 Open Access Article
Open Access ArticleBiological properties and DNA nanomaterial biosensors of exosomal miRNAs in disease diagnosis
Zhikun
Zhang
a,
Md. Ahasan
Ahamed
d and
Dayong
Yang
 *bc
*bc
aSchool of Chemical and Pharmaceutical Engineering, Hebei University of Science and Technology, Shijiazhuang, 050018, PR China
bDepartment of Chemistry, State Key Laboratory of Molecular Engineering of Polymers, Shanghai Key Laboratory of Molecular Catalysis and Innovative Materials, College of Chemistry and Materials, Fudan University, Shanghai, 200438, PR China. E-mail: dayongyang@fudan.edu.cn
cBioinformatics Center of AMMS, 100850, Beijing, PR China
dDepartment of Electrical Engineering, Pennsylvania State University, University Park, 16802, USA
First published on 7th February 2025
Abstract
MicroRNAs (miRNAs) regulate gene expression and are important biomarkers in molecular diagnostics, prognosis, and personalized medicine. The miRNAs that are found within exosomes, also known as exo-miRs, have been shown to demonstrate increased levels of both abundance and stability. Thus, exo-miRs show potential as a reliable biomarker for further investigation. Due to the programmable nanostructures, biocompatibility, and excellent molecular recognition ability, biosensing platforms based on DNA nanomaterial are considered promising for detecting exo-miRs in clinical analysis, including cancer, neurodegenerative disorders, and infectious diseases. Although considerable advancements have been achieved in exo-miR-based testing, there are ongoing challenges in accurately detecting and analyzing multiple targets concurrently at low concentrations in complex biological samples. The primary focus of our research is to thoroughly analyze the biogenesis of exo-miRs, carefully assess their levels of expression in various clinical diseases, and comprehensively investigate their correlations with a wide range of diseases, including cancer, infection, and neurodegenerative disorders. We also examined recent progress in DNA nanomaterial-based detection methods for exo-miRs. This study explores the challenges and intricacies faced during the creation and execution of exo-miR tests within a clinical setting to diagnose diseases. The successful development and implementation of DNA nanomaterials for exo-miR detection can significantly revolutionize the early detection, monitoring, and management of various medical conditions, leading to enhanced healthcare outcomes.
1 Introduction
MicroRNAs (miRNAs or miRs) are a group of endogenous, small-molecule (typically 22 nucleotides long), non-coding, and structurally functional RNAs.1,2 miRs have become increasingly recognized as essential biomarkers in the fields of molecular diagnostics, disease progression monitoring, and individualized medical treatment because of their notable impact on the regulation of gene expression.3 Dysregulated miRNAs are strongly associated with the development of various diseases,4,5 such as tumor,5,6 cardiovascular diseases,7,8 central nervous system diseases,9 kidney injury,10 and other conditions. Different miRNAs may be present in bodily fluids, including freely circulating miRNAs, miRNAs bound to ribonucleoproteins, and miRNAs enclosed within various exosomes. Exosomes are extracellular vesicles released by all types of cells, serving a crucial function in transporting biomolecules, including proteins, metabolites, and assorted nucleic acids in biological processes.3,11,12 miRs contained within exosomes (so-called exo-miRs) exhibit higher levels of abundance and stability under the protection of a lipid bilayer membrane when contrasted with freely circulating miRs in the bloodstream.13 They exhibit greater value in liquid biopsy due to enhanced preservation when exposed to enzymes commonly found in bodily fluids. Therefore, exo-miRs have been the subject of thorough investigation in the past few years as they emerge as a promising biomarker for liquid biopsy, attracting considerable attention in disease diagnostics.The conventional methods for analyzing exo-miRs are reverse transcription-polymerase chain reaction (RT-PCR),14,15 digital PCR,16 and next-generation sequencing (NGS) technology.17 Despite their excellent analytical performance, these methods still exhibit limitations for clinical applications. RT-PCR as the gold standard technique can be utilized to detect exo-miRs; however, it exhibits a comparatively restricted need for costly equipment, involving multiple manual steps that prolong the assay time.18 Digital PCR can detect very low levels of miRNAs. However, it requires a significant financial investment and involves a complex operational process.16 Next-generation sequencing typically involves extensive pretreatment and multiple procedural steps.19 Therefore, the direct and accurate detection of exo-miRs derived from tumors presents a significant challenge, requiring the precise identification and quantitative assessment of exo-miRs amidst complex background interference, diverse subtypes of extracellular vesicles (EVs), and varying expression levels of various exo-miRs. Therefore, developing a sensitive, accurate, and simplified bioassay and platform to quantify disease-derived exo-miRs in complicated biofluid samples is highly desirable to promote the development of clinically viable miRNA biomarkers of disease diagnostics.
Currently, there has been a significant focus on the advancement of multiplexed, sensitive, and straightforward techniques for exo-miR detection, including surface-enhanced Raman scattering (SERS),20 surface plasmon resonance (SPR),21 microfluidics,22 fluorescence,23,24 and electrochemistry.25 DNA nanotechnology generates a wide array of nanostructures due to its precise design, unique ability for programming, and natural compatibility with biological systems.26–28 DNA nanomaterial-based biosensors have garnered significant interest as potential diagnostic tools within this field. The attributes of DNA nanostructures, such as their diminutive scale, precise molecular recognition abilities, and inherent self-assembly stability, render them a feasible biosensing platform fabrication for facilitating the integration of nanotechnology and diagnostics. This allows DNA nanoassemblies to fulfill a crucial function in advancing the development of highly specific and sensitive biosensors with improved performance for exo-miR detection. This review offers an in-depth analysis of the biogenesis of exo-miRs, emphasizing the linkages between miRs and various diseases. We examined the latest developments in DNA nanomaterials, focusing on their design and mechanisms in exo-miR detection (Fig. 1). This review article also explores the challenges faced and offers suggestions for future progress. The goal is to bridge the gap between research on exo-miRs and their application in disease diagnostics to facilitate the development of novel, clinically useful diagnostic tools.
 | ||
| Fig. 1 Various DNA nanomaterials for detecting exo-miRs associated with diseases by fluorescence, electrochemical, SERS, and UV-vis sensing platforms. | ||
2 Biosynthesis of exo-miRs
Exo-miR biosynthesis starts with the traditional miRNA biogenesis pathway inside the cell.29 Their size ranges from 40 to 120 nm, which provides valuable information for cancer diagnosis, brain diseases, infectious diseases, and therapeutic treatment. Their origins can be traced back to the intraluminal budding of multivesicular bodies (MVBs), a fusion of MVBs with cell membranes, and endolysosomal pathways. Generally, it contains a variety of membranes, cytosolic metabolites, DNA, proteins, circular RNA (circRNA), messenger RNAs (mRNAs), and microRNAs (miRNAs), as shown in Fig. 2a. Among these, miRNAs are non-coding RNAs that regulate genetic expression, which is transcribed from DNA and converted to primary miRNAs, processed into precursor forms, and mature miRNAs.30 Generally, miRNAs bind with the 3′ untranslated region (UTR) of a gene target, which causes miRNA degradation or translation inhibition. They can also interact with the 5′ UTR and gene promoters.29 Normally, miRNA activity depends on the subcellular target site, quantity, and interaction strength.31 One way of intraluminal vesicles (ILVs) with miRNAs is secretion in extracellular fluids via vesicles like exosomes or bound to proteins like argonaute 2 (AGO2).32–34 In other studies, exo-miRs can be found in biological fluids such as saliva and serum, which are now widely used for nucleic acid extraction.35–38 Primarily, miRNAs are transcribed in the nucleus as primary miRNAs (pri-miRNAs) by RNA polymerase II followed by cleavage activity by Drosha-DGCR8 and create precursor miRNAs (pre-miRNAs), which exportin-5 exports to the cytoplasm.31,35 Then, the Dicer enzyme comes into play to process pre-miRNAs to mature miRNA duplexes in the cytoplasm.39 One strand of the duplex can regulate a gene's expression by inhibiting or degrading the translation of target mRNAs after being loaded into the RNA-induced silencing complex (RISC).40 On the other hand, various miRNAs are specifically packed into exosomes, which facilitate intercellular communication.41 Following general miRNA synthesis, the packaging of exo-miRs is a selective process.35 miRNA sorting and packaging is a very complex process where heterogeneous nuclear ribonucleoproteins (hnRNPs), endosomal sorting complex required for transport (ESCRT) and AGO2 proteins are responsible for sorting, identifying and managing specific miRNAs for exosome packaging.42 Finally, these exosomes and microvesicles are released from the MVBs into extracellular space, and lysosomes are formed inside the cell through the process of Golgi apparatus, as shown in Fig. 2b.43 | ||
