman, M. Aspelund, A. Hunter, G. Kilby and X. Chen, J. Pharm. Sci., 2021, 110, 2904–2915 CrossRef CAS PubMed.
- L. Duhamel, Y. Gu, G. Barnett, Y. Tao, S. Voronov, J. Ding, N. Mussa and Z. J. Li, Anal. Bioanal. Chem., 2019, 411, 5617–5629 CrossRef CAS PubMed.
- M. Fei, Q. Zhang, L. Zhang, Y. Zhang, L. Wang, Y. Zhao and Z. Zhang, Electrophoresis, 2024, 45, 1325–1338 CrossRef PubMed.
- J. Giorgetti, A. Beck, E. Leize-Wagner and Y.-N. François, J. Pharm. Biomed. Anal., 2020, 182, 113107 CrossRef CAS PubMed.
- T. Xu, F. Zhang, D. Chen, L. Sun, D. Tomazela and L. Fayadat-Dilman, mAbs, 2023, 15, 2229102 CrossRef PubMed.
- J. Dugal-Tessier, S. Thirumalairajan and N. Jain, J. Clin. Med., 2021, 10, 838 CrossRef CAS PubMed.
- Q. Liu, J. Hong, Y. Zhang, Q. Wang, Q. Xia, M. D. Knierman, J. Lau, C. Dayaratna, B. Negron, H. Nanda and H. P. Gunawardena, Sci. Rep., 2024, 14, 20239 CrossRef CAS PubMed.
- M. Li, X. Zhao, D. Shen, G. Wu, W. Wang, C. Yu, J. Sausen, H. Xu and L. Wang, J. Chromatogr. A, 2022, 1684, 463560 CrossRef CAS PubMed.
- J. Römer, A. Stolz, S. Kiessig, B. Moritz and C. Neusüß, J. Pharm. Biomed. Anal., 2021, 201, 114089 CrossRef PubMed.
- O. Höcker, M. Knierman, J. Meixner and C. Neusüß, Electrophoresis, 2021, 42, 369–373 CrossRef PubMed.
- ICH, Q2(R2) Validation of Analytical Procedures, Step 5 Version, 2024.
- G. Bou-Assaf, D. Tribby and M. Santos, ChemRxiv, 2025, Preprint, DOI:10.26434/chemrxiv-2025-x5fh2.
- Q. Liu, J. Hong and Y. Zhang, et al. , Sci. Rep., 2024, 14, 20239 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |


