Open Access Article
Open Access ArticleHierarchical biofilm models using sodium alginate beads containing bacteria embedded in a cellulose–chitosan hydrogel matrix†
Shiwei
Li
 a,
Yuxuan
Ji
a,
Xuxiang
Xue
a,
Yaohao
Yang
b,
Yi
Huang
a,
Yuxuan
Ji
a,
Xuxiang
Xue
a,
Yaohao
Yang
b,
Yi
Huang
 b,
Shuiqing
Yang
c and
Xianfeng
Chen
b,
Shuiqing
Yang
c and
Xianfeng
Chen
 *a
*a
aSchool of Engineering, Institute for Bioengineering, University of Edinburgh, The Kings Buildings, EH9 3JL Edinburgh, UK. E-mail: michael.chen@ed.ac.uk; Tel: +44 (0)131 6502784
bSchool of Engineering, Institute for Materials & Processes, University of Edinburgh, The Kings Buildings, EH9 3JL Edinburgh, UK
cJiangsu Dingying New Materials Co., Ltd. Jiangsu Dingying New Materials Co., Ltd., Changzhou, Jiangsu 213031, China
First published on 17th February 2025
Abstract
In biofilm studies, a stable model is crucial for exploring infection mechanisms, antibiotic resistance, and evaluating materials’ antibiofilm performance. Cultured biofilms often face challenges, such as slow maturation or rapid bacteria dispersion. Therefore, developing a stable, mature-stage biofilm model is critical for effective biofilm research. In this study, we report a beads-in-hydrogel biofilm model, in which sodium alginate (SA) hydrogel beads that contain bacteria are embedded within a chitosan–cellulose hydrogel film to simulate natural bacterial biofilms. This model can retain bacteria for a relatively long period of time, preventing their dispersion to the surrounding areas while keeping them viable. The reliability of the model was validated by measuring functional molecules, including extracellular DNA and biofilm-forming related proteins. Overall, this study presents a stable 3D beads-in-hydrogel biofilm model that effectively replicates natural biofilms, providing a reliable platform for exploring infection mechanisms, antibiotic resistance, and evaluating antibiofilm strategies.
1 Introduction
A biofilm is a type of bacterial community characterized by strong antibiotic resistance, vertical gene transfer, and a self-producing polymer matrix.1 Research studies on biofilms are crucial across various fields, including the food industry,2 environmental protection2 and medical research.3 Biofilm infections typically cause varying degrees of harm to the human body. Consequently, developing innovative strategies to combat biofilms is essential, utilizing both in vitro and in vivo models. These models often involve bacteria like E. coli, P. aeruginosa, and S. aureus, known for their ability to form biofilms and cause chronic diseases.4In vitro models have become increasingly valuable compared to in vivo models due to their controllability, lower costs, and ability to yield precise data in a controlled environment. In vitro biofilm models include both two-dimensional (2D) and three-dimensional (3D) reactor systems.5 2D models are typically utilized to investigate fundamental mechanisms such as biofilm adhesion and growth.5,6 However, these models are limited in their ability to replicate the complex microenvironmental changes that occur during biofilm formation. Consequently, 3D models have gained prominence. These models allow for the study of dynamic biofilm formation, including the development of extracellular polymeric substance (EPS) architecture and bacterial dynamics within the biofilm.7,8
To develop 3D biofilm models, researchers choose appropriate scaffolds or substrates, such as hydrogels, alginate beads, or microfluidic devices, to simulate the natural environments of biofilms. These substrates are inoculated with bacterial suspensions to initiate biofilm formation, under carefully controlled conditions, including nutrient flow and shear forces. Systems such as drip flow reactors or rotating disk reactors are employed to maintain these conditions.7
Hydrogels are particularly advantageous for constructing 3D biofilm models due to their high water content and adjustable mechanical properties, which closely replicate the natural extracellular matrix of biofilms.9 Made from materials like alginate, agarose, and polyacrylamide, these hydrogels provide a versatile platform for biofilm cultivation.10 The inclusion of nutrients, signaling molecules, and other environmental factors within the hydrogels facilitates the study of biofilm dynamics, including formation, maturation, and resistance mechanisms.11 These models are essential for testing antimicrobial agents, developing new treatment strategies, and exploring the fundamental biology of biofilms. Furthermore, they can accommodate multiple bacterial strains, thereby reflecting the polymicrobial nature of many biofilms associated with chronic infections.12
