Near-IR nanolignin sensitizers based on pyrene-conjugated chlorin and bacteriochlorin for ROS generation, DNA intercalation and bioimaging†
Kunal
Gogde
ab,
Seema
Kirar
a,
Anil Kumar
Pujari
ac,
Devesh
Mohne
ac,
Ashok Kumar
Yadav
b and
Jayeeta
Bhaumik
 *a
*a
aCenter of Innovative and Applied Bioprocessing (CIAB), Department of Biotechnology (DBT), Government of India, Sector 81 (Knowledge City), S. A. S. Nagar 140306, Punjab, India. E-mail: jayeeta@ciab.res.in; jbhaumi@gmail.com
bUniversity Institute of Pharmaceutical Sciences, Panjab University, Sector 14, Chandigarh 160306, India
cIndian Institute of Sciences Education and Research (IISER), Sector 81 (Knowledge City), S. A. S. Nagar 140306, Punjab, India
First published on 21st October 2024
Abstract
Near-infrared (NIR) fluorescent agents are extensively used for biomedical imaging due to their ability for deep tissue penetration. Tetrapyrrole-based photosensitizers are promising candidates in this regard. Further, the extended conjugation of such macromolecules with chromophores can enhance their fluorescence efficiency and DNA intercalation ability. Herein, pyrene-conjugated NIR photosensitizers, such as chlorin (PyChl) and bacteriochlorin (PyBac), were synthesized from the corresponding pyrene–porphyrin (PyP). The correlation between the theoretical and experimental optical properties (absorption and fluorescence spectroscopy results) was determined using the DFT/TD-DFT computational approach. Next, studies on the photophysical properties, reactive oxygen species (ROS) production, and DNA binding were conducted on these macrocycles to study the effect of pyrene conjugation on the pyrrolic ring. Furthermore, each photosensitizer was loaded into lignin nanoparticles (LNPs) using the solvent–antisolvent method to accomplish fluorescence-guided imaging. The developed near-IR chlorin- and bacteriochlorin-doped lignin nanocarriers (PyChl-LNCs and PyBac-LNCs) exhibited significant in vitro singlet oxygen generation upon red LED light exposure. Moreover, these macrocycle-loaded nanolignin sensitizers showed good fluorescence-guided bioimaging with fungal cells (Candida albicans). Further, the nanoprobes exhibited pH-dependent release profiles for biological applications. These nanolignin sensitizers demonstrated promising potential to be utilized in near-IR image-guided photodynamic therapy.
Introduction
Near-infrared-assisted photodynamic therapy (PDT) and bioimaging are effective treatments for eradicating microbes.1 Near-IR photosensitizers could be useful owing to their unique optical properties that utilize the phototherapeutic window (i.e. 650–850 nm).1,2 Since the last decade, near-IR (NIR) fluorophores have attracted much attention for the non-invasive imaging of microbial cells.1 Among natural mimics, bacteriochlorins, which are present in bacteriochlorophylls, are attracting interest as bacteriochlorophyll-based pigments can harvest far-red to near-infrared light.3,4 Chemically, they feature the 18-π-electron conjugation inside the porphyrin ring connected by four methylene bridges and two reduced pyrroles on the opposite sides.5 Bacteriochlorin structure does not require two peripheral double bonds to maintain aromaticity.3,5 Thus, reducing one or both bonds of porphyrin leads to chlorin and bacteriochlorin, respectively. They have unique optical properties that make them suitable for absorption in the phototherapeutic window.5,6 These near-IR light-absorbing photodynamic agents can penetrate deep tissue during PDT.4–6 Various π-electron chromophores are being utilized to develop novel probes with unique photophysical characteristics, such as a high quantum yield and longer fluorescence lifetime.7 For example, the substitution of aromatic chromone, naphthalene, pyrene, and pyrazole can be used for the study.7 Among these, pyrene is a versatile, well-studied, and highly fluorescent polycyclic aromatic ring, and its attachment to near-IR photosensitizer can provide intramolecular charge transfer, which could enhance optical properties.8 Thus, pyrene attachment can make better analytical and diagnostic probes for bioimaging and therapy. The pyrene ring could also help to bind with DNA. Further, it can assist in microbial and cancerous DNA intercalation during photodynamic therapy.9,10 Hence, we designed pyrene-conjugated near-IR photosensitizers that could serve as unique pharmacophores for near-IR imaging.Bacteriochlorin-based NIR photosensitizers have been reported for photodynamic and bioimaging applications in the past two decades. Being second-generation, they are considered promising agents for photodynamic therapy and bioimaging compared to natural bacteriochlorophyll.2,9 Recent examples are platinated bacteriochlorins, tetrafluorophenyl bacteriochlorin, cationic bacteriochlorins, and 5,10,15,20-tetrakis[4-(3-N,N-dimethylaminopropoxy)phenyl] chlorin for PDT and bioimaging applications.2,11–14 However, they have a few drawbacks, such as a lower aqueous solubility, self-aggregation, and lack of targeting, which limit the utilization of porphyrin, chlorin, and bacteriochlorin for biological applications.14 A nanotechnological approach can be utilized to deliver the photosensitizer using a drug-delivery system to improve these properties.15–17 Diverse ranges of nano-photosensitizers, such as metal nanoclusters, carbon dots, colloidal metal photosensitizers based on a silica shell, and gold nanocarriers have been reported for NIR bioimaging and ROS-enhanced photodynamic therapy.18 One such example is NIR-absorbing platinum(II)–acetylide conjugated polymers (CPs) containing BODIPY, which exhibited potential as phototheranostic agents in cancer treatment.19
Agri-biomass-derived lignin, a non-toxic, biodegradable polymer, has been preferred to prepare nanoparticles that can enhance the efficacy and targeted delivery of hydrophobic photosensitizers.20–22 The aromatic structure of lignin can be beneficial in phototheranostic applications due to the presence of phenyl propane units, such as sinapyl, coniferyl, and p-coumaric alcoholic hydroxyl, and carboxyl groups on the surface of lignin.23–26 It provides unique photoluminescent properties, with utility in photodynamics and bioimaging, and some studies have shown lignin's biocompatibility with cells.27,28 Thus, lignin nanoparticles could be a promising platform for the delivery of NIR photosensitizers. The highly polyphenolic chemical structure of lignin provides the probability of non-covalent interactions, such as π–π stacking and H-bonding, with the hydrophobic photosensitizer(s).20–22 Conclusively, these properties enhance the loading efficiency of photosensitizer drugs.26,29 The biocompatibility of lignin nanoparticles has been tested in both in vitro cell viability studies as well as in in vivo studies.26 These studies revealed that they formed non-toxic byproducts.26 Also, lignin is the most abundant biopolymer after cellulose, and its ease of availability makes it widely accessible.23,24
In this study, pyrene-conjugated near-IR photosensitizers with chlorin and bacteriochlorin were synthesized using porphyrin. Furthermore, they were characterized by various spectroscopic techniques. An in silico TD-DFT study was conducted to correlate the theoretical study with the optical properties, and it also provided the molecular orbital electron-density distributions and energies. Due to the lack of water solubility and self-aggregation, their use in in vitro and in vivo biological studies has been limited to date.12,20 Hence, we loaded photodynamic agents (PyP, PyChl, and PyBac) in the lignin nanoparticles. Analysis of singlet oxygen generation and ROS production by the photosensitizers (PyP, PyChl, and PyBac) and their nanoformulations was performed using a green/red-light-emitting diode (LED) light (12 W). The NIR fluorescence bioimaging potential of the nanoformulations was validated using Candida albicans fungal cells. The photosensitizers (8, 9a, and 9b) with a fluorophore group, i.e. pyrene, displayed a higher DNA binding affinity and enhanced fluorescence properties.
Results and discussion
Synthesis and characterization of pyrene-conjugated near-IR photosensitizers
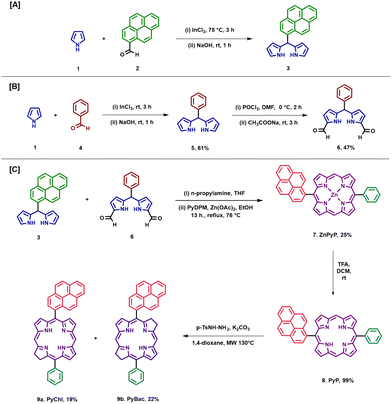
The hydrogenation or reduction of porphyrins is one of the common approaches to scaling up bacteriochlorin.5,30,31 Whitlock et al. reported the first diimide reduction of porphyrin under mild conditions.31 To reduce the long reaction process, Nascimento et al. modified and optimized the diimide reduction of porphyrin under microwave irradiation and managed to scale up the bacteriochlorins with higher yields (65% and 85%).30 Here, pyrene–porphyrin was synthesized by modifying previously reported methods.22,32–36 The starting aldehyde (benzaldehyde or 2-pyrene carboxaldehyde) and pyrrole were reacted under solvent-free conditions in the presence of indium chloride to obtain meso-substituted dipyrromethanes (DPMs) [Scheme 1(A)].32 Pyrene-dipyrromethane (3) was characterized by 1H and 13C-NMR and mass spectroscopy (Fig. S2, S3, and S7 ESI†). Furthermore, diformylated phenyl dipyrromethane was synthesized by adding the Vilsmeier reagent (a mixture of POCl3 in DMF solvent) to phenyl dipyrromethane (5) [Scheme 1(B)]. Phenyl dipyrromethane (5) and diformyl phenyl dipyrromethane (6) were characterized as per our previous work.36 Zn-metallated porphyrin (ZnPyP; 7) was synthesized by the condensation of pyrene DPM (3) and diformylated DPM (6) in ethanol via a McDonald-type reaction, and the yield obtained was up to 25%.32 Zn(II) pyrene–porphyrin (7) was characterized by mass spectroscopy and UV-vis spectroscopy (Fig. S8 and S16 (i), ESI† respectively). With this ZnPyP (7), the demetallation reaction was performed to obtain free-base pyrene–porphyrin, PyP (8) in a 99% yield [Scheme 1 (C)]. The pyrene–porphyrin (8) was confirmed by UV–vis, fluorescence spectroscopy, 1H NMR, and mass spectroscopy [Fig. 1 (i), (ii) and ESI,† Fig. S4, S9]. The targeted near-IR photosensitizers pyrene-substituted bacteriochlorin (PyBac) and chlorin (PyChl) were then synthesized via the diimide reduction of pyrene–porphyrin, PyP (8), under microwave irradiation with a slight modification.30 The main advantage of using microwave irradiation to synthesize near-IR photosensitizers over conventional heating was to reduce the reaction time and volume largely. Notably, PyChl (9a) and PyBac (9b) were obtained by the reduction of the precursor pyrene–porphyrin (8) and further reacted in the presence of p-toluene sulfonyl hydrazide and K2CO3 using a 1,4-dioxane solvent. The final products PyChl (9a) and PyBac (9b) were obtained with moderate yields of 19% and 22%, respectively.The reaction parameters, such as temperature, duration, and concentration of the reactants, and work-up procedures were optimized for maximizing the yields of the near-IR photosensitizers (Table S1 in the ESI†). Pyrene–chlorin and pyrene–bacteriochlorin were isolated by column chromatography (Fig. S1, ESI†), and characterized by UV-vis, fluorescence, and mass spectroscopy [Fig. 1 (i), (ii), and Fig. S8, S9 in the ESI†]. The purity of all the photosensitizers (7, 8, 9a, and 9b) was confirmed by HPLC analysis (Fig. S10–S14, ESI†).
