Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Creating taxonomically-informed metabolome libraries for any species using the pubchem.bio R package
Corey D. Broeckling
Colorado State University, Analytical Resources Core – Bioanalysis and Omics Center, Fort Collins, CO 80523, USA. E-mail: cbroeckl@colostate.edu; Tel: +1 970-491-2273
First published on 15th December 2025
Abstract
Annotation remains a significant challenge in metabolomics, in large part due to the enormous structural diversity of small molecules. PubChem represents one of the largest curated chemical structure databases, with more than 122![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structures, supplemented by extensive biological metadata provided by numerous external sources. While many of these structures are relevant to metabolomics, a majority are unlikely to be measured in a typical metabolomics experiment. This article describes the R package, pubchem.bio, which enables users to: (1) download the metabolomics-centric subset of PubChem onto their local computer, (2) build a metabolomic structured library of biological compounds in PubChem, (3) develop custom metabolite structure libraries for any species or collection of species using selected or all available taxonomic data in PubChem and (4) define a core biological metabolome, comprising metabolites plausibly found in any species. Species-specific metabolomes are enabled through the adoption of a lowest-common-ancestor chemotaxonomy approach, which is implemented by associating PubChem CIDs into the NCBI Taxonomy database hierarchy, enabling extrapolation of the taxonomic range beyond the species reported. This package is available via CRAN, and can be used to simplify the annotation process and embed biological metadata into the annotation process.
000 structures, supplemented by extensive biological metadata provided by numerous external sources. While many of these structures are relevant to metabolomics, a majority are unlikely to be measured in a typical metabolomics experiment. This article describes the R package, pubchem.bio, which enables users to: (1) download the metabolomics-centric subset of PubChem onto their local computer, (2) build a metabolomic structured library of biological compounds in PubChem, (3) develop custom metabolite structure libraries for any species or collection of species using selected or all available taxonomic data in PubChem and (4) define a core biological metabolome, comprising metabolites plausibly found in any species. Species-specific metabolomes are enabled through the adoption of a lowest-common-ancestor chemotaxonomy approach, which is implemented by associating PubChem CIDs into the NCBI Taxonomy database hierarchy, enabling extrapolation of the taxonomic range beyond the species reported. This package is available via CRAN, and can be used to simplify the annotation process and embed biological metadata into the annotation process.
Introduction
A critical step in any mass spectrometry-based metabolomics workflow is ‘annotation’ – the process of assigning chemical structures to chromatographically coupled mass spectrometry signals. The annotation task has been considered as one of the largest challenges in metabolomics since the field's inception,1 and remains problematic today.2–4There have been innumerable approaches designed to improve annotation accuracy. One of the major challenges in metabolomics annotation is appropriately defining the chemical search space for any given untargeted metabolomics experiment. The full theoretical chemical search space has been estimated to be on the order of 1060 potential structures at a molecular weight of 1000 or less.5 PubChem is one of the largest publicly available repositories for small molecule structures, with more than 122![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. This size makes it one of the most comprehensive structure databases available, but there are many structures in PubChem that are unlikely to ever be observed in most metabolomics experiments. PubChem Lite6 was designed to extract the compounds that are most likely to be observed, with a focus toward support for non-target analysis or exposomics experiments.
000. This size makes it one of the most comprehensive structure databases available, but there are many structures in PubChem that are unlikely to ever be observed in most metabolomics experiments. PubChem Lite6 was designed to extract the compounds that are most likely to be observed, with a focus toward support for non-target analysis or exposomics experiments.
Numerous biological databases have been developed for metabolomics research as a way of restricting the chemical structure search space, each with a different focus. If a study is focused on clinical samples, the Human Metabolome Database7 is an invaluable resource. For natural products, Coconut,8 Lotus,9 NPASS,10 and others are available. These databases are extremely valuable in cataloging the existing known chemical space, particularly for specialized metabolites. However, there are relatively few species for which a comprehensive database exists, due to the effort necessary to compile databases from the literature and the incredible diversity of life. Some of the natural product databases explicitly link chemical structures to taxonomy, enabling taxonomy filtering, but these databases are biased toward natural products and therefore not inclusive of more highly conserved metabolic pathways.
Taxonomy has been shown to be a useful piece of metadata to include in annotation approaches.11,12 An ideal metabolomics library for a given biological sample would include all the potential small molecules that would plausibly be found in a given sample, with as few extraneous compounds as possible. This is feasible through manual effort, but remains a cumbersome task.13 PubChem14 has become not only a vast repository of chemical structure data, but also a vast repository of associated biological metadata, incorporating metadata from metabolomics, metabolic pathway, and natural product databases. The pubchem.bio R package described in this manuscript is an informatic resource which streamlines the building of biological and taxonomically informed metabolite libraries from PubChem in support of metabolomics, and other applications that can benefit from a comprehensive list of plausible small molecules found in a given species.
Approach
In this article, the term ‘metabolite’ is used to be inclusive of all small molecule structures that are linked to biology, including traditional metabolites, but also lipids, polysaccharides, and even exogenous compounds such as pesticides, which are frequently found in biological metabolome libraries and pathways.PubChem is an accessible and freely available data source, which incorporates a wealth of metadata. The pubchem.bio functions were executed on August 22, 2025, to generate summary statistics presented here. There are several biological classes of metadata built into PubChem, including:
1. Data source: PubChem can be subset based on which organizations deposited structures. 885 different data sources have contributed. Suitable biological data sources include HMDB, CheBI, Metabolomics Workbench, Lipid Maps, and others. Any data source can be selected, but the default values are those considered to clearly fit the category of ‘biological’.
2. Pathways: 10 organizations have deposited pathway data. The presence of a chemical in a pathway means that there is biological transformation which either produces or consumes it. These chemicals will be either native biological metabolites, or metabolic products of enzymatic processes acting on well-characterized exogenous compounds.