Despite the advantages of 3D hydrogel-based biofilm models, challenges remain in fully replicating the natural conditions of biofilm formation and maintaining the stability of hydrogel matrices over extended periods. Various methods have been developed to address specific research needs. One approach used alginate beads to culture biofilms within an alginate matrix, allowing growth in three-dimensional space that mimics deep tissue colonization seen in clinical settings.12,13 However, the stability of alginate hydrogels is limited, and maintaining biofilm structures over 2 days is difficult. Another method involves using hydrogels composed of polymers like 3-sulfopropyl acrylate potassium salt and polyethylene glycol diacrylate, polymerized under UV light to create a stable matrix suitable for studying microbial interactions and testing the efficacy of antimicrobial agents.11 Additionally, 3D bioprinting techniques, such as those explored by Aliyazdi et al., have been used to fabricate E. coli MG1655 biofilms using a gelatin–alginate hydrogel as a bioink.14 This method ensures that biofilm properties are retained post-printing, as evidenced by various analyses, including antibiotic susceptibility assays and metabolic profiling, which demonstrate a high similarity to native biofilms. However, the poor mechanical stability causes challenges in biofilm transfer and shape retention. Especially, for long-time cultivation, the printed hydrogel scaffold will be easily broken down.
Additionally, wound infection models are frequently utilized in the development of 3D hydrogel biofilm models. For instance, Thaarup et al. developed a collagen-based, layered chronic wound biofilm model to simulate the dynamic nutrient influx and waste exchange of chronic wounds.15 This model uses a gelatin–alginate hydrogel matrix incorporating Staphylococcus aureus and Pseudomonas aeruginosa, cast in transwell inserts within wound-simulating media. The setup allows for continuous nutrient and waste exchange, maintaining bacterial stability and more accurately mimicking the in vivo environment. However, this model was built based on a semi-solid environment which may impact the biofilm architecture construction and accuracy of treatment efficacy assessments.
In this research, a long-term bead-in-hydrogel platform is designed to closely mimic mature-phase biofilms. The design involves a hierarchy structure achieved by embedding sodium alginate (SA) hydrogel beads containing bacteria within a chitosan and cellulose gel film, which effectively inhibits bacterial migration into the surrounding culture medium. In the beads-in-hydrogel platform, bacterial hydrogel beads are formed through ionic crosslinking between SA and calcium chloride (CaCl2), while the hydrogel films are stabilized via hydrogen bonding between chitosan and cellulose in an acidic environment.16 Typically, the interaction between calcium ions (Ca2+) and SA lacks long-term stability, leading to bead degradation and subsequent bacterial leakage. However, the introduction of a double-crosslink gel film significantly strengthens the microstructure,17 including ionic and hydrogen bonds between CaCl2 and SA, as well as between chitosan and cellulose, and enables the model to retain bacteria for extended periods compared to isolated hydrogel beads.18,19 In this work, extracellular DNA (eDNA) and secreted protein composition of this model align with those of natural biofilms. Additionally, this setup not only replicates the natural distribution process of bacteria prior to dispersion but also enhances the model's longevity to keep its architecture for 9 days.
2 Methods and materials
2.1 Materials
SA (Sigma-Aldrich), chitosan (low molecular weight, Sigma-Aldrich), calcium chloride (anhydrous, BioReagent, suitable for insect cell culture, suitable for plant cell culture, ≥96.0%, Sigma-Aldrich), cellulose (microcrystalline, powder, Sigma-Aldrich), hydrochloric acid solution (1.0 N, BioReagent, suitable for cell culture), the LB medium (Sigma-Aldrich), the LB agar medium (Sigma-Aldrich), SimplyBlue™ SafeStain (Thermo Fisher scientific), 20X Bolt™ MES SDS running buffer (Thermo Fisher scientific), PageRuler™ Plus Prestained Protein Ladder (10 to 250 kDa, Thermo Fisher scientific), Novex™ Tris–Glycine SDS Sample Buffer (2×, Thermo Fisher scientific), Novex™ Tris–Glycine Mini Protein Gels (10–20%, 1.0 mm, WedgeWell™ format, Thermo Fisher scientific), 1X phosphate-buffered saline (PBS, Sigma-Aldrich), TE buffer (20X, RNase-free, Thermo Fisher scientific), DEPC-treated water (Thermo Fisher scientific), ladder, Quant-iT™ PicoGreen™ dsDNA Assay Kits and dsDNA Reagents (Thermo Fisher scientific), and QuantiPro™ BCA Assay Kit (Sigma-Aldrich) were used in this study.2.2 Methods
For the separation of EPS proteins, the culture media were removed from the biofilm, which was then washed once with PBS. The biofilm was scraped from the surface using a sterile spatula and collected in a suitable container. The collected biofilm was resuspended in PBS, and the cells were disrupted using a vortex mixer to release the EPS. The resulting suspension was centrifuged at 4 °C and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 minutes to pellet the cells and debris. The supernatant was carefully collected and filtered through a 0.22 μm filter to remove any remaining cellular debris, followed by dialysis against PBS to remove small molecules and salts.
