Open Access Article
Open Access ArticleOrgan-targeted drug delivery systems (OTDDS) of poly[(N-acryloylglycine)-co-(N-acryloyl-L-phenylalanine methyl ester)] copolymer library and effective treatment of triple-negative breast cancer†
Sukanya
Patra
a,
Jyotirmayee
b,
Krishan
Kumar
c,
Divya
Pareek
a,
Prem Shankar
Gupta
a,
Anjali Ramsabad
Mourya
 a,
Taniya
Das
a,
Kirti
Wasnik
a,
Malkhey
Verma
b,
Ruchi
Chawla
c,
Tarun
Batra
d and
Pradip
Paik
a,
Taniya
Das
a,
Kirti
Wasnik
a,
Malkhey
Verma
b,
Ruchi
Chawla
c,
Tarun
Batra
d and
Pradip
Paik
 *a
*a
aSchool of Biomedical Engineering, Indian Institute of Technology, (Banaras Hindu University), Varanasi, Uttar Pradesh 221 005, India. E-mail: paik.bme@iitbhu.ac.in
bSchool of Biotechnology, Institute of Science, (Banaras Hindu University), Varanasi, Uttar Pradesh 221 005, India
cDepartment of Pharmaceutical Engineering and Technology, Indian Institute of Technology (Banaras Hindu University), Varanasi, Uttar Pradesh 221 005, India
dDepartment of Oncology, Institute of Medical Sciences (IMS), Banaras Hindu University, Varanasi, Uttar Pradesh 221 005, India
First published on 12th February 2025
Abstract
Organ-targeted drug delivery systems (OTDDS) are essential for the effective treatment of complicated diseases. Triple-negative breast cancer (TNBC) is an aggressive cancer with high mortality and requires targeted therapeutics. This work was aimed at designing a library of polymeric OTDDS with N-acryloyl-glycine (NAG) and N-acryloyl-L-phenylalanine methyl ester (NAPA) [p(NAG-co-NAPA)(x:y)] and screening its in vivo organ-targeting specificity. Among this library, the best p(NAG-co-NAPA)(x:y) NPs with an x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y ratio of 1
y ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and size of 160–210 nm targeted breasts to a high extent compared to other organs and thus were optimized for TNBC treatment. A network pharmacology study was performed, which revealed that 14 genes were responsible for TNBC, and a combination of DHA (targets 6 genes) and piperine (targets 8 genes) drugs was used to optimize the formulation, achieving the maximum therapeutic efficiency against TNBC with an IC50 value of 350 μg mL−1. The designed organ-specific polymeric nanoparticle (NP) library, identification of target genes and proteins responsible for TNBC, and the optimized formulation for effective combination therapy established an effective therapeutic option for TNBC. The findings of this work further demonstrate that this polymeric library of NPs shows exciting therapeutic potential for treating TNBC and presents innovative treatment options for critical diseases of the liver, heart, lungs and kidney.
4 and size of 160–210 nm targeted breasts to a high extent compared to other organs and thus were optimized for TNBC treatment. A network pharmacology study was performed, which revealed that 14 genes were responsible for TNBC, and a combination of DHA (targets 6 genes) and piperine (targets 8 genes) drugs was used to optimize the formulation, achieving the maximum therapeutic efficiency against TNBC with an IC50 value of 350 μg mL−1. The designed organ-specific polymeric nanoparticle (NP) library, identification of target genes and proteins responsible for TNBC, and the optimized formulation for effective combination therapy established an effective therapeutic option for TNBC. The findings of this work further demonstrate that this polymeric library of NPs shows exciting therapeutic potential for treating TNBC and presents innovative treatment options for critical diseases of the liver, heart, lungs and kidney.
1. Introduction
The precise organ-specific delivery of therapeutic biomolecules requires interdisciplinary research among scientists in chemistry and biology, engineers, and clinicians for successful treatment outcomes, given that the surface chemistry of organs is very complicated.1 The development of effective organ-targeted drug delivery systems (OTDDS) necessitates a clear understanding of how the physical and chemical properties of OTDDS influence biological processes at the molecular, cellular and organ levels. OTDDS can deliver precise drug doses for the desired therapeutic outcomes rather than delivering an arbitrary amount of free drug at the targeted site.2 However, for effective treatment, OTDDS must overcome challenges such as poor biodistribution and biological fate.1 Generally, three targeting mechanisms, i.e., passive, active, and endogenous, are considered to engineer OTDDS.3–5 The passive and active targeting mechanisms have been widely studied to date. Alternatively, endogenous targeting is emerging as a promising approach based on how the chemical versatility of nanoparticles (NPs) leads to binding with distinct plasma protein subsets of target organs, directing NPs to specified organs and enhancing their cellular uptake.6,7 However, for specific cellular uptake, controlled chemical modification of NPs is necessary. For instance, 7C1, an oligomer derived from lipidated PEI600, was used for gene silencing in lung epithelial cells, compared to kidney and heart endothelial cells. However, the standardization of the lipidation process is necessary for improved silencing across different organs.8,9 IDD-3, a terpolymer formulated with lipid components, enables lung-selective mRNA delivery. Interestingly, compounds sharing the same polymerization degree as IDD-3 failed to target the lungs when their amino-alcohol building block was altered.10 Furthermore, PEI modified with vitamin A was reported together with enhanced retinol binding protein 4 in its protein corona, targeting hepatic stellate cells for liver fibrosis treatment.11 Similarly, phospholipidation of cationic polymers could facilitate mRNA delivery to the spleen and lymph nodes.12 However, identifying NPs that can target inaccessible tissues such as muscles and the heart, central nervous system, and gastrointestinal tract is challenging.1 In this case, although chemical modification can enhance the target specificity of the delivery vehicles, can also increase the toxicity and side effects in the long run. These insights motivated us to synthesize copolymer NPs with varying ratios (x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y) of biologically safe monomers (amino acid based) without using any endogenous or exogenous ligands for enhanced organ-specificity.
y) of biologically safe monomers (amino acid based) without using any endogenous or exogenous ligands for enhanced organ-specificity.
Several OTDDS have been approved by the FDA for clinical use including viral vectors, molecular conjugates, antibody–drug conjugates, and chemically modified polymer NPs. Among them, polymer NPs are particularly useful due to their ability to load various drugs separately or in combination with minimal immunogenicity, enhanced capacity of endosomal escape,12 cell and tissue tropism10 and easy modulation of their properties through controlled chemical synthesis.1 Amphiphilic copolymer-based nanocarriers are gaining attention for hydrophobic drug delivery in cancer treatment because of their synthetic versatility, favourable size, biocompatibility, and stability.13–15 Compared to free drugs, nano-scaled polymeric carriers can enhance their accumulation in solid tumor cells via the enhanced permeability and retention (EPR) effect.16 Furthermore, OTDDS can improve the drug delivery to metastatic cancer cells and potentially bypass permeability barriers. However, despite the development of numerous polymeric nanovehicles for targeted or receptor-mediated (exogenous ligands) drug delivery,17 achieving the expected anticancer efficacy remains a challenge due to off-target effects.18 The key feature of polymeric OTDDS is their organ-targeted delivery, which can be engineered by varying the ratios of the selected monomers, which is the main focus of this work. By varying the x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y ratio, a library of copolymer NPs i.e. organ-targeted drug delivery nanoparticles (OTDDN) can be generated, and subsequently the selection of a particular OTDDN can become dominant for the treatment of invasive cancers such as triple-negative breast cancer (TNBC).
y ratio, a library of copolymer NPs i.e. organ-targeted drug delivery nanoparticles (OTDDN) can be generated, and subsequently the selection of a particular OTDDN can become dominant for the treatment of invasive cancers such as triple-negative breast cancer (TNBC).
Considering different-organ based diseases, TNBC is the most prevalent malignancy and the leading cause of cancer-related mortality (lifetime is 18 months from detection) among women worldwide, accounting 15%–20% of breast cancer cases.19–21 It has the worst prognosis regarding tumor relapse and patient survival, making it a clinically aggressive cancer with limited therapeutic options.22,23 This highlights the urgent need for developing combinatorial therapies for TNBC using suitable copolymer-based drug delivery system OTDDN to eliminate chemoresistance issues.24–26 To address this, a combination delivery of DHA and piperine can be used for better efficacy. Dihydroartemisinin (DHA), a hydrophobic metabolite of artemisinin, shows significant potential in treating TNBC.27,28 Also, piperine, a hydrophobic alkaloid from black pepper (Piper nigrum), has demonstrated anticancer effects.29–31 Thus, treatment with a combination of DHA and piperine can be an effective strategy for TNBC through targeted drug delivery systems (TDDS) from a designed copolymeric NP library. This option can potentially delay TNBC adaptation, while minimizing the severe side effects associated with free drug combinations.32–39 Previously, we also reported the development of biocompatible acrylate-amino acid-based polymers for various therapeutic applications.14,40–44 We reported that poly(N-acryloyl-L-phenylalanine methyl ester) [p(NAPA)] NPs exhibit adjuvant properties on the innate immune system14 and porous poly(N-acryloyl-glycine) [p(NAG)] NPs serve as an excellent drug delivery carrier with enhanced anti-inflammatory effects.40,43 Presently, our idea of designing a library of copolymer-based OTDDN without depending on organ-targeting mechanisms can be a further game-changing option for the treatment of TNBC.
Accordingly, this work reports the synthesis of an OTDDN library of porous amphiphilic copolymer NPs, specifically [p(NAG-co-NAPA)(x:y)], using N-acryloyl glycine and N-acryloyl-L-phenylalanine methyl ester as monomers in varying molar ratios (x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y). Furthermore, the physicochemical properties of OTDDNs were studied to confirm their structural and chemical functionality and further evaluate their efficiency to target different organs. The targeting efficiency of these copolymer NPs was assessed based on in vivo organ-based biodistribution studies. The [p(NAG-co-NAPA)(1
y). Furthermore, the physicochemical properties of OTDDNs were studied to confirm their structural and chemical functionality and further evaluate their efficiency to target different organs. The targeting efficiency of these copolymer NPs was assessed based on in vivo organ-based biodistribution studies. The [p(NAG-co-NAPA)(1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4)] NPs demonstrated the maximum targeting efficiency to breast tissue, which facilitated the loading of DHA and piperine individually or in combination, and exhibited pH-based drug release behavior. Also, in silico studies, including network pharmacology and molecular docking, were performed to predict the gene targets for TNBC. Additionally, the in vitro IC50 values of nanoformulations (NFs) on the MDA-MB-231 cell line were analyzed to assess the TNBC inhibition efficiency and mechanisms through quantitative estimation of the reactive oxygen species (ROS), mitochondrial membrane potential assay, cell cycle arrest, and apoptosis via flow cytometry. Finally, the future direction is proposed for the use of various OTDDNs from this library, which can be effective for several organ-targeted therapies.
4)] NPs demonstrated the maximum targeting efficiency to breast tissue, which facilitated the loading of DHA and piperine individually or in combination, and exhibited pH-based drug release behavior. Also, in silico studies, including network pharmacology and molecular docking, were performed to predict the gene targets for TNBC. Additionally, the in vitro IC50 values of nanoformulations (NFs) on the MDA-MB-231 cell line were analyzed to assess the TNBC inhibition efficiency and mechanisms through quantitative estimation of the reactive oxygen species (ROS), mitochondrial membrane potential assay, cell cycle arrest, and apoptosis via flow cytometry. Finally, the future direction is proposed for the use of various OTDDNs from this library, which can be effective for several organ-targeted therapies.