Photophysical analysis of the pyrene-conjugated porphyrin, chlorin and bacteriochlorin
| PS | Stock solution (Colour) | λ max/Molar extinction coefficient (ε) (mM L)−1 cm−1 | Fluorescence (at 365 nm) | λ em (nm) | Fluorescence quantum yield (ΦF)* | |||
|---|---|---|---|---|---|---|---|---|
| B-bands | Soret | Q y -band | Q x -band | |||||
| PyP (8) | Red | 327 nm (2263.91) | 418 nm (2140.25) | 571 nm (2633.08) | 625 nm (2553.87) | Pink | 632, 699, 810 | 0.55 ± 0.07 |
| 341 nm (2563.63) | ||||||||
| PyChl (9a) | Green | 323 nm (7388.04) | 406 nm (6897.83) | — | 624 nm (7906.23) | Dark pink | 630 | 0.85 ± 0.12 |
| 340 nm (7286.86) | ||||||||
| PyBac (9b) | Pink | 349 nm (3432.79) | 374 nm (3430.79) | 507 nm (3455.99) | 730 nm (3438.33) | Light pink | 645, 735 | 0.66 ± 0.03 |
| 387 nm (3928.57) | ||||||||
The fluorescence study of 8, 9a, and 9b was conducted at different excitation wavelengths. After analysis, they all showed emission within the phototherapeutic window (NIR-I window). Pyrene–porphyrin (8) displayed emission peaks at 632, 699, and 810 nm, while pyrene–chlorin showed a single emission peak at 630 nm, and pyrene–bacteriochlorin showed two peaks at 645 and 735 nm. Hence, all the photosensitizers emitted fluorescence around 630–750 nm, which is good for NIR fluorescence bioimaging.
Fluorescence quantum yield (ΦF)
The fluorescence quantum yield is the key optical parameter to quantify a photosensitizer's ability to emit fluorescence upon excitation. The fluorescence quantum yields (ΦF) of pyrene-conjugated porphyrin (8), chlorin (9a), and bacteriochlorin (9b) were determined by a steady-state comparative method, with reference to zinc phthalocyanine (ZnPc; in DMF). All the photosensitizers exhibited greater fluorescence quantum yields (ΦF) than ZnPc (ΦF = 0.3) due to the attachment of a fluorophore, i.e. pyrene. Among them, PyChl (9a) and PyBac (9b) showed higher values (ΦF = 0.85 ± 0.12 and 0.66 ± 0.03) compared to porphyrin (ΦF = 0.55 ± 0.07) (Table 1). Generally, the reduced double bond of the one and two pyrrole rings of the PyChl and PyBac structures alters the distortion and non-planarity in the macrocycle structure compared to the planar extensive π-conjugated porphyrin. Consequently, this reduces the non-radiative relaxation pathways, such as vibrational relaxation and internal conversion, which enhances the fluorescence efficiency of the NIR photosensitizers.Computational analysis of the pyrene-conjugated porphyrin, chlorin, and bacteriochlorin
Density functional theory (DFT) and time-dependent DFT (TD-DFT) based computational calculations were carried out using the Gaussian 16 package.37 The Gouterman four-orbital model (CAM-B3LYP) method and LANL2DZ basis set were utilized to study the electronic properties of the photosensitizers [PyP (8), PyChl (9a), and PyBac (9b)].22,36,38 The optical and electronic properties of pyrene-conjugated porphyrin (8), chlorin (9a), and bacteriochlorin (9b) were compared. The frontier molecular orbitals, and HOMO and LUMO energy diagrams were observed to establish the correlation between the experimental and theoretical study findings. The delocalization of π-orbitals, i.e. HOMO and LUMO electrons, were situated mainly over the whole of the pyrrolic backbone. This did not change the general characteristics of the parent porphyrin, chlorin, and bacteriochlorin, but the pyrene ring donates electrons to the pyrrolic macrocycle, which results in the delocalization of HOMO electrons throughout the macrocycle.36,38–40 Interestingly, electron density transfer from the bacteriochlorin macrocycle (PyBac) to the pyrene ring was seen in the LUMO+1 (Fig. 2). Pyrene–porphyrin was compared with the corresponding chlorin and bacteriochlorin. A previous study suggested that the reduction in the number of π-electrons in bacteriochlorin from 22 to 18 changes and narrows the HOMO–LUMO energy gap.41 Consequently, the energy gap (HOMO–LUMO transition) of pyrene–porphyrin (8) was found to be higher (0.165 eV) than that of pyrene–chlorin (9a; 0.153 eV) and pyrene–bacteriochlorin (9b; 0.136 eV). The electronic transition (from the HOMO to LUMO+1) of pyrene–porphyrin (8) required higher energy (0.181 eV) compared to the corresponding chlorin (9a, 0.167 eV) and bacteriochlorin (9b, 0.173 eV). Pyrene-substitution in porphyrin exhibited the lowest energy level of LUMO+1, providing a longer wavelength of the Q-band at up to 625 nm, as could be observed in the UV-vis spectrum of pyrene–porphyrin (Fig. 1).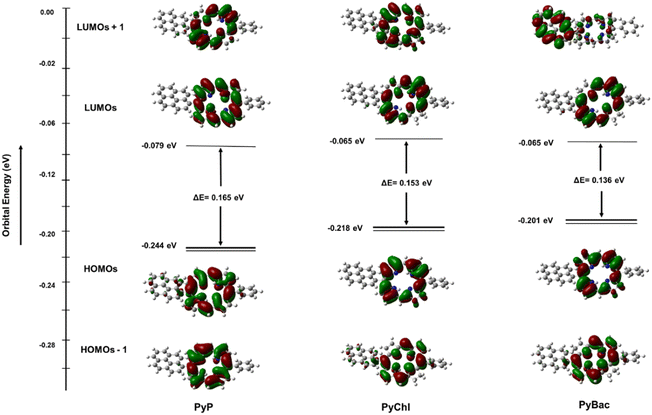 | ||
| Fig. 2 Excited state geometries of pyrene-conjugated PyP (8), PyChl (9a), and PyBac (9b) obtained from TD-DFT calculations at the CAM-B3LYP/LANL2DZ level of theory. | ||
As reported in the previous literature, the origin of the Q-bands involves only HOMO−1, HOMO, LUMO, and LUMO+1 level transitions.39 Herein, the present study revealed that the Qx band (λmax = 735 nm) of pyrene–bacteriochlorin (9b) mainly occurred due to the HOMO → LUMO transition and a small part of the next-HOMO → next-LUMO transition. The Qy band of PyBac, which appeared at 507 nm, equally corroborated the HOMO−1 → LUMO and HOMO → LUMO+1 transitions. The Bx (Soret band) rose due to the HOMO → LUMO+1 electronic transition. Whereas the next By (next band) corresponded to the HOMO−1 to LUMO+1 transition. This induced a bathochromic (red-shift) and hyperchromic effect on the PyBac (9b) spectra (Fig. 1). In the pyrene–chlorin (9a) counterpart, the Qx band was displayed at 624 nm (i.e. red region of the electromagnetic spectrum). The Qx and Qy bands of porphyrin were also observed at 625 and 571 nm, respectively. These electronic changes also raised the energy of the HOMO by changing its nature.41 The photophysical changes of the photosensitizers had a strong effect on the oscillator strengths. PyBac (9b) showed the highest oscillator strength (f; 0.37) compared to pyrene–porphyrin (8; 0.03) and pyrene–chlorin (9a; 0.17), which was because a change in the transition dipole moment leads to an enhanced oscillator strength (f) [Table 2].
| Parameters | PyP | PyChl | PyBac |
|---|---|---|---|
| Electronic energy (Hartree) | −1833.62 | −1834.80 | −1836 |
| HOMO−1 | −0.247 | −0.243 | −0.241 |
| HOMO | −0.244 | −0.218 | −0.201 |
| LUMO | −0.079 | −0.065 | −0.065 |
| LUMO+1 | −0.063 | −0.051 | −0.028 |
| Energy gap, (Eg) (HOMO–LUMO+1) | 0.181 | 0.167 | 0.173 |
| Energy gap, (Eg) (HOMO–LUMO) | 0.165 | 0.153 | 0.136 |
| Excited state 1 | 590 nm | 637 nm | 732 nm |
| Oscillator strength | 0.0305 | 0.1757 | 0.370 |
Conclusively, the TD-DFT study of the photosensitizers (8, 9a, and 9b) established the correlation between the experimental and theoretical calculations based on the photophysical parameters. The computed results also provided insights into the distribution of electrons within the HOMOs and LUMOs (Fig. 2).39,41 In the case of pyrene–porphyrin (8), it was observed that the distinctive electronic transitions (from HOMO to LUMO; HOMO to LUMO+1) led to a shift of the Qx band to the red region at 625 nm [Fig. 1(i)].