| Fig. 2 Exosome structure and biosynthesis. (a) Exosome size and functional components such as miRNA, mRNA, circRNA, proteins, and lipids.44 (b) miRNA is transcribed in the nucleus, processed by Drosha and Dicer, and loaded into RISC. Exosomes are released from MVBs and carry miRNA to recipient cells.40 | ||
Exosomes travel toward the recipient cells by bodily fluids such as blood and serum to interact with the cell and outside exo-miRs. Recipient cells can take exosomes and reflect on their unique miRNAs, which control gene expression and facilitate intercellular communication.45 Then, miRNAs fuses inside the cell membrane or receptor–ligand interaction called endocytosis.46 After endocytosis, the exosomal cargo miRNA is trapped. Once the extracellular microRNAs are located within an early endosome that contains intraluminal vesicles, which are small vesicles found in multivesicular bodies and contain microRNA, DNA, RNA, proteins, and lipids. They are created through the inward budding of the MVBs' membrane, and the membrane fusion process facilitates the release of miRNA to the cytosol.35,47 Moving forward, RISC interacts with miRNA, and ESCRT facilitates the sorting process.48 On the other hand, the ILVs are released into the extracellular environment as exosomes when the MVBs and plasma membrane fuse. To create diagnostic instruments and treatment plans, it is essential to find out the miRNA type that is primarily responsible for early disease detection and the amount of miRNA present at that stage, where exo-miRs act as significant biomarkers.39
3 Correlation between exo-miRs and various diseases
Exo-miRs have emerged as potential biomarkers for disease detection and monitoring and can be found in various non-invasive biological fluids such as blood, saliva, and urine. Previously, we provided deep insight into miRNA biogenesis, where they serve as a primary cargo content of exosomes in early diagnosis of cancer, neurological diseases such as Parkinson's disease (PD) and Alzheimer's disease (AD), cardiovascular diseases such as heart diseases, pulmonary arterial hypertension, and aortic aneurysms, infectious diseases, and respiratory conditions. Modulating immune responses and inflammation of infectious and respiratory diseases, potential drug resistance for cancer and neurological disease, and miRNA detection are valuable tools for early diagnosis and setting up specific treatment plans. Their role in disease pathogenesis makes them attractive targets for developing diagnostic and therapeutic interventions.3.1 Cancer
Exo-miRs are crucial in regulating cancer drug resistance through gene expression modulation, drug efflux, metabolic reprogramming, DNA damage repair, apoptosis, and epithelial-to-mesenchymal transition (EMT).40 MicroRNAs are integral to an intricate network that includes their target genes (RhoA, FIH, ACOX1, etc.) and signalling cascades (the EGF-AKT signalling axis, the ERK-MMP9 cascade, the Hippo signalling pathway, etc.), which plays a crucial role in the progression of cancer.49 These miRNAs are transferred between cancer cells and the tumor microenvironment, influencing tumor growth, metastasis, and therapeutic resistance. For example, Guo et al. explained in their study that exo-miRs like miR-1246, secreted by ovarian cancer (OC) cells, regulate drug efflux transporters like ATP-binding cassette subfamily B member 1 (ABCB1), contributing to chemotherapy resistance by decreasing drug efficacy.40,50 Similarly, miR-365 induces resistance to gemcitabine by increasing cytidine deaminase activity in pancreatic cancer (PC) cells.51 Exo-miRs like miR-21 and miR-155 also play the most significant roles in cancer progression by modulating apoptosis, promoting tumor survival and drug resistance when carried by exosomes.52 miR-21, often overexpressed in breast cancers (BC), colorectal cancers, and OC, is transferred by cancer-associated fibroblasts (CAFs) to nearby cancer cells. It reduces the expression of pro-apoptotic factors like phosphatase and tensin homolog (PTEN) and allows cells to survive chemotherapy.40 The transfer of miR-21 via exosomes has explicitly been noted to reduce the expression of pro-apoptotic factors such as PTEN, leading to enhanced cell survival and drug resistance.53 On the other hand, miR-155 reprograms tumor-associated macrophages (TAMs) into a pro-tumorigenic phenotype, aiding in tumor proliferation and immune evasion.54 These macrophages, in turn, secrete factors that promote tumor cell proliferation and immune evasion. In addition, exosomal miR-155 contributes to chemotherapy resistance by downregulating genes involved in DNA repair and apoptosis, allowing cancer cells to thrive even in chemotherapeutic agents, as shown in Fig. 3A. This dynamic interaction between exo-miRs and immune cells in the tumor microenvironment highlights the sophisticated mechanisms by which tumors evade therapeutic interventions.40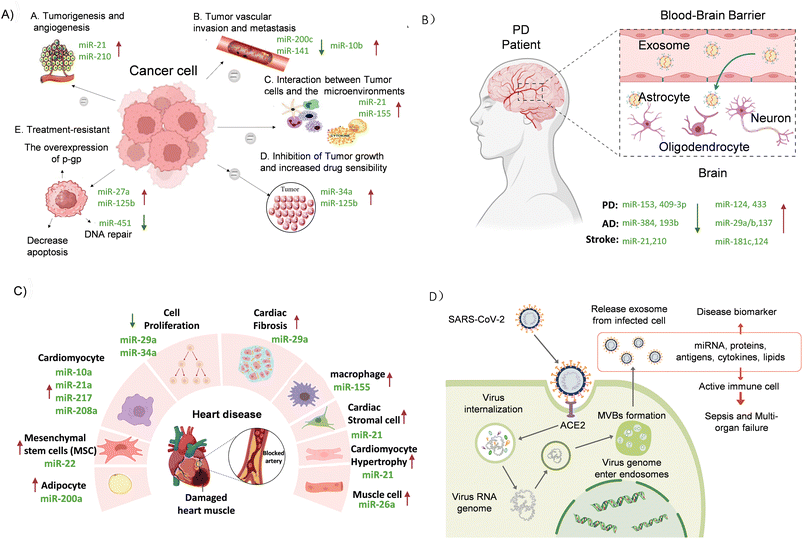 | ||
| Fig. 3 Exo-miRs' role in diseases. (A) Cancer: miR-21 and miR-210 (upregulated) promote tumor growth and angiogenesis, while miR-27a and miR-125b enhance treatment resistance. Downregulated miR-451 impairs DNA repair.52 (B) Brain disease: miR-153 and miR-409-3p (upregulated) are associated with neuroinflammation and α-synuclein accumulation, while miR-124 and miR-133b (downregulated) impair neuronal repair. In AD, miR-384 and miR-193b are upregulated, contributing to disease progression, while miR-29a/b and miR-137 (downregulated) affect synaptic function.55 (C) Cardiovascular disease: miR-29a and miR-34a (upregulated) drive cell proliferation and fibrosis, while miR-155 promotes inflammation. miR-21 contributes to cardiac fibrosis and hypertrophy.56–58 (D) Infectious disease (SARS-CoV-2): exosomes from infected cells carry miRNAs like miR-21, triggering immune responses that lead to severe inflammation and potentially multi-organ failure.59 | ||
Exo-miRs also significantly impact cancer metastasis. In BC, for instance, Zhang et al. showed that miR-9 induces fibroblast cells to acquire cancer-associated fibroblast-like properties, supporting tumor growth and metastasis. At the same time, miR-200b promotes proliferation and invasion in colorectal cancer by downregulating tumor suppressors.60 Modani et al. note that miR-10b achieves this by targeting genes that regulate cell adhesion and migration, thus facilitating the spread of cancer to distant organs.39 miR-10b, secreted by highly metastatic BC cells, enhances recipient cells' migratory and invasive capabilities, facilitating cancer spread to distant organs.40 These exo-miRs drive metastasis and create a supportive tumor microenvironment by influencing gene expression and facilitating communication between cancer and stromal cells. In addition to miRNAs, these exosomes transfer proteins, lipids, and other molecules that modify the behavior of both cancerous and non-cancerous cells in the microenvironment. Additionally, exo-miRs are crucial for maintaining cancer stem cell (CSC) populations, which are inherently resistant to conventional therapies.61 For example, Santos et al. noted that miR-155, secreted by CSCs, triggers the EMT process, making recipient cells more resistant to chemotherapy.62 Meanwhile, miR-210, transferred by exosomes in gemcitabine-resistant PC stem cells, inhibits apoptosis and promotes survival.39
Furthermore, circulating exo-miRs offer a minimally invasive method for early cancer detection and monitoring treatment response. For example, miR-1290 and miR-375 elevated in prostate cancer patients potentially serve as reliable biomarkers for early detection.63 Another example is that miR-122 reduces glucose uptake in liver cells, providing metabolic advantages to cancer cells, while miR-126-5p alters lipid metabolism, essential for cancer cell proliferation. This metabolic reprogramming can exacerbate conditions such as cachexia, diabetes, and obesity, highlighting the intricate relationship between cancer, exosomes, and systemic metabolic dysfunction.64 Exo-miRs, such as miR-19, linked to brain cancer metastasis, and miR-25-3p promoting liver metastasis in colorectal cancer via the CXCL12/CXCR4 pathway, are key diagnostic markers.65 Since exo-miRs are available in saliva, Zhang et al. highlight the critical role of salivary miRNAs in the early diagnosis and progression of various cancers, including oral squamous cell carcinoma (OSCC), esophageal cancer (EC), gastric cancer (GC), pancreatic cancer (PC), and breast cancer (BC).66 The miRNAs, such as miR-200 and miR-21, are significantly altered in OSCC, making them promising biomarkers for early detection.67,68 Similarly, elevated levels of miR-10, miR-144, and miR-451 are associated with EC,69–71 while miR-140-5p and miR-301a are linked to GC. For PC, miR-1246, miR-4644, miR-103, and miR-107 in salivary exosomes show promise for differentiating patients from normal controls.66,72 In BC, miR-21 is a significant marker, showing reduced levels post-treatment.73 Understanding the roles of exo-miRs in cancer drug resistance, metastasis, and tumor–stroma interactions can help develop new therapeutic strategies to improve cancer diagnosis and treatment outcomes.