2. Results and discussion
2.1 Synthesis and physicochemical characterization of monomers, homopolymers and library of OTDDNs
To develop a library of OTDDNs of amino acid-based amphiphilic copolymers, N-acryloyl glycine (NAG) and N-acryloyl-L-phenylalanine methyl ester (NAPA) monomers were used. The monomers were synthesized (yield 60–65%) and their structure confirmed through 1H (Fig. S2a and S3a, ESI†), 13C NMR (Fig. S2b and S3b, ESI†) and FTIR (Fig. S4, ESI†), respectively. The monomer synthesis process followed the Shorten–Baumann reaction approach, where the amine group of the monomers reacts with acryloyl chloride in basic medium to form an amide bond. In the final stage of washing or extraction, ethyl acetate was used as an organic phase to improve the yield percentage of monomers. Further, the free radical miniemulsion polymerization method (Fig. S1, ESI†) was used to synthesize a library of [p(NAG-co-NAPA)(x:y)] copolymers. p(NAG) and p(NAPA) homopolymers were synthesized and used as references. Here, hexadecane (HD) was used as a co-stabilizer of the droplets formed during the emulsion process, which prevented quiescent crystallization and grain growth. Initially, the monomers were dispersed in the oil phase (toluene), forming monomer droplets. AIBN, a radical initiator, was used to initiate the polymerization reaction at ∼70–75 °C. The oily core surrounded by a thin polymeric membrane was stabilized by crosslinking the monomer droplets with DVB, which helped generate pores and provide mechanical strength in the NPs. To generate a library of copolymers, the NAG to NAPA wt% ratio was varied (Table 1) and a series of reactions was performed. Additionally, SDS was used as a surfactant to avoid aggregation during the emulsion process. AIBN initiated the polymerization process by interacting with the terminal sp2-hybridized carbon of NAPA, NAG, and DVB, forming radicals. This led to chain propagation and crosslinking, resulting in the formation of the amphiphilic copolymer [p(NAG-co-NAPA)(x:y)] NPs. The carboxylic acid groups of the NAG monomer formed a hydrophilic shell, while its phenyl rings created a hydrophobic core, potentially forming dense-core NPs with pores.| Nanoparticle | Size (d. nm) | Zeta potential (mV) | PDI | EE% | LE% |
|---|---|---|---|---|---|
| P4 | 538.8 | −34.40 | 0.730 | — | — |
| PP4 | 429.7 | −31.36 | 0.722 | 48.60 | 32.69 |
| DP4 | 492.6 | −37.50 | 0.736 | 93.85 | 48.09 |
The chemical functionalities of the [p(NAG-co-NAPA)(x:y)] NPs were confirmed through 1H NMR, 13C NMR, FTIR, and UV-Vis spectroscopic analysis. It can be clearly noted that for simplicity and better understanding, the data points mentioned in this section are particularly for the P1 copolymer, which was already published in another report. All the other copolymers present in the library (P2–P5) are quite similar to P1 with a variation in a considerable range and their spectra are shown in their respective images for reference. The number of protons with probable positions was determined using 1H NMR spectroscopy (Fig. 1a). The distinct bands identified for p(NAG) and p(NAPA) in their 13C NMR spectra are presented in the ESI† (Fig. S4, ESI†). As mentioned in our earlier works, the δ (1H NMR) for the P1 copolymer are as follows: 8.21 (1H, d, secondary amine of glycine monomer and phenylalanine monomer), 7.16 (5H, m, aromatic protons), 4.1-3.2 (m, backbone H of monomers), 3.29 (3H, alkoxide of phenylalanine monomer), 2.4 (–CH2, m, DVB), 1.07 (–CH2, m, monomer). In the 13C NMR, the δ corresponding to the monomers disappeared and a single band in the range of 40.27 to 39.18 appeared, indicating the presence of a splitting of 2° alkanes (Fig. S4, ESI†). Further, in the FTIR spectra, the distinct bands observed for the p(NAG) and p(NAPA) homopolymers are shown in Fig. 1b and coincide well with the library of copolymers (Fig. 1b). The major characteristic bands present in the copolymers are (ν in cm−1): 3413 (secondary –NH, s), 1739 (ester C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, s) and 1659 (–NH–C
O, s) and 1659 (–NH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, s). Similarly, the UV-vis spectra of the copolymers confirmed the formation of p(NAG-co-NAPA) NPs, showing two distinct bands at 213 nm and 265 nm, which correspond to the π–π* transition and phenyl rings, respectively (Fig. 1c). Furthermore, based on the XRD results (Fig. 1d), it is clearly noticed that OTDDNs are amorphous in nature. Compared to the homopolymers, the copolymers exhibited higher thermal stability (Fig. 1e). The P1 to P5 NPs showed three degradation steps, where the first step observed is due to the loss of surface moisture, followed by polymer chain degradation (Fig. 1e). According to DSC (exo up and endo down), it was revealed besides the homopolymers, all five copolymers showed a phase transition at ∼70–75 °C with an endothermic peak (Fig. 1f). The MALDI-ToF spectra were recorded for all the samples (Fig. S6–S12, ESI†) and tabulated in Fig. 1g. The results clearly showed the presence of heterogeneous fragments in the copolymers, indicating the formation of random copolymers. The linearity in the PDI index represents the uniformity of the chains. It was observed that with an increase in the monomer ratio, there was a change in
O, s). Similarly, the UV-vis spectra of the copolymers confirmed the formation of p(NAG-co-NAPA) NPs, showing two distinct bands at 213 nm and 265 nm, which correspond to the π–π* transition and phenyl rings, respectively (Fig. 1c). Furthermore, based on the XRD results (Fig. 1d), it is clearly noticed that OTDDNs are amorphous in nature. Compared to the homopolymers, the copolymers exhibited higher thermal stability (Fig. 1e). The P1 to P5 NPs showed three degradation steps, where the first step observed is due to the loss of surface moisture, followed by polymer chain degradation (Fig. 1e). According to DSC (exo up and endo down), it was revealed besides the homopolymers, all five copolymers showed a phase transition at ∼70–75 °C with an endothermic peak (Fig. 1f). The MALDI-ToF spectra were recorded for all the samples (Fig. S6–S12, ESI†) and tabulated in Fig. 1g. The results clearly showed the presence of heterogeneous fragments in the copolymers, indicating the formation of random copolymers. The linearity in the PDI index represents the uniformity of the chains. It was observed that with an increase in the monomer ratio, there was a change in ![[M with combining macron]](https://www.rsc.org/images/entities/i_char_004d_0304.gif) w and
w and ![[M with combining macron]](https://www.rsc.org/images/entities/i_char_004d_0304.gif) n; however, the PDI was calculated for all the NPs to be ∼1.0 (table in Fig. 1). This may be due to the fact that the overall weight of the two monomers always remained fixed (500 mg). Overall, the chemical functionality and MALDI-ToF studies confirmed the successful formation of the library of NPs [p(NAG-co-NAPA)(x:y)]. All the library NPs were checked for their different organ-target specificity, as shown in the subsequent section.
n; however, the PDI was calculated for all the NPs to be ∼1.0 (table in Fig. 1). This may be due to the fact that the overall weight of the two monomers always remained fixed (500 mg). Overall, the chemical functionality and MALDI-ToF studies confirmed the successful formation of the library of NPs [p(NAG-co-NAPA)(x:y)]. All the library NPs were checked for their different organ-target specificity, as shown in the subsequent section.
The morphology and colloidal stability of the library of NPs were studied through HRTEM, DLS, zeta potential analysis and CD spectroscopy. The HRTEM images of p(NAG) and p(NAPA) NPs are shown in Fig. S13 (ESI†) with their respective SAED patterns. The HRTEM image of p(NAG) suggested the formation of porous NPs with a size of ∼130–190 nm (Fig. S13a1 and a2, ESI†), while the p(NAPA) NPs showed a dense core with a porous shell and size of ∼100–120 nm (Fig. S13b1 and b2, ESI†). In this series, it can be observed that from P1 to P5 a smooth change in porous spherical structure with a change in size was observed (Fig. 2a1–e3). In the case of P1, the particles with a size of ∼130 nm agglomerated (Fig. 2a1–a3), while in the case of P2–P4, the particles were monodispersed in nature, mimicking the structure of p(NAPA) NPs with an increase in the concentration of NAPA (Fig. 2b1–d3), respectively. Alternatively, in the case of P5, spherical co-connected NPs with a chain-type structure were observed (Fig. 2e1–e3). This chain-like structure was formed with an increase in the concentration of NAPA, i.e., with a higher content of phenyl rings, they tended to form a linearly arranged structure with less steric hindrance. Further, the hydrodynamic size and ζ of the NPs were studied, as shown in Fig. 2(f). It was clearly noticed that with an increase in NAPA concentration, the hydrodynamic size and obtained ζ increased. The higher hydrodynamic size of p(NAG) is due to the spongy porous structure of the NPs, as observed from HRTEM (Fig. S13a1 and a2, ESI†). Furthermore, CD spectroscopy was performed to study the chiral and optical properties of the NPs. It was noticed that due to the presence of a phenyl ring in the copolymer system, for P1–P4, the intensity of the CD band increased and shifted towards the left. This shifting can be attributed to the increase in the number of phenyl rings in the system.45 In the case of P5, no 2° arrangement was observed. The absence of folding in P5 is due to the formation of chains at this ratio rather than dispersed NPs (Fig. 2e1 and e2). The corresponding α-helix and β-sheet turns and random coil % are provided in ESI† (Table S2). These porous structures, spherical-shaped morphology, and negative zeta potential with chiro-optical properties of the OTDDNs offer valuable insights into designing specific drugs or vaccines with greater stability and shelf-life in biological media.
2.2 Biocompatibility, hemocompatibility and biodistribution of library OTDDNs
For the therapeutic application of the OTDDNs, they should be biocompatible, hemocompatible and target specific in nature. Initially, the cell viability of the designed OTDDNs was checked against L929 cells, i.e., mouse fibroblast cells. Fig. S14a and d (ESI†) and Fig. 3a–d represent the cell viability results of p(NAG), p(NAPA), P1 NPs and P2–P5 NPs, respectively, for concentrations in the range of 1 to 500 μg mL−1 against L929 cells. It was observed that for the p(NAG) NPs, the cell viability was ∼100% at 500 μg mL−1, while at a lower concentration starting from 50 to 1 μg mL−1, the cell viability % varied from 110% to 101%. Alternatively, for the p(NAPA) NPs, the cell viability decreased from 74% (100 μg mL−1) to 69% (500 μg mL−1). In the case of P1 NP, at a higher concentration, the cell viability was observed to be 78% and 89% for 500 μg mL−1 and 250 μg mL−1, respectively. In the range of 100 to 25 μg mL−1, the cell viability varied from 114% to 102%. The cell viability % observed for P2 was ∼85% to 75% from lower to higher concentrations. In contrast, for P4, the achieved viability was better with respect to P3 and P5 for all the concentration ranges used here. In the case of P5, at 500 μg mL−1, it exhibited toxicity (viability ∼60%) due to the higher content of NAPA monomer. Furthermore, the cytotoxicity of the OTDDNs was tested against two cancer cell lines, i.e., A549 and MDA-MB-231 (Fig. S14b, c and e (ESI†) and Fig. 3e–h), at concentrations varying from 7.8125 to 1000 μg mL−1. The p(NAG) NPs were incubated with both cancer cells and the viability observed to range from 100% to 80% from lower to higher concentration (Fig. S14b and c, ESI†), respectively.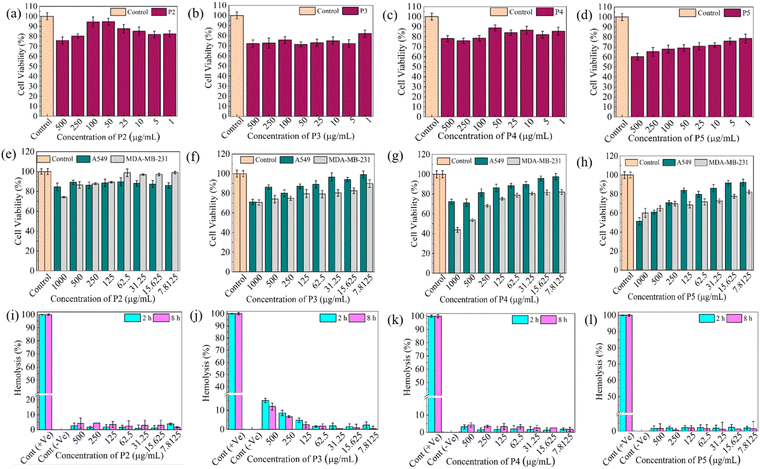 | ||
| Fig. 3 Cytocompatibility and hemocompatibility of OTDDNs. (a)–(d) Cytocompatibility against L929 cells, (e)–(h) A549 and MDA-MB-231 cell lines and (i)–(l) hemocompatibility of P2, P3, P4 and P5 NPs at different concentrations. For (a)–(l), the statistical significance values are provided in the ESI† (Table S3–S6). | ||
In the case of p(NAPA), the cell viability % against A549 was determined to be ∼ 100% for all concentrations, whereas MDA-MB-231 exhibited ∼80% viability at 1000 μg mL−1. Further, P1 NPs were also incubated with A549 and MDA-MB-231 cell lines and the cell viability % observed to be 85% and 67% for 1000 μg mL−1, respectively. Considering P2 to P5, other than P2 in all three OTDDNs, a similar trend of cell viability %, i.e. with an increase in concentration, a smooth decrease in cell viability % was observed. In the case of P2, at all the tested concentrations, the cell viability against A549 was determined to be ∼90%, while against MDA-MB-231, at a lower concentration it exhibited ∼100% viability. Among P3, P4 and P5, P4 showed the lowest % cell viability at 1000 μg mL−1 (∼45%). Thus, it can be concluded that all the NPs in the library are quite toxic against MDA-MB-231 cells with respect to A549.