Fabrication and characterization of nanolignin sensitizers
Lignin, an aromatic macromolecule with a negative charge due to the presence of polyphenolic groups and their nanoparticles can be used as a nanocarrier to deliver hydrophobic photosensitizers.21,42 The highly aromatic PyP (8) and NIR sensitizers [PyChl (9a) and PyBac (9b)] were doped in lignin nanoparticles via the solvent–antisolvent method.21 Kraft lignin powder (20 mg mL−1) was prepared in ethanol. Then, pyrene–porphyrin (PyP) and the NIR sensitizers (PyChl and PyBac) were dissolved in a saturated solution of ethanol–acetone (1:1). Both solutions were the mixed, and added to deionized water at a flow rate of 0.1 mL min−1. The resulting mixture was subjected to sonication for 10 min. Then, the reaction was removed and centrifuged. The final pellet was diluted with deionized water and lyophilized to obtain a dried powder, which was utilized for further analysis (Fig. 3).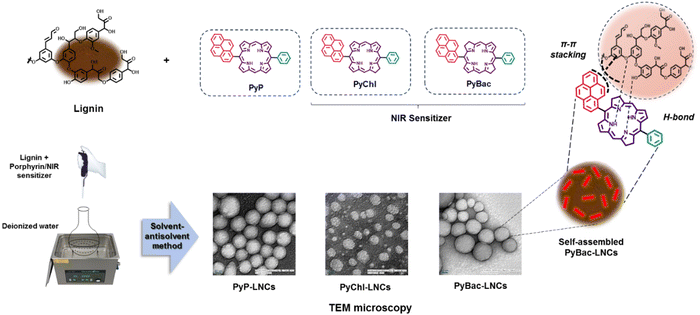 | ||
| Fig. 3 Fabrication and characterization of pyrene–porphyrin, pyrene–chlorin, and pyrene–bacteriochlorin-loaded lignin nanoparticles (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs). | ||
Mechanistically, the highly aromatic photosensitizer can attach to the lignin surface by electrostatic, hydrogen bonding, and π–π stacking, which provides certain advantages, including good stability and aqueous solubility.21,43
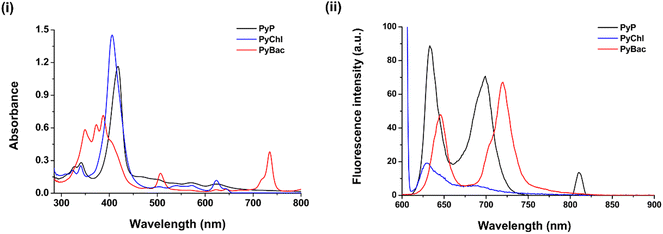
The mean particle size and surface zeta potential (for PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) of the nanoformulations were initially measured using dynamic light scattering (DLS). The average particle size of the bare LNPs was ∼74 nm with a good polydispersity index of 0.17. After the encapsulation of each photosensitizer [PyP (8)/PyChl (9a)/PyBac (9b)], the average size of the nanoparticles (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) increased by ∼45 nm. The pyrene–porphyrin-loaded lignin nanoparticles (PyP-LNCs) were found to be ∼116 nm. With the encapsulation of the NIR sensitizers, the histogram of pyrene–chlorin-loaded LNCs (PyChl-LNCs) and pyrene–bacteriochlorin-loaded LNCs (PyBac-LNCs) exhibited ∼123 nm and ∼114 nm Z-averages, respectively (Fig. 4 and Table S2 in the ESI†). The surface charges of PyP-LNCs, PyChl-LNCs, and PyBac-LNCs nanoparticles were found to be ∼−31, ∼−29, and ∼−34 eV, respectively (Fig. S3 in the (ESI†) and Table 3). TEM images were obtained to provide information regarding the morphology and size of the nanoparticles. The nanolignin sensitizers (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) were found to be spherical in the size range of 100–130 nm (Fig. 4).
| Sample | Standard equation | Φ Δ |
|---|---|---|
| a All the experiments were performed in triplicate. | ||
| PyP (8) | y = −0.001x + 1.039 | 0.30 ± 0.02 |
| PyChl (9a) | y = −0.0014x + 0.916 | 0.41 ± 0.02 |
| PyBac (9b) | y = −0.0012x + 1.121 | 0.36 ± 0.01 |
| PyP-LNCs | y = −0.0008x + 1.0097 | 0.23 ± 0.01 |
| PyChl-LNCs | y = −0.0011x + 1.0201 | 0.32 ± 0.02 |
| PyBac-LNCs | y = −0.001x + 0.9756 | 0.29 ± 0.04 |
| ZnPc | y = −0.0019x + 0.9217 | 0.56 |
Spectroscopic analysis of the nanoformulations (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) was done to confirm the characteristics of the Soret peak and Q-bands. The pyrene–porphyrin-loaded lignin nanoparticles (PyP-LNCs) exhibited a Soret peak at 408 nm; while the pyrene–chlorin-loaded nanoparticles (PyChl-LNCs) showed the Soret peak at 421 nm. The spectroscopy analysis also confirmed the NIR peak of PyBac-LNCs at 753 nm [Fig. S16 (ii), ESI†]. These results indicated that the near-IR sensitizers retained their photophysical properties as intact post-nano entrapment.
Percentage (%) loading, pH-dependent release kinetics, and stability studies of the nanolignin sensitizers
In order to determine the efficacy of the nanolignin sensitizers as imaging probes, percentage loading, pH-dependence, and stability studies were performed. In this line, the percentage loading of the developed pyrene–porphyrin and NIR sensitizers (pyrene–chlorin and pyrene–bacteriochlorin)-loaded lignin nanoparticles was determined using a previously reported method.36,44 The loading efficiency of the nanoprobes, namely PyP-LNCs, PyChl-LNCs, and PyBac-LNCs, was found to be ∼95%, ∼90%, and ∼91%, respectively (Fig. S17 and Table S3 in the ESI†).The release kinetics pattern depends on the surrounding conditions of the diseased area, which could alter the efficiency and distribution of the photosensitizer molecule.20 The release pattern, i.e. rapid or sustained release, plays a key role in systemic and cellular toxicity.20 Here, a biological buffer system in acidic pH 5 and slightly basic 7.4 pH conditions was employed to assess the release patterns of PyP, PyChl, PyBac from PyP-LNCs, PyChl-LNCs, and PyBac-LNCs, respectively. The % cumulative release was determined using a UV-vis spectroscopic technique. The data revealed that PyP, PyChl, and PyBac had a rapid and sustained release over 24 h with percentage cumulative drug releases of 78 ± 3%, 79 ± 5%, and 84 ± 2%, respectively, at pH 5 (Fig. 5 and ESI,† Fig. S18). The cumulative drug release at pH 7.4 was 64 ± 7% for PyP-LNCs, 63 ± 6% for PyChl-LNCs, and 62 ± 2% for PyBac-LNCs (Fig. 5). Overall, it could be concluded that all the nanoparticles showed pH-responsive release kinetics, mimicking the acid nature of Candida fungal cells.
The stability study of the nanoformulations PyP-LNCs, PyChl-LNCs, and PyBac-LNCs was performed in terms of the mean particle size, polydispersity index, and surface charge of the nanoparticles. The nanoformulations were immersed in a buffer solution of pH 7, which was stored at 4 °C.21 The above parameters were monitored at different time intervals and the nanolignin sensitizers were found to be stable for up to 3 months (Fig. S19, ESI†). Furthermore, the stability of nanoparticles was observed using TEM morphology analysis and their corresponding DLS data after one month. The size and morphology of each of the nanoformulations PyP-LNCs, PyChl-LNCs, and PyBac-LNCs were stable with Z-average sizes of 116, 129, and 115 nm, respectively (Fig. S20, ESI†).