3. Taxonomy: 11 organizations have submitted taxonomy/structure relationship datasets to PubChem in the form of ‘Annotations’. For example, the Natural Product Activity and Species Sources (NPASS) has 540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 494 annotations in PubChem. The Lotus database has submitted 434
494 annotations in PubChem. The Lotus database has submitted 434![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081 annotations, where an ‘annotation’ is defined as a relationship between a taxonomy identifier and a pubchem structure.
081 annotations, where an ‘annotation’ is defined as a relationship between a taxonomy identifier and a pubchem structure.
The pubchem.bio package is designed to utilize all these data sources to enable efficient and comprehensive custom metabolome library creation through PubChem centralization. The vast majority of the functionality of pubchem.bio is arranged as five R functions. These functions retrieve, organize, and subset PubChem data to enable the generation of custom taxonomically informed libraries.
get.pubchem.ftp: The puchem.bio R package accesses NCBI's PubChem and Taxonomy databases programatically, downloading data primarily through the FTP interface. The downloaded files are stored in a temporary directory, unzipped, and parsed, retaining only the portions of the data needed. Several derivative datasets are generated, each indexed by PubChem CID or taxonomy ID, and stored internally as data.table15 formatted data frames, enabling fast searching and filtering. These files are saved and retained to a local drive determined by the user. This function only downloads and parses data into smaller, more easily managed chunks, to enable further downstream handling.
build.cid.lca: This function utilizes all selected sources of taxonomy data, which can include both pathway data such as WikiPathways,16 which is sometimes built for a specific species, and the taxonomic annotations from PubChem's data sources, such as the Lotus database.9 To make full use of these taxonomy data, pubchem.bio organizes each PubChem CID into the nested NCBI Taxonomy17 hierarchy. This network structure (also stored as a data.table) enables the extrapolation of metabolite (CID) presence even in species for which the metabolite has not been reported, through the notion of lowest common ancestor (LCA). The LCA approach has been adopted for metaproteomics studies as a mechanism to deal with redundancy in peptide sequence within a given taxonomic clade.18 The pubchem.bio package adapts this logic for small molecule structures, enabling inference on the plausible metabolites for any given species.
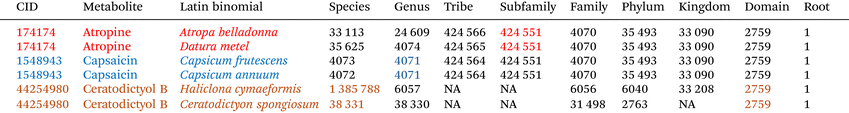
Table 1 provides an example of how LCA is assigned. Note that not all taxonomic levels are displayed, for simplicity. If one wished to determine the lowest common ancestor for capsaicin (in blue, Table 1), the metabolite in peppers which provides their spicy heat, all biological species that are known to contain capsaicin are catalogued (only a small subset is shown in Table 1). The full taxonomic hierarchy is then generated for each species, and the lowest taxonomic level that contains all examples of capsaicin is assigned at the LCA of 4071, the genus Capsicum. Alternatively, consider atropine, in red. Atropine is found in both Atropa and Datura species, and the lowest common ancestor is Taxonomy ID 424551, the subfamily Solanoideae, since each of the two species have a distinct genus and tribe. The Ceratodictyol B example is more complex, and will be discussed in the results.
This output cid.lca dataset generated by the build.cid.lca function is both saved with results from get.pubchem.ftp and returned to the R console, and is used for downstream library creation. This function is separated from get.pubchem.ftp only for practical reasons, as the function takes a bit of time to run.
build.pubchem.bio: With all of the data now organized from the first two functions, the build.pubchem.bio function will generate a data.table containing only metabolites that are found from the selected data sources, which may include structure databases, pathway databases, and/or taxonomy–structure databases. The returned dataset will include CID, compound name, molecular formula, monoisotopic mass, SMILES structure, InChIKey, etc. Additionally, this function enables the calculation of all physicochemical properties available through the rcdk package.18 The dataset is both saved in the local directory and returned as an R data.table.
build.taxon.metabolome: The pubchem.bio dataset created in the above step contains all biological compounds from PubChem. The build.taxon.metabolome function uses all selected taxonomy data to filter and/or score all biological compounds that have an assigned LCA. The user supplies one or more target taxa, using a NCBI Taxonomy number. For each taxon identifier, the full taxonomic hierarchy is extracted, and all CIDs that have a lowest common ancestor that matches any of the taxon's hierarchy levels are assigned a score of ‘1’. All CIDs that have taxonomy data assigned, but do not fall within the selected taxonomic hierarchy are assigned a score between ‘0’ and ‘1’, based on how many taxonomic levels separate the metabolite lowest common ancestor from the ‘root’ of the taxonomic tree. Put another way, parent taxa up to the lowest common ancestor inherits the metabolome of child taxa, and then traverse the tree using the proposed scoring system. In this way, highly specialized metabolites from Streptomyces, for example, are assigned a low taxonomy score if the user is building a Solanum metabolome library. All CIDs that have no taxonomic data are left with a taxonomy score of NA.
Table 1 demonstrates the cid.lca theory. If we build a taxon metabolome for Datura metel, our exported metabolome will contain both atropine and capsaicin. However, atropine will have a taxonomy score of ‘1’, while capsaicin will have a taxonomy score between 0 and 1. Since capsaicin is found in a genus (Capsicum) that is relatively closely related to Datura metel – they share a common subfamily – the taxonomy score will be larger, but less than 1. In practice, the assigned score for capsaicin, when building a library for Datura metel, is 0.76, reflecting the fractional number of taxonomic levels that separate the two, relative to the total number of taxonomic levels.
Note also that if one were to build a taxon metabolome library for the genus Capsicum, the metabolome would contain atropine with a taxonomy score of 1, since the lowest common ancestor for atropine – Taxonomy ID 424551 – is a direct taxonomic ancestor of the Capsicum genus. This may not, at first glance, appear to be a desirable result. However, this result captures evolutionary concepts. Atropine is found in multiple genera within the subfamily Solanoideae. We must either assume that (1) the only species atropine is found in are those listed or (2) that the species that have been found to contain atropine are some subset of all species that contain atropine, and that there are missing data in PubChem. These missing data are due to some combination of (a) unpublished or uncatalogued studies that demonstrate the absence of atropine from other Datura species, and (b) studies looking at atropine in other species which have yet to be performed. If no one has published on the presence or absence of atropine in Datura ferox, for example, we have no evidence that atropine is absent in that species.