Hemocompatibility evaluation for biomaterials is crucial for all therapeutic purposes given that they enter the systemic circulation and cause harmful effects by limiting therapeutic benefits. A material considered to be an effective biomaterial if its hemolysis value is below 5%. Therefore, the hemolytic activity of all the samples was studied in both dose- and time-dependent manners in the range of 7.8125 to 500 μg mL−1 (Fig. 3i–l). A variation in the degree of hemolysis % was observed for selected concentrations at 2 h and 8 h. It was seen that for p(NAG), p(NAPA) and P1, the hemolysis % was below 5% up to 8 h (Fig. S14, ESI†). In the case of P3, at higher concentrations (500 and 250 μg mL−1) the hemolysis % was determined to be 15% and 8%, respectively, whereas for all the other samples, the hemolysis results were determined to be in a considerable range. The images of the hemolytic tubes are presented in the ESI† (Fig. S15). Therefore, OTDDNs are suitable biomaterials and can be used for different therapeutic uses. To evaluate the organ selectivity of OTDDNs, in vivo biodistribution studies were conducted following the process mentioned in the Experimental section (Fig. 4a). Five major organs (lungs, heart, liver, kidney, and breast) were considered to quantify the target efficiency of OTDDNs by measuring the fluorescence intensity and compared with the control p(NAG) and p(NAPA) NPs. According to the in vivo biodistribution study, the diverse properties of the library of NPs were observed. According to the intensity, it is obvious that p(NAG), P3, and P5 are target specific to the lungs (Fig. 4b). The control sample p(NAG) targeted the lungs and heart, whereas p(NAPA) targeted the heart and liver. P1 was target specific to the heart, kidney and liver efficiently. P2 was only target specific to the heart. P3 preferably targeted the lungs. Interestingly, it is exciting to report that P4 is very target specific to the breast (700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 p s−1 cm−2 sr−1). However, P5 is target specific to the lungs, heart, liver and kidney. The detailed target specificity is mentioned in Table (Fig. 4g). Therefore, the obtained results of cell viability, hemolysis and in vivo biodistribution are clear evidence that P4 can be a suitable OTDDS for the targeted delivery of drugs for the treatment of TNBC. Further, to use the P4 NPs for TNBC treatment, a biodegradation study was performed in simulated body fluid (SBF) (pH 7.4), as discussed in the Method section. The morphology of the degraded P4 NPs was scrutinized through HRTEM after 7 days of post-incubation with SBF (Fig. S16, ESI†). The results exhibited that the P4 NPs (of size 180 to 200 nm) were degraded into small fragments (10 to 20 nm), as shown in the supporting HRTEM images (Fig. S16a1 and b1, ESI†). Therefore, it can be concluded that the P4 NPs are degradable.
000 p s−1 cm−2 sr−1). However, P5 is target specific to the lungs, heart, liver and kidney. The detailed target specificity is mentioned in Table (Fig. 4g). Therefore, the obtained results of cell viability, hemolysis and in vivo biodistribution are clear evidence that P4 can be a suitable OTDDS for the targeted delivery of drugs for the treatment of TNBC. Further, to use the P4 NPs for TNBC treatment, a biodegradation study was performed in simulated body fluid (SBF) (pH 7.4), as discussed in the Method section. The morphology of the degraded P4 NPs was scrutinized through HRTEM after 7 days of post-incubation with SBF (Fig. S16, ESI†). The results exhibited that the P4 NPs (of size 180 to 200 nm) were degraded into small fragments (10 to 20 nm), as shown in the supporting HRTEM images (Fig. S16a1 and b1, ESI†). Therefore, it can be concluded that the P4 NPs are degradable.
2.3 In silico assessment of DHA and piperine as targeted drugs for TNBC
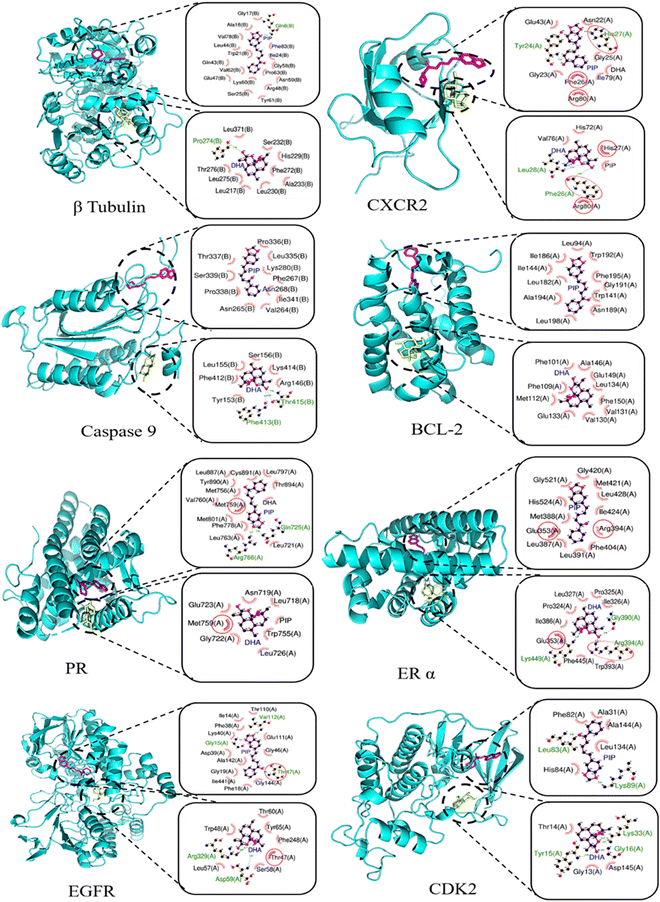
The in silico assessment of selected anticancer drugs, i.e.; DHA and piperine, was conducted using network pharmacology and molecular docking, which can predict gene targets and the binding affinity of selected ligands with them, respectively. The results exhibited that both drugs are effective as anticancer agents. The Swiss Target prediction tool was used to determine the targeted enzymes of TNBC (seven for piperine and two for DHA). These enzymes were run through DisGeNET and the targeted proteins were listed and further analysed as a PPI network using Cytoscape. A network of common genes and target compounds with enzymes was constructed (Fig. 5a and b). Additionally, more gene targets were found from GEPIA2 and used to construct a Venn diagram (Fig. 5c), which represents that 860 genes are responsible for TNBC. Piperine and DHA can target 94 and 92 genes, respectively. Among the 860 genes responsible for TNBC, piperine and DHA can target 6 and 8 genes, respectively. However, none of these common genes of TNBC have been reported, which can be targeted by both piperine and DHA. Furthermore, the gene targets were collected from NCBI and recent literature on TNBC46–49 and the top 40 genes with the highest connectivity screened. A network-like structure was constructed with different colour gradients, which represents the degree of relatedness of genes with TNBC (Fig. 5d). The identified genes were further explored to understand their pathways related to the targeted genes responsible for TNBC (Fig. 5e). A lollipop plot was drawn to represent the top 15 pathways. It was clearly observed that among the various cancers, breast cancer GO enrichment is ∼50 fold, and these targeted genes can be highly expressed in breast cancer following various pathways. According to the network pharmacology study, 24 receptor proteins responsible for TNBC were selected for molecular docking with paclitaxel as an FDA approved anti-TNBC drug. Then, single docking was performed for the 24 receptor proteins with piperine and DHA separately. The obtained binding energies, inhibition constant (Ki) values and amino acid residues are listed in Table S7 (ESI†). It is clearly evident that piperine and DHA interact differently with different amino acid residues. Further, among the 24 proteins, seven proteins (CXCR2, Caspase 9, BCL-2, PR, ER α, EGFR and CDK2) dominantly responsible for TNBC are selected according to their highest negative binding energy with lowest Ki. Their binding energy and inhibition constant values are compared with paclitaxel (Fig. 5f and g). Here, β-tubulin is also considered a potential target, given that paclitaxel inhibits the growth of TNBC by binding with the β-subunit of tubulin.50–52Fig. 5f and g reveal that compared to paclitaxel, both piperine and DHA show a greater binding affinity with lower Ki. | ||
| Fig. 5 Network pharmacology and molecular docking results of piperine and DHA against TNBC. (a) and (b) Target protein network for piperine and DHA. (c) Venn diagram showing the common gene targets of piperine and DHA in TNBC. (d) Interrelated network map of top 40 gene targets of piperine and DHA. (e) Gene expression responsible for TNBC pathways, (f) binding energy, and (g) Ki for target proteins of piperine and DHA. (h) Binding energy and (i) Ki for the combination of piperine and DHA with paclitaxel as reference. The colors in Fig. 5e show the p-values for each term, with lower p-values having darker colours. The dot size demonstrates the number of genes associated with each pathway. | ||
Given that one of the important objectives of this work is to determine the synergistic anti-TNBC effects of piperine and DHA, combination docking has been performed to find the binding pockets with the selected 8 receptor proteins in two steps, (i) piperine followed by DHA and (ii) DHA followed by piperine. The binding energy and Ki values are shown in Fig. 5c and d. The docking scores are shown in Table S8 and S9 (ESI†), which clearly show that for the combination study, the binding energy is highly negative, and Ki is very low with respect to paclitaxel. Therefore, a combination of piperine and DHA can be used for TNBC treatment with better efficacy. The detailed binding energy, Ki values and amino acid residues are listed in Table S8 and S9 (ESI†) for β-tubulin, CXCR2, Caspase 9, BCL-2, PR, ER α, EGFR and CDK2. Further, their 3D structures and corresponding 2D Lig Plots were extracted (Fig. 6), as mentioned in the Method section, which clearly represents both polar (H-bonds) and nonpolar interactions with specific amino acid residues. According to the figures and tables, it is clearly visible that piperine and DHA form H-bonds at Gln98 and Pro274 for β-tubulin, at Try24, His27 and Leu28, and Phe26 for CXCR2, whereas for caspase 9, DHA only forms H-bonds at Phe413 and Thr415. In the case of piperine, PR formed H-bonds with Arg766 and Gln725 residues. In the case of ER α, DHA formed strong H-bonding with three amino acid residues (Lys449, Gly390 and Arg394). In the case of EGFR, piperine established three polar interactions (Val112, Gly15 and Thr47) and DHA showed two polar interactions (Arg329 and Asp59) with amino acid residues. Similarly, in CDK2, DHA forms three hydrogen bonds (Lys33, Try15 and Gly16), whereas it is limited to two polar interactions for piperine (Leu83 and Lys89). The presence of these common interacting amino acid residues suggests that piperine and DHA can inhibit TNBC more efficiently by targeting these four proteins (CXCR2, PR, ER α and EGFR) among the selected eight proteins. The network pharmacology and molecular docking results further revealed that a number of protein targets exist for piperine and DHA to inhibit TNBC, and among them the major potential targets can be selected for in vitro and in vivo TNBC studies.