3.2 Concussion and Parkinson's disease
Exo-miRs are also implicated in the early stages of concussion, neurodegenerative diseases, Parkinson's disease (PD), and Alzheimer's disease (AD). Specific miRNAs, like miR-124 and miR-132, are found to be dysregulated in exosomes derived from neural cells, contributing to disease pathogenesis.74 Concussions, especially prevalent in children and adolescents, represent about two-thirds of mild traumatic brain injury (TBI) cases and involve the regulation of gene expression through miRNA. While most children recover within two weeks, around one-third may experience prolonged concussion symptoms (PCS). miR-27 has been identified as a potential biomarker for concussions due to its accuracy in distinguishing concussed individuals. Additionally, miRNAs such as miR-320, miR-133, and let-7a have been linked to prolonged symptoms, while specific miRNAs (miR-320, miR-629, let-7b-5p) correlate with symptoms like memory issues, headaches, and fatigue.66 In the central nervous system (CNS), exosomes facilitate communication between neurons and glial cells, contributing to the progression of various diseases. They carry miRNAs, such as miR-150, that regulate neuroinflammatory responses and contribute to disease progression.75 Exosomal release from various CNS cells, including neurons, astrocytes, and microglial cells, influences synaptic communication and neuro-glial interactions.76 miR-150, miR-223, and miR-23a have been identified in the serum and plasma of multiple sclerosis (MS) patients. These biomarkers have shown high diagnostic accuracy, with miR-150 demonstrating 85% sensitivity and 90% specificity, making them valuable tools for MS diagnosis.77 Other examples are miR-126, miR-223, miR-9, and miR-124, which have shown potential in diagnosing ischemic stroke with higher sensitivity and specificity compared to free circulating miRNAs. For instance, miR-223 demonstrated an AUC of 0.859 with 84.0% sensitivity and 78.8% specificity for stroke diagnosis.78 miR-21, miR-222, and miR-124-3p are significantly elevated in the serum of GBM patients and have shown high diagnostic accuracy in distinguishing GBM from healthy controls. ROC curve analysis for miR-21 revealed an AUC of 0.927, demonstrating its strong potential as a biomarker for GBM.79 Additionally, miR-301a levels were found to correlate with tumor recurrence and surgical resection outcomes, highlighting their role in monitoring disease progression.77Parkinson's disease (PD) is a progressive neurodegenerative disorder that primarily affects individuals over the age of 65 and is the second most common neurodegenerative disorder after Alzheimer's disease (AD).66 Its diagnosis is typically based on clinical symptoms such as tremors, rigidity, and non-motor issues like depression and dementia. Vital pathological features of PD include the loss of dopaminergic neurons and the accumulation of α-synuclein.76 Recent studies have highlighted alterations in miRNA levels in saliva, cerebrospinal fluid (CSF), and serum of PD patients, which affect genes linked to the disease.80 Research has indicated that miRNAs can regulate genes linked to Parkinson's disease, such as the SNCA gene, PRKN gene, PARK7 gene, etc.81,82 Notably, miR-153, miR-409-3p, miR-375, miR-1468-5p, miR-19b, miR-195, and miR-24, found in CSF and plasma, have emerged as potential diagnostic biomarkers.83 Among these, miR-153 and miR-223 have been associated with α-synuclein accumulation, showing moderate diagnostic accuracy.84 Other miRNAs, including miR-29, miR-28, miR-19b-3p, and elevated levels of miR-409-3p and let-7g-3p, exhibit high sensitivity and specificity as diagnostic biomarkers.77 Additionally, miRNAs such as miR-7, miR-34b/c, miR-124, miR-221/222, miR-137, and miR-433 play roles in PD pathology, contributing to neuroinflammation, mitochondrial dysfunction, and dopamine metabolism, as depicted in Fig. 3B. Exo-miRs like miR-193b, miR-135a, and miR-384 have been identified as potential biomarkers for early AD detection. miR-384 has been particularly effective in distinguishing AD from other neurological diseases like PD with dementia and vascular dementia.77 Furthermore, studies report that while miR-1 and miR-19b-3p are reduced, miR-153 and miR-409-3p are elevated in PD patients' CSF exosomes. Exo-miRs may also help detect mild cognitive impairment (MCI), an early stage of AD, underscoring their potential as noninvasive tools for early diagnosis and treatment strategies in both PD and AD.55,83,85–89
3.3 Infectious diseases
Exo-miRs serve as a potential biomarker for diagnosing various infectious diseases, significantly influencing cell signaling and directing the host immune response. Changes in their expression profiles often appear early, even before the pathogen is detectable. These small, non-coding RNAs encapsulated within exosomes are protected from degradation and can be transported to recipient cells, which modulate gene expression and immune responses. Below, we discuss the role of exo-miRs in various infectious diseases, focusing on their functions and mechanisms of action in SARS-CoV-2, HIV, HCV, enterovirus, Epstein–Barr virus (EBV), HBV, and Ebola virus.90Several exo-miRs play a role in inhibiting viral replication and modulating immune responses in SARS-CoV-2 infection. For example, Drury et al. noted their significance in diseases caused by various pathogens, including Helicobacter pylori and SARS-CoV-2.91 Zhou and Xu found that downregulation of miR-204 in pulp tissue, serum, and saliva indicated H. pylori infection, highlighting the role of miRNA in regulating inflammatory pathways and their potential for early gastric cancer detection.92 Hicks et al. observed altered salivary miRNA levels, such as decreases in miR-4495, miR-296, miR-548, and miR-1273, in children with severe SARS-CoV-2 infection.93 Saulle et al. identified significant downregulation of let-7a, let-7b, and let-7c, and upregulation of miR-23a, miR-23b, and miR-29c, alongside three immunomodulatory miRNA in SARS-CoV-2.94 miR-223-3p, miR-24-3p, miR-145-5p, and miR-75p have been shown to inhibit SARS-CoV-2 replication by targeting viral proteins and mediating membrane fusion and viral entry into host cells. Additionally, miR-148a and miR-590 modulate the immune response during SARS-CoV-2 infection by targeting IRF9. miR-148a inhibits USP33, reducing IRF9 protein expression and impacting the immune response to viral infection. Similarly, miR-590 directly targets IRF9, regulating pro-inflammatory pathways. Both miRNAs are key in lowering viral load and controlling immune reactions.95–97Fig. 3D depicts the mechanism of exosomal miRNA involvement in SARS-CoV-2 infection. Infected cells release exosomes containing viral RNA, miRNAs, proteins, and other molecules, facilitating immune system activation. These exosomes contribute to inflammation and can lead to severe immune responses, including sepsis and multi-organ failure, while also serving as potential biomarkers for disease progression. However, challenges remain, such as the lack of consensus on specific miRNAs to be used as biomarkers, the influence of factors like age, gender, and lifestyle on miRNA expression, and variations in laboratory practices for detecting these miRNAs. The specificity issue can be resolved by integrating a clustered, regularly interspaced short palindromic repeats (CRISPR) assay.18,98–100 Despite these challenges, exosomal miRNA offers excellent potential for improving early diagnosis and therapeutic interventions in infectious diseases.90,91 Influenza infection is characterized by the modulation of miRNAs like miR-155 and miR-323, which influence the host's antiviral response and inhibit viral replication.101
For HIV and HIV-1, vmiR-TAR reduces Cdk9 and Bcl-2, delaying apoptosis and boosting viral production. vmiR88 and vmiR99 activate TLR8 in macrophages, increasing pro-inflammatory cytokines. Exosomal miR-155-5p from HIV-infected T cells activates NF-κB in cervical cancer cells, promoting invasion.102,103 In HIV infection, miRNAs like miR-223, miR-21, and miR-29b are involved in modulating viral replication and immune responses, with specific miRNAs downregulated during infection, affecting disease progression and immune function. Hepatitis viruses, including HBV and HCV, deregulate miRNAs like miR-122 and miR-29b, linked to liver fibrosis and viral replication.101 In hepatitis B virus (HBV) infections, exosomal miR-122 promotes viral replication, while miR-574-5p inhibits HBV polymerase, reducing replication. These miRNAs serve as potential targets for antiviral therapies and disease monitoring.104,105 miR-122 promotes anti-hepatitis C virus (HCV) antibody production by stimulating B cell proliferation. Exosomal let-7b and miR-206 activate macrophages via TLR7, enhancing BAFF release. These miRNAs help control viral replication and support the immune response during HCV infection.106,107 During enterovirus 71 (EV71) infection, miR-146a suppresses type I interferon responses, boosting viral replication. Similarly, miR-30a from EV71-infected cells targets MyD88 in macrophages, inhibiting immune responses and enhancing replication.108,109 In Epstein–Barr Virus (EBV) infection, miR-BHRF1-1 plays a role in immune evasion. At the same time, miR-BART3 is known for modulating gene expression linked to apoptosis and inflammation.110 miR-BART3 targets IPO7 to increase IL-6 expression, while miR-BHRF1 down-regulates p53, enhancing cell survival and viral replication.111,112 In Ebola virus (EBOV) infections, exosomal miR-21 and miR-222-3p enhance cell survival and viral replication by promoting anti-apoptotic and pro-inflammatory responses, making them potential therapeutic targets for controlling EBOV infections.113 miR-VP-3p and miR-155 are linked to immune response modulation and interferon signaling, and their presence in serum can be used for early diagnosis of infection.101 Chikungunya virus infection leads to differential expression of miRNAs in mosquito saliva, such as miR-184 and miR-125, which regulate inflammation and immune responses, influencing the transmission and severity of the disease in humans.101 Bayer et al. found that infection with varicella zoster virus could be attenuated in cells that had been pre-exposed to exosome-packaged miRNA clusters from chromosome 19; this mechanism offers broader antiviral resistance, especially against rubella and togavirus and HIV-1.114 Kaposi sarcoma-associated herpesvirus (KSHV), human papillomavirus (HPV), and BK polyomavirus (BKV) use exo-miRs to manipulate host cells. KSHV promotes the Warburg effect via miR-K12-11,115 shifting metabolism to aerobic glycolysis and enhancing angiogenesis and cell migration. HPV infection leads to the upregulation of miR-20a-5p, miR-423-3p, and let-7d-5p, which promote cancer progression by influencing cell survival and proliferation. BKV-associated bkv-miR-B1-3p and bkv-miR-B1-5p are detected in urine and blood as biomarkers for diagnosing BK virus nephropathy and monitoring viral replication in kidney transplant patients.110 In tuberculosis, miRNAs such as miR-144 and miR-361-5p are differentially expressed, mediating immune response modulation and disease progression.116 Filariasis involves miRNAs like miR-223 and miR-71, which regulate immune responses and are critical in the development and persistence of the infection. These miRNAs offer insights into the pathophysiological conditions associated with filariasis and could be used as diagnostic markers to evaluate disease severity and treatment efficacy.101