000g for 15 minutes to pellet the cells and debris. The supernatant was carefully collected and filtered through a 0.22 μm filter to remove any remaining cellular debris, followed by dialysis against PBS to remove small molecules and salts.
To separate proteins from a bead-in-hydrogel biofilm model, the culture media were removed and the biofilm was washed once with PBS. The bead-in-hydrogel biofilm model was scraped from the surface with a sterile spatula and collected in a suitable container. The biofilm was resuspended in PBS and 100 mM citrate acid buffer, allowed to stand for 10 minutes, and then disrupted using a vortex mixer to break the hydrogel crosslinks in the model. The suspension was centrifuged at 4 °C and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 minutes to pellet the cells and debris. The supernatant was collected, filtered through a 0.22 μm filter to remove any remaining cellular debris, and dialyzed against PBS to remove salts and small molecules.
000g for 15 minutes to pellet the cells and debris. The supernatant was collected, filtered through a 0.22 μm filter to remove any remaining cellular debris, and dialyzed against PBS to remove salts and small molecules.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The mixture was stirred until a uniform color was achieved. An equal volume of QuantiPro working solution and each sample (1
1. The mixture was stirred until a uniform color was achieved. An equal volume of QuantiPro working solution and each sample (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added to the wells of a 96-well plate. The plate was then incubated at 37 °C for 2 hours. Following incubation, the absorbance was measured at 562 nm using a plate reader (CLARIOstar Plus, BMG LABTECH, UK).
1) were added to the wells of a 96-well plate. The plate was then incubated at 37 °C for 2 hours. Following incubation, the absorbance was measured at 562 nm using a plate reader (CLARIOstar Plus, BMG LABTECH, UK).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The mixture was then heated at 85 °C for 4 minutes to denature the proteins, followed by a brief spin in a microcentrifuge to collect the sample at the bottom of the tube. The inner chamber of the electrophoresis apparatus was filled with running buffer. A protein ladder was loaded into the first well, followed by the loading of 30 μL of each protein sample into the subsequent wells. The voltage was set to 100–120 V for the stacking phase, which was then increased to 250 V for running the gel. The gel was run until the dye front reached the bottom of the gel. After electrophoresis, the gel was carefully removed from the apparatus and placed in a staining solution, where it was gently agitated for 1 hour. Following staining, the gel was removed from the staining solution and placed in deionized (DI) water, with gentle agitation until the background became clear.
1 ratio. The mixture was then heated at 85 °C for 4 minutes to denature the proteins, followed by a brief spin in a microcentrifuge to collect the sample at the bottom of the tube. The inner chamber of the electrophoresis apparatus was filled with running buffer. A protein ladder was loaded into the first well, followed by the loading of 30 μL of each protein sample into the subsequent wells. The voltage was set to 100–120 V for the stacking phase, which was then increased to 250 V for running the gel. The gel was run until the dye front reached the bottom of the gel. After electrophoresis, the gel was carefully removed from the apparatus and placed in a staining solution, where it was gently agitated for 1 hour. Following staining, the gel was removed from the staining solution and placed in deionized (DI) water, with gentle agitation until the background became clear.