2.4 Preparation, characterization, in vitro cytotoxicity and migratory effect of piperine or DHA-loaded P4 NPs (PP4 and DP4)
P4 was selected to load piperine or DHA for targeting TNBC according to the method discussed in the Experimental section. The amphiphilic nature of the copolymers enhanced the loading of hydrophobic piperine and DHA in the core and shell of the P4 NPs. Piperine and DHA were loaded separately in P4 NPs and designated as PP4 and DP4, respectively. The LE% and EE% were calculated for both drugs (Table 1) and it was observed that the encapsulation efficiencies were very good for both piperine (48.60%) and DHA (93.85%). Due to the difference in hydrophobicity, different extents of loading were observed. It can be noted that the increased EE% in DHA is high due to its more hydrophobic complex ring structure with respect to piperine.The thermal stability of all the samples was studied from 25 °C to 600 °C and it was observed that they were quite stable up to 50 °C. The minor weight loss (2–3%) observed up to 100 °C is due to the surface moisture. Beyond 160 °C, the weight loss of the polymer particles occurred due to the breaking of the polymer bonds. After loading drugs, PP4 and DP4 showed greater thermal stability compared to P4 (Fig. 7a). The in vitro drug release behavior of PP4 and DP4 was studied using the dialysis bag approach in 1× PBS buffer (pH 7.4) acidic medium (pH 6.8) for up to 30 days and the results are shown in Fig. 7b. In the case of neutral medium, ∼5% and ∼15% release were observed at 24 h for PP4 and DP4, respectively. It was clearly observed that the release occurred stepwise in a stimuli-responsive nature53 for all the samples at all pH. However, it is interesting to mention that DP4 at pH 6.8 released DHA very fast compared to the other samples. It can also be mentioned that up to 30 day, DP4 released the maximum of ∼40% DHA. This can be explained by pH acting as a stimulus and there is a possibility of breaking the network of copolymer chains periodically, which influenced the stepwise release of piperine/DHA.53 To assess the effects of NFs against TNBC, the MTT assay was performed (Fig. 7c–e) by varying the doses of PP4 and DP4 from 25 to 2500 μg mL−1. According to Fig. 7c–e, it is clearly evident that the effective treatment of TNBC is dose dependent. The IC50 values for PP4 and DP4 were calculated to be 450 μg mL−1 and 409.5 μg mL−1, respectively. Interestingly, with a variation in the ratio of DP4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PP4 for combination therapy, synergetic TNBC inhibition was observed (Fig. 7d and Fig. S17, ESI†). In the case of combination therapy (DP4 + PP4), the IC50 obtained for a 7
PP4 for combination therapy, synergetic TNBC inhibition was observed (Fig. 7d and Fig. S17, ESI†). In the case of combination therapy (DP4 + PP4), the IC50 obtained for a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio (Fig. 7d) is promising. Further, different concentrations of mixed DP4
3 ratio (Fig. 7d) is promising. Further, different concentrations of mixed DP4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PP4 at 7
PP4 at 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 were employed to study the TNBC inhibition and the IC50 was found to be 350 μg mL−1. According to this study, it can be concluded that for each composition ratio, different IC50 values were obtained. By selecting the IC50, based on the severity of TNBC in patients, their dose-dependent treatment can be programmed. Furthermore, the IC50 values for PP4 and DP4 were used to calculate CI and it was observed that the CI value was less than ‘1’, which implies the synergetic inhibition effect of DHA and piperine against TNBC. The Ki value for each NF was determined by using the Dixon plot54 (Fig. 7h), where the Ki values were obtained by extrapolating the cytotoxicity results and found to be 0.941 μg mL−1, 0.571 μg mL−1 and 0.45 μg mL−1 for PP4, DP4 and Co-NP(7
3 were employed to study the TNBC inhibition and the IC50 was found to be 350 μg mL−1. According to this study, it can be concluded that for each composition ratio, different IC50 values were obtained. By selecting the IC50, based on the severity of TNBC in patients, their dose-dependent treatment can be programmed. Furthermore, the IC50 values for PP4 and DP4 were used to calculate CI and it was observed that the CI value was less than ‘1’, which implies the synergetic inhibition effect of DHA and piperine against TNBC. The Ki value for each NF was determined by using the Dixon plot54 (Fig. 7h), where the Ki values were obtained by extrapolating the cytotoxicity results and found to be 0.941 μg mL−1, 0.571 μg mL−1 and 0.45 μg mL−1 for PP4, DP4 and Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), respectively. Thus, the effectiveness of the designed NFs follows the order of Co-NP(7
3), respectively. Thus, the effectiveness of the designed NFs follows the order of Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) > DP4 > PP4. All the designed NFs show competitive inhibition, which matches well with the earlier mentioned docking results.
3) > DP4 > PP4. All the designed NFs show competitive inhibition, which matches well with the earlier mentioned docking results.
To effectively prevent metastasis caused by tumor cells, it is crucial to study the behavior of MDA-MB-231 cell migration in the presence of our designed NFs. An in vitro wound scratch assay was performed to study the proliferation and migratory properties of the cells in presence of P4, PP4, DP4 and Co-NP (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for 24 h (Fig. 7f and g). The change in width (%) results revealed that compared to the control (22.82%), for the P4 NPs (15.94%), a minor change was observed. Alternatively, compared to P4 or the control with NFs (for PP4, DP4 and Co-NP), the change in width (%) values were below 5%, which is negligible (Fig. 7f). Similarly, according to the rate of cell migration results, it was found that although the rate of migration for the control and P4 was 0.95% and 0.66%, respectively, for PP4, DP4 and Co-NP(7
3) for 24 h (Fig. 7f and g). The change in width (%) results revealed that compared to the control (22.82%), for the P4 NPs (15.94%), a minor change was observed. Alternatively, compared to P4 or the control with NFs (for PP4, DP4 and Co-NP), the change in width (%) values were below 5%, which is negligible (Fig. 7f). Similarly, according to the rate of cell migration results, it was found that although the rate of migration for the control and P4 was 0.95% and 0.66%, respectively, for PP4, DP4 and Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), the obtained values were 0.06%, 0.08% and 0.06%, respectively (Fig. 7g). The rate of migration by NFs decreased by 90% of that of the control. Thus, by considering the above-mentioned results, further studies were conducted with 350 μg mL−1 for all the NFs including P4 NPs as the control.
3), the obtained values were 0.06%, 0.08% and 0.06%, respectively (Fig. 7g). The rate of migration by NFs decreased by 90% of that of the control. Thus, by considering the above-mentioned results, further studies were conducted with 350 μg mL−1 for all the NFs including P4 NPs as the control.
2.5. Effect of PP4, DP4, and Co-NPs(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 3) on programmed cell death, cell cycle and TNBC inhibition mechanism
3) on programmed cell death, cell cycle and TNBC inhibition mechanism
The IC50 values were obtained from the MTT assays for the individual and Co-NPs(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for the MDA-MB-231 cells, and to confirm whether NFs induced apoptotic or necrotic pathway, flow cytometric assays have been performed (Fig. 8a). Herein, Annexin-V/PI was used to deduce the various stages of apoptosis.55 P4, PP4 and DP4 together with Co-NPs(7
3) for the MDA-MB-231 cells, and to confirm whether NFs induced apoptotic or necrotic pathway, flow cytometric assays have been performed (Fig. 8a). Herein, Annexin-V/PI was used to deduce the various stages of apoptosis.55 P4, PP4 and DP4 together with Co-NPs(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) were treated with MDA-MB-231 cells at 350 μg mL−1 and their cell death pathway was studied. As observed earlier, P4 induced cytotoxicity in the cells (Fig. 3g). The flow cytometry results revealed that P4 induced necrotic cell death (51.26%). Comparing PP4 and DP4, PP4 induced more necrotic cell death (extent of live cells: 13.68%, late apoptotic cells: 1.14% and necrotic cells: 85.12%). However, DP4 promoted late apoptotic cell death (live cell: 41.63%, late apoptotic: 28.48% and necrotic cells: 29.24%), which further justified the considered target ratio of DP4
3) were treated with MDA-MB-231 cells at 350 μg mL−1 and their cell death pathway was studied. As observed earlier, P4 induced cytotoxicity in the cells (Fig. 3g). The flow cytometry results revealed that P4 induced necrotic cell death (51.26%). Comparing PP4 and DP4, PP4 induced more necrotic cell death (extent of live cells: 13.68%, late apoptotic cells: 1.14% and necrotic cells: 85.12%). However, DP4 promoted late apoptotic cell death (live cell: 41.63%, late apoptotic: 28.48% and necrotic cells: 29.24%), which further justified the considered target ratio of DP4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PP4, i.e., 7
PP4, i.e., 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 for TNBC treatment. The use of DHA surged the late apoptotic stage from 1.14% to 28.48%, which is highly remarkable. Furthermore, Co-NPs(7
3 for TNBC treatment. The use of DHA surged the late apoptotic stage from 1.14% to 28.48%, which is highly remarkable. Furthermore, Co-NPs(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) demonstrated a decrease in necrotic cells (9.21%), with a significant increase in live cells (56.44%), early apoptotic cells (13.31%), and slight decrease in late apoptotic cells (21.04%). This study further revealed that instead of P4, PP4 and DP4, Co-NP(7
3) demonstrated a decrease in necrotic cells (9.21%), with a significant increase in live cells (56.44%), early apoptotic cells (13.31%), and slight decrease in late apoptotic cells (21.04%). This study further revealed that instead of P4, PP4 and DP4, Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) can be potentially used as a TNBC target, given that it showed programmed cell death (apoptosis) at the IC50 concentration (350 μg mL−1).
3) can be potentially used as a TNBC target, given that it showed programmed cell death (apoptosis) at the IC50 concentration (350 μg mL−1).