The pubchem.bio package uses the LCA approach to infer metabolome content for any species, whereby one can assume that the data listed in PubChem are a subset of the species in which atropine can be found. Rephrased, we do not currently have complete data, so we must infer the taxonomic true range of each given metabolite. Given that all occurrences of atropine are found within the subfamily Solanoideae, we infer that any member of this subfamily may plausibly have evolved the capacity to synthesize atropine. There is of course also a practical reason for wanting to be inclusive in our metabolome search space – if we are measuring the metabolome of peppers, and atropine is present in some samples, we want to identify it as such, as it can have toxic properties.
build.primary.metabolome: In theory, the PubChem metabolites that have been assigned an LCA of ‘1’ – the root of all life – can be considered a list of primary metabolites contained within PubChem. This function enables users to extract all highly conserved metabolites from the full pubchem.bio output.
Additionally, there are currently four ‘export’ functions, for exporting the full dataset in .csv format, or in formats compatible with Sirius or MSFinder.
Results
The pubchem.bio package was demonstrated on August 22, 2025. All processing reflects performance on a Windows 11 PC, with 64 GB RAM, a 1.0 TB SSD hard drive, and a 13th Gen Intel Core i9-13900K 3.00 GHz processor. The get.pubchem.bio function was run to retrieve and store data for later use. The downloaded content occupies approximately 18 GB on the disk, although users should ensure there is at least triple that to accommodate temporary files during processing. The get.pubchem.ftp function took approximately 2.5 hours to run.There are 885 total sources of PubChem data. For this example, five were considered as biological databases: Metabolomics Workbench, Human Metabolome Database (HMDB),7 ChEBI,19 LIPID MAPS,20 and MassBank of North America (MoNA).21 Users can select any of the 885 databases as source databases. There are ten sources of pathway data, with PathBank,22 PlantCyc,23 Plant Reactome,24 BioCyc,25 Reactome,26 and WikiPathways16 each listing more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 records. All pathway sources were used, and any taxonomy-assigned pathway was incorporated into the CID–LCA assignment. Eleven sources provide taxonomy–structure (CID) pairs, with FooDB,27 NPASS,10 LOTUS,9 HMDB,7 KNApSAcK,28 and NPA29 each supplying more than 10
000 records. All pathway sources were used, and any taxonomy-assigned pathway was incorporated into the CID–LCA assignment. Eleven sources provide taxonomy–structure (CID) pairs, with FooDB,27 NPASS,10 LOTUS,9 HMDB,7 KNApSAcK,28 and NPA29 each supplying more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 records. For this example, only LOTUS was used, but the user is free to use any or all of the sources. CID–taxonomy associations within PubChem amount to 3
000 records. For this example, only LOTUS was used, but the user is free to use any or all of the sources. CID–taxonomy associations within PubChem amount to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 857
857![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 records, including 434
536 records, including 434![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081 from LOTUS. Adding taxonomic associations from pathway sources added an additional 375
081 from LOTUS. Adding taxonomic associations from pathway sources added an additional 375![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 375 CID–taxonomy pairs, bringing the total to 809
375 CID–taxonomy pairs, bringing the total to 809![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 456. The build.cid.lca function took approximately 1.1 hours to run, resulting in 247
456. The build.cid.lca function took approximately 1.1 hours to run, resulting in 247![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 050 CID–LCA pairs. Some pathways and biological data sources do include non-biological compounds, such as pesticide degradation pathways in plants. It should be noted that this package does not remove those compounds from the resulting structure library.
050 CID–LCA pairs. Some pathways and biological data sources do include non-biological compounds, such as pesticide degradation pathways in plants. It should be noted that this package does not remove those compounds from the resulting structure library.
There are numerous ‘NA’ values in the taxonomy hierarchy – not all species have assignments at each level. In fact, the only two taxonomic levels that have no missing values are species and domain. This can be seen in Table 1, specifically for the Ceratodictyol B example.
In preliminary CID–LCA assignments, there were nearly 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 metabolites that were demonstrated to have a listed lowest common ancestor of ‘1’ – the root of all biology. These could be interpreted as being the most highly conserved, and therefore classified as ‘primary’ metabolites. It was, however, observed that sparse and spurious CID–taxonomy associations can generate false positive assignments with unexpectedly high LCA taxonomic assignments. For example, Ceratodictyol B was assigned an LCA of ‘2759’, as depicted in Table 1. Taxonomy ID 2759 is ‘Eukaryota’, or eukaryotes. Assignment to this level would result in assignment of Ceratodictyol B to every eukaryote metabolome library. This occurred due to Ceratodictyol B being reported from precisely two species, each listed by two sources: a marine sponge, Haliclona cymaeformis, and a red algae, Ceratodictyon spongiosum. Ceratodictyon spongiosum can form symbioses with sponges, calling into question whether Haliclona is producing Ceratodictyol B, or harboring Ceratodictyon, which is producing Ceratodictyol B. In fact, the paper describing these results does not distinguish between the two.30 This result can therefore be interpreted as a false positive assignment of LCA at a much higher taxonomic level than is warranted. The LCA concept is built on a premise that each taxon–CID association reflects phylogeny, while, in practice, the LCA approach implemented is dependent on taxonomy as a surrogate, and the case in question reflects complex cross-taxa symbioses that taxonomy doesn't capture well.