3.4 Other diseases
In cardiovascular diseases, exo-miRs like miR-1 and miR-133a are associated with early cardiac dysfunction, regulate gene expression between cells and myocardial injury, and facilitate communication between cells involved in heart function.117 Exo-miRs have been linked to the progression of atherosclerosis, myocardial infarction, and heart failure, influencing inflammation, cell death, and tissue repair.118Fig. 3C illustrates cell communication involving exo-miRs in the context of heart disease and their interaction with damaged heart muscle. The figure shows how different cell types, including cardiomyocytes, mesenchymal stem cells (MSCs), adipocytes, macrophages, and cardiac stromal cells, release or respond to exo-miRs that regulate critical processes like cell proliferation (e.g., miR-29a, miR-34a), fibrosis (e.g., miR-29a), and inflammation (e.g., miR-155). These miRNAs communicate between cells and influence damaged heart tissue's repair and response mechanisms, contributing to pathological processes like fibrosis, hypertrophy, and inflammatory responses in heart disease. Atherosclerosis is a leading cause of cardiovascular disease, characterized by the thickening and hardening of arterial walls due to the accumulation of lipids and fibrous tissues. Exo-miRs, such as miR-143 and miR-145, regulate vascular smooth muscle cells (VSMCs) and endothelial cells, mediating intercellular communication to prevent or promote atherosclerotic plaque formation.119 These miRNAs help reduce atherosclerosis by modulating inflammatory responses and promoting the repair of damaged endothelial cells.120,121 Acute coronary syndrome (ACS), which includes myocardial infarction and unstable angina, is marked by the rupture or erosion of atherosclerotic plaques. ACS patients, miRNAs such as miR-133a/b, the miR-30 family, miR-19, miR-20, miR-499, miR-1, miR-146a-5p, miR-208a, miR-126, and miR-21 are elevated, correlating with ACS and coronary artery disease (CAD).122,123 Ischemia–reperfusion injury (MIRI) occurs when blood flow is restored to a previously ischemic myocardial tissue, exacerbating oxidative stress and inflammation damage. miR-126 and miR-423-3p have reduced MIRI by targeting specific signaling pathways to inhibit cardiomyocyte apoptosis and promote cardiac cell survival. These exo-miRs offer therapeutic potential for reducing myocardial damage after ischemic events.56 In heart failure (HF), exo-miRs contribute to myocardial remodeling, leading to heart muscle thickening and dysfunction. Elevated levels of miRNAs such as miR-425 and miR-744 are linked to cardiac fibroblast activation and collagen synthesis, both critical in myocardial fibrosis and the progression of HF.124Pulmonary arterial hypertension (PAH) is characterized by increased pulmonary vascular resistance due to remodeling of the pulmonary arteries. miR-143 and miR-145 establish the communication between pulmonary artery smooth muscle cells and endothelial cells, promoting vascular remodeling.125 Aortic aneurysm, an abnormal enlargement of the aorta, is caused by miR-106a and miR-24, which have been implicated in the pathogenesis of aortic aneurysm by promoting the apoptosis of vascular smooth muscle cells and inflammation.126 Vascular calcification (VC) involves abnormal calcium deposition in blood vessels linked to atherosclerosis and increased cardiovascular risk. miR-324-3p regulates calcification pathways, offering potential therapeutic targets to reverse calcification and stabilize plaques.127 In rheumatic valvular disease (RVD), miR-155-5p modulates inflammation and fibrosis in heart valves via the S1PR1/STAT3 pathway, providing potential treatment options for valve dysfunction.118
4 DNA nanomaterial-based sensing platform for exo-miR detection
Exo-miRs present in saliva, blood, plasma, and urine have been identified at detectable levels, offering potential benefits for disease diagnosis using liquid biopsy.4,128–130 The efficient and prompt monitoring of exo-miRs allows for the convenient diagnosis of diseases in patients. Nevertheless, there are various challenges in detecting exo-miRs in situ for molecule biosensors,131 stemming from their complex compositions, small size, membrane obstruction, and low concentration.131,132 The designed molecular biosensors must be capable of entering a confined space to identify the target and maintain a consistent and stable signal output within a nanoscale environment in exosomes. Biosensors incorporating nucleic acid nanomaterials possess distinctive qualities that allow them to effectively address the limitations associated with detecting exo-miRs, owing to their small size, unsurpassed programmability, natural biocompatibility, and superior molecular recognition capacity.133,134 In current state of the art, various well-established DNA nanomaterials are utilized as biosensors for detecting exo-miRs, such as tetrahedral DNA nanostructures (TDNs), branched DNA, spherical DNA, and hairpin DNA.135,136 This selection explores the recent advancements in nucleic acid synthesis and their application in detecting exo-miRs, providing valuable insights for precise biological analysis and clinical diagnostics.4.1 Tetrahedral DNA nanomaterials
Tetrahedral DNA nanostructures (TDNs) were initially developed by Turberfield in 2004,137,138 exhibiting structural stability, easy internalization, biocompatibility, and versatile functionality.137,139–143 Briefly, TDNs could be prepared by four specifically designed single DNA strands using the complementary base pairing rules. In exosomes, TDN scaffold biosensors are utilized to prevent nucleases from degrading them. Maintaining a consistent spatial distance can help minimize the entanglement and local overcrowding among neighboring probes.144 As a result of these benefits, biosensors based on TDN can effectively penetrate exosomes spatially oriented, which enhances the ability to detect miRs by accurately adjusting the lateral distance between the biosensors and miRs.145,146 Furthermore, they have exhibited potential as biosensors for accurately identifying extracellular microRNAs.To attain simple, quick detection abilities and highly sensitive detection, methods utilizing TDN scaffold for electrochemical and fluorescence detection of exo-miRs were developed.147,148 Yang et al. utilized TDN nanomaterials to form a sandwich structure with exo-miRs and the capture DNA on an electronic chemosensor for ultrasensitive detection exo-miRs (Fig. 4A).149 In the detection process, TDN nanomaterials containing the spatial structure were capable of linking with multiple electroactive compounds in stoichiometry on electrochemical sensors, which increased the sensitivity. The recommended sensing system illustrated remarkable anti-interference characteristics, specificity, and sensitivity, capable of determining a low level of 34 aM in serum derived from patients with breast cancer without the need for enzymes. Motivated by this mechanism, Chao et al. utilized flowered nickel–iron layered double hydroxide@AuNPs and TDNs to design a reusable electrochemiluminescence biosensor for miR-27a.150 TDN-Ru(bpy)32+ functions as an electrochemiluminescence sensor, establishes a steady sandwich structure with miR-27a and hairpin DNA through complementary base pairing, enabling miRNA detection. This platform exhibited heightened sensitivity, exceptional selectivity, and consistent reproducibility. The limit of detection (LoD) decreased to 11.7 aM with a broad linear dynamic range. Furthermore, Miao et al. designed TDN nanomaterials with a walker strand on top and multiple track strands around it by assembling a triplex-forming oligonucleotide for miR-141 in cell and serum samples.151 The platform enabled the regeneration of the detecting interface by manipulating pH conditions. Using the tetrahedral DNA-supported walking nanomachine allowed for a highly sensitive miR-141 assay by electrochemical method. The LoD was found to be as low as 4.9 aM, indicating a relatively low value. Simultaneously, the designed platform was effectively utilized for miRNA detection in cell and serum samples, allowing for differentiation between pathological data and those of healthy individuals. Based on the aforementioned reports, the TDN-based electronic chemosensors exhibited high detection sensitivity, selectivity, and specificity, along with ease of operation. However, combining these methods requires a two-step process that includes the extraction of exosomes and the detection of miRNA, ultimately making it overly complex and impractical for practical applications. Within exosomes, TDNs were able to efficiently attach to specific microRNAs as a result of their spatial configuration, enabling the identification of target molecules. Furthermore, TDNs can readily be linked with electroactive compounds to generate electrochemical signals, which effectively amplified the detection signal. Currently, TDN-based various electrochemical detection systems for exo-miRs have been developed based on the mechanism described above, with the successful detection of aM-level exo-miRs (Table 1).