One of the key steps in developing potential anticancer NFs is controlling the cell cycle.56 Based on the cell cycle study (Fig. 8b), the developed NFs possessed potential to initiate apoptosis in MDA-MB-231 cells. In the case of P4, the activation of the G0–G1 phase (65.72%) is responsible for reduction in cell viability %. In contrast, in the case of PP4 and DP4, the G0–G1 phase percentage was in a similar in range; however, drastic differences were observed for the S and G2-M phases, e.g., the S phase decreased from 20.80% to 13.52%, while the G2-M phase increased from 11.73% to 22.74%. This indicates that the G2-M phase is dominant for DP4 and the value almost doubles compared to that for PP4. In the case of Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), the extent of S phase decreased, whereas the G2-M phase increased compared to the P4, PP4 and DP4 NPs. It can be stated that Co-NP(7
3), the extent of S phase decreased, whereas the G2-M phase increased compared to the P4, PP4 and DP4 NPs. It can be stated that Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) inhibited cell growth by arresting the cell cycle at the G2-M phase. Subsequently, the TNBC inhibition mechanism was established by quantifying the reactive oxygen species (ROS) and mitochondrial membrane potential (MMP) (Fig. 8c and d). ROSs are strong oxidants that can trigger cells by increasing the intracellular reactive oxygen species, further causing DNA, protein or lipid degradation and diminishing their activities. Here, DCFDA is a green fluorescent dye used to quantify the ROS generated by the treatment conditions. The fluorescence intensity is directly proportional to the amount of ROS present in the sample. Similarly, apoptosis is related to the change in mitochondrial membrane potential and mitochondrial damage. The mitochondrial potential is the determinant for cell death. Rhodamine-123 (Rh-123) was used to estimate MMP. It enters the mitochondria when the membrane potential is intact. However, when the mitochondria lose their potential, the excess Rh-123 leaks out, resulting in a detectable reduction in fluorescence. This decrease in fluorescence is directly related to the loss of mitochondrial potential. In the case of ROS, all the NFs including Co-NP(7
3) inhibited cell growth by arresting the cell cycle at the G2-M phase. Subsequently, the TNBC inhibition mechanism was established by quantifying the reactive oxygen species (ROS) and mitochondrial membrane potential (MMP) (Fig. 8c and d). ROSs are strong oxidants that can trigger cells by increasing the intracellular reactive oxygen species, further causing DNA, protein or lipid degradation and diminishing their activities. Here, DCFDA is a green fluorescent dye used to quantify the ROS generated by the treatment conditions. The fluorescence intensity is directly proportional to the amount of ROS present in the sample. Similarly, apoptosis is related to the change in mitochondrial membrane potential and mitochondrial damage. The mitochondrial potential is the determinant for cell death. Rhodamine-123 (Rh-123) was used to estimate MMP. It enters the mitochondria when the membrane potential is intact. However, when the mitochondria lose their potential, the excess Rh-123 leaks out, resulting in a detectable reduction in fluorescence. This decrease in fluorescence is directly related to the loss of mitochondrial potential. In the case of ROS, all the NFs including Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) exhibited greater ROS-based cell inhibition than the control, and the inhibition efficiency followed the order of DP4 > P4 > PP4> Co-NP(7
3) exhibited greater ROS-based cell inhibition than the control, and the inhibition efficiency followed the order of DP4 > P4 > PP4> Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). In the case of Co-NP(7
3). In the case of Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), the maximum fluorescent intensity value approached the order of ∼105. Further, between ROS and MMP, MMP was dominant, and compared to the control, P4, PP4, and DP4 shifted more towards the right side of the fluorescence intensity plot with a range in the order of 106 (Fig. 8c). Further, Co-NP(7
3), the maximum fluorescent intensity value approached the order of ∼105. Further, between ROS and MMP, MMP was dominant, and compared to the control, P4, PP4, and DP4 shifted more towards the right side of the fluorescence intensity plot with a range in the order of 106 (Fig. 8c). Further, Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) shifted more, showing a broad spectrum with an increase in the cell count, which depicts that the underlying cell killing mechanism of Co-NP(7
3) shifted more, showing a broad spectrum with an increase in the cell count, which depicts that the underlying cell killing mechanism of Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) is majorly due to the change in the membrane potential of TNBC (Fig. 8d). Therefore, it can be stated that Co-NP(7
3) is majorly due to the change in the membrane potential of TNBC (Fig. 8d). Therefore, it can be stated that Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) exhibited late apoptotic cell death by inducing MMP-based TNBC killing. Therefore, all the above-mentioned results corroborate that Co-NP(7
3) exhibited late apoptotic cell death by inducing MMP-based TNBC killing. Therefore, all the above-mentioned results corroborate that Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) can be a potential agent for the treatment of TNBC. It can also be noted that all the combinations of PP4 and DP4 can be used according to the cancer severity.
3) can be a potential agent for the treatment of TNBC. It can also be noted that all the combinations of PP4 and DP4 can be used according to the cancer severity.
3. Conclusions
The current challenges in targeted therapy are associated with the fact that none of the reported nanocapsules are 100% target specific, where most of the existing drug delivery devices miss the target, leading to side effects. Considering this, the present work focused on constructing a library of organ-targeting amino acid-based amphiphilic copolymers, i.e., p(NAG-co-NAPA)(x:y), for therapy. Physicochemical characterization of the synthesized p(NAG-co-NAPA)(x:y) NPs was conducted to ensure their successful synthesis, and subsequently screened for their potential to act as drug delivery vehicles to different organs. The in vivo organ-based biodistribution was studied to reveal the best OTDDNs targeting the breast, i.e., p(NAG-co-NAPA)(1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) NP (P4). In addition, an in silico study was also performed to justify the selection of piperine and DHA, given that they target 14 genes responsible for TNBC. The prepared piperine (PP4) and DHA (DP4)-loaded NFs exhibited an excellent drug loading and pH-dependent drug release, which enhanced the targeted delivery in the TNBC site by minimizing systemic side effects. The low IC50 values in combination treatment, i.e. with Co-NPs(7
4) NP (P4). In addition, an in silico study was also performed to justify the selection of piperine and DHA, given that they target 14 genes responsible for TNBC. The prepared piperine (PP4) and DHA (DP4)-loaded NFs exhibited an excellent drug loading and pH-dependent drug release, which enhanced the targeted delivery in the TNBC site by minimizing systemic side effects. The low IC50 values in combination treatment, i.e. with Co-NPs(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), revealed the synergetic treatment efficiency of TNBC rather than antagonism behaviour. The flow cytometry results revealed that the inhibition of TNBC by the Co-NP(7
3), revealed the synergetic treatment efficiency of TNBC rather than antagonism behaviour. The flow cytometry results revealed that the inhibition of TNBC by the Co-NP(7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) formulation was due to mitochondrial membrane potential-mediated apoptosis by arresting the G2-M phase in the cell cycle. It can be noted that block-co-polymers with varying mers have different extents of target efficiency to various organs, which has never been reported. This finding is similar to that based on the treatment of interest, given that any member of the library of nanocapsules can be selected for the optimum delivery of drugs and to achieve the maximum therapeutic efficiency, which may not be restricted to the major organs such as liver, heart, lungs and kidney, rather it can be used to treat other organs too. It can further be noted that the used polymeric NPs can be subsequently degraded and eliminated from the body. However, to comprehend the efficacies of the NFs and progress towards future clinical trials, in vivo studies are under consideration and the results will be reported as a continuation of this work.
3) formulation was due to mitochondrial membrane potential-mediated apoptosis by arresting the G2-M phase in the cell cycle. It can be noted that block-co-polymers with varying mers have different extents of target efficiency to various organs, which has never been reported. This finding is similar to that based on the treatment of interest, given that any member of the library of nanocapsules can be selected for the optimum delivery of drugs and to achieve the maximum therapeutic efficiency, which may not be restricted to the major organs such as liver, heart, lungs and kidney, rather it can be used to treat other organs too. It can further be noted that the used polymeric NPs can be subsequently degraded and eliminated from the body. However, to comprehend the efficacies of the NFs and progress towards future clinical trials, in vivo studies are under consideration and the results will be reported as a continuation of this work.
4. Experimental
4.1 Materials
All chemicals were used as received without further purification unless otherwise stated. Glycine (98%, Qualigens), L-phenylalanine methyl ester (Sigma), triethylamines (≥99.5%, Merck), 1,4-dioxane (99%) extra pure (Merck), potassium hydroxide (KOH), 2,2′-azo-bis-isobutyronitrile (AIBN, 98%, SRL), SDS (sodium dodecyl sulfate) (90%, Merck), magnesium sulphate anhydrous (SRL), hexadecane anhydrous (HD) (99%, Sigma–Aldrich), acryloyl chloride stab. with 400 ppm phenothiazine (96%, Alfa Aesar), divinyl benzene (DVB), diethyl ether, dichloromethane, sodium bisulphate, potassium chloride, potassium hydrogen phosphate, magnesium chloride, sodium sulphate, sodium chloride, toluene, DMSO d6, CDCl3, phosphate buffered saline (pH 7.4), phosphate buffered saline (pH 6.8), isopropanol, piperine (Sigma), dihydroartemisinin (TCI), phosphotungstic acid (Sigma), Triton-X, DNS (sodium chloride and dextrose injection IP (0.9% & 5% w/v)) (Jedux), calcium chloride, methyl thiazoltetrazolium (MTT, >99.9%, Himedia), Dulbecco's modified Eagle medium (DMEM, Cell Clone), trypsin-EDTA, thiopentone sodium (Abbott), 1,1′-dioctadecyl-3,3,3′,3′- tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD) dye, 4%, Triton-X (Thermo), Fetal Bovine serum (FBS, Gibco), FACS Kit (Invitrogen), DCFH-DA (Cymann), Rhodamine 123 (Sigma), penicillin–streptomycin cocktail (Himedia), and DMSO (Merck). L929 (mouse fibroblast cells), A549 (lung carcinoma epithelial cells) and MDA-MB-231 (triple-negative breast cancer, TNBC) cell lines were acquired from NCCS-Pune Repository, India. A Pure Lab Ultra water system (ELGA, High Wycombe, United Kingdom) was used to obtain Ultrapure water (18.2 M) for sample processing.4.2 Synthesis and characterization of NAG, NAPA, p(NAG), p(NAPA), and library of OTDDNs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NAPA, as mentioned in Table 2, were dispersed in toluene. Then, HD and DVB were added and sonicated for 5 min at RT. The mixture was stirred for 30 min. Then, a water-based SDS solution and defined amount of AIBN were added to the above-mentioned reaction mixture and stirred at RT for 1 h. Then, the reaction mixture was sonicated (Sonics, Vibra Cell, 750 Watt, 45
NAPA, as mentioned in Table 2, were dispersed in toluene. Then, HD and DVB were added and sonicated for 5 min at RT. The mixture was stirred for 30 min. Then, a water-based SDS solution and defined amount of AIBN were added to the above-mentioned reaction mixture and stirred at RT for 1 h. Then, the reaction mixture was sonicated (Sonics, Vibra Cell, 750 Watt, 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 s, 40% power) for a few minutes in an ice bath, followed by polymerization under continuous stirring until the reaction finished.
15 s, 40% power) for a few minutes in an ice bath, followed by polymerization under continuous stirring until the reaction finished.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y)] with different monomer ratios
y)] with different monomer ratios
| Polymer code | Composition | Monomer's ratio (x![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y) y) |
|---|---|---|
| p(NAG) | NAG | NA |
| p(NAPA) | NAPA | NA |
| P1 | p(NAG-co-NAPA) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| P2 | p(NAG-co-NAPA) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| P3 | p(NAG-co-NAPA) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
| P4 | p(NAG-co-NAPA) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
| P5 | p(NAG-co-NAPA) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
Finally, the residual toluene was evaporated. The prepared NPs were washed thoroughly with a (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) ethanol and water mixture 8–10 times, followed by centrifugation. Then, the samples were freeze dried and stored for further studies. The p(NAG) and p(NAPA) homopolymers were synthesized following the same procedure and used as control samples. The different ratios of monomers used are mentioned in Table 2. The details synthesis method of this polymeric NP library has been filed for an Indian patent.
1) ethanol and water mixture 8–10 times, followed by centrifugation. Then, the samples were freeze dried and stored for further studies. The p(NAG) and p(NAPA) homopolymers were synthesized following the same procedure and used as control samples. The different ratios of monomers used are mentioned in Table 2. The details synthesis method of this polymeric NP library has been filed for an Indian patent.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of CDCl3 and DMSO-D6 (co-solvency approach) for p(NAG), p(NAPA), and [p(NAG-co-NAPA)(x:y)], respectively, and plotted using MestReNova 14.1. UV-vis spectroscopy (Lambda 750 spectrophotometer, PerkinElmer) was performed (λ = 180–300 nm). Matrix-assisted laser desorption/ionization-time of flight (MALDI-ToF) mass spectra were acquired using a Bruker Autoflex instrument using dithranol in THF as the matrix and the method followed for analysis was RN_900-4500_Da.par. The thermal stability and phase transition of the NPs were studied through TGA (TGA-50, M/s Shimadzu (Asia Pacific)) from 25 °C to 600 °C at a heating rate of 10 °C min−1 with 100 mL min−1 of N2 gas flow and DSC (DSC-60 Plus, M/s Shimadzu (Asia Pacific)) from 25 °C to 300 °C at a heating rate of 10 °C min−1 with 50 mL min−1 of N2-gas flow, respectively. The crystallinity of the NPs was evaluated using high-resolution X-Ray diffraction (Rigaku SmartLab 9 kW Powder type (without χcradle) at 2θ = 10°–90° equipped with a Cu Kα X-Ray source with λ = 1.54
1 mixture of CDCl3 and DMSO-D6 (co-solvency approach) for p(NAG), p(NAPA), and [p(NAG-co-NAPA)(x:y)], respectively, and plotted using MestReNova 14.1. UV-vis spectroscopy (Lambda 750 spectrophotometer, PerkinElmer) was performed (λ = 180–300 nm). Matrix-assisted laser desorption/ionization-time of flight (MALDI-ToF) mass spectra were acquired using a Bruker Autoflex instrument using dithranol in THF as the matrix and the method followed for analysis was RN_900-4500_Da.par. The thermal stability and phase transition of the NPs were studied through TGA (TGA-50, M/s Shimadzu (Asia Pacific)) from 25 °C to 600 °C at a heating rate of 10 °C min−1 with 100 mL min−1 of N2 gas flow and DSC (DSC-60 Plus, M/s Shimadzu (Asia Pacific)) from 25 °C to 300 °C at a heating rate of 10 °C min−1 with 50 mL min−1 of N2-gas flow, respectively. The crystallinity of the NPs was evaluated using high-resolution X-Ray diffraction (Rigaku SmartLab 9 kW Powder type (without χcradle) at 2θ = 10°–90° equipped with a Cu Kα X-Ray source with λ = 1.54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Å).