000 metabolites that were demonstrated to have a listed lowest common ancestor of ‘1’ – the root of all biology. These could be interpreted as being the most highly conserved, and therefore classified as ‘primary’ metabolites. It was, however, observed that sparse and spurious CID–taxonomy associations can generate false positive assignments with unexpectedly high LCA taxonomic assignments. For example, Ceratodictyol B was assigned an LCA of ‘2759’, as depicted in Table 1. Taxonomy ID 2759 is ‘Eukaryota’, or eukaryotes. Assignment to this level would result in assignment of Ceratodictyol B to every eukaryote metabolome library. This occurred due to Ceratodictyol B being reported from precisely two species, each listed by two sources: a marine sponge, Haliclona cymaeformis, and a red algae, Ceratodictyon spongiosum. Ceratodictyon spongiosum can form symbioses with sponges, calling into question whether Haliclona is producing Ceratodictyol B, or harboring Ceratodictyon, which is producing Ceratodictyol B. In fact, the paper describing these results does not distinguish between the two.30 This result can therefore be interpreted as a false positive assignment of LCA at a much higher taxonomic level than is warranted. The LCA concept is built on a premise that each taxon–CID association reflects phylogeny, while, in practice, the LCA approach implemented is dependent on taxonomy as a surrogate, and the case in question reflects complex cross-taxa symbioses that taxonomy doesn't capture well.
To enable customization in the assignment of LCA, the build.cid.lca function was provided an option, ‘min.taxid.table.length’, which can prevent such spurious LCA assignments. In the case of Ceratodictyol B, it can be seen that there are 2 taxonomic identifiers at each level – species, genus, family, phylum, and kingdom, and only when we arrive at domain do we find that the number of unique taxonomy identifiers drops to 1. There are very few levels of unique taxonomy ID count across taxonomy levels, which is used as an indicator that there are few records that are taxonomically (and presumably phylogenetically) isolated from each other. For each taxonomic level, the number of unique taxonomy identifiers that map to the CID is calculated, and the frequency examined. All instances with a frequency of ‘0’ (all NA) are removed. If the length of the resulting tables is less than or equal to min.taxid.table.length, then the LCA is assigned within the lowest taxonomic level with the most frequently observed n.taxa, where n.taxa is the number of taxa within that taxonomic level. The default value for min.taxid.table.length is ‘3’. In this way, metabolites which are observed in very few taxa that are very disparate in their taxonomic distance from each other can be assigned to two (or infrequently more) LCAs.
In the example depicted in Table 1, Ceratodictyol B is found in two species, genera, families, and phyla. As such, the most frequently observed n.taxa is ‘2’. The lowest common taxonomic level for all taxonomic levels with n.taxa = 2 is species, so the LCA is assigned within the level ‘species’. In this case, there are two species, and Ceratodictyol B is assigned two LCA values, one for each species. Hypothetically, if there were three Ceratodictyon spp. linked to Ceratodictyol B, the LCA would be assigned within the level ‘genus’ instead. Since there is still only one species within the genus Haliclona, the first LCA would still be assigned to the species Haliclona cymaeformis (taxonomy ID = 1385788). For the second taxa set, there are three species of Ceratodictyon, the LCA for which would then be assigned at the genus level (taxonomy ID = 38330).
After running the build.cid.lca function, setting the min.taxid.table.length equal to three, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859 metabolites were assigned an LCA equal to 1. When considering the construction of in-house libraries of metabolites, this list can be used to ensure that the investment in metabolite standards is spent on metabolites most likely to be observed across all sample types. The full table of assigned primary metabolites can be regenerated at any time by using the pubchem.bio package, ensuring that as the data in PubChem grow, so can the list of primary metabolites.
859 metabolites were assigned an LCA equal to 1. When considering the construction of in-house libraries of metabolites, this list can be used to ensure that the investment in metabolite standards is spent on metabolites most likely to be observed across all sample types. The full table of assigned primary metabolites can be regenerated at any time by using the pubchem.bio package, ensuring that as the data in PubChem grow, so can the list of primary metabolites.
The build.pubchem.bio function was used to create a biological metabolome library, using default values, which include the use of the biologically associated datasource including Metabolomics Workbench, Human Metabolome Database (HMDB), ChEBI, LIPID MAPS, and MassBank of North America (MoNA). XLogP, AcidicGroupCount, BasicGroupCound, and TPSA for each metabolite were predicted using rcdk.18 This biological subset of PubChem took approximately 1.1 hours to build, and resulted in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 268
268![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 778 metabolites. The molecular weight distribution of metabolites is clearly altered between PubChem (Fig. 1a) and the pubchem.bio dataset (Fig. 1b).
778 metabolites. The molecular weight distribution of metabolites is clearly altered between PubChem (Fig. 1a) and the pubchem.bio dataset (Fig. 1b).
Fig. 1c plots the relationship between the number of taxa that map to a given metabolite and the number of metabolites that also have an LCA of ‘1’ (root). A strong log-linear relationship is observed at taxa counts above 25, before dropping between 25 and 28 and then falling rapidly above 28. These inflection points in the curve represent some combination of undersampling of taxa–structure relationships, and the loss of metabolic conservation across taxa – we cannot disentangle the two from the available data. The default value for assignment of a metabolite as ‘primary’ in this function is therefore conservatively set to 25, although this value is a variable in the function that can be further refined as more and more data are incorporated into PubChem. At a minimum frequency of 25 taxonomic occurrences, there are 1069 metabolites considered as primary. This value falls between the ∼200 reported recently in a survey of mammalian metabolomics data31 and the ∼6000 reported in a genome-informed approach.32 As can be seen from Fig. 1c, even within the pubchem.bio package, the value can range from several hundred to over 2000. This package can help to inform on core metabolic functionalities, but is not going to provide an unambiguous answer to this important evolutionary question.