 | ||
| Fig. 4 (A) Illustrative diagram of TDN synthesis for nanolabel supercharge-based electrochemical platform for exo-miR analysis.149 (B) The diagrammatic representation of the designed DTNP-based FRET detecting sensor for miRNA analysis in living cells.142,152 (C) Synthesis of FDTp sensors for two types of miR-21 and miR-155 imaging in living cells.153 | ||
| DNA nanomaterials | Methods | Target miRNA | Linear range | LoD | Diseases | Ref. |
|---|---|---|---|---|---|---|
| TDNs | Electrochemical sensor | miR-27a | 100 aM–1 nM | 11.7 aM | Tumors | 150 |
| Electrochemical sensor | miR-21 | 100 aM–1 nM | 34.0 aM | Tumors | 149 | |
| Fluorescence sensor | miR-21, miR-141 | — | 100.0 fM | Cancer | 154 | |
| Branched DNA | Fluorescence sensing | miR-21, miR-27a, miR-375 | 5–140 nM, 2–100 nM, 7–70 nM | 462.0 pM, 301.0 pM, 154.0 pM | Cancer | 155 |
| Electrochemical sensor | miR-21 | 10–70 fM | 2.3 fM | MCF-7 cell | 156 | |
| Fluorescence sensor | miR-141 | 100–900 pM | 57.6 pM | Cancer | 157 | |
| Cubic DNA | Fluorescence sensor | miR-21, miR-221 | 10–100 nM, 10–120 nM | 3.4 nM, 2.3 nM | U251 cells | 140 |
| Fluorescence sensor | miR-21 | 100 pM–25 nM | 77.4 pM | No | 158 | |
| Fluorescence sensor | miR-44438 | No | No | Sleep disorder | 159 | |
| Spherical DNA | Fluorescence sensor | miR-1246 | 0–30 nM | 0.68 nM | Breast cancer | 160 |
| SERS | miR-10b | No | 0.21 fM | Pancreatic ductal adenocarcinoma | 161 | |
| Hairpin DNA | Fluorescence sensor | miR-21 | 103–108 particles per μL | 103 particles per μL | Tumor | 162 |
| Electrochemical sensor | miR-181 | 10 fM–100 nM | 7.94 fM | Cardiovascular disease | 163 |
To simplify and expedite the detection process, TDN-based optical platforms were developed for exo-miR detection in situ for diagnostic purposes. Fluorescence resonance energy transfer (FRET) is one approach that enables sensitive, real-time monitoring of molecular interactions, enhancing the specificity of diagnostic assays.164 Wang et al. created a TDN-based FRET sensing platform to sensitively detect tumor-related miR-146b by DNA-assisted cyclic amplification (Fig. 4B).152 When there was no hsa-miR-146b as target, the 5′ terminal and the 3′ terminal of fluorescent DNA (HP) were functionalized with FAM and TAMRA did not change the hairpin structure due to the hybridization with the extended DNA strand of the TDNs. Under that condition, the detection system exhibited a diminished FRET signal. The target hsa-miR-146b existed and induced the hybridization with the extended DNA strand of the DNA tetrahedron, inducing the release of HP and producing the significant FRET. Therefore, the level of hsa-miR-146b can be determined through the alteration in the fluorescent value. Furthermore, another DNA was designed to replace hsa-miR-146b for cyclic signal amplification, increasing the sensitivity of the assay in various cells. The LoD for hsa-miR-146b-5p was found to be 6 pM under optimal experimental conditions. Meanwhile, Hu et al. created a fluorescent TDNs probe using catalytic hairpin assembly (CHA) to enable the sensitive and specific detection of miR-21 and miR-155 in living cells through fluorescence signaling, as depicted in Fig. 4C.153 The DNA tetrahedron underwent functionalization with elements such as H1, H2, and protector, each incorporating two pairs of fluorophores and quenching molecules. Exo-miR-21 triggered the chain displacement effect and emitted Cy3 fluorescence. The amplification of the signal in the CHA between H1 and H2 on FDTp was observed in the presence of miR-155, resulting in increased sensitivity of FAM fluorescence to miR-155. The miRNAs miR-21 and miR-155 were effectively visualized within viable cells, with a LoD of 5 pM. Meanwhile, Electrochemical and fluorescent sensors can detect exo-miRs in TDN-based sensors. Electrochemical methods have excellent sensitivity. When comparing electrochemical methods to fluorescence, it is important to note that fluorescence possesses the unique ability to identify exo-miRs precisely in situ, making it a straightforward and practical approach for application. The optical approaches of TDNs usually requires the utilization of fluorescent molecules, demonstrating sensitivity typically within the picomolar range. The detection sensitivity is lower compared to electrochemical methods. One of the primary benefits is that the fluorescence detection method enables the in vivo detection of exosomal microRNAs through a simple procedure.