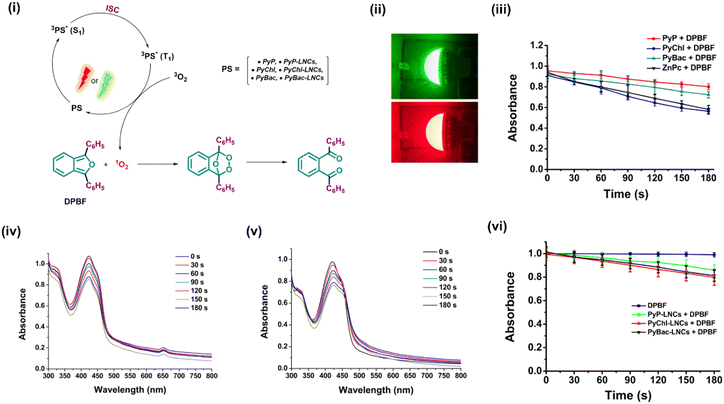
Singlet oxygen generation analysis
The reactive oxygen species (ROS) generation ability of a photosensitizer is a key parameter to determine its efficacy towards destroying microbes when activated by light. Therefore, the singlet oxygen quantum yield (ΦΔ) of all the samples [PyP (8), PyChl (9a), PyBac (9b), PyP-LNCs, PyChl-LNCs, and PyBac-LNCs] was determined using the DPBF assay, with zinc phthalocyanine used as the standard. The highest singlet oxygen quantum yield was observed in the case of PyChl (0.41 ± 0.02) compared with the corresponding PyP (0.30 ± 0.02), PyBac (0.36 ± 0.01) under a green LED light (12 W; 500–570 nm, 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 J cm−2)/red LED light (12 W; 620–750, 32
400 J cm−2)/red LED light (12 W; 620–750, 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 J cm−2), and using DMF as the solvent [Fig. S21 and Table 3]. Similarly, the NIR nanoprobes: PyChl-LNCs (0.32 ± 0.02) and PyBac-LNCs (0.29 ± 0.04) showed good quantum yields compared to the pyrene–porphyrin-loaded LNCs (PyP-LNCs; 0.23 ± 0.01) [Fig. 6 and Table 3]. Notably, the ΦΔ values of the nanoprobes were relatively lower compared to their respective photosensitizers due to the restriction of light access to the photosensitizers in the aqueous medium. Other factors, such as polarity and the access to molecular oxygen by the nanoparticle matrix, may also play a key role in reducing the ΦΔ values. The DPBF assay used in determining the ΦΔ values involved a chemical probe named 1,3-diphenylisobenzofuran (DPBF). DPBF is a fluorescent molecule that acts as a singlet oxygen quencher, which reacts specifically with singlet oxygen and forms the non-fluorescent endoperoxide 1,2-dibenzoyl benzene irreversibly.20,36,45
400 J cm−2), and using DMF as the solvent [Fig. S21 and Table 3]. Similarly, the NIR nanoprobes: PyChl-LNCs (0.32 ± 0.02) and PyBac-LNCs (0.29 ± 0.04) showed good quantum yields compared to the pyrene–porphyrin-loaded LNCs (PyP-LNCs; 0.23 ± 0.01) [Fig. 6 and Table 3]. Notably, the ΦΔ values of the nanoprobes were relatively lower compared to their respective photosensitizers due to the restriction of light access to the photosensitizers in the aqueous medium. Other factors, such as polarity and the access to molecular oxygen by the nanoparticle matrix, may also play a key role in reducing the ΦΔ values. The DPBF assay used in determining the ΦΔ values involved a chemical probe named 1,3-diphenylisobenzofuran (DPBF). DPBF is a fluorescent molecule that acts as a singlet oxygen quencher, which reacts specifically with singlet oxygen and forms the non-fluorescent endoperoxide 1,2-dibenzoyl benzene irreversibly.20,36,45
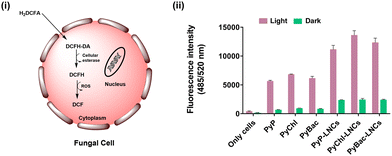
Intracellular reactive oxygen species (ROS) determination
The generation of reactive oxygen species (ROS) by photosensitizers/nanophotosensitizers is crucial under light treatment during photodynamic and bioimaging applications.46 Therefore, ROS generation quantification of the photosensitizers (8, 9a, 9b) and nanolignin sensitizers (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) was done using a DCFDA assay with a green LED (12 W; 500–570 nm, 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 J cm−2) and red LED (12 W; 620–750 nm, 32
400 J cm−2) and red LED (12 W; 620–750 nm, 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 J cm−2) respectively. The oxidation-sensitive DCFDA fluorescent probe was used to stain the fungal cells, followed by incubation with each sample treatment for 30 min (at 100 rpm; 37 °C). After incubation, the DCFDA was converted into non-fluorescent 2,7-dichlorofluorescein (DCFH2) in the presence of cellular esterase enzyme. DCFH2 further reacts with the ROS produced by the light-responsive photosensitizers/nanolignin sensitizers, which leads to the formation of highly fluorescent dichlorofluorescein (DCF) [Fig. 7(i)]. All the treatments were performed with different concentrations of 5, 10, and 15 μg mL−1. The 5 and 10 μg mL−1 treatments were compared with their dark treatments, producing 2–5-fold higher ROS [Fig. S23, ESI†]. However, when the nanolignin sensitizers with 15 μg mL−1 dose were chosen, they showed up to a 12-fold higher ROS than their light-treated counterpart against fungal cells (Candida albicans) [Fig. 7(ii)]. Further, morphological deformities of the fungal cell walls were observed due to ROS production by the nanolignin sensitizers, as depicted in the SEM analysis (Fig. S22, ESI†). Overall, the nanolignin sensitizers are promising ROS generators for the eradication of microbes as exemplified by their effect on fungal cells.
400 J cm−2) respectively. The oxidation-sensitive DCFDA fluorescent probe was used to stain the fungal cells, followed by incubation with each sample treatment for 30 min (at 100 rpm; 37 °C). After incubation, the DCFDA was converted into non-fluorescent 2,7-dichlorofluorescein (DCFH2) in the presence of cellular esterase enzyme. DCFH2 further reacts with the ROS produced by the light-responsive photosensitizers/nanolignin sensitizers, which leads to the formation of highly fluorescent dichlorofluorescein (DCF) [Fig. 7(i)]. All the treatments were performed with different concentrations of 5, 10, and 15 μg mL−1. The 5 and 10 μg mL−1 treatments were compared with their dark treatments, producing 2–5-fold higher ROS [Fig. S23, ESI†]. However, when the nanolignin sensitizers with 15 μg mL−1 dose were chosen, they showed up to a 12-fold higher ROS than their light-treated counterpart against fungal cells (Candida albicans) [Fig. 7(ii)]. Further, morphological deformities of the fungal cell walls were observed due to ROS production by the nanolignin sensitizers, as depicted in the SEM analysis (Fig. S22, ESI†). Overall, the nanolignin sensitizers are promising ROS generators for the eradication of microbes as exemplified by their effect on fungal cells.
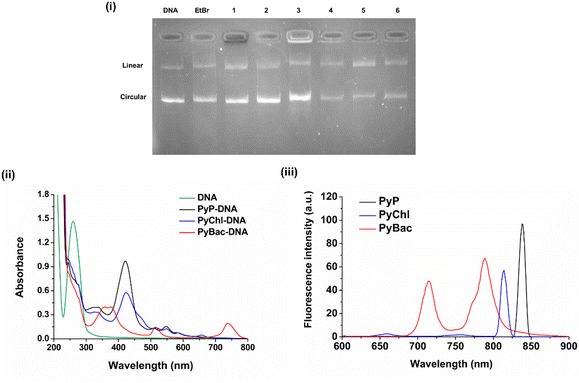
DNA-intercalation study using the nanolignin sensitizers
The DNA binding analysis is crucial for studying preliminary biological responses. Determining DNA intercalation helps with the initial drug-development process. It was previously reported that the pyrene group binds with DNA and shows excellent DNA intercalation properties.46,47 The planar structure of pyrene helps its insertion inside DNA base pairs, i.e. intercalation. The polypyrrole macrocycle could also help the non-covalent interactions, such as H-bonding and van der Waals forces, stabilize the photosensitizer–DNA complex.44 Herein, to study DNA intercalation by the photosensitizers [PyP (8), PyChl (9a), and PyBac (9b)] and their nanoformulations (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs), gel electrophoresis was performed using plasmid DNA pUC19. The results indicated that the photosensitizers bind/intercalate with DNA, as depicted by the hindrance in the movement of the DNA plasmid pUC19 (Fig. 6). Interestingly, all the pyrene-conjugated pyrrolic photosensitizers [PyP (8), PyChl (9a), and PyBac (9b)] showed the DNA intercalation property. Among these, pyrene–bacteriochlorin (9b) was found to be a very efficient DNA intercalator compared to EtBr. All the nanoformulations (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) also demonstrated DNA-intercalation capabilities, as shown in Fig. 8, Lanes 4, 5, and 6, respectively. Hence, the NIR fluorescent dyes (8, 9a, 9b) and their nanoformulations (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) can be used for DNA intercalation. Moreover, the effect of DNA binding on the photosensitizers was also examined by UV-vis and fluorescence spectroscopy. DNA pUC19 showed absorption around 260 nm.48,49 After binding with each photosensitizer, a hypochromic shift of the DNA peak was observed. After DNA binding, the UV-vis spectrum of pyrene–porphyrin showed a hypochromic shift and red-shift by 3 nm from 418 nm to 421 nm. Pyrene–chlorin also exhibited hypochromic and red-shifted peaks: the Soret peak from 406 nm to 423 nm and Q-band from 630 nm to 657 nm. Meanwhile, pyrene–bacteriochlorin also showed hypochromic and red-shifts from 349 to 357 nm and 374 to 376 nm. The DNA intercalation of the photosensitizers (8, 9a, and 9b) was also monitored by the fluorescence intensity (Fig. 8(ii) and (iii)). In this study, PyP exhibited a fluorescence emission peak at 837 nm upon excitation at 418 nm. PyChl showed the emission at 814 nm upon 406 nm excitation. Meanwhile, PyBac displayed two emission peaks at 715 and 789 nm through 507 nm excitation. Conclusively, the spectroscopic study suggested that the DNA binding of each photosensitizer (8, 9a, and 9b) could bring photophysical changes (bathochromic and hypochromic shifts) due to intercalation.Ethidium bromide (EtBr) and 4′,6-diamidino-2-phenyl-indole (DAPI) displacement study