To demonstrate the utility of taxonomy filtering and scoring, a food example is considered. Salsa is comprised largely of tomato, pepper, cilantro, onion and garlic. These foods map to Taxonomy database IDs of 4081, 4072, 4047, 4679, and 4682. Building the salsa metabolome from these five ingredients took 6.5 minutes. For the ingredients listed above, there were 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 162, 17
162, 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058, 16
058, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 895, 17
895, 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062, and 17
062, and 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 metabolites mapped, respectively. In total, 18
027 metabolites mapped, respectively. In total, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 116 metabolites are mapped to these five species, from four different genera. Tomato and pepper are closely related, and 98% of all tomato metabolites are also in the pepper metabolome. Garlic and onion are also closely related to each other, sharing the same genus, Allium. 99.7% of garlic metabolites are also present in the onion metabolome. Tomato and garlic are more distantly related, with a common taxonomic ancestor at the kingdom level, Viridiplantae. 97% of all tomato metabolites are also present in the garlic metabolome. A total of 243
116 metabolites are mapped to these five species, from four different genera. Tomato and pepper are closely related, and 98% of all tomato metabolites are also in the pepper metabolome. Garlic and onion are also closely related to each other, sharing the same genus, Allium. 99.7% of garlic metabolites are also present in the onion metabolome. Tomato and garlic are more distantly related, with a common taxonomic ancestor at the kingdom level, Viridiplantae. 97% of all tomato metabolites are also present in the garlic metabolome. A total of 243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 792 metabolites are assigned an aggregate taxonomy score, with scores ranging from 0 (extremely unlikely to be found given the five species listed) to 1 (highly plausible, as they are metabolites that map to at least one of the species). Intermediate scores represent increasing probability that a metabolite may be present, given imperfect knowledge of species-specific metabolism (Fig. 2). Of course, the small molecule composition of salsa will ultimately be additionally impacted by cooking and preservation, so the reported library should be viewed as the potential small molecule components of salsa which are derived directly from the ingredients, not comprehensive of, for example, Maillard products that may form.
792 metabolites are assigned an aggregate taxonomy score, with scores ranging from 0 (extremely unlikely to be found given the five species listed) to 1 (highly plausible, as they are metabolites that map to at least one of the species). Intermediate scores represent increasing probability that a metabolite may be present, given imperfect knowledge of species-specific metabolism (Fig. 2). Of course, the small molecule composition of salsa will ultimately be additionally impacted by cooking and preservation, so the reported library should be viewed as the potential small molecule components of salsa which are derived directly from the ingredients, not comprehensive of, for example, Maillard products that may form.
Usage discussion
The pubchem.bio package is performing tasks that would take humans much longer to do. The process of assigning the lowest common ancestor to PubChem structures is relatively novel, but based on the same principles as chemotaxonomy – a field with decades of applications,33 and highly similar to that used in metaproteomics studies.34 The LCA approach turns a limitation of chemotaxonomy – that even specialized metabolites are often not unique to a given species – into a useful trait, enabling generalization of metabolite distributions for creating more comprehensive metabolome libraries for any species, even those poorly studied in the past. However, it must be noted that the LCA approach is difficult to validate and optimize, given the incomplete knowledge of any given biological sample's metabolic components. Default values are a reasonable starting point for users, and should provide relatively conservative results – a database that utilizes all available data to generate a comprehensive list of analytes that may plausibly be observed.Taxonomic and biological data in PubChem are both incomplete (false negative) and can contain errors (false positive). Each of these types of errors can result in inaccurate taxon-specific metabolome data and assigned LCA. For example, capsaicin is widely considered to be a specialized metabolite of peppers, genus Capsicum. The ‘taxonomy’ section of the PubChem webpage for capsaicin lists numerous Capsicum species, but also the prokaryote Streptomyces roseofulvus, submitted by the NPASS database. The original reference supporting this association is not apparent, so it is difficult to evaluate this claim. That said, if the user selects both Lotus and NPASS databases when building the LCA, the LCA for capsaicin may be ‘1’ – the root of all biology, depending on the assigned ‘min.taxid.table.length’ value used. However, if the users opt to use only the more selective Lotus database (the current default value), the LCA is returned as ‘4071’, the Taxonomy ID for the Capsicum genus.
Biological databases and pathway databases may each incorporate exogenous compounds into their data in a taxonomy-informed manner. Capsaicin can be degraded by E. coli, for example, and therefore can be reported as an E. coli metabolite, despite the fact that E. coli is not known to be able to synthesize capsaicin. In theory, if all reaction data for a given species were available in computer readable format, one could remove metabolites that are not considered metabolic products of a reaction – this is an opportunity for additional development in the future.
The taxonomy scoring applied here is based on taxonomic relationships as described by decades of taxonomic research. It should be noted that the taxonomic approaches are not necessarily consistent across all clades. While taxonomy is a representation of phylogeny, the scoring algorithm of pubchem.bio is based on what must be considered an imperfect taxonomic representation of phylogenic time and distance between species.
The work here demonstrates that the pubchem.bio package is able to return meaningful results to users, enabling users to create taxonomy-informed libraries in an automated manner. This does not mean that the process is completely objective – the user will need to select which sources to use and to assign an appropriate ‘min.taxid.table.length’ value, for example. The ‘correct’ metabolome library is one that serves the need of the user, whether that be the minimal most confident metabolome, the maximum plausible metabolome, or some intermediate level. The package is designed to enable users to build the library they need, depending on the circumstance. As the resources in PubChem grow, so does the ability for pubchem.bio to convert that collected knowledge into metabolome libraries.
The pubchem.bio package fills a functional void. There are no other resources to enable rapid generation of customizable metabolome libraries for any species. While individual researchers have manually done this in the past, pubchem.bio enables what would have taken hours, at best, to be done in minutes. The most similar tool available, an R package called tima, available via zenodo,35 performs comparable tasks to pubchem.bio, but does so using more limited resources, and does not appear to utilize a lowest common ancestor extrapolation approach. There are also tools for retrieving and handling existing structure libraries. The CompoundDB R package, which pubchem.bio can export metabolite libraries to, is useful for retrieving and storing structure data from a few select well-curated database sources, such as HMDB, and provides an extensible data structure to integrate into other RforMassSpectrometry packages.36
Conclusion
Using freely available data from PubChem, derived through many invaluable sources that have been contributed by individual database efforts, the pubchem.bio package enables the creation of metabolomics-ready libraries fully informed by biological and taxonomic digital resources. After an initial setup phase requiring several hours of computer time, each new species metabolome library can be generated in minutes. These species-specific metabolome libraries have the potential to dramatically improve annotation accuracy.Future directions
Additional metadata are downloaded as part of the get.pubchem.ftp function, notably MeSH data, full CID-pathway membership details, CID–substance relationships, synonyms, and CAS numbers. These metadata have potential utility in, for example, pathway analysis.37 The pubchem.bio package makes no effort to incorporate any additional exogenous compounds beyond what is already incorporated into biological databases such as CheBI or HMDB. Future work may incorporate the data from PubChem Lite, which is focused on these exogenous analytes. Additionally, the use of reactions rather than pathways may enable the removal of exogenous catabolic reactants from the species-specific metabolome output, when desired.Conflicts of interest
There are no conflicts to declare.Data availability
This manuscript describes no new analytical data. Rather it describes a new informatic resource, the R pubchem.bio package. https://github.com/cbroeckl/pubchem.bioSoftware availability: The pubchem.bio package is available via CRAN under a GPL-3 license.