3 Results and discussion
3.1 Bacterial retention
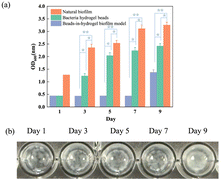
An E. coli culture solution was mixed with SA and then added to the CaCl2 solution to prepare hydrogel beads. Subsequently, these beads were embedded within a film of cellulose crosslinked with chitosan. This bead-in-hydrogel model was fabricated using a dual crosslinking system: one involves ionic bonding between SA and Ca2+, while the other relies on hydrogen bonding between cellulose and chitosan in an acidic environment. The hydrogen bonds are formed between the hydroxyl groups of cellulose and the protonated amino groups of chitosan.16 To evaluate the long-term bacterial retention of a beads-in-hydrogel biofilm model, the biofilm was immersed in a LB medium within a 96-well plate, ensuring that the biofilm model covered the entire bottom surface of each well, and the OD600 of the supernatant was measured on days 1, 3, 5, 7, and 9 to assess bacterial presence; as shown in Fig. 1(a), during this period, the medium was not exchanged to ensure precise results. The two control groups are hydrogel beads (SA beads) group and a natural biofilm group cultured using E. coli and LB media in a 96-well plate. Digital photographs of the bead-in-hydrogel biofilm model at different time points are displayed in Fig. 1(b). Natural biofilms are reported to typically mature on solid surfaces within a few days, with some beginning to disperse as early as 24 hours after reaching maturation,20 as indicated by dead bacteria detachment and dispersion into the surrounding environment. The bacteria are encapsulated in the hydrogel beads on day 1 and day 3, indicated by the high transparency of the solution. Later, when bacteria swim into the gel film layer, the model becomes less transparent on day 5 and day 7. By the 9th day, the culture medium becomes cloudy, indicating that the bacteria have entered the medium. In contrast, the bead-in-hydrogel biofilm model can effectively entrap bacteria for 7 days, which show significant differences compared with the other two groups (p < 0.05 compared with the bacterial hydrogel beads group, and p < 0.01 compared with the natural biofilm group). Beyond this period, bacteria reach the maximum capacity of the model and begin to enter the surrounding medium, causing an increase in the OD value, but still much lower than the control groups. The digital images of the bead-in-hydrogel biofilm model of days 1, 3, 5, 7, and 9 (Fig. 1(b)) present the transparent level and reflect bacterial density. This observation suggests that the bead-in-hydrogel biofilm model substantially delays bacterial migration. The OD changes of the pure LB medium, bacterial solution, and the suspension medium of the beads-in-hydrogel model can also indicate our biofilm model stability during this period (Fig. S1, ESI†). The OD value of our bead-in-hydrogel suspension (orange line) began increasing on day 7, whereas the bacterial hydrogel bead suspension showed an OD increase as early as day 3. This further indicates that our bead-in-hydrogel platform has significantly enhanced stability compared to the SA hydrogel alone.3.2 Bacterial density and distribution
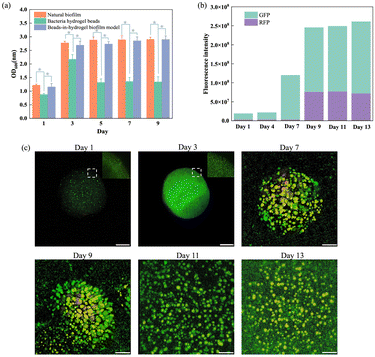
To validate the artificial beads-in-hydrogel biofilm model as a replicate of natural biofilms, changes in bacterial density were meticulously investigated over specific intervals – days 1, 3, 5, 7, and 9. The experiment involved extracting bacteria from natural biofilms, hydrogel beads, and the beads-in-hydrogel biofilm model. The optical density (OD600) of these extracted bacterial solutions was then measured (Fig. 2(a)). During the initial phase (from day 1 to day 3), as the natural biofilm matures, there is a notable increase in OD600, indicating a rise in bacterial density. After this phase, the density stabilizes, demonstrating the biofilm's maturity.In the hydrogel bead group, there was an increase in bacterial density during the first three days, followed by a gradual decline. This decrease correlates with bacteria leaking from the beads into the surrounding environment, consequently reducing the number of bacteria recoverable from the beads.
In contrast, the bead-in-hydrogel biofilm model exhibited changes in bacterial density that closely mirrored those observed in natural biofilms (p > 0.05, between the beads-in-hydrogel biofilm model and the natural biofilm group from day 1 to day 9). The difference is that the bacterial density in the beads-in-hydrogel biofilm model continued to increase after day 3 and maintained the high bacterial count until the end of the testing period of 9 days.