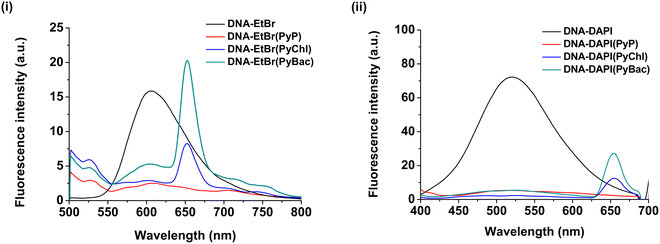
Furthermore, the DNA-intercalation study findings were validated by an EtBr and DAPI displacement study.50,51 The basic mechanism of the study was the competition between the DAPI/EtBr and the photosensitizer samples (8, 9a, and 9b).50 The decrease in the fluorescence intensity of DAPI/EtBr after displacement by the photosensitizer sample confirmed their DNA-intercalation properties. Naturally, DNA does not possess fluorescence.50,51 Therefore, two fluorophores, namely EtBr and DAPI, were combined to create pUC19(DNA)-EtBr (intercalator/binder) and pUC19(DNA)-DAPI (groove binder) adducts, respectively. After the addition of each of PyP, PyChl, and PyBac to the EtBr-DNA/DAPI-DNA adduct, the fluorescence intensity was found to be significantly reduced [Fig. 9 (i) and (ii)]. This fluorescence quenching indicated that each photosensitizer strongly displaced the EtBr and DAPI from the binding site present in the DNA (pUC19). Consequently, it provided strong evidence of the DNA-binding abilities of the designed photosensitizers: PyP, NIR-absorbing PyChl, and PyBac. The quenching constant (Kq) was calculated using the Stern–Volmer equation for each sample (Table 4):| F0/F = Kq[Q] + 1 | (1) |
| Sample | K q[Q] for EtBr | K q[Q] for DAPI |
|---|---|---|
| PyP (8) | 6.10 M−1 | 12.46 M−1 |
| PyChl (9a) | 4.51 M−1 | 35.11 M−1 |
| PyBac (9b) | 2.06 M−1 | 12.79 M−1 |
In vitro fluorescence imaging using the nanolignin sensitizers
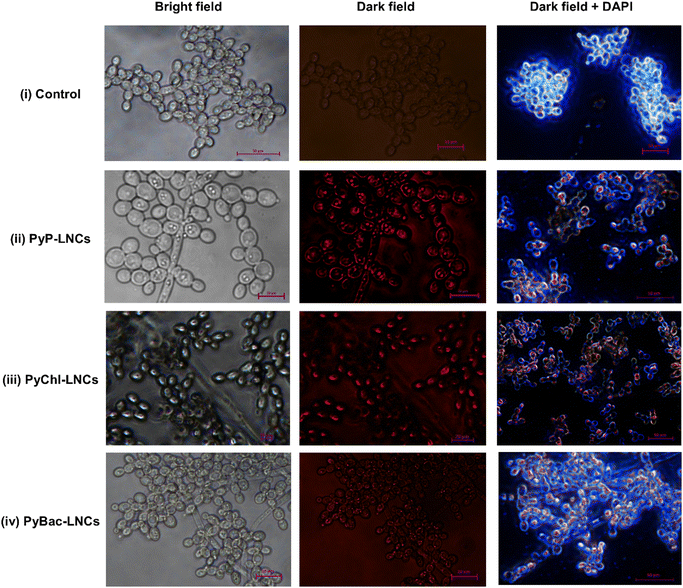
Fungus, such as Candida albicans, can cause skin infections, and the overgrowth of this species causes candidiasis.52,53 The development of resistance to antifungal medications has been found to be ineffective.54,55 The major reason for the inefficacy in the treatment is due to the lack of detection of such fungi. In this line, bioimaging studies of the nanolignin sensitizers, namely PyP-LNCs, PyChl-LNCs, and PyBac-LNCs, were conducted on Candida albicans fungal cells. The Candida cells were stained by 4′,6-diamidino-2-phenyl-indole (DAPI, blue fluorescent stain) and were then incubated with each nanoprobe (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs; Fig. 10). The fluorescence imaging was performed using an inverted fluorescence microscope (Fig. 8). In the study, the treated cells were excited with a wavelength of 525 nm, and the dark field images showed the red-light emission due to the pyrene–porphyrin, pyrene–chlorin, and pyrene–bacteriochlorin, as shown in Fig. 10 (ii)–(iv) respectively. The nanosensitizers were also compared with untreated/control cells [Fig. 10 (i) bright field, (ii) control cells in the dark field, and (iii) DAPI-treated control in the dark field] to prove their efficacy. To note, the pyrene–porphyrin, pyrene–chlorin, and pyrene–bacteriochlorin-based nanosensitizers entered the cells, possibly via passive diffusion.48,49 Interestingly, the pyrene–porphyrin LNCs and pyrene–chlorin LNCs exhibited greater red emission compared to PyBac-LNCs. The nanosensitizers showed the binding with the Candida albicans cell membrane and the cytoplasm (Fig. 8). This indicated that the lignin nanoparticles could be a good drug-delivery carrier for the photosensitizers [PyP (8), PyChl (9a), and PyBac (9b)] for bioimaging purposes.In vitro photoinactivation study of Candida albicans by PyP, PyChl, PyBac and their nanoformulations
The photodynamic inactivation of Candida albicans was tested to investigate the antimicrobial PDT potential of the photosensitizers [PyP (8), PyChl (9a), and PyBac (9b)] and their nanoformulations (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs). The cell viability of these samples was evaluated based on the colony-forming units (CFUs) on the Yeast Peptone Dextrose (YPD) Agar Plates. After the incubation of each sample with C. albicans, they were subjected to dark and light (LED) treatments. The treated cells [with pyrene–porphyrin (PyP) and their nanoformulation (PyP-LNCs)] were irradiated under a green LED (12 W; 500–570 nm, 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 J cm−2) light, while pyrene–chlorin (PyChl), pyrene–bacteriochlorin (PyBac), and their nanoformulations (PyChl-LNCs and PyBac-LNCs) were irradiated under a red LED (12 W; 620–750 nm) light. Each nanoformulation was taken as 0, 5, 10, 15, 20, and 25 μg doses. After 36 h incubation, the CFUs were counted after incubation and the % cell viability [Fig. 11] was calculated. Each sample set produced the IC50, as demonstrated using a standard calibration plot drawn at 5, 10, 15, 20 and 25 μg mL−1 (Fig. 11 and Table 5).
400 J cm−2) light, while pyrene–chlorin (PyChl), pyrene–bacteriochlorin (PyBac), and their nanoformulations (PyChl-LNCs and PyBac-LNCs) were irradiated under a red LED (12 W; 620–750 nm) light. Each nanoformulation was taken as 0, 5, 10, 15, 20, and 25 μg doses. After 36 h incubation, the CFUs were counted after incubation and the % cell viability [Fig. 11] was calculated. Each sample set produced the IC50, as demonstrated using a standard calibration plot drawn at 5, 10, 15, 20 and 25 μg mL−1 (Fig. 11 and Table 5).
| Sample | IC50 in dark condition (dose in μg) | IC50 in dark condition (dose in μM) | IC50 in light condition (dose in μg) | IC50 in light condition (dose in μM) | Fold reduction of doses (dark vs. light) |
|---|---|---|---|---|---|
| a All the experiments were conducted in triplicate. | |||||
| PyP (8) | 28.97 ± 3.31 | 0.038 ± 0.05 | 14.74 ± 0.6 | 0.025 ± 0.001 | 2 |
| PyChl (9a) | 21.52 ± 0.41 | 0.036 ± 0.001 | 10.21 ± 0.22 | 0.017 ± 0.001 | 2.1 |
| PyBac (9b) | 20.56 ± 0.35 | 0.034 ± 0.001 | 7.26 ± 0.17 | 0.012 ± 0.001 | 2.8 |
| PyP-LNCs | 13.84 ± 0.42 | — | 9.42 ± 0.92 | — | 1.5 |
| PyChl-LNCs | 14.27 ± 2.82 | — | 5.19 ± 0.14 | — | 2.7 |
| PyBac-LNCs | 13.72 ± 1.37 | — | 5.35 ± 0.6 | — | 2.5 |
The light treatment of each photosensitizer [PyP (8), PyChl (9a) and PyBac (9b)] produced up to 62–87% inhibition at 25 μg mL−1 concentration (P > 0.001) compared to the positive control, as shown in Fig. 11 (i). The IC50 of the PyBac (9b) photosensitizer was found to be very effective (IC50:0.012 μM) compared to the IC50 of PyP (8) and PyChl (9a) with 0.025 μM and 0.017 μM doses, respectively (Table 5). Consequently, PyBac was found to have 2 and 1.4 times more potential than PyP and PyChl under light conditions [Fig. 11 (iii) and Fig. S21, ESI†]. Similarly, under the dark conditions for the treatments, they could inhibit the Candida cells by 47–51% (Fig. 11 (i)). Furthermore, their nanolignin sensitizers (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) increased the % inhibition by 81–92% at a 25 μg mL−1 concentration (P > 0.001) under light treatment [Fig. 11 (iii)]. The % cell viability data revealed that the nanolignin sensitizers of the respective photosensitizer were very effective against Candida albicans cells. Hence, the IC50 values of the nanolignin sensitizers PyP-LNCs, PyChl-LNCs, and PyBac-LNCs were observed at 9.42, 5.19, and 5.35 μg mL−1, which were 1.3, 1.5, and 2 times less compared with PyP, PyChl, and PyBac, respectively (Table 5). Moreover, the antifungal PDT study using the nanolignin sensitizers demonstrated, improved hydrophilicity, leading to the enhanced effectiveness of the hydrophobic photosensitizers during antimicrobial photodynamic activity (Fig. 11), and the destruction of Candida cells could also be observed after each nanolignin sensitizer treatment (Fig. S22, ESI†).