Acknowledgements
I would like to acknowledge the scientists who compiled datasets and submitted them to PubChem,7–10,14,16,17,22–29 without which there would be no pubchem.bio. I would also like to acknowledge Paul Theissen with PubChem, who patiently provided valuable guidance for accessing PubChem programmatically. CDB works in the Colorado State University Analytical Resources Core, Research Resource ID (RRID: SCR_021758).References
- R. J. Bino, R. D. Hall, O. Fiehn, J. Kopka, K. Saito, J. Draper, B. J. Nikolau, P. Mendes, U. Roessner-Tunali, M. H. Beale, R. N. Trethewey, B. M. Lange, E. S. Wurtele and L. W. Sumner, Potential of Metabolomics as a Functional Genomics Tool, Trends Plant Sci., 2004, 9(9), 418–425, DOI:10.1016/j.tplants.2004.07.004.
- N. F. de Jonge, K. Mildau, D. Meijer, J. J. R. Louwen, C. Bueschl, F. Huber and J. J. J. van der Hooft, Good Practices and Recommendations for Using and Benchmarking Computational Metabolomics Metabolite Annotation Tools, Metabolomics, 2022, 18(12), 103, DOI:10.1007/s11306-022-01963-y.
- A. Delabrière, C. Gianfrotta, S. Dechaumet, A. Damont, T. Hautbergue, P. Roger, E. L. Jamin, O. Puel, C. Junot, F. Fenaille and E. A. Thévenot, mineMS2: Annotation of Spectral Libraries with Exact Fragmentation Patterns, J. Cheminf., 2025, 17(1), 111, DOI:10.1186/s13321-025-01051-y.
- C. H. Chang, S. C. Schwartz, A. K. Im, K. J. Bloodsworth, B.-J. M. Webb-Robertson, R. G. Ewing, T. O. Metz and D. H. Ross, Assessing the Impact of Measurement Precision on Metabolite Identification Probability in Multidimensional Mass Spectrometry-Based, Reference-Free Metabolomics, Anal. Chem., 2025, 97(26), 13861–13871, DOI:10.1021/acs.analchem.5c01067.
- R. S. Bohacek, C. McMartin and W. C. Guida, The Art and Practice of Structure-Based Drug Design: A Molecular Modeling Perspective, Med. Res. Rev., 1996, 16(1), 3–50, DOI:10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6.
- A. Elapavalore, D. H. Ross, V. Grouès, D. Aurich, A. M. Krinsky, S. Kim, P. A. Thiessen, J. Zhang, J. N. Dodds, E. S. Baker, E. E. Bolton, L. Xu and E. L. Schymanski, PubChemLite Plus Collision Cross Section (CCS) Values for Enhanced Interpretation of Nontarget Environmental Data, Environ. Sci. Technol. Lett., 2025, 12(2), 166–174, DOI:10.1021/acs.estlett.4c01003.
- D. S. Wishart, A. Guo, E. Oler, F. Wang, A. Anjum, H. Peters, R. Dizon, Z. Sayeeda, S. Tian, B. L. Lee, M. Berjanskii, R. Mah, M. Yamamoto, J. Jovel, C. Torres-Calzada, M. Hiebert-Giesbrecht, V. W. Lui, D. Varshavi, D. Varshavi, D. Allen, D. Arndt, N. Khetarpal, A. Sivakumaran, K. Harford, S. Sanford, K. Yee, X. Cao, Z. Budinski, J. Liigand, L. Zhang, J. Zheng, R. Mandal, N. Karu, M. Dambrova, H. B. Schiöth, R. Greiner and V. Gautam, HMDB 5.0: The Human Metabolome Database for 2022, Nucleic Acids Res., 2022, 50(D1), D622–D631, DOI:10.1093/nar/gkab1062.
- V. Chandrasekhar, K. Rajan, S. R. S. Kanakam, N. Sharma, V. Weißenborn, J. Schaub and C. Steinbeck, COCONUT 2.0: A Comprehensive Overhaul and Curation of the Collection of Open Natural Products Database, Nucleic Acids Res., 2025, 53(D1), D634–D643, DOI:10.1093/nar/gkae1063.
- A. Rutz, M. Sorokina, J. Galgonek, D. Mietchen, E. Willighagen, A. Gaudry, J. G. Graham, R. Stephan, R. Page, J. Vondrášek, C. Steinbeck, G. F. Pauli, J.-L. Wolfender, J. Bisson and P.-M. Allard, The LOTUS Initiative for Open Knowledge Management in Natural Products Research, eLife, 2022, 11, e70780, DOI:10.7554/eLife.70780.
- H. Zhao, Y. Yang, S. Wang, X. Yang, K. Zhou, C. Xu, X. Zhang, J. Fan, D. Hou, X. Li, H. Lin, Y. Tan, S. Wang, X.-Y. Chu, D. Zhuoma, F. Zhang, D. Ju, X. Zeng and Y. Z. Chen, NPASS Database Update 2023: Quantitative Natural Product Activity and Species Source Database for Biomedical Research, Nucleic Acids Res., 2023, 51(D1), D621–D628, DOI:10.1093/nar/gkac1069.