4.2 Branched DNA nanomaterials
Branched DNA with multi-arm junctions was first created by Seeman, including 3-, 4-, 5-, 6-, 8-, and 12-arm junctions.165–167 These nanomaterials have shown promise in serving as a flexible element in producing multifunctional nanomaterial biosensors by strategically adjusting their locations.26,28,168 Luo's group utilized branched DNA labeled with fluorescent dyes (green and red) to create a system capable of detecting multiple targets.169,170 Um et al. designed a fluorescence-coded DNA nanostructure probe system to detect intracellular miR-21 and miR-22 in cells (Fig. 5A).171 The system is capable of detecting multiple miRNAs across various breast cancer cell lines. Flow cytometry results illustrated the feasibility of directly comparing miRNA expression levels among different breast cancer cell lines with distinct characteristics. Additionally, Yang et al. detailed various branched DNA nanostructures used as scaffolds for creating super-branched silver nanoclusters (super-AgNCs), which exhibited adjustable spatial structures and fluorescence characteristics for biosensing applications.172,173 Branched DNA AgNCs displayed remarkable biocompatibility with living organisms and minimal harmful effects, which allows for their versatile use in a wide range of biomedical purposes. Meanwhile, Yang et al. designed thermostable DNA nanostructures to construct a nucleic acid detection platform,174–176 which was used to ultra-sensitively detect miR-223 on the surface-enhanced Raman spectroscopy (SERS) platform (Fig. 5B).176 This work successfully detected multiple liver cancer biomarkers involving miR-223 and protein. The intricately structured branched DNA utilized various complementary single-strand DNA as captures for detecting miR-223, along with a double-stranded steady branched centre to enhance the distance of captures and expose a greater number of DNA captures to minimize lateral interactions for improved sensitivity.177 The incorporation of branched DNA results in 2 orders of magnitude increase in sensitivity for miRNA detection compared to using single-stranded DNA. The LoDs for miR-223 and AFP were 10 aM and 10−12 M (S/N = 3), respectively. Lin et al. created a localized branched catalytic hairpin assembly (bCHA) strategy for miRNAs in living cells by a spatial-confinement approach of branched DNA structure without enzymes (Fig. 5C).178 This platform utilized three separate nanoprobes, each containing a distinct bCHA hairpin structure. The specific miRNA triggers bCHA among the LRN probes, resulting in fluorescent signal amplification. Improvements in detection sensitivity and speed were achieved by anchoring each bCHA hairpin to individual nanospheres while maintaining accuracy through the reduction of non-specific interactions. The LoD of miR-21 in vitro can reach as low as 3.66 pM in live cells and clinical tissues.179 One target-induced connection of probes orderly to each other enhances the contrast in a cell by avoiding signal dispersion in amplification. This strategy was first programmed to implement miR-21 detection of MCF-7 cells by target-triggered hairpin-free chain-branching assembly (HFCBA). Following the introduction of probes, the signal in cells exhibited a gradual increase in intensity in tandem with the growth of DNA dendrimers. This led to a significant enhancement in contrast within HFCBA through the accumulation of in situ fluorescence while also preventing signal dispersion during amplification. In 2020, Wang et al. developed an all-in-one biosensor to detect multiplexed exo-miRs in serum in situ for breast cancer diagnosis.155 They utilized a branched DNA to construct a simple all-in-one biosensor for simultaneous determination of several miRNAs in situ by strand displacement reaction. Branched DNA included three oligonucleotides, which were extended at their 5′ ends by incorporating three single-stranded recognition sequences with quenchers, involving BHQ1, BHQ2, and BHQ2, respectively. Following this, three reporter sequences were attached to the corresponding recognition sequences using different fluorophores (FAM, Cy3, and Cy5) to create a multicolor DNA biosensor capable of emitting self-quenched fluorescence. The biosensor is capable of entering exosomes, hybridizing with complementary miRNA targets to create longer duplexes, and releasing reporter sequences to activate significant fluorescence signals for the simultaneous assay of several miRNAs in exosomes. MiR-21, miR-27a, and miR-375 were selected as representative targets due to their elevated expression levels in breast cancer cells (MCF-7). The fluorescence characteristics of the detecting platform in the presence of MCF-7 exosomes illustrated the positive correlations to their level, and the LoDs were found to be 0.116 μg mL−1, 0.125 μg mL−1, and 0.287 μg mL−1 for exosomes through detection of miR-21, miR-27a and miR-375, respectively. Conversely, no apparent correlations were found between fluorescence intensities and controlled concentrations of MCF-10A exosomes. Chen developed a ratiometric electrochemical DNA biosensor for the detection of exo-miR-21 (Fig. 5D).156,179 They created a locked nucleic acid (LNA)-modified branched DNA nanostructure to construct a ratiometric electrochemical DNA platform for exo-miR-21 detection. miR-21 triggered the LNA-assisted strand displacement reaction on the branched DNA, resulting in a structure alteration and signal ratio change. This indicates the varying distances between the electrode surface and the two electroactive molecules attached to the branched DNA. The biosensor demonstrated high accuracy and sensitivity using a dual-signal ratiometric method, achieving a limit of detection as low as 2.3 fM. Furthermore, due to the linear correlation between the logarithm of the signal ratio and the logarithm of the miR-21 concentration, the biosensor demonstrated sufficient stability in detecting miR-21 in MCF-7 cell-derived exosomes with effective selectivity. Branched DNA presents multiple modification sites that uphold a specific spatial arrangement. Various signaling molecules can be linked with different sites to enable multiplexing of exo-miRs. Currently, the integration of branched DNA with SERS, electrochemical, and fluorescence sensors has led to the development of various detection platforms for exo-miRs in situ, achieving a high sensitivity at the attomole (aM) level. | ||
| Fig. 5 (A) Branched DNA nanomaterials with fluorescent dyes for intracellular miR-22 and miR-21 detection.171 (B) Branched DNA-based AgNF for detecting AFP and miRNA on SERS.176 (C) Synthesis of branched DNA nanomaterials for miR-21 detection in living cells.178 (D) Schematic diagram of target triggered synthesis of DNA dendrimers for contrast-increased imaging of miRNA in living cells.179 | ||
4.3 Cubic DNA nanocage sensors
The cubic DNA nanocage exhibits improved structural stability against enzymatic degradation, remarkable cellular permeability, high biocompatibility, and size-specific discrimination capability through the spatial confinement phenomenon.28 Based on cubic DNA nanocages, the sensing platform primarily uses a fluorescence beacon to create an optical detection assay directly on the samples. For example, Ye et al. created a cubic DNA nanocage probe designed to infiltrate EVs originating from MDA-MB-231 cells to monitor miRNA-10b in situ using a combination of fluorescence and quenching.180 Within the exosome, miRNA-10b selectively binds with the quencher, restoring fluorescence for the cubic DNA nanocage. A strong linear relationship was found between fluorescent value and the concentration of extracellular vesicles (EVs) within the range of 1.11 × 108 to 5.70 × 1010 particles per mL in 120 min. Meanwhile, Mi et al. developed a cubic DNA nanocage three-dimensional molecular beacon to accurately detect multiplexed exo-miRs in confined spaces (Fig. 6A).140 They devised a precise and adaptable cubic DNA nanostructure-based three-dimensional molecular beacon (ncMB), effectively overcoming the aforementioned challenge and achieving the detection of exo-miRNA. In this detection platform, steric hindrance and electrostatic repulsion caused by the distinctive three-dimensional arrangement of the ncMB created a barrier between fluorescent probes, preventing unintended fluorescence quenching in single exo-miR detection and unexpected FRET in dual exo-miR detection. In comparison to conventional molecular beacons used for detecting exosomal miRNA in both single and dual modes, the larger DNA nanocage of the ncMB was found to shield the fluorophore from interactions with the quencher and acceptor fluorophore by employing steric hindrance and electrostatic repulsion. Additionally, Li et al. utilized a spatial confinement strategy to design an accelerated DNA nanoprobe for fast, in situ monitoring of exo-miRs (Fig. 6B).181 This nanoprobe is capable of detecting exo-miRs and differentiating tumor-derived exosomes from those originating from normal cells to a high degree of accuracy, thereby establishing a foundation for bioimaging exo-miRs and diagnosing diseases. Sun et al. developed DNA cage-based exo-miR detection for diagnostics of various stages in cancer progression using the characteristics of the size-selective ability and polyethylene glycol (PEG)-enhanced thermophoretic accumulation.182 This method was a highly sensitive, selective, and in situ detection of exo-miRs with a LoD of 2.05 fM in serum samples. Exo-miR-21 or miR155 demonstrated a discrimination accuracy of 90% in clinical tests when distinguishing between BC patients and healthy people, surpassing the performance of conventional molecular probes capable of detecting miRs. The cubic DNA nanocage combined with a fluorescence beacon can effectively transfer into the exosome and detect the miRs on site. The main focus of the detection process of the cubic DNA nanostructure primarily centers on the use of the optical detection system with a cage structure. This structure serves to efficiently bind fluorescent molecules, safeguard them within the confines of the cage, and effectively shield them from interference by the fluorescence of neighboring molecules within the surrounding exosome. This analysis strategy increases the accuracy and precision of the detection system, allowing for the in situ detection of exo-miRs with fM levels of LoD.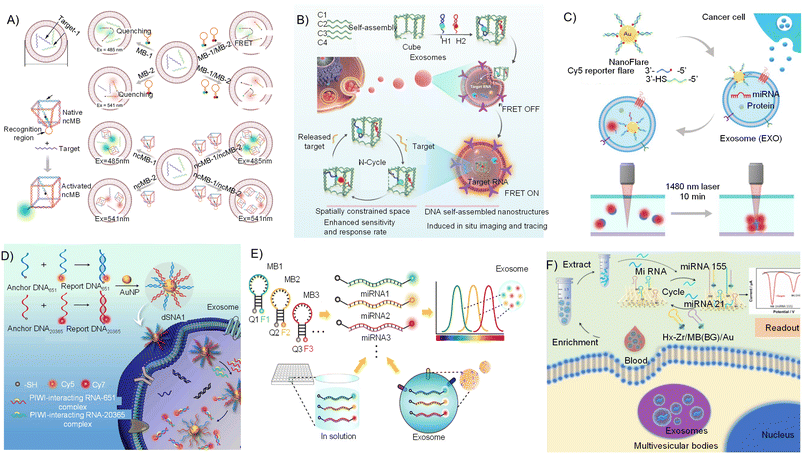 | ||
| Fig. 6 (A) Molecular beacon-based cubic DNA nanocage for exosomal miRNA detection in confined spaces.140 (B) Spatial confinement-based cubic DNA nanocage exosomal miRNA detection in situ.181 (C) Functional components and equivalent SSDC for the cyclic amplification detection of miRNAs.183 (D) Thermophoretic sensor based on NanoFlare (TSN) for in situ determination of exo-miRs.184 (E) In situ simultaneous determination of several miRNAs in exosomes using MBs for diagnosis. F: fluorescent dye. Q: quencher.185 (F) Simultaneous exosomal miR-21 and miR-155 detection by nucleic acid-functionalized disposable paper-based sensors.186 | ||
4.4 Spherical DNA nanomaterials