Experimental section
Synthesis and characterization of pyrene-conjugated porphyrin, chlorin, and bacteriochlorin and their intermediates
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (1
n-hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) solvent system and silica gel (100–200 mesh size). The yields of compounds 3 and 5 were calculated to be 68% and 61%, respectively. (Compound 3) Yellow solid crystals: 1H NMR. (500 MHz, DMSO-d6) δ: 5.64 (d, J = 10 Hz, 2H), 5.94 (d, J = 5 Hz, 2H), 6.53 (s, 1H, R3H), 6.66 (d, J = 5 Hz, 2H), 7.72 (s, 1H, ArH), 8.04 (d, 15 Hz, 2H, ArH), 8.17 (d, 5 Hz, 2H, ArH), 8.20 (d, 10 Hz, 2H, ArH), 8.26 (s, 1H, ArH), 8.47 (s, 1H, ArH), 9.97 (d, 2H, NH) ppm; 13C NMR. (125 MHz, CDCl3) δ: 106.85, 106.97, 116.88, 123.41, 124.17, 124.74, 124.83, 125.07, 126.08, 126.70, 127.14, 127.39, 127.80, 129.38, 130.20, 130.82, 133.16, 137.83. HPLC-MS: (C25H18N2) m/z [M]+ calc. 346.15; found 346.15, m/z [M – 1]+ found 345.16.
9) solvent system and silica gel (100–200 mesh size). The yields of compounds 3 and 5 were calculated to be 68% and 61%, respectively. (Compound 3) Yellow solid crystals: 1H NMR. (500 MHz, DMSO-d6) δ: 5.64 (d, J = 10 Hz, 2H), 5.94 (d, J = 5 Hz, 2H), 6.53 (s, 1H, R3H), 6.66 (d, J = 5 Hz, 2H), 7.72 (s, 1H, ArH), 8.04 (d, 15 Hz, 2H, ArH), 8.17 (d, 5 Hz, 2H, ArH), 8.20 (d, 10 Hz, 2H, ArH), 8.26 (s, 1H, ArH), 8.47 (s, 1H, ArH), 9.97 (d, 2H, NH) ppm; 13C NMR. (125 MHz, CDCl3) δ: 106.85, 106.97, 116.88, 123.41, 124.17, 124.74, 124.83, 125.07, 126.08, 126.70, 127.14, 127.39, 127.80, 129.38, 130.20, 130.82, 133.16, 137.83. HPLC-MS: (C25H18N2) m/z [M]+ calc. 346.15; found 346.15, m/z [M – 1]+ found 345.16.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane solvent system (1
n-hexane solvent system (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9). The brownish solid product was obtained (680 mg, 49%). The product was characterized by 1H-NMR spectroscopy and the data were found to be consistent with a previous report.36
9). The brownish solid product was obtained (680 mg, 49%). The product was characterized by 1H-NMR spectroscopy and the data were found to be consistent with a previous report.36
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (1
n-hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) as the solvent system. Red solid crystal was found, ESI-MS: m/z [M + 1]+ calc. for C42H24N4Zn: 649.13; found 649.16. HPLC analysis, tR = 10.86 [the analysis was performed isocratically in 0.1% TFA in an acetonitrile and water composition (90:10) run over 20 min, purity >99%].
9) as the solvent system. Red solid crystal was found, ESI-MS: m/z [M + 1]+ calc. for C42H24N4Zn: 649.13; found 649.16. HPLC analysis, tR = 10.86 [the analysis was performed isocratically in 0.1% TFA in an acetonitrile and water composition (90:10) run over 20 min, purity >99%].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 588.24; found 588.23 and [M+1]+ found 589.28. HPLC analysis, tR = 2.89 [the analysis was performed isocratically in 0.1% TFA in an acetonitrile and water composition (90:10) run over 20 min, purity >99%].
588.24; found 588.23 and [M+1]+ found 589.28. HPLC analysis, tR = 2.89 [the analysis was performed isocratically in 0.1% TFA in an acetonitrile and water composition (90:10) run over 20 min, purity >99%].
| ε = A/c·l | (2) |
Fluorescence quantum yield (ΦF)
The ΦF yields were recorded by a fluorescence spectrophotometer using a comparative method with zinc phthalocyanine in DMF solvent as a reference (ΦF = 0.3; Table S4, ESI†). The following standard equation was used for the calculation of ΦF:| ΦF(u) = ΦF(ref) × Iu/Iref × Aref/Au × ηu2/ηref2 | (3) |
Molecular orbital calculations
Time-dependent density functional theory (TD-DFT) calculations were performed with the CAM-B3LYP method and LANL2DZ basis set, and the equilibrium geometries were fully optimized using the same parameters.37,38,40Development of lignin nanoparticles (LNPs) and pyrene-based porphyrin and NIR photosensitizers-loaded lignin nanoparticles
The PyP-LNCs, PyChl-LNCs and PyBac-LNCs were prepared according to previous publications with minor modifications.21 A stock solution of Kraft lignin (20 mg mL−1) was prepared in ethanol. On the other side, the pyrene–porphyrin (PyP) and NIR sensitizers (PyChl and PyBac) [1 mg mL−1] were taken in an ethanol–acetone solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 saturated solution). Furthermore, stock solutions (0.5 mL of each) were mixed and sonicated to completely dissolve the mixture. The resulting mixture (1 mL) was added dropwise in 10 mL of deionized water in RBF via a peristaltic pump and then sonicated for 10 min. Then, the reaction was removed and centrifuged at 10 000 rpm for 8 min for further purification. The final pellet was diluted with deionized water and lyophilized to obtain a dried solid, which was utilized for further analysis.
1 saturated solution). Furthermore, stock solutions (0.5 mL of each) were mixed and sonicated to completely dissolve the mixture. The resulting mixture (1 mL) was added dropwise in 10 mL of deionized water in RBF via a peristaltic pump and then sonicated for 10 min. Then, the reaction was removed and centrifuged at 10 000 rpm for 8 min for further purification. The final pellet was diluted with deionized water and lyophilized to obtain a dried solid, which was utilized for further analysis.
Determination of the particle size, zeta potential, and morphology of the nanoparticles
The Z-average size and surface potential charge of PyP-LNCs, PyChl-LNCs, and PyBac-LNCs were determined using a zeta sizer (Malvern Zetasizer Nano ZS1211332, Malvern, UK). Also, the morphology of these nanoparticles was observed by high-resolution transmission electron microscopy (HR-TEM), whereby a dispersed sample of the nanoparticles (in water) was placed on a copper grid and allowed to air dry. Then, they were observed by TEM microscopy.Determination of the % loading efficiency of the nanoparticles
The loading efficiency (%) was calculated using a reported protocol and quantified by optical density using UV–vis spectroscopy.44 After loading the pyrene–porphyrin and NIR sensitizers, the unreacted photosensitizer supernatant was removed and diluted with acetone–ethanol solution. The absorbance was recorded at 418 nm (PyP), 406 nm (PyChl), and 735 nm (PyBac) using UV–vis spectroscopy. The amount of free sensitizer drugs was determined by the standard calibration curves of each sample. The experiment was performed in triplicate for each sample and the results presented as the mean ± standard deviation. The loading efficiency (%) of the nanoformulations was determined by the following equation, and standard curves were also calibrated for the analysis:| Loading efficiency % = [(Total drug added − free drug in supernatant)/Total drug added] × 100 | (4) |
Spectroscopic analysis
The pyrene–porphyrin (PyP-LNCs), and NIR sensitizers (PyChl-LNCs, and PyBac-LNCs) were analyzed using a UV-Vis spectrophotometer. The analysis was performed in deionized water.Singlet oxygen quantification of PyP, PyChl, PyBac, and their nanoformulations
Following a reported protocol,56 the singlet oxygen quantum yield (ΦΔ) was determined by 1,3-diphenylisobenzofuran (DPBF) dye. Stock solutions of DPBF (1 mg mL−1) were prepared in DMF and DMSO. Furthermore, stock solutions of the photosensitizer molecules [PyP (8), PyChl (9a), PyBac (9b); 1 mg mL−1] and each nanoprobe [(PyP-LNCs, PyChl-LNCs and PyBac-LNCs)] were prepared in DMF and water, respectively. They were then diluted with the respective solvents to maintain a solution with an OD ≤ 1. The DPBF dye (10 μg mL−1) and each photosensitizer molecule (8, 9a, and 9b) at a 10 μg mL−1 concentration, were then irradiated with a green LED light (12 W; 500–570 nm, 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 J cm−2) for pyrene–porphyrin (8) and red light (12 W; 620–750 nm, 32
400 J cm−2) for pyrene–porphyrin (8) and red light (12 W; 620–750 nm, 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 J cm−2) for the PyChl (9a) and PyBac (9b) samples. At the same time, the OD for DPBF degradation was recorded at different time intervals (0, 30, 60, 90, 120, 150, and 180 s) at 410 nm. Similarly, the same procedure was followed for the nanoformulations (20 μg mL−1) to record DPBF decay at 417 nm. The SOQY calculations were performed in triplicate for each experiment, and the obtained data were calculated as the mean ± standard deviation using zinc phthalocyanine (ZnPc) as a standard (ΦΔ, 0.56). The singlet oxygen quantum yield (ΦΔ) was quantified by the following formula:
400 J cm−2) for the PyChl (9a) and PyBac (9b) samples. At the same time, the OD for DPBF degradation was recorded at different time intervals (0, 30, 60, 90, 120, 150, and 180 s) at 410 nm. Similarly, the same procedure was followed for the nanoformulations (20 μg mL−1) to record DPBF decay at 417 nm. The SOQY calculations were performed in triplicate for each experiment, and the obtained data were calculated as the mean ± standard deviation using zinc phthalocyanine (ZnPc) as a standard (ΦΔ, 0.56). The singlet oxygen quantum yield (ΦΔ) was quantified by the following formula:| ΦΔ = ΦΔS × ms/mu | (5) |
Microorganisms and culture conditions
The fungal strain C. albicans was employed for the study. The fungal cells were grown and incubated in YPD broth (yeast, peptone, and dextrose). The growth of the cells was continued till the OD600 reached 0.6.In vitro ROS generation analysis
The C. albicans suspension with an OD600 up to 0.6 was reproduced and revived for the analysis. To stain it with dye, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) was mixed with the suspension of the C. albicans cells and underwent incubation at 100 rpm (at 37 °C) for 30 min. The suspension was removed and centrifuged at 3700 rpm for 5 min. The pellet was removed and washed three times in deionized water. The cells were treated with different doses: 5, 10, and 15 μg mL−1 under light (source green/red LED of 12 W) and dark conditions. The fluorescence intensity of each condition was measured at the excitation wavelength of 485 nm and emission wavelength of 520 nm.DNA-intercalation study
The DNA-intercalation study of the samples was carried out using a DNA plasmid (pUC19) by agarose gel electrophoresis. A 10 μM solution of each sample PyP, PyChl, and PyBac was prepared in DMSO-tris buffer, and 1 mg mL−1 of each nanoformulation (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) was prepared in distilled water. Before the electrophoresis, they were incubated at 37 °C for 1 h. Next, 0.25 μg of DNA plasmid pUC19 was taken, mixed with each sample, and incubated at 37 °C for 1 h, and the total reaction volume was 14 μL. The agarose gel-electrophoresis of PyP (8), PyChl (9a), PyBac (9b) and each nanoprobe (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs) (tris buffer, 1 h) was done to prove the ability to intercalate the DNA.EtBr and DAPI displacement study
For the displacement of DAPI/EtBr, 2.5 μg DNA in 500 μL molecular biology grade water containing EtBr/DAPI was incubated for 1 h. After the incubation, 100 μM PyP/PyChl/PyBac was added and incubated to displace the DAPI/EtBr. The fluorescence intensities were recorded from 400 to 800 nm with excitation at 480 and 358 nm wavelength for DAPI and EtBr, respectively.Fluorescence imaging
The C. albicans cells were incubated with 20 μg mL−1 of each nanoprobe PyP-LNCs, PyChl-LNCs, and PyBac-LNCs at 37 °C for 1 h. They were washed in distilled water, pelleted, and resuspended in 50 μL distilled water and chemically fixed using 5% glutaraldehyde in distilled water. Also, the fluorescence imaging was validated by staining the Candida cells with 20 μg mL−1 of 4′,6-diamidino-2-phenyl-indole (DAPI, blue fluorescent stain) in distilled water. They were also washed in distilled water, pelleted, and resuspended in 50 μL distilled water and chemically fixed using 5% glutaraldehyde in distilled water. Next, 10 μL of each sample was placed on a microscope slide and covered with a coverslip. Then, they were observed by a fluorescence microscope (Nikon; TS2FL) with a 2880 × 2048 pixel resolution. An excitation wavelength of 525 nm was used to capture the dark field images and a 650 to 755 nm bandpass filter for the red-light absorbing pyrene–chlorin and near-IR pyrene–bacteriochlorin.In vitro photoinactivation study of Candida albicans by PyP, PyChl, PyBac, and their nanoformulations
The Candida albicans were aerobically grown in YPD broth (yeast, peptone, and dextrose) for 12 h at 37 °C and 100 rpm in an orbital shaking incubator. The broth was then centrifuged and washed three times with deionized water. The OD600 of the broth was maintained at 0.6, corresponding to ca. 103 CFU per mL. Then, 1 mL diluted suspension of the C. albicans cells was taken and treated with the synthesized photosensitizers (PyP, PyChl, and PyBac), and the developed nanolignin sensitizers (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs). The common dose concentrations were 0, 5, 10, 15, 20, and 25 μg mL−1 for all the samples. These concentrations of pyrene–porphyrin (PyP) and their nanolignin sensitizers (PyP-LNCs) were individually incubated with C. albicans for 45 min and further irradiated for about 40 min with a green LED (12 W; 500–570 nm, 32 400 J cm−2) light. Whereas, pyrene–chlorin (PyChl), pyrene–bacteriochlorin (PyBac), and their nanolignin sensitizers (PyChl-LNCs, and PyBac-LNCs) were incubated with C. albicans for 45 min and irradiated for about 40 min with a red LED (12 W; 620–750 nm, 32 400 J cm−2) light. After each treatment, 100 μL suspension was withdrawn and plated on the YPD Agar culture plates. Then, they were incubated at 37 °C and examined after 24 h. The grown colonies were counted, and the colony-forming unit (CFU) method was used to determine the % cell viability after each set of treatments. The IC50 value of all the samples was determined using a standard linear calibration equation drawn at 0, 5, 10, 15, 20, and 25 μg mL−1 doses of PyP, PyChl, PyBac, and nanolignin sensitizers (PyP-LNCs, PyChl-LNCs, and PyBac-LNCs).Conclusion
In summary, we developed pyrene-substituted NIR photosensitizers, namely chlorin and bacteriochlorin, from their respective porphyrin derivatives. TD-DFT calculations with these NIR fluorescence imaging agents were performed to correlate the experimental photophysical properties with the in silico optical studies results. Owing to their hydrophobicity, the NIR photosensitizers were loaded in lignin nanoparticles to form hydrophilic nanolignin sensitizers, which facilitated fluorescence imaging. Due to the presence of pyrene fluorophores, the native near-IR photosensitizers and their corresponding nanoformulations displayed higher DNA binding affinities and enhanced fluorescence properties than porphyrin. Furthermore, the nanolignin sensitizers were found to be promising ROS generators. Conclusively, it was demonstrated that the pyrene-based near-IR photosensitizers (chlorin and bacteriochlorin) are promising candidates for red and near-IR light-assisted photodynamic therapy and bioimaging applications.Data availability
All data supporting the findings of this study are available within the paper and its ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
JB thanks the Department of Biotechnology (DBT), Govt. of India for funding the lignin valorization flagship project. K. G. acknowledges CIAB for a senior research fellowship and University Institute of Pharmaceutical Sciences, Panjab University, for PhD registration. S. K. acknowledges the Women Scientists Scheme A (WOS-A), DST, and Govt. of India. A. K. P. thanks CIAB for a senior research fellowship and IISER Mohali for PhD registration. D. M. thanks DBT for a junior research fellowship and IISER Mohali for PhD registration. The authors want to acknowledge the Center of Innovative and Bioprocessing (CIAB), Department of Biotechnology (DBT), Govt. of India for funding and infrastructural support.References
- Y. Zhang, P. Huang, D. Wang, J. Chen, W. Liu, P. Hu, M. Huang, X. Chen and Z. Chen, Nanoscale, 2018, 10, 15485–15495 RSC.
- L. Huang, Y. Y. Huang, P. Mroz, G. P. Tegos, T. Zhiyentayev, S. K. Sharma, Z. Lu, T. Balasubramanian, M. Krayer, C. Ruzié, E. Yang, H. L. Kee, C. Kirmaier, J. R. Diers, D. F. Bocian, D. Holten, J. S. Lindsey and M. R. Hamblin, Antimicrob. Agents Chemother., 2010, 54, 3834–3841 CrossRef CAS PubMed.
- D. M. George, A. S. Vincent and H. R. Mackey, Biotechnol. Rep., 2020, 28, e00563 CrossRef PubMed.
- M. K. Peters, F. Röhricht, C. Näther and R. Herges, Org. Lett., 2018, 20, 7879–7883 CrossRef CAS PubMed.
- W. Zhu, Y. H. Gao, P. Y. Liao, D. Y. Chen, N. N. Sun, P. A. Nguyen Thi, Y. J. Yan, X. F. Wu and Z. L. Chen, Eur. J. Med. Chem., 2018, 160, 146–156 CrossRef CAS.
- Z. Yu and M. Ptaszek, Org. Lett., 2012, 14, 3708–3711 CrossRef CAS.
- X.-D. Jiang, D. Xi, C.-L. Sun, J. Guan, M. He and L.-J. Xiao, Tetrahedron Lett., 2015, 56, 4868–4870 CrossRef CAS.
- H. S. Lee, J. H. Han, J. H. Park, M. E. Heo, K. Hirakawa, S. K. Kim and D. W. Cho, Phys. Chem. Chem. Phys., 2017, 19, 27123–27131 RSC.
- P. A. Panchenko, M. A. Grin, O. A. Fedorova, M. A. Zakharko, D. A. Pritmov, A. F. Mironov, A. N. Arkhipova, Y. V. Fedorov, G. Jonusauskas, R. I. Yakubovskaya, N. B. Morozova, A. A. Ignatova and A. V. Feofanov, Phys. Chem. Chem. Phys., 2017, 19, 30195–30206 RSC.
- J. H. Han, H. Y. Cho, D. Y. Kim, Y. J. Jang, Y.-A. Lee and S. K. Kim, J. Mol. Struct., 2020, 1215, 128264 CrossRef CAS.