To further explore bacterial distribution and structural changes, confocal laser microscopy was employed to observe these samples. Images were captured on days 1, 3, 7, 9, 11, and 13 after culturing, with bacteria in the model stained for better visualization (Fig. 2(c)). At the beginning of the beads-in-hydrogel biofilm model, bacteria were already densely packed within the beads (the sphere on days 1, 3, and 7 represents a SA bead), with density further increasing until the 4th day. From day 4 to day 9, bacteria began to migrate from SA beads into the cellulose–chitosan hydrogel film and formed new colonies.
Subsequently, the live/dead ratio of bacteria were quantitatively analyzed by measuring fluorescence intensity, which helped track bacterial viability over time (Fig. 2(b)). The total fluorescence intensity (including both live and dead fluorescence signals) increased from day 1 to day 13, corresponding to changes in the total bacterial population during this period. During the initial 4 days, GFP (living bacteria) gradually increased, while RFP (dead bacteria) did not change much. Later, bacteria migrated into the gel film and formed colonies from day 7. The amount of dead bacteria on the 9th day significantly increased, possibly due to nutrient deprivation, accumulation of waste products, and programmed cell death in the biofilm.21
3.3 Protein components
Previous research has shown that proteins extracted from biofilms exhibit 40% different types of proteins compared to those from planktonic bacteria.22 To demonstrate that our beads-in-hydrogel biofilm model closely mimics natural biofilms, the protein concentrations of a 9-day cultured beads-in-hydrogel biofilm model, natural biofilms, and planktonic bacteria were measured using both the BCA assay and SDS-PAGE.The BCA assay results (Fig. 3(a)) indicated that the protein concentrations in the natural E. coli biofilm and the bead-in-hydrogel biofilm model increased to more than double, reaching 55.7 ± 1.5 μg mL−1 and 52.0 ± 1.0 μg mL−1, respectively, compared to 19.7 ± 1.4 μg mL−1 secreted by planktonic bacteria. The secreted protein concentration in planktonic bacteria shows a significant difference with the natural biofilm and beads-in-hydrogel model (p < 0.05). This difference is likely due to the requirement of numerous functional proteins for the structural integrity of biofilms, adherence to surfaces, interaction with extracellular DNA,23,24 and communication with other bacteria.25 To further explore the differences in protein composition, SDS-PAGE was employed to separate proteins by their molecular weights from the mixed samples (Fig. 3(b)). The SDS-PAGE results highlighted distinct protein profiles for each sample. Proteins secreted by planktonic bacteria showed three bands at 100 kDa, 35 kDa, and 25 kDa. In contrast, the natural biofilm EPS displayed a broader range of proteins, with bands at 100 kDa, 55–70 kDa, 55 kDa, 15–25 kDa, and 10–15 kDa. The protein extract from the beads-in-hydrogel biofilm model was similar to that of the natural biofilm, featuring bands at 100 kDa, 55 kDa, 15–25 kDa, and 10–15 kDa. Notably, additional bands appeared in the beads-in-hydrogel biofilm model sample in the ranges of 55–70 kDa and 15–20 kDa. This could be attributed to the unique polysaccharide components (SA and chitosan) used in the model, potentially altering the E. coli metabolism and resulting in the production of different proteins.26 This analysis underscores the beads-in-hydrogel biofilm model effectiveness in mimicking natural biofilm protein profiles and supports its utility in biofilm research.