Spherical nucleic acids (SNAs) consist of a core nanoparticle surrounded by a densely packed layer of oligonucleotides. In 1996, Mirkin et al. first developed SNA, which has since demonstrated notable effects in diverse disciplines.187,188 First, regarding the sandwich structure for miRNA detection, Liu et al. developed a “sandwich” structure of SNA and peptide nucleic acid (PNA) to construct an electrochemical exo-miR detection platform.189 PNA with a neutrally charged peptide skeleton serves as a capture probe, leading to a reduced charge and increased the binding efficiency with SNA-miRNA on the electrode surface. RuHex, being an electroactive compound, has the capability to interact with PNA-miRNA-SNA “sandwich” structures that have been assembled on the electrode surface via electrostatic adsorption. Chronocoulometry was utilized to measure the charges of RuHex, indicating the level of exo-miR. The LoD can reach as low as 49 aM with a detection range from 100 aM to 1 nM. The method demonstrates good sensitivity, but its complexity can be attributed to the initial lysis step. For achieving the exo-miR detection on site, in 2023, Yang et al. created a precise and reliable one-step method named dual-surface-protein-guided orthogonal recognition of tumor by profiling of miRs, which effectively increased the detection accuracy of prostate cancer.190 This platform employed two allosteric aptamers against exosomal marker CD63 and tumor marker EpCAM to build barcode for tumor-specific exosome subtypes. Meanwhile, the tumor-derived exosomes with barcode triggered targeted membrane fusion with liposome probes to introduce miRNA detection reagents, allowing for in situ sensitive detection of tumor-derived exo-miRs. With a signature of six miRs, this sensor differentiated PCa and benign prostatic hyperplasia with an accuracy of 100%. Additionally, the diagnostic accuracy was found to be 90.6% in classifying metastatic and nonmetastatic PCa. Wu et al. developed spherical nucleic acid-mediated spatial matching-guided nonenzymatic DNA circuits for miRNA detection to predict and prevent malignant tumor (Fig. 6C).183 In this detection system, matching-guided nonenzymatic DNA circuits (SSDCs) were created using AuNP-based spherical nucleic acid to mediate spatial matching for effective detection of miRNAs. Because of the significantly improved ability to resist nuclease degradation, the occurrence of false positive signals is prevented even when present in a complex biological environment. The miRNA target has the capability to align the hairpin DNA probe into a linear structure, enabling the probe to enter the inner section of a core/shell DNA-functionalized signal nanoamplifier and trigger a strand displacement reaction that leads to the production of an enhanced fluorescence signal. The LoD was measured to be as low as 17 pM, and the imaging of microRNA was found to be well aligned with the widely accepted gold standard polymerase chain reaction (PCR) technique. The capability to image intracellular miRNAs is greatly improved compared to traditional fluorescence in situ hybridization methods, allowing in vivo SSDC-based imaging to be proficient in accurately predicting the development of tumors. The use of intratumoral chemotherapy, directed by SSDC-based imaging, effectively managed the growth and advancement of tumors prior to the presentation of noticeable clinical symptoms. Sun et al. created a thermophoretic sensor known as NanoFlare (TSN) to detect exo-miRNAs in situ without the need for RNA extraction or target amplification (Fig. 6D).184 The accumulation of NanoFlare treated exosomes through thermophoresis results in an increased fluorescence signal when exo-miRs bind to NanoFlare, enabling the direct and quantitative measurement of exo-miRs at concentrations as low as 0.36 fM in 0.5 μL serum samples within a 15 min time frame. One of the most reliable indicators, exo-miR-375, demonstrated an accuracy rate of 85% in identifying estrogen receptor-positive breast cancer in its early stages (stages I and II). This study offers a practical tool for enhancing cancer diagnosis. Duan et al. synthesized an Au-cored spherical nucleic acid biosensor to detect multiplex exosomal RNAs in situ.191 The probes' structures have been specifically tailored for the large targets of the PIWI-interacting RNA complex to prevent any steric hindrance in the analytical process, ensuring reliable sensitivity. Moreover, the strategic adjustment of the input during the synthesis of the double-strand DNA containing functional DNA and capture DNA with substantial concentration variations promotes the consistency of the product probes in both structure and detection sensitivity. The probes demonstrate the ability to detect piRNAs in exosomes in real human plasma samples with high sensitivity and specificity, allowing for differentiation between breast cancer patients and normal controls. This indicates that they may have the potential to serve as a straightforward and precise liquid biopsy tool for cancer diagnostics. Although the methods based on SNAs have high sensitivity, they exhibit significant noise in detecting exo-miRs. Spherical DNA interacts with various metal nanoparticles to synthesize sizable ball structures with the nucleic acid surface, allowing for the efficient capture of target miRNAs for high sensitivity; however, this technique remains vulnerable to the impact of external interferences, resulting in the production of non-specific signals. This method allows for the detection of exosomal microRNA in its natural environment with attomolar sensitivity.4.5 Hairpin DNA-based sensors
Hairpin DNA sensors (HDS) were designed with a single-stranded DNA sequence that features a stem structure created by base pairs and a loop containing unpaired nucleotides.192 The stem–loop of HDS demonstrated notable recognizable properties of hybridization specificity and distinct selectivity towards target miRs. HDS is extensively utilized as a biosensor for exo-miRs, predominantly employing HDS-based molecular beacons (MBs) and hybridization chain reaction (HCR).193,194MBs are hairpin DNA structures with fluorophores and a quencher specifically engineered to monitor exo-miRs.195 Also, it is noteworthy that the choice of fluorophores for multiplexing biomarkers in an exosome is crucial because of its small size. Rhee et al. documented the in vivo single-step identification of particular miR-21 within intact exosomes using a nano-scale molecular beacon probe.130 Permeabilization by streptolysin treatment further enhanced the delivery of MBs into exosomes, giving significantly increased signals from miR-21. Rhee et al., for the first time, further developed simultaneous and multiplexed detection exo-miRs (from breast cancer cell line MCF-7) using MBs (Fig. 6E).185 MBs successfully hybridized with multiple miR-21, miR-375, and miR-27a in the cancer cell-derived exosomes even in the presence of high human serum concentration. The proposed method is beneficial to high-throughput analysis for disease diagnosis, prognosis, and response to treatment because it is a time-, labor-, and cost-saving technique. Zhang et al. developed accurate and rapid detection of exo-miR-21 by a DNA zipper-mediated membrane fusion approach for cancer diagnosis.196 In this detection platform, a lipid vesicle biosensor with MBs was synthesized and subsequently functionalized with zipper DNA nanostructures (ZDCs) on its surface. Additionally, complementary zipper DNA nanostructures (cZDCs) are incorporated on the exosome. Upon co-mixing ZDCs and cZDCs, the process activates membrane fusion between exosomes and vesicle probes, leading to the identification of exo-miR-21 by encapsulated MBs and subsequent fluorescence emission that can be detected within a 30 min time frame. Significantly, flow cytometry allows for the differentiation of miR-21-overexpressed tumor exosomes derived from cell culture medium or clinical patient serums from exosomes secreted by normal cells.
Additionally, hybridization chain reaction (HCR) proved to be yet another effective technique for isothermal amplification that operates based on the opening of functional hairpin DNA structures.197 Once the specific target miRNA was identified, it resulted in the formation of lengthy double-helix structures composed of multiple repeated units through the process of hybridization between two DNA hairpins.198 Zou et al. utilized an HCR-based platform to construct a sensitive imaging method for intracellular miRNA in living cells.199 For the purpose of delivery and recruitment, exosomes were altered by incorporating three different cholesterol-modified hairpins (designated as H1, H2, and H3), with H1 specifically intended for anchoring the target miRNA, while H2 and H3 were equipped with photocleavable linkers (PC linkers) to facilitate spatiotemporal HCR. By strategically releasing probes with concentrated quantities in specific locations to effectively initiate HCR and subsequently gathering the resultant double-stranded DNA polymers rather than allowing them to disperse throughout the cytoplasm, the LCR-HCR technique has the potential to greatly enhance imaging contrast by confining all of the chemical substances involved on exosome carriers. The comparison of low- and high-contrast reverse immunosensor systems measured the microRNA-21 at a limit of detection as low as 3.3 pM (equivalent to 165 attomoles per 50 microliters) under laboratory conditions and exhibited a response in vivo that was four times greater than that achieved by conventional high-contrast reverse immunosensors. The LCR-HCR method offers a highly effective tool for the visualization of microRNAs in living cells and for the detection of cancer. Li and colleagues created a method that combines HCR and DNAzyme technology to amplify signals without the need for enzymes. This method allows for the detection of specific exo-miRs in cancer cell culture media and serum samples from cancer patients through the self-assembly of polymer DNAzyme nanostructures triggered by the target.200 He et al. synthesized hairpins that loaded DNAzyme subunits in the 3′ and 5′ ends. When exo-miRs existed, long DNA double helixes were produced, leading to various activated DNAzymes by two nearby units. After that, the DNAzymes prompted the cleavage of fluorescent reporter probes, resulting in the emission of fluorescence signals. Based on this mechanism, quantifying miRNA assays were conducted in circulating exosomes. Choi et al. developed hydrogel-based HCR for multiplex signal amplification to detect urinary exo-miRs (hsa-miR-6090 and hsa-miR-3665) from human clinical samples.201 They identified small amounts (∼amol) of exo-miRs from 600 μL of urine with up to ∼35-fold amplification. Furthermore, they created a ratiometric detection platform using a reference miRNA, which indicated that this analysis platform has a great potential for differentiating prostate cancer patients from healthy controls. HDS-based molecular beacons (MBs) and the HCR-based detection platform primarily rely on the target molecule initiating the opening of a hairpin, leading to fluorescence signal enhancement or amplification reactions. The assay necessitates carefully designed DNA sequences that specifically target regions within hairpins to minimize non-specific binding. Currently, this method can detect at the femtomolar level of sensitivity.