Å).
The colloidal stability of the OTDDNs were evaluated by measuring their zeta potential (ζ) using a Zeta sizer (Malvern Zeta Sizer) at 25 °C. For this, suspensions of 500 μg mL−1 of NPs in MilliQ were prepared and three acquisitions were recorded for each sample and the average hydrodynamic diameter and ζ values were recorded. Circular dichroism (CD) experiments using a Jasco J-1500 CD spectrometer were performed in MilliQ at 250 μg mL−1 at RT from λ = 190 to 300 nm with a 0.5 nm data pitch and bandwidth of 1.0 nm. For the HRTEM analysis, ∼0.1 mg OTDDNs were taken in 500 μL of isopropanol with two drops of phosphotungstic acid and sonicated at regular intervals up to 8 h to form a uniform dispersion. Then, a few drops of suspension were added to a C-coated Cu grid (200 mesh size). Images and EDAX patterns were acquired using a Tecnai G2 20 TWIN (FEI Company of USA (S.E.A.) PTE, LTD) equipped with an EDS SYSTEM and Octane Plus SDD detector.
4.3 Cell viability study of p(NAG), p(NAPA), and library of OTDDN [p(NAG-co-NAPA)(x:y)] NPs
The cell viability of the homopolymers and OTDDNs was determined using one healthy cell line (L929) and two cancer cell lines, A549 and MDA-MB-231. DMEM supplemented with 10% of FBS and 100 U of penicillin–streptomycin antibiotic was used to culture the cells and maintained in incubator at 37 °C with 5% CO2. 1 × 104 cells per well were cultured in a 96-well plate for 24 h, followed by the addition of OTDDNs (1 to 500 μg mL−1 and 7.8125 to 1000 μg mL−1). After incubation, the samples were removed, and 5 μg mL−1 of MTT reagent added to each well and kept for 4 h. Further, MTT reagent was removed and 100 μL of DMSO was added per well and incubated for 20 min in the dark. Then, the cell viability was estimated by recording the absorbance at λmax = 570 nm using a microplate reader (Biotek, SYNERGY H1M).4.4 Hemocompatibility of p(NAG), p(NAPA), and library of OTDDN [p(NAG-co-NAPA)(x:y)] NPs
Hemolysis was conducted on RBCs collected from rat whole blood. Initially, RBCs were collected and suspended in DNS. To determine the hemolytic effect of the copolymer NPs, 5 × 106 RBCs were incubated with all the samples separately at different concentrations (7.8125 to 500 μg mL−1) for 2 h and 8 h at 37 °C with 100 rpm. Incubation with distilled water and DNS were considered as the positive and negative control, respectively. After incubation, the samples were centrifuged at 3500 rpm for 5 min at RT. Then, the supernatants were collected, and hemolysis was calculated by recording the absorbance at λmax = 540 nm. Experiments were performed with the same rat blood in three distinct tubes for each concentration of NPs.4.5 In vivo biodistribution of p(NAG), p(NAPA), and library of OTDDN [p(NAG-co-NAPA)(x:y)] NPs
The in vivo biodistribution of all the NPs was studied using DiD dye. Initially, 20 μL of dye (1 mg mL−1 in DMSO) was incubated with a NP suspension (1× PBS) for 24 h in the dark with constant stirring at 100 rpm. The NP suspension was centrifuged for 5 min at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, followed by removal of the supernatant, washing of the pellet, and re-suspending it in PBS for bio-imaging studies. Swiss albino mice (female) were divided into seven groups with n = 3 and anaesthetized by IP injection with thiopentone sodium at a dose of 0.2 mg kg−1 of body weight, followed by injection of 200 μL of DiD-loaded NPs through their tail vein. After the administration of NPs, images were acquired at 1, 6, 12 and 24 h by using an IVIS imaging system (PHOTONIMAGER OPTIMA, Biospace Lab, France) at the excitation/emission maxima of 637/672 nm in the near infrared (IR) region.
000 rpm, followed by removal of the supernatant, washing of the pellet, and re-suspending it in PBS for bio-imaging studies. Swiss albino mice (female) were divided into seven groups with n = 3 and anaesthetized by IP injection with thiopentone sodium at a dose of 0.2 mg kg−1 of body weight, followed by injection of 200 μL of DiD-loaded NPs through their tail vein. After the administration of NPs, images were acquired at 1, 6, 12 and 24 h by using an IVIS imaging system (PHOTONIMAGER OPTIMA, Biospace Lab, France) at the excitation/emission maxima of 637/672 nm in the near infrared (IR) region.
4.6 Biodegradation of p(NAG-co-NAPA)(1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 4) (P4) NPs
4) (P4) NPs
The biodegradation study was only performed57 for P4 NPs in simulated body fluid (SBF) at pH 7.4. In brief: 4 mg of P4 NPs was suspended in 8 mL of SBF solution, and then shaken at 37 °C and 100 rpm for 7 days. The degraded NPs were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 15 min and lyophilized further for morphology observations through HRTEM. 0.2 mg mL−1 of sodium azide was used to prevent any microbial contamination. The biodegradation study was only conducted for P4 NPs given that were finally used for the TNBC treatment.
000 rpm at 4 °C for 15 min and lyophilized further for morphology observations through HRTEM. 0.2 mg mL−1 of sodium azide was used to prevent any microbial contamination. The biodegradation study was only conducted for P4 NPs given that were finally used for the TNBC treatment.
4.7 Network pharmacology and molecular docking assessment for finding targets of DHA and piperine
4.7.2.1 Preparation of target proteins and ligands. 3D crystal structures of the targeted proteins were retrieved from the Protein Data Bank (PDB) and pre-processed by removing water molecules, and the ligand present, identical chains, and other heteroatoms listed in Table S1 (ESI†). After that, polar H-atoms and Kollman charges were added, followed by saving in.pdbqt format. Ligand structures (DHA (3000518), piperine (638024) and paclitaxel (36314)) were downloaded from the PubChem database in SDF format and energy minimization was performed using the Avogadro Software (Universal force field) to ensure the optimal conformation for the docking study and saved in PDB format.
4.7.2.2 Docking procedure. The docking studies were conducted in two steps, as follows: (1) single docking (protein with ligand) and (2) sequential docking (proteins with piperine followed by DHA and proteins with DHA followed by piperine). Blind docking was performed using AutoDock Tools 1.5.6. For single docking, the grid box centered on a protein with dimensions to cover the complete protein. For sequential docking, one ligand was docked with the protein and saved as a complex in.pdbqt format and further docked with another ligand following the same steps. The parameters set here were GA run of 50, population size of 300 and saving the output file by selecting the Lamarckian Genetic Algorithm (4.2) in “protein.dpf” format. Further, 3D structures of the protein–ligand complex were visualized in Pymol and all types of interactions (polar hydrogen bonding and hydrophobic nonpolar interactions) between the ligands and proteins were obtained from the Lig Plot.
4.8 Preparation of DHA or piperine-loaded p(NAG-co-NAPA) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 4) NPs (DP4 and PP4)
4) NPs (DP4 and PP4)
DHA and piperine were encapsulated in the pores of P4 NPs via the vacuum loading method. Briefly, 200 mg of drugs (DHA and piperine) and 200 mg of NPs (P4) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were separately dispersed in 8 mL of ethanol, followed by the dropwise addition of the formed to the latter under continuous stirring at 4 °C. After complete addition, two different mixtures of (1) DHA in P4 (DP4) and (2) piperine in P4 (PP4) were placed in a rocker and shaken at 300 rpm at 4 °C for 24 h. Within 24 h, 4 cycles of vacuum (20
1) were separately dispersed in 8 mL of ethanol, followed by the dropwise addition of the formed to the latter under continuous stirring at 4 °C. After complete addition, two different mixtures of (1) DHA in P4 (DP4) and (2) piperine in P4 (PP4) were placed in a rocker and shaken at 300 rpm at 4 °C for 24 h. Within 24 h, 4 cycles of vacuum (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 s, i.e. on–off cycle) was performed to achieve the maximum loading efficiency. Further, the NFs were centrifuged at 14
10 s, i.e. on–off cycle) was performed to achieve the maximum loading efficiency. Further, the NFs were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM for 5 min at 4 °C, and then the supernatants were collected and stored for calculating the drug entrapment efficiency (EE (%)) and loading efficiency (LE (%)). Further, the pellets were washed with MilliQ twice and centrifuged at 14
000 RPM for 5 min at 4 °C, and then the supernatants were collected and stored for calculating the drug entrapment efficiency (EE (%)) and loading efficiency (LE (%)). Further, the pellets were washed with MilliQ twice and centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min at 4 °C to remove the unloaded drug molecules, then lyophilized and stored in airtight amber colour tubes at 4 °C. The supernatant was assessed for DHA (λmax= 205 nm) and piperine (λmax= 343 nm) loaded in P4 by UV-vis spectroscopy. The following equations (Eqn 1 and 2) were used to calculate EE (%) and LE (%) for the respective NFs. Further, TGA was studied to confirm the loading of drugs in the NFs.
000 rpm for 5 min at 4 °C to remove the unloaded drug molecules, then lyophilized and stored in airtight amber colour tubes at 4 °C. The supernatant was assessed for DHA (λmax= 205 nm) and piperine (λmax= 343 nm) loaded in P4 by UV-vis spectroscopy. The following equations (Eqn 1 and 2) were used to calculate EE (%) and LE (%) for the respective NFs. Further, TGA was studied to confirm the loading of drugs in the NFs. | (1) |
 | (2) |
4.9 In vitro drug release study of DP4 and PP4 NFs
The dialysis bag method was followed to study the in vitro drug release % of NFs. Briefly, 4 mg of prepared NFs was added to 2 mL of 1× PBS (pH 7.4) and the solution was placed in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kDa dialysis bag and immersed in a beaker containing 50 mL of 1 × PBS (pH 7.4). The beaker was placed in an orbital shaker at 100 rpm and 37 °C ± 2 °C for a period of 30 days (720 h). At predefined intervals, 3 mL of supernatant was collected from the beaker for UV-vis analysis at λ = 205 and 343 nm for DHA and piperine, respectively. Simultaneously, the beaker was replenished with an equal amount of fresh 1 × PBS (pH 7.4). The release studies were also performed at pH = 6.8, given that the pH of the tumor microenvironment is at a similar level. All the release studies were performed in triplicate for statistical significance.
000 kDa dialysis bag and immersed in a beaker containing 50 mL of 1 × PBS (pH 7.4). The beaker was placed in an orbital shaker at 100 rpm and 37 °C ± 2 °C for a period of 30 days (720 h). At predefined intervals, 3 mL of supernatant was collected from the beaker for UV-vis analysis at λ = 205 and 343 nm for DHA and piperine, respectively. Simultaneously, the beaker was replenished with an equal amount of fresh 1 × PBS (pH 7.4). The release studies were also performed at pH = 6.8, given that the pH of the tumor microenvironment is at a similar level. All the release studies were performed in triplicate for statistical significance.