- N. A. Le, V. Babu, M. Kalt, L. Schneider, F. Schumer and B. Spingler, J. Med. Chem., 2021, 64, 6792–6801 CrossRef CAS.
- M. Wu, Z. Liu and W. Zhang, Chem. Sci., 2021, 12, 1295–1301 RSC.
- L. Huang, M. Krayer, J. G. S. Roubil, Y. Y. Huang, D. Holten, J. S. Lindsey and M. R. Hamblin, J. Photochem. Photobiol., B, 2014, 141, 119–127 CrossRef CAS PubMed.
- B. Pucelik, A. Sułek and J. M. Dąbrowski, Coord. Chem. Rev., 2020, 416, 213340 CrossRef CAS.
- X. Zhang, Y. Ma, X. Zhang, X. Pang and Z. Yang, Biomed. Pharmacother., 2023, 165, 115014 CrossRef CAS PubMed.
- N. Dragicevic, J. Predic-Atkinson, B. Nikolic, S. B. Pajovic, S. Ivkovic and M. Adzic, Expert Opin. Drug Delivery, 2024, 21, 279–307 CrossRef CAS PubMed.
- V. M. Desai, M. Choudhary, R. Chowdhury and G. Singhvi, Mol. Pharmacol., 2024, 21, 1591–1608 CrossRef CAS.
- M. Tavakkoli Yaraki, B. Liu and Y. N. Tan, Nano-Micro Lett., 2022, 14, 1–49 CrossRef PubMed.
- G. Li, R. Zhou, W. Zhao, B. Yu, J. Zhou, S. Liu, W. Huang and Q. Zhao, Research, 2020, 2020, 1–14 Search PubMed.
- S. Paul, A. De, S. Kirar, N. S. Thakur, A. K. Pujari, K. Gogde and J. Bhaumik, ACS Appl. Nano Mater., 2024, 7(9), 10984–10997 CrossRef CAS.
- S. Paul, N. S. Thakur, S. Chandna, Y. N. Reddy and J. Bhaumik, J. Mater. Chem. B, 2021, 9, 1592–1603 RSC.
- A. K. Pujari, R. Kaur, Y. N. Reddy, S. Paul, K. Gogde and J. Bhaumik, J. Med. Chem., 2024, 67, 2004–2018 CrossRef CAS PubMed.
- J. H. Lee, K. Kim, X. Jin, T. M. Kim, I. G. Choi and J. W. Choi, Int. J. Biol. Macromol., 2021, 183, 660–667 CrossRef CAS PubMed.
- R. Kumar, A. Butreddy, N. Kommineni, P. Guruprasad Reddy, N. Bunekar, C. Sarkar, S. Dutt, V. K. Mishra, K. R. Aadil, Y. K. Mishra, D. Oupicky and A. Kaushik, Int. J. Nanomed., 2021, 16, 2419–2441 CrossRef CAS PubMed.
- D. A. Ali and M. M. Mehanna, Int. J. Biol. Macromol., 2022, 221, 934–953 CrossRef CAS PubMed.
- J. H. Lee, J. S. Im, X. Jin, T. M. Kim and J. W. Choi, ACS Sustainable Chem. Eng., 2022, 10, 5792–5802 CrossRef CAS.
- M. Mishra, Lignin-based Materials, 2023, pp. 223–246 Search PubMed.
- M. Arcentales, R. Martín-Sampedro, R. Santos-Oliveira, M. F. Attia, J. N. Anker, D. C. Whitehead, A. Debut, L. Deng, Y. Zheng, B. Hu, M. P. Romero, F. López and F. Alexis, ACS Appl. Mater. Interfaces, 2023, 15, 31320–31329 CrossRef CAS.
- A. Boarino, H. Wang, F. Olgiati, F. Artusio, M. Özkan, S. Bertella, N. Razza, V. Cagno, J. S. Luterbacher, H. A. Klok and F. Stellacci, ACS Sustainable Chem. Eng., 2022, 10, 14001–14010 CrossRef CAS PubMed.
- B. F. O. Nascimento, A. M. d A. Rocha Gonsalves and M. Pineiro, Inorg. Chem. Commun., 2010, 13, 395–398 CrossRef CAS.
- H. W. Whitlock, R. Hanauer, M. Y. Oester and B. K. Bower, J. Am. Chem. Soc., 1969, 91, 7485–7489 CrossRef CAS.
- J. Bhaumik, Z. Yao, K. E. Borbas, M. Taniguchi and J. S. Lindsey, J. Org. Chem., 2006, 71, 8807–8817 CrossRef CAS PubMed.
- J. K. Laha, S. Dhanalekshmi, M. Taniguchi, A. Ambroise and J. S. Lindsey, Org. Process Res. Dev., 2003, 7, 799–812 CrossRef CAS.
- R. Paolesse, A. Marini, S. Nardis, A. Froiioa, F. Mandoj, D. J. Nurco, L. Prodi, M. Montalti and K. M. Smith, J. Porphyrins Phthalocyanines, 2012, 7, 25–36 CrossRef.
- J. Bhaumik, N. S. Thakur, P. K. Aili, A. Ghanghoriya, A. K. Mittal and U. C. Banerjee, ACS Biomater. Sci. Eng., 2015, 1, 382–392 CrossRef CAS.
- K. Gogde, S. Paul, A. K. Pujari, A. K. Yadav and J. Bhaumik, J. Med. Chem., 2023, 66, 13058–13071 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, C.01, 2016 Search PubMed.
- M. Gouterman, J. Mol. Spectrosc., 1961, 6, 138–163 CrossRef CAS.
- O. Cramariuc, T. I. Hukka and T. T. Rantala, J. Phys. Chem. A, 2004, 108, 9435–9441 CrossRef CAS.
- Z. Melissari, H. C. Sample, B. Twamley, R. M. Williams and M. O. Senge, ChemPhotoChem, 2020, 4, 601–611 CrossRef CAS.
- S. K. Sharma, M. Krayer, F. F. Sperandio, L. Huang, Y. Y. Huang, D. Holten, J. S. Lindsey and M. R. Hamblin, J. Porphyrins Phthalocyanines, 2013, 17, 73–85 CrossRef CAS PubMed.
- Y. N. Reddy, K. Gogde, S. Paul and J. Bhaumik, in Biomass for Bioenergy and Biomaterials, ed. N. Adlakha, R. Bhatnagar and S. S. Yazdani, CRC Press, Boca Raton, Florida, 2021, pp. 31–64.
- D. Yang, S. Gao, Y. Fang, X. Lin, X. Jin, X. Wang, L. Ke and K. Shi, Nanomedicine, 2018, 13, 3159–3177 CrossRef CAS PubMed.
- A. Shalviri, G. Raval, P. Prasad, C. Chan, Q. Liu, H. Heerklotz, A. M. Rauth and X. Y. Wu, Eur. J. Pharm. Biopharm., 2012, 82, 587–597 CrossRef CAS PubMed.
- P. Carloni, E. Damiani, L. Greci, P. Stipa, F. Tanfani, E. Tartaglini and M. Wozniak, Res. Chem. Intermed., 1993, 19, 395–405 CrossRef CAS.
- S. Kirar, N. S. Thakur, J. K. Laha and U. C. Banerjee, ACS Appl. Bio Mater., 2019, 2, 4202–4212 CrossRef CAS PubMed.
- M. Haeubl, L. M. Reith, B. Gruber, U. Karner, N. Müller, G. Knör and W. Schoefberger, J. Biol. Inorg. Chem., 2009, 14, 1037–1052 CrossRef CAS PubMed.
- B. P. de Oliveira, N. U. de, C. Bessa, J. F. do Nascimento, C. S. de Paula Cavalcante, R. O. dos, S. Fontenelle and F. O. M. da S. Abreu, Int. J. Biol. Macromol., 2023, 227, 805–814 CrossRef.
- J. Pfister, A. Lichius, D. Summer, H. Haas, T. Kanagasundaram, K. Kopka and C. Decristoforo, Sci. Rep., 2020, 10, 1–9 CrossRef PubMed.
- M. Bera, M. Das, M. Dolai, S. Laha, M. M. Islam, B. C. Samanta, A. Das, I. Choudhuri, N. Bhattacharyya and T. Maity, ACS Omega, 2023, 8, 636–647 CrossRef CAS.
- M. Das, S. Mukherjee, M. M. Islam, I. Choudhuri, N. Bhattacharyya, B. C. Samanta, B. Dutta and T. Maity, ACS Omega, 2022, 7, 23276–23288 CrossRef CAS.
- E. M. Kojic and R. O. Darouiche, Clin. Microbiol. Rev., 2004, 17, 255 CrossRef PubMed.
- M. Nitz, B. W. Purse and D. R. Bundle, Org. Lett., 2000, 2, 2939–2942 CrossRef CAS PubMed.
- K. M. Sakita, P. C. V. Conrado, D. R. Faria, G. S. Arita, I. R. G. Capoci, F. A. V. Rodrigues-Vendramini, N. Pieralisi, G. B. Cesar, R. S. Gonçalves, W. Caetano, N. Hioka, E. S. Kioshima, T. I. E. Svidzinski and P. S. Bonfim-Mendonça, Future Microbiol., 2019, 14, 519–531 CrossRef CAS PubMed.
- N. Martins and C. F. Rodrigues, J. Clin. Med, 2020, 9, 722 CrossRef PubMed.
- Y. N. Reddy, N. Singh Thakur and J. Bhaumik, ChemNanoMat, 2020, 6, 239–247 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available Instruments and methods, materials ad chemicals, tables. See DOI: https://doi.org/10.1039/d4tb01627k |
| This journal is © The Royal Society of Chemistry 2025 |