The analytical performance of the various strategies for exo-miRNA detection is summarized in Table 1. Even though there is a recent emergence of exo-miRNAs as potential biomarkers, their clinical applications continue to be constrained by their low concentration in small quantities of clinical samples. Consequently, the endeavor to create a diagnostic tool that is non-invasive and specific, and to profile markers of exosomal microRNAs from urine, presents a considerable and pressing challenge. Even though the identification of terminal stem–loop/hairpin DNA-based exo-miRs brings various benefits due to its straightforward structure and exceptional sensitivity, it is also confronted with obstacles linked to elevated background signals and potential external disruptions.
5 Importance and challenges
Exo-miRs are emerging as powerful early biomarkers, forging new directions in molecular diagnostics, prognosis, and personalized disease management, which represents a notable advancement, yielding quicker diagnostic results and greater accessibility for patients. In situ detection facilitates the simultaneous detection of exo-miRs, which is crucial for examining complex miRNA-involved regulatory networks. DNA nanomaterials have excellent cell permeability, biocompatibility, low toxicity, and resistance to enzyme degradation for their exceptional sensitivity, enabling the detection of low levels of exo-miR in confined spaces. Despite the many advancements in technologies for exo-miR detection, we must stress a few critical points about exo-miR diagnostics, establish ultrasensitive biosensing on site, reliable biomarkers, and multiplexed and sensitive assays.The concentration of exo-miRs in exosomes is low, and these molecules' exclusive expression or downregulation content was not significant in specific human diseases. Therefore, it is essential that the detection method be both accurate and highly sensitive to target miRs effectively. The integration of DNA nanomaterials with both electrochemical and optical detection systems has now facilitated the detection of exo-miRs at unprecedentedly low attomolar levels, thereby representing a notable breakthrough in the field of clinical disease diagnostics. However, numerous factors influence miRNA expression levels in real complex samples, including age, gender, lifestyle, and an individual's multiple factors collaboratively influence the expression level of exo-miRs. The expression levels of target miRNAs were reportedly upregulated or downregulated in patients with various diseases. Considering these factors, certain miRNA and concentrations as standards for disease detection need to be first found. Further research is needed to explore the regulation of miRNA expression levels and its correlation with disease in relation to interfering factors and identify the precise target exosomal microRNA molecule for reliable disease diagnostics. As mentioned above, in specific diseases, multiplexed exo-miRs should be used as biomarkers simultaneously for accurate disease diagnostics. Therefore, it is important to create DNA nanomaterials that feature several target molecular junction sites, with each site corresponding to a unique signal to enable recognition of multiple targets. The above-mentioned challenges of DNA nanomaterial-based detection platforms still need to be further studied for exo-miRs as biomarkers of disease diagnostics.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
Zhikun Zhang: conceptualization (equal); writing – original draft (equal); writing – review and editing (equal). Md. Ahasan Ahamed: writing – original draft (equal); writing – review and editing (equal). Dayong Yang: conceptualization (equal); funding acquisition (lead); supervision (lead); writing – original draft (equal); writing – review and editing (equal).Conflicts of interest
All authors declare no conflict of interests.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No.: 22225505).References
- R. Shang, S. Lee, G. Senavirathne and E. C. Lai, Nat. Rev. Genet., 2023, 24, 816–833 Search PubMed.
- F. Wahid, A. Shehzad, T. Khan and Y. Y. Kim, Biochim. Biophys. Acta, Mol. Cell Res., 2010, 1803, 1231–1243 CrossRef CAS PubMed.
- Z. Sun, Y. Wu, F. Gao, H. Li, C. Wang, L. Du, L. Dong and Y. Jiang, Acta Biomater., 2023, 155, 80–98 Search PubMed.
- S. Shin, Y. H. Park, S.-H. Jung, S.-H. Jang, M. Y. Kim, J. Y. Lee and Y. Chung, npj Genomic Med., 2021, 6, 45 Search PubMed.
- M. Jang, G. Choi, Y. Y. Choi, J. E. Lee, J.-H. Jung, S. W. Oh, D. H. Han, H. Lee, J.-H. Park, J.-H. Cheong and P. Kim, NPG Asia Mater., 2019, 11, 79 Search PubMed.
- L. Saadatpour, E. Fadaee, S. Fadaei, R. Nassiri Mansour, M. Mohammadi, S. M. Mousavi, M. Goodarzi, J. Verdi and H. Mirzaei, Cancer Gene Ther., 2016, 23, 415–418 Search PubMed.
- N. Jafari, P. Llevenes and G. V. Denis, Nat. Rev. Endocrinol., 2022, 18, 327–328 CrossRef PubMed.
- Z. Zhang, Y. Zou, C. Song, K. Cao, K. Cai, S. Chen, Y. Wu, D. Geng, G. Sun, N. Zhang, X. Zhang, Y. Zhang, Y. Sun and Y. Zhang, J. Adv. Res., 2023, S2090123223004022 Search PubMed.
- Y. Yu, K. Hou, T. Ji, X. Wang, Y. Liu, Y. Zheng, J. Xu, Y. Hou and G. Chi, Mol. Cell. Biochem., 2021, 476, 2111–2124 CrossRef CAS PubMed.
- N. Mahtal, O. Lenoir, C. Tinel, D. Anglicheau and P.-L. Tharaux, Nat. Rev. Nephrol., 2022, 18, 643–662 CrossRef CAS PubMed.
- B. Mohan, S. Kumar, S. Kumar, K. Modi, D. Tyagi, D. Papukashvili, N. Rcheulishvili and A. J. L. Pombeiro, Sens. Diagn., 2023, 2, 78–89 RSC.
- J. Tang, X. Jia, Q. Li, Z. Cui, A. Liang, B. Ke, D. Yang and C. Yao, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2303822120 CrossRef CAS PubMed.
- N. P. Hessvik and A. Llorente, Cell. Mol. Life Sci., 2018, 75, 193–208 CrossRef CAS PubMed.
- M. A. U. Khalid, Md. A. Ahamed, M. Dong, A. Kshirsagar and W. Guan, Biosens. Bioelectron., 2025, 268, 116895 CrossRef CAS PubMed.
- Y. Xue, K. Wang, Y. Jiang, Y. Dai, X. Liu, B. Pei, H. Li, H. Xu and G. Zhao, Biosens. Bioelectron., 2024, 247, 115927 Search PubMed.
- H. Zhang, L. Cao, J. Brodsky, I. Gablech, F. Xu, Z. Li, M. Korabecna and P. Neuzil, TrAC, Trends Anal. Chem., 2024, 174, 117676 Search PubMed.
- H. Wang, Cells, 2024, 13, 1277 CrossRef CAS PubMed.
- M. A. Ahamed, A. J. Politza, T. Liu, M. A. U. Khalid, H. Zhang and W. Guan, Nanotechnology, 2025, 36, 042001 Search PubMed.
- S. Tam, R. De Borja, M.-S. Tsao and J. D. McPherson, Lab. Invest., 2014, 94, 350–358 CrossRef CAS PubMed.
- H.-S. Tan, T. Wang, H.-N. Sun, A. Liu and S.-S. Li, TrAC, Trends Anal. Chem., 2023, 167, 117253 CrossRef CAS.
- W. Wu, X. Yu, J. Wu, T. Wu, Y. Fan, W. Chen, M. Zhao, H. Wu, X. Li and S. Ding, Biosens. Bioelectron., 2021, 175, 112835 CrossRef CAS PubMed.
- J. Peng, B. Li, Z. Ma, Z. Qiu, H. Hu, Y. Jiang and D. Gao, Talanta, 2025, 281, 126838 Search PubMed.
- Y. Xia, Z. Huang, T. Chen, L. Xu, G. Zhu, W. Chen, G. Chen, S. Wu, J. Lan, X. Lin and J. Chen, Biosens. Bioelectron., 2022, 209, 114259 Search PubMed.
- T