4.10 Evaluation of IC50 and combination index (CI) for DP4, PP4 and DP4:PP4 (Co-NPs)
The IC50 values of the developed NFs were determined using the MTT assay. In the case of Co-NPs, a ratio-based concentration was considered for the MTT study. Further, the combination index for Co-NPs was determined using the Chou and Talalay method.58 The synergistic or antagonist effect of Co-NPs on MDA-MB-231 was studied using eqn (3). | (3) |
4.11 In vitro inhibition of MDA-MB-231 cell migration
The migration inhibitory effect of P4, DP4, PP4 and Co-NPs was investigated using MDA-MB-231 cells by considering no treatment as the negative control. 2 × 105 cells per well was cultured with 800 μL of complete media in a 12-well plate and kept for 24 h for 70%–80% confluency with adhesion. Then, a linear scratch wound was generated in each well using a 20 μL sterile tip, followed by washing with 1 × PBS three times. Then, the desired concentration of NPs (350 μg mL−1) was added to the wells. Further, images were acquired at 0 and 24 h. The Fiji ImageJ software was used to quantify the change in width (%) and rate of migration (%).4.12 Apoptosis study by annexin-V/propidium iodide
The apoptosis study was performed using an annexin-v/PI kit (Cat. No. BMS500F1-100, Invitrogen). 2 × 105 cells per well were cultured in a 6-well plate and incubated for 24 h. Then, P4, DP4, PP4, and Co-NPs at a final concentration of 350 μg mL−1 each were separately added to the cells and incubated for 24 h. Then, the cells were trypsinized in PBS, followed by centrifugation at 200×g for 5 min. After discarding the supernatant, 5 μL of annexin-V and 10 μL of PI were added to each tube, mixed thoroughly and incubated in the dark for 30 min. Finally, 400 μL of binding buffer was added to each tube and flow cytometry performed for the apoptosis study (CytoFlex B53000, Beckmann Coulter).4.13 Cell cycle analysis
The cell cycle analysis was performed according to an earlier study with slight modification.59 2 × 105 cells per well were cultured in a 6-well plate and kept for 24 h. Then, P4, DP4, PP4, and Co-NPs at a final concentration of 350 μg mL−1 each were separately added to the cells and incubated for 24 h. Then, the cells were trypsinized in PBS, followed by centrifugation at 200×g for 5 min. After discarding the supernatant, the cells were fixed in 70% ethanol (−20 °C) and incubated for 20 min. Further, the cells were centrifuged at 200×g for 5 min and the supernatant discarded. 500 μL of RNase A (200 μg mL−1 stock) was added to each tube and incubated for 30 min. Then, 500 μL of PI (50 μg mL−1 stock) was added to each tube and incubated for 30 min in the dark (in ice). Finally, the samples were mixed thoroughly and analyzed through flow cytometry.4.14 Quantification of intracellular ROS generation and mitochondrial membrane potential (MMP)
Flow cytometry analysis was also conducted to quantify the intracellular ROS and change in mitochondrial membrane potential. After 24 h of treatment with NPs (P4) and NFs (DP4, PP4, and Co-NP) at 350 μg mL−1 and untreated as the control, the adhered cells were collected (1200 rpm) and washed with 1 × PBS 3 times. Then, the cells were stained with 50 μM solution of DCFH-DA and incubated at 37 °C for 30 min in the dark. Further, the supernatant was removed and washed with 1 × PBS two times and suspended in 500 μL of 1 × PBS. The shift in the bands and intracellular ROS generation were analyzed. Similarly, for the mitochondrial membrane potential measurement, instead of DCFH-DA, Rhodamine-123 was used and all other steps were followed, similar to the procedure for the measurement of ROS.4.15 Animal ethical approval
All animal experiments were carried out in accordance with the guidelines of CPCSEA and approved by the Institutional Animal Ethical Committee (IAEC) of IIT (BHU), Varanasi, Uttar Pradesh, India (regd. no. 2123/GO/Re/S/21/CPCSEA). The animal ethical approval reference No. is IIT(BHU)/IAEC/2024/L049.4.16 Statistical significance
One-way ANOVA with independent Student's t-test was used to evaluate the in vitro and in vivo data statistically. The significance was calculated by considering p < 0.05 and represented as mean (±) SD, unless otherwise stated.Author contributions
P. P. was the principal investigator (PI) of this project. Ideation, experimental designs, result analysis and manuscript writing were mainly carried out by S. P. and P. P. MTT and hemolysis study were performed by D. P., K. W. and P. S. G. Network pharmacology and cell-based study were performed by Jyotirmayee, T. B. and M. V. Docking study was done by A. M. and T. D. In vivo study was performed by K. K. and R. C. Manuscript was written and finalized by S. P. and P. P. All the authors have checked and given approval to the final version of the manuscript.Data availability
The data supporting this study are available in the ESI† of this article.Conflicts of interest
There is no conflict of interest to report.Acknowledgements
The authors acknowledge financial support awarded to Prof. Paik by I-DAPT (File no. I-DAPT/IT (BHU)/2023-24/Project Sanction/47), Indian Council of Medical Research (ICMR), India (Ref: EMDR/SG/12/2023-4724), STARS-IISc Bangalore (Ref. MoE-STARS/STARS-2/2023-0318) and Anusandhan National Research Foundation (ANRF), India (Ref: CRG/2023/005576). Sukanya acknowledges the awarded DST-INSPIRE fellowship (IF180185) and financial support from ICMR to carry out the PhD work. The authors acknowledge Dr Monika Singh for giving access of Biorender to draw ToC, Dr Avnish Parmar (Department of Physics, IIT (BHU)) for Malvern Zeta Analyser and Dr Alakh Niranjan Sahu (Department of Pharmaceutical Engineering and Technology, IIT (BHU)) for UV-vis facility.References
- S. A. Dilliard and D. J. Siegwart, Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs, Nat. Rev. Mater., 2023, 8(4), 282–300, DOI:10.1038/s41578-022-00529-7.
- N. Bertrand and J.-C. Leroux, The journey of a drug-carrier in the body: An anatomo-physiological perspective, J. Controlled Release, 2012, 161(2), 152–163, DOI:10.1016/j.jconrel.2011.09.098.
- W. Sun, Q. Hu, W. Ji, G. Wright and Z. Gu, Leveraging Physiology for Precision Drug Delivery, Physiol. Rev., 2017, 97(1), 189–225, DOI:10.1152/physrev.00015.2016.
- J. Li and K. Kataoka, Chemo-physical Strategies to Advance the in Vivo Functionality of Targeted Nanomedicine: The Next Generation, J. Am. Chem. Soc., 2021, 143(2), 538–559, DOI:10.1021/jacs.0c09029.
- S. A. Dilliard, Q. Cheng and D. J. Siegwart, On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(52), e2109256118, DOI:10.1073/pnas.2109256118.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Cellular uptake of nanoparticles: journey inside the cell, Chem. Soc. Rev., 2017, 46(14), 4218–4244, 10.1039/C6CS00636A.
- S. Peng, Y. Wang, N. Li and C. Li, Enhanced cellular uptake and tumor penetration of nanoparticles by imprinting the “hidden” part of membrane receptors for targeted drug delivery, Chem. Commun., 2017, 53(81), 11114–11117, 10.1039/C7CC05894B.
- W. Xue, J. E. Dahlman, T. Tammela, O. F. Khan, S. Sood, A. Dave, W. Cai, L. M. Chirino, G. R. Yang and R. Bronson, et al., Small RNA combination therapy for lung cancer, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(34), E3553–E3561, DOI:10.1073/pnas.1412686111.
- O. F. Khan, P. S. Kowalski, J. C. Doloff, J. K. Tsosie, V. Bakthavatchalu, C. B. Winn, J. Haupt, M. Jamiel, R. Langer and D. G. Anderson, Endothelial siRNA delivery in nonhuman primates using ionizable low–molecular weight polymeric nanoparticles, Sci. Adv., 2018, 4(6), eaar8409, DOI:10.1126/sciadv.aar8409.
- P. S. Kowalski, U. Capasso Palmiero, Y. Huang, A. Rudra, R. Langer and D. G. Anderson, Ionizable Amino-Polyesters Synthesized via Ring Opening Polymerization of Tertiary Amino-Alcohols for Tissue Selective mRNA Delivery, Adv. Mater., 2018, 30(34), 1801151, DOI:10.1002/adma.201801151 (acccessed 2024/09/22).
- Z. Zhang, C. Wang, Y. Zha, W. Hu, Z. Gao, Y. Zang, J. Chen, J. Zhang and L. Dong, Corona-directed nucleic acid delivery into hepatic stellate cells for liver fibrosis therapy, ACS Nano, 2015, 9(3), 2405–2419 CrossRef CAS PubMed.
- S. Liu, X. Wang, X. Yu, Q. Cheng, L. T. Johnson, S. Chatterjee, D. Zhang, S. M. Lee, Y. Sun and T.-C. Lin, et al., Zwitterionic Phospholipidation of Cationic Polymers Facilitates Systemic mRNA Delivery to Spleen and Lymph Nodes, J. Am. Chem. Soc., 2021, 143(50), 21321–21330, DOI:10.1021/jacs.1c09822.
- B. Pandey, N. G. Patil, G. S. Bhosle, A. V. Ambade and S. S. Gupta, Amphiphilic Glycopolypeptide Star Copolymer-Based Cross-Linked Nanocarriers for Targeted and Dual-Stimuli-Responsive Drug Delivery, Bioconjugate Chem., 2019, 30(3), 633–646, DOI:10.1021/acs.bioconjchem.8b00831.
- A. K. Yamala, V. Nadella, Y. Mastai, H. Prakash and P. Paik, Poly-N-acryloyl-(l-phenylalanine methyl ester) hollow core nanocapsules facilitate sustained delivery of immunomodulatory drugs and exhibit adjuvant properties, Nanoscale, 2017, 9(37), 14006–14014, 10.1039/C7NR03724D.
- S. Kim, Y. Shi, J. Y. Kim, K. Park and J.-X. Cheng, Overcoming the barriers in micellar drug delivery: loading efficiency, in vivo stability, and micelle–cell interaction, Expert Opin. Drug Delivery, 2010, 7(1), 49–62, DOI:10.1517/17425240903380446.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nanocarriers as an emerging platform for cancer therapy, Nat. Nanotechnol., 2007, 2(12), 751–760, DOI:10.1038/nnano.2007.387.
- H. Y. Choi and J.-E. Chang, Targeted Therapy for Cancers: From Ongoing Clinical Trials to FDA-Approved Drugs, Int. J. Mol. Sci., 2023, 24(17), 13618 CrossRef CAS PubMed.
- N. Salari, F. Faraji, F. M. Torghabeh, F. Faraji, K. Mansouri, F. Abam, S. Shohaimi, H. Akbari and M. Mohammadi, Polymer-based drug delivery systems for anticancer drugs: A systematic review, Cancer Treat. Res. Commun., 2022, 32, 100605, DOI:10.1016/j.ctarc.2022.100605.
- E. T. Sedeta, B. Jobre and B. Avezbakiyev, Breast cancer: Global patterns of incidence, mortality, and trends, Am. Soc. Clin. Oncol., 2023, 10528 CrossRef.
- Breast Cancer. World Health Organization. World Health Organization. 13-03-2024. https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed) Search PubMed.
- X. Li, J. Yang, L. Peng, A. A. Sahin, L. Huo, K. C. Ward, R. O’Regan, M. A. Torres and J. L. Meisel, Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer, Breast Cancer Res. Treat., 2017, 161(2), 279–287, DOI:10.1007/s10549-016-4059-6.
- B. Mamnoon, J. Loganathan, M. I. Confeld, N. De Fonseka, L. Feng, J. Froberg, Y. Choi, D. M. Tuvin, V. Sathish and S. Mallik, Targeted Polymeric Nanoparticles for Drug Delivery to Hypoxic, Triple-Negative Breast Tumors, ACS Appl. Bio Mater., 2021, 4(2), 1450–1460, DOI:10.1021/acsabm.0c01336.
- S. Tripathy, S. Haque, S. Londhe, S. Das, C. C. Norbert, Y. Chandra, B. Sreedhar and C. R. Patra, ROS mediated Cu[Fe(CN)5NO] nanoparticles for triple negative breast cancer: A detailed study in preclinical mouse model, Biomater. Adv., 2024, 160, 213832, DOI:10.1016/j.bioadv.2024.213832.
- G. Bianchini, C. De Angelis, L. Licata and L. Gianni, Treatment landscape of triple-negative breast cancer—expanded options, evolving needs, Nat. Rev. Clin. Oncol., 2022, 19(2), 91–113, DOI:10.1038/s41571-021-00565-2.