3.4 Extracellular DNA measurements
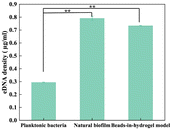
eDNA plays a vital role in biofilm formation, contributing to the structural integrity of the EPS and facilitating bacterial communication.27 We measured the concentration of eDNA extracted from the beads-in-hydrogel biofilm model, natural biofilm, and planktonic bacteria using the Quant-iT™ PicoGreen™ dsDNA Assay Kits and dsDNA Reagents (Fig. 4). The results revealed distinct differences in eDNA concentrations among the samples. Specifically, the planktonic bacterial solution contained only 0.290 ± 0.002 μg mL−1 of eDNA, whereas much higher concentrations of 0.790 ± 0.016 μg mL−1 and 0.740 ± 0.003 μg mL−1 were observed in the natural biofilm and the bead-in-hydrogel biofilm model, respectively. The extracellular DNA concentration in planktonic bacteria shows a significant difference with the natural biofilm and beads-in-hydrogel model (p < 0.05).These findings indicate that the bead-in-hydrogel biofilm model maintains a high eDNA concentration when most bacteria are viable.28 Thus, eDNA within the beads-in-hydrogel biofilm model serves a structural function similar to that in natural biofilms. In the beads-in-hydrogel model, eDNA forms DNA fibers29 that preserve the biophysical properties of the biofilm30 and promote biofilm growth and dispersion.27
4 Conclusions
In this study, we developed a three-dimensional (3D) hydrogel-based bead-in-hydrogel biofilm model by entrapping bacteria in SA/CaCl2 hydrogel beads and embedding these beads in chitosan and cellulose hydrogel films. This design aims to closely replicate the mature phase of natural biofilms formed by bacteria with LB media, which can be cultured in 96-well plates. This innovative model can sustain bacterial populations for at least 7 days under appropriate culture conditions, exhibiting remarkable similarity to natural biofilms in terms of cell density and bacterial distribution dynamics. The hydrogel matrix effectively encapsulates bacteria, supporting sustained growth and biofilm formation. The molecular composition of the model, including eDNA and proteins, closely mirrors that of natural biofilms, validating its use as an artificial replicate. This alignment in molecular components ensures that the bead-in-hydrogel biofilm model accurately mimics the behavior and structural characteristics of natural biofilms.The long-term stability and consistent composition of proteins and eDNA in this beads-in-hydrogel biofilm model highlight its significant potential for medical research applications. It offers a reliable platform for testing the efficacy of anti-biofilm agents, providing a robust tool for evaluating new therapeutic strategies. Additionally, the model affords valuable insights into the mechanisms underlying biofilm growth and dispersion, which are critical for understanding and managing biofilm-associated infections.
Overall, the 3D hydrogel-based beads-in-hydrogel biofilm model represents a significant advancement in biofilm research, offering a robust and accurate tool for studying biofilm dynamics and testing therapeutic interventions. Its ability to replicate the natural biofilm environment makes it an invaluable resource for developing more effective strategies to combat biofilm-related challenges in both medical and industrial contexts.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.Conflicts of interest
The authors declare that they have no known conflicts of interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors thank Jiangsu Dingying New Materials Co. Ltd. for funding support.References
- P. Shree, C. K. Singh, K. K. Sodhi, J. N. Surya and D. K. Singh, Med. Microecol., 2023, 16, 100084 Search PubMed.
- M. Papale, S. Fazi, M. Severini, R. Scarinci, O. Dell’Acqua, M. Azzaro, V. Venuti, B. Fazio, E. Fazio and V. Crupi, Sci. Total Environ., 2024, 943, 173773 Search PubMed.
- J. Yu, W. Han, Y. Xu, L. Shen, H. Zhao, J. Zhang, Y. Xiao, Y. Guo and F. Yu, BMC Microbiol., 2024, 24, 241 CrossRef CAS PubMed.
- S. S. Ramachandra, P. Wright, P. Han, A. Abdal-hay, R. S. Lee and S. Ivanovski, MicrobiologyOpen, 2023, 12, e1377 CrossRef PubMed.
- S. S. Ramachandra, A. Abdal-hay, P. Han, R. S. B. Lee and S. Ivanovski, Biomater. Adv., 2023, 145, 213251 CrossRef CAS PubMed.
- J. Chandra, P. K. Mukherjee and M. A. Ghannoum, Nat. Protoc., 2008, 3, 1909–1924 CrossRef CAS PubMed.
- P. Pearce, B. Song, D. J. Skinner, R. Mok, R. Hartmann, P. K. Singh, H. Jeckel, J. S. Oishi, K. Drescher and J. Dunkel, Phys. Rev. Lett., 2019, 123, 258101 CrossRef CAS PubMed.
- L. Shang, D. Deng, B. P. Krom and S. Gibbs, Crit. Rev. Microbiol., 2024, 50, 397–416 CrossRef CAS PubMed.
- M. Diba, S. Spaans, S. I. Hendrikse, M. M. Bastings, M. J. Schotman, J. F. van Sprang, D. J. Wu, F. J. Hoeben, H. M. Janssen and P. Y. Dankers, Adv. Mater., 2021, 33, 2008111 Search PubMed.
- L. H. P. Pham, K. L. Ly, M. Colon-Ascanio, J. Ou, H. Wang, S. W. Lee, Y. Wang, J. S. Choy, K. S. Phillips and X. Luo, Biofilm, 2023, 5, 100103 Search PubMed.
