Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Development of a tannic acid- and silicate ion-functionalized PVA–starch composite hydrogel for in situ skeletal muscle repairing
Longkang
Li
a,
Huipeng
Li
b,
Zhentian
Diao
a,
Huan
Zhou
*b,
Yanjie
Bai
*bc and
Lei
Yang
b
aSchool of Materials Science and Engineering, Hebei University of Technology, Tianjin, 300130, China
bCenter for Health Science and Engineering, Hebei Key Laboratory of Biomaterials and Smart Theranostics, School of Health Sciences and Biomedical Engineering, Hebei University of Technology, Tianjin, 300130, China. E-mail: zhouhuan@hebut.edu.cn
cDepartment of Chemical Engineering, Hebei University of Technology, Tianjin, 300130, China. E-mail: 2021100@hebut.edu.cn
First published on 6th February 2025
Abstract
Correction for ‘Development of a tannic acid- and silicate ion-functionalized PVA–starch composite hydrogel for in situ skeletal muscle repairing’ by Longkang Li et al., J. Mater. Chem. B, 2024, 12, 3917–3926, https://doi.org/10.1039/D3TB03006G.
The authors note a mistake in Fig. 5 in which the image of PSTS3 in day 3 in Fig. 5C was mistakenly duplicated as Con in day 3. The corrected Fig. 5 is shown here. Meanwhile, corresponding original images are provided in an additional supplementary information file.
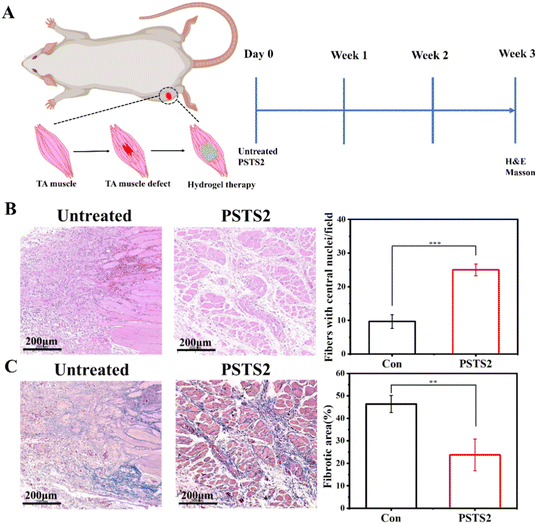
The authors note the sample size used for quantitative analysis in Fig. 7 was not enough. Therefore, the section staining data of all the mice in the Con group and the PSTS group (n = 3) in Fig. 7B and C was reanalyzed with three same-sized regions of the section from the same batch of mice taken into processing. In general, the initial localization of the damaged area was firstly selected based on the difference between normal tissue and damaged tissue. The damaged tissue exhibited a more disorganized structure with a large amount of scar tissue production and a less distinct cellular structure when compared with normal muscle tissue. After that, representative images located in the entire injury region were chosen for analysis, and they were selected according to the following criteria: (1) areas should completely cover the injury region and possess injury characteristics; (2) areas should represent the regions with healing tendency as much as possible; (3) the selected area should be maintained at a consistent size.
The corrected Fig. 7 is shown here. Meanwhile, corresponding original images of all processed sections are provided in an additional supplementary information file.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2025 |