- L. Zhang, Z. Ren, J. Lü, X. Mo, J. Lin, Y. Li, W. Ma, P. Liu, Y. Shen and Q. Zhao, et al., Nanoparticles carrying paclitaxel and anti-miR-221 for breast cancer therapy triggered by ultrasound, Cell Death Discovery, 2023, 9(1), 298, DOI:10.1038/s41420-023-01594-9.
- S. Londhe, S. Haque, S. Tripathy, S. Bojja and C. R. Patra, Silver nitroprusside nanoparticles for breast cancer therapy: in vitro and in vivo approach, Nanoscale, 2023, 15(23), 10017–10032, 10.1039/d3nr00221g.
- S. Kongpatanakul, S. Chatsiricharoenkul, A. Khuhapinant, S. Atipas and J. Kaewkungwal, Comparative study of dihydroartemisinin and artesunate safety in healthy Thai volunteers, Int. J. Clin. Pharmacol. Ther., 2009, 47(9), 579–586 CrossRef CAS PubMed.
- J.-J. Lu, L.-H. Meng, Y.-J. Cai, Q. Chen, L.-J. Tong, L.-P. Lin and J. Ding, Dihydroartemisinin induces apoptosis in HL-60 leukemia cells dependent of iron and p38 mitogen-activated protein kinase activation but independent of reactive oxygen species, Cancer Biol. Ther., 2008, 7(7), 1017–1023, DOI:10.4161/cbt.7.7.6035.
- S. Kumar, C. Bhandari, P. Sharma and N. Agnihotri, Role of piperine in chemoresistance, in Role of nutraceuticals in cancer chemosensitization, Elsevier, 2018, pp. 259–286 Search PubMed.
- S. Abdelhamed, S. Yokoyama, A. Refaat, K. Ogura, H. Yagita, S. Awale and I. Saiki, Piperine Enhances the Efficacy of TRAIL-based Therapy for Triple-negative Breast Cancer Cells, Anticancer Res., 2014, 34(4), 1893 CAS.
- C. R. Quijia and M. Chorilli, Piperine for treating breast cancer: A review of molecular mechanisms, combination with anticancer drugs, and nanosystems, Phytother. Res., 2022, 36(1), 147–163 CrossRef CAS PubMed.
- T. Chen, J. Xu, L. Zhu and D. Yan, Cancer-cell-membrane-camouflaged supramolecular self-assembly of antisense oligonucleotide and chemodrug for targeted combination therapy, Nanoscale, 2023, 15(4), 1914–1924, 10.1039/D2NR05669K.
- L. Lin, Z. Schneiderman, A. Venkatraman and E. Kokkoli, Formation of ssDNA nanotubes from spherical micelles and their use as a delivery vehicle for chemotherapeutics and senolytics to triple negative breast cancer cells, Nanoscale, 2023, 15(22), 9801–9812, 10.1039/d3nr00196b.
- Y. Liu, H. Liu, L. Wang, Y. Wang, C. Zhang, C. Wang, Y. Yan, J. Fan, G. Xu and Q. Zhang, Amplification of oxidative stress via intracellular ROS production and antioxidant consumption by two natural drug-encapsulated nanoagents for efficient anticancer therapy, Nanoscale Adv., 2020, 2(9), 3872–3881, 10.1039/D0NA00301H.
- A. K. Rajora, E. D. Ahire, M. Rajora, S. Singh, J. Bhattacharya and H. Zhang, Emergence and impact of theranostic-nanoformulation of triple therapeutics for combination cancer therapy, Smart Med., 2024, 3(1), e20230035, DOI:10.1002/SMMD.20230035.
- W. Zhou, X. Ma, J. Wang, X. Xu, O. Koivisto, J. Feng, T. Viitala and H. Zhang, Co-delivery CPT and PTX prodrug with a photo/thermo-responsive nanoplatform for triple-negative breast cancer therapy, Smart Med., 2022, 1(1), e20220036, DOI:10.1002/SMMD.20220036.
- Y. Gong, W. Chen, X. Chen, Y. He, H. Jiang, X. Zhang, L. Pan, B. Ni, F. Yang and Y. Xu, et al., An Injectable Epigenetic Autophagic Modulatory Hydrogel for Boosting Umbilical Cord Blood NK Cell Therapy Prevents Postsurgical Relapse of Triple-Negative Breast Cancer, Adv. Sci., 2022, 9(23), 2201271, DOI:10.1002/advs.202201271 (acccessed 2025/02/02).
- S. K. Nethi, S. Kothadiya, B. M. White, S. Rachagani, R. Bardhan and S. K. Mallapragada, Polyanhydride Copolymer-Based Niclosamide Nanoparticles for Inhibiting Triple-Negative Breast Cancer: Metabolic Responses and Synergism with Paclitaxel, ACS Appl. Mater. Interfaces, 2024, 16(51), 70362–70377, DOI:10.1021/acsami.4c17961.
- S. Zheng, M. Li, W. Xu, J. Zhang, G. Li, H. Xiao, X. Liu, J. Shi, F. Xia and C. Tian, et al., Dual-targeted nanoparticulate drug delivery systems for enhancing triple-negative breast cancer treatment, J. Controlled Release, 2024, 371, 371–385, DOI:10.1016/j.jconrel.2024.06.012.
- P. S. Gupta, K. Wasnik, S. Patra, D. Pareek, G. Singh, D. D. Yadav, S. Maity and P. Paik, Nitric oxide releasing novel amino acid-derived polymeric nanotherapeutic with anti-inflammatory properties for rapid wound tissue regeneration, Nanoscale, 2024, 16(4), 1770–1791, 10.1039/D3NR03923D.
- K. Wasnik, P. S. Gupta, G. Singh, S. Maity, S. Patra, D. Pareek, S. Kumar, V. Rai, R. Prakash and A. Acharya, et al., Neurogenic and angiogenic poly(N-acryloylglycine)-co-(acrylamide)-co-(N-acryloyl-glutamate) hydrogel: preconditioning effect under oxidative stress and use in neuroregeneration, J. Mater. Chem. B, 2024, 12(25), 6221–6241, 10.1039/D4TB00243A.
- K. Wasnik, P. S. Gupta, S. Mukherjee, A. Oviya, R. Prakash, D. Pareek, S. Patra, S. Maity, V. Rai and M. Singh, et al., Poly(N-acryloylglycine-acrylamide) Hydrogel Mimics the Cellular Microenvironment and Promotes Neurite Growth with Protection from Oxidative Stress, ACS Appl. Bio Mater., 2023, 6(12), 5644–5661, DOI:10.1021/acsabm.3c00807.
- P. S. Gupta, K. Wasnik, G. Singh, S. Patra, D. Pareek, D. D. Yadav, M. S. Tomar, S. Maiti, M. Singh and P. Paik, In vivo potential of polymeric N-acryloyl-glycine nanoparticles with anti-inflammatory activities for wound healing, Mater. Adv., 2023, 4(20), 4718–4731, 10.1039/D3MA00378G.
- A. K. Yamala, V. Nadella, Y. Mastai, H. Prakash and P. Paik, P-LME polymer nanocapsules stimulate naïve macrophages and protect them from oxidative damage during controlled drug release, J. Appl. Polym. Sci., 2020, 137(6), 48363, DOI:10.1002/app.48363.
- L. Werber, L. C. Preiss, K. Landfester, R. Muñoz-Espí and Y. Mastai, Isothermal Titration Calorimetry of Chiral Polymeric Nanoparticles, Chirality, 2015, 27(9), 613–618, DOI:10.1002/chir.22473 (acccessed 2024/09/15).
- H. Wang, Network pharmacology- and molecular docking-based approaches to unveil the pharmacological mechanisms of dihydroartemisinin against esophageal carcinoma, Front. Genet., 2022, 13, 1017520, DOI:10.3389/fgene.2022.1017520 Original Research.
- P. M. Paarakh, D. C. Sreeram, Shruthi S D and S. P. S. Ganapathy, In vitro cytotoxic and in silico activity of piperine isolated from Piper nigrum fruits Linn, In Silico Pharmacol., 2015, 3(1), 9, DOI:10.1186/s40203-015-0013-2.
- A. A. El-Shehawy, A. Elmetwalli, A. H. El-Far, S. A. E.-R. Mosallam, A. F. Salama, A. O. Babalghith, M. A. Mahmoud, H. Mohany, M. Gaber and T. El-Sewedy, Thymoquinone, piperine, and sorafenib combinations attenuate liver and breast cancers progression: epigenetic and molecular docking approaches, BMC Complement. Med. Ther., 2023, 23(1), 69 CrossRef CAS.
- X. Dai, X. Zhang, W. Chen, Y. Chen, Q. Zhang, S. Mo and J. Lu, Dihydroartemisinin: A Potential Natural Anticancer Drug, Int. J. Biol. Sci., 2021, 17(2), 603–622, DOI:10.7150/ijbs.50364 Review.
- H. M. Nawara, S. M. Afify, G. Hassan, M. H. Zahra, A. Seno and M. Seno, Paclitaxel-Based Chemotherapy Targeting Cancer Stem Cells from Mono- to Combination Therapy, Biomedicines, 2021, 9(5), 500 CrossRef CAS.
- O. Wattanathamsan, R. Thararattanobon, R. Rodsiri, P. Chanvorachote, C. Vinayanuwattikun and V. Pongrakhananon, Tubulin acetylation enhances lung cancer resistance to paclitaxel-induced cell death through Mcl-1 stabilization, Cell Death Discovery, 2021, 7(1), 67, DOI:10.1038/s41420-021-00453-9.
- X. Wang, B. Gigant, X. Zheng and Q. Chen, Microtubule-targeting agents for cancer treatment: Seven binding sites and three strategies, MedComm: Oncol., 2023, 2(3), e46, DOI:10.1002/mog2.46.
- P. Zhang, D. Gao, K. An, Q. Shen, C. Wang, Y. Zhang, X. Pan, X. Chen, Y. Lyv and C. Cui, et al., A programmable polymer library that enables the construction of stimuli-responsive nanocarriers containing logic gates, Nat. Chem., 2020, 12(4), 381–390, DOI:10.1038/s41557-020-0426-3.
- C. Ruggeri, N. Drinkwater, K. K. Sivaraman, R. S. Bamert, S. McGowan and A. Paiardini, Identification and Validation of a Potent Dual Inhibitor of the P. falciparum M1 and M17 Aminopeptidases Using Virtual Screening, PLoS One, 2015, 10(9), e0138957, DOI:10.1371/journal.pone.0138957.
- A. G. Porter and R. U. Jänicke, Emerging roles of caspase-3 in apoptosis, Cell Death Differ., 1999, 6(2), 99–104, DOI:10.1038/sj.cdd.4400476.
- A. Boroumand Moghaddam, M. Moniri, S. Azizi, R. Abdul Rahim, A. Bin Ariff, M. Navaderi and R. Mohamad, Eco-Friendly Formulated Zinc Oxide Nanoparticles: Induction of Cell Cycle Arrest and Apoptosis in the MCF-7 Cancer Cell Line, Genes, 2017, 8(10), 281, DOI:10.3390/genes8100281.
- J. Zhu, H. Han, F. Li, X. Wang, J. Yu, C.-C. Chu and D. Wu, Self-assembly of amino acid-based random copolymers for antibacterial application and infection treatment as nanocarriers, J. Colloid Interface Sci., 2019, 540, 634–646, DOI:10.1016/j.jcis.2018.12.091.
- T.-C. Chou and P. Talalay, Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors, Adv. Enzyme Regul., 1984, 22, 27–55, DOI:10.1016/0065-2571(84)90007-4.
- P. K. Gupta, S. Pappuru, S. Gupta, B. Patra, D. Chakraborty and R. S. Verma, Self-assembled dual-drug loaded core-shell nanoparticles based on metal-free fully alternating polyester for cancer theranostics, Mater. Sci. Eng.: C, 2019, 101, 448–463, DOI:10.1016/j.msec.2019.03.041.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb02445a |
| This journal is © The Royal Society of Chemistry 2025 |