- A. Rutz, M. Dounoue-Kubo, S. Ollivier, J. Bisson, M. Bagheri, T. Saesong, S. N. Ebrahimi, K. Ingkaninan, J.-L. Wolfender and P.-M. Allard, Taxonomically Informed Scoring Enhances Confidence in Natural Products Annotation, Front. Plant Sci., 2019, 10 DOI:10.3389/fpls.2019.01329.
- S. Zuffa, R. Schmid, A. Bauermeister, P. W. Gomes, A. M. Caraballo-Rodriguez, Y. El Abiead, A. T. Aron, E. C. Gentry, J. Zemlin, M. J. Meehan, N. E. Avalon, R. H. Cichewicz, E. Buzun, M. C. Terrazas, C.-Y. Hsu, R. Oles, A. V. Ayala, J. Zhao, H. Chu, M. C. M. Kuijpers, S. L. Jackrel, F. Tugizimana, L. P. Nephali, I. A. Dubery, N. E. Madala, E. A. Moreira, L. V. Costa-Lotufo, N. P. Lopes, P. Rezende-Teixeira, P. C. Jimenez, B. Rimal, A. D. Patterson, M. F. Traxler, R. d. C. Pessotti, D. Alvarado-Villalobos, G. Tamayo-Castillo, P. Chaverri, E. Escudero-Leyva, L.-M. Quiros-Guerrero, A. J. Bory, J. Joubert, A. Rutz, J.-L. Wolfender, P.-M. Allard, A. Sichert, S. Pontrelli, B. S. Pullman, N. Bandeira, W. H. Gerwick, K. Gindro, J. Massana-Codina, B. C. Wagner, K. Forchhammer, D. Petras, N. Aiosa, N. Garg, M. Liebeke, P. Bourceau, K. B. Kang, H. Gadhavi, L. P. S. de Carvalho, M. Silva Dos Santos, A. I. Pérez-Lorente, C. Molina-Santiago, D. Romero, R. Franke, M. Brönstrup, A. Vera Ponce de León, P. B. Pope, S. L. La Rosa, G. La Barbera, H. M. Roager, M. F. Laursen, F. Hammerle, B. Siewert, U. Peintner, C. Licona-Cassani, L. Rodriguez-Orduña, E. Rampler, F. Hildebrand, G. Koellensperger, H. Schoeny, K. Hohenwallner, L. Panzenboeck, R. Gregor, E. C. O'Neill, E. T. Roxborough, J. Odoi, N. J. Bale, S. Ding, J. S. Sinninghe Damsté, X. L. Guan, J. J. Cui, K.-S. Ju, D. B. Silva, F. M. R. Silva, G. F. da Silva, H. H. F. Koolen, C. Grundmann, J. A. Clement, H. Mohimani, K. Broders, K. L. McPhail, S. E. Ober-Singleton, C. M. Rath, D. McDonald, R. Knight, M. Wang and P. C. Dorrestein, microbeMASST: A Taxonomically Informed Mass Spectrometry Search Tool for Microbial Metabolomics Data, Nat. Microbiol., 2024, 9(2), 336–345, DOI:10.1038/s41564-023-01575-9.
- F. S. Bragagnolo, E. Ibáñez, A. Cifuentes and M. A. Rostagno, Building Your Own Database for Foodomics: Tips and Tricks, Food Chem. Int., 2025, 1(2), 183–186, DOI:10.1002/fci2.70009.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, PubChem 2023 Update, Nucleic Acids Res., 2023, 51(D1), D1373–D1380, DOI:10.1093/nar/gkac956.
- T. Barrett, M. Dowle, A. Srinivasan, J. Gorecki, M. Chirico, T. Hocking, B. Schwendinger and I. Krylov, Data.Table: Extension of 'data.Frame‘, 2025. DOI:10.32614/CRAN.package.data.table.
- A. Agrawal, H. Balcı, K. Hanspers, S. L. Coort, M. Martens, D. N. Slenter, F. Ehrhart, D. Digles, A. Waagmeester, I. Wassink, T. Abbassi-Daloii, E. N. Lopes, A. Iyer, J. M. Acosta, L. G. Willighagen, K. Nishida, A. Riutta, H. Basaric, C. T. Evelo, E. L. Willighagen, M. Kutmon and A. R. Pico, WikiPathways 2024: Next Generation Pathway Database, Nucleic Acids Res., 2024, 52(D1), D679–D689, DOI:10.1093/nar/gkad960.
- C. L. Schoch, S. Ciufo, M. Domrachev, C. L. Hotton, S. Kannan, R. Khovanskaya, D. Leipe, R. Mcveigh, K. O'Neill, B. Robbertse, S. Sharma, V. Soussov, J. P. Sullivan, L. Sun, S. Turner and I. Karsch-Mizrachi, NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools, Database, 2020, 2020, baaa062, DOI:10.1093/database/baaa062.
- R. Guha, Chemical Informatics Functionality in R, J. Stat. Softw., 2007, 18(5), 1–16 Search PubMed.
- J. Hastings, G. Owen, A. Dekker, M. Ennis, N. Kale, V. Muthukrishnan, S. Turner, N. Swainston, P. Mendes and C. Steinbeck, ChEBI in 2016: Improved Services and an Expanding Collection of Metabolites, Nucleic Acids Res., 2016, 44(D1), D1214–D1219, DOI:10.1093/nar/gkv1031.
- V. B. O'Donnell, E. A. Dennis, M. J. O. Wakelam and S. Subramaniam, LIPID MAPS: Serving the next Generation of Lipid Researchers with Tools, Resources, Data, and Training, Sci. Signaling, 2019, 12(563), eaaw2964, DOI:10.1126/scisignal.aaw2964.
- G. Wohlgemuth, S. S. Mehta, R. F. Mejia, S. Neumann, D. Pedrosa, T. Pluskal, E. L. Schymanski, E. L. Willighagen, M. Wilson, D. S. Wishart, M. Arita, P. C. Dorrestein, N. Bandeira, M. Wang, T. Schulze, R. M. Salek, C. Steinbeck, V. C. Nainala, R. Mistrik, T. Nishioka and O. Fiehn, SPLASH, a Hashed Identifier for Mass Spectra, Nat. Biotechnol., 2016, 34(11), 1099–1101, DOI:10.1038/nbt.3689.