- E. M. Townsend, L. Sherry, R. Rajendran, D. Hansom, J. Butcher, W. G. Mackay, C. Williams and G. Ramage, Biofouling, 2016, 32, 1259–1270 CrossRef CAS PubMed.
- H. R. Ali, P. Collier and R. Bayston, Microorganisms, 2024, 12, 203 CrossRef CAS PubMed.
- Y. Zhang, P. Young, D. Traini, M. Li, H. X. Ong and S. Cheng, Biotechnol. J., 2023, 18, 2300074 Search PubMed.
- S. Aliyazdi, S. Frisch, A. Hidalgo, N. Frank, D. Krug, R. Müller, U. F. Schaefer, T. Vogt, B. Loretz and C.-M. Lehr, Biofabrication, 2023, 15, 035019 Search PubMed.
- I. C. Thaarup, M. Lichtenberg, K. T. Nørgaard, Y. Xu, J. Lorenzen, T. R. Thomsen and T. Bjarnsholt, Wound Repair Regen., 2023, 31, 500–515 CrossRef.
- W. E. J, R. Solanki, M. Dhanka, P. Thareja and D. Bhatia, Mater. Adv., 2024, 5, 5365–5393 Search PubMed.
- S. A. A. Ghavimi, E. S. Lungren, J. L. Stromsdorfer, B. T. Darkow, J. A. Nguyen, Y. Sun, F. M. Pfieffer, C. L. Goldstein, C. Wan and B. D. Ulery, AAPS J., 2019, 21, 41 CrossRef PubMed.
- I. A. Udoetok, L. D. Wilson and J. V. Headley, ACS Appl. Mater. Interfaces, 2016, 8, 33197–33209 CrossRef.
- S. Tang, J. Yang, L. Lin, K. Peng, Y. Chen, S. Jin and W. Yao, Chem. Eng. J., 2019, 393, 124728 CrossRef.
- W. Xinyu, L. Ming, Y. Chuanjiang, L. Jing and Z. Xikun, Mol. Biomed., 2023, 4, 49 CrossRef.
- P. S. S. Mark and E. Roberts, Antimicrob. Agents Chemother., 2004, 48, 48–52 CrossRef.
- D. Ren, L. A. Bedzyk, S. M. Thomas, R. W. Ye and T. K. Wood, Appl. Microbiol. Biotechnol., 2004, 64, 515–524 CrossRef.
- J. Nobile Clarissa, E. Nett Jeniel, R. Andes David and P. Mitchell Aaron, Eukaryot. Cell, 2006, 5, 1604–1610 CrossRef.
- S. Kavanaugh Jeffrey, E. Flack Caralyn, J. Lister, B. Ricker Erica, B. Ibberson Carolyn, C. Jenul, E. Moormeier Derek, A. Delmain Elizabeth, W. Bayles Kenneth and R. Horswill Alexander, mBio, 2019, 10, e01137 Search PubMed.
- M. Rybtke, J. Berthelsen, L. Yang, N. Høiby, M. Givskov and T. Tolker-Nielsen, MicrobiologyOpen, 2015, 4, 917–930 Search PubMed.
- L. C. Gomes and F. J. Mergulhão, FEMS Microbiol. Lett., 2017, 364, fnx042 CrossRef.
- K. Sharma Dhirendra and S. Rajpurohit Yogendra, J. Bacteriol., 2024, 206, e00006 Search PubMed.
- M. Alhede, M. Alhede, K. Qvortrup, K. N. Kragh, P. Ø. Jensen, P. S. Stewart and T. Bjarnsholt, Pathog. Dis., 2020, 78, ftaa018 CrossRef.
- T. Seviour, F. R. Winnerdy, L. L. Wong, X. Shi, S. Mugunthan, Y. H. Foo, R. Castaing, S. S. Adav, S. Subramoni, G. S. Kohli, H. M. Shewan, J. R. Stokes, S. A. Rice, A. T. Phan and S. Kjelleberg, npj Biofilms Microbiomes, 2021, 7, 27 CrossRef PubMed.
- L. Ferguson Danielle, S. Gloag Erin, R. Parsek Matthew and J. Wozniak Daniel, J. Bacteriol., 2023, 205, e00238 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb02015d |
| This journal is © The Royal Society of Chemistry 2025 |