- D. S. Wishart, R. Kruger, A. Sivakumaran, K. Harford, S. Sanford, R. Doshi, N. Khetarpal, O. Fatokun, D. Doucet, A. Zubkowski, H. Jackson, G. Sykes, M. Ramirez-Gaona, A. Marcu, C. Li, K. Yee, C. Garros, D. Y. Rayat, J. Coleongco, T. Nandyala, V. Gautam and E. Oler, PathBank 2.0—the Pathway Database for Model Organism Metabolomics, Nucleic Acids Res., 2024, 52(D1), D654–D662, DOI:10.1093/nar/gkad1041.
- C. Hawkins, B. Xue, F. Yasmin, G. Wyatt, P. Zerbe and S. Y. Rhee, Plant Metabolic Network 16: Expansion of Underrepresented Plant Groups and Experimentally Supported Enzyme Data, Nucleic Acids Res., 2025, 53(D1), D1606–D1613, DOI:10.1093/nar/gkae991.
- P. Gupta, S. Naithani, J. Preece, S. Kim, T. Cheng, P. D'Eustachio, J. Elser, E. E. Bolton and P. Jaiswal, Plant Reactome and PubChem: The Plant Pathway and (Bio)Chemical Entity Knowledgebases, in Plant Bioinformatics: Methods and Protocols, ed. D. Edwards, Springer US, New York, NY, 2022, pp. 511–525. DOI:10.1007/978-1-0716-2067-0_27.
- P. D. Karp, R. Billington, R. Caspi, C. A. Fulcher, M. Latendresse, A. Kothari, I. M. Keseler, M. Krummenacker, P. E. Midford, Q. Ong, W. K. Ong, S. M. Paley and P. Subhraveti, The BioCyc Collection of Microbial Genomes and Metabolic Pathways, Briefings Bioinf., 2019, 20(4), 1085–1093, DOI:10.1093/bib/bbx085.
- K. Rothfels, M. Milacic, L. Matthews, R. Haw, C. Sevilla, M. Gillespie, R. Stephan, C. Gong, E. Ragueneau, B. May, V. Shamovsky, A. Wright, J. Weiser, D. Beavers, P. Conley, K. Tiwari, B. Jassal, J. Griss, A. Senff-Ribeiro, T. Brunson, R. Petryszak, H. Hermjakob, P. D'Eustachio, G. Wu and L. Stein, Using the Reactome Database, Curr. Protoc., 2023, 3(4), e722, DOI:10.1002/cpz1.722.
- D. S. Wishart, Knowledge Translation and Knowledge Mobilization from the FoodBAll Project, Appl. Physiol., Nutr., Metab., 2024, 49(9), 1279–1285, DOI:10.1139/apnm-2023-0573.
- Y. Nakamura, F. Mochamad Afendi, A. Kawsar Parvin, N. Ono, K. Tanaka, A. Hirai Morita, T. Sato, T. Sugiura, M. Altaf-Ul-Amin and S. Kanaya, KNApSAcK Metabolite Activity Database for Retrieving the Relationships Between Metabolites and Biological Activities, Plant Cell Physiol., 2014, 55(1), e7, DOI:10.1093/pcp/pct176.
- J. A. van Santen, E. F. Poynton, D. Iskakova, E. McMann, T. A. Alsup, T. N. Clark, C. H. Fergusson, D. P. Fewer, A. H. Hughes, C. A. McCadden, J. Parra, S. Soldatou, J. D. Rudolf, E. M.-L. Janssen, K. R. Duncan and R. G. Linington, The Natural Products Atlas 2.0: A Database of Microbially-Derived Natural Products, Nucleic Acids Res., 2022, 50(D1), D1317–D1323, DOI:10.1093/nar/gkab941.
- T. Akiyama, R. Ueoka, R. W. M. van Soest and S. Matsunaga, Ceratodictyols, 1-Glyceryl Ethers from the Red Alga–Sponge Association Ceratodictyon Spongiosum/Haliclona Cymaeformis, J. Nat. Prod., 2009, 72(8), 1552–1554, DOI:10.1021/np900355m.
- O. Liska, G. Boross, C. Rocabert, B. Szappanos, R. Tengölics and B. Papp, Principles of Metabolome Conservation in Animals, Proc. Natl. Acad. Sci. U. S. A., 2023, 120(35), e2302147120, DOI:10.1073/pnas.2302147120.
- J. M. Peregrín-Alvarez, C. Sanford and J. Parkinson, The Conservation and Evolutionary Modularity of Metabolism, Genome Biol., 2009, 10(6), R63, DOI:10.1186/gb-2009-10-6-r63.
- P. M. Smith, The Chemotaxonomy of Plants, 1976 Search PubMed.
- T. Vande Moortele, B. Devlaminck, S. Van de Vyver, T. Van Den Bossche, L. Martens, P. Dawyndt, B. Mesuere and P. Verschaffelt, Unipept in 2024: Expanding Metaproteomics Analysis with Support for Missed Cleavages and Semitryptic and Nontryptic Peptides, J. Proteome Res., 2025, 24(2), 949–954, DOI:10.1021/acs.jproteome.4c00848.
- A. Rutz, Tima: Taxonomically Informed Metabolite Annotation, 2024. DOI:10.5281/zenodo.14515116.
- M. Witting and J. Rainer, Bio- and Chemoinformatic Approaches for Metabolomics Data Analysis, Methods Mol. Biol., 2025, 2891, 67–89, DOI:10.1007/978-1-0716-4334-1_4.
- C. Wieder, C. Frainay, N. Poupin, P. Rodríguez-Mier, F. Vinson, J. Cooke, R. P. Lai, J. G. Bundy, F. Jourdan and T. Ebbels, Pathway Analysis in Metabolomics: Recommendations for the Use of over-Representation Analysis, PLoS Comput. Biol., 2021, 17(9), e1009105, DOI:10.1371/journal.pcbi.1009105.
| This journal is © The Royal Society of Chemistry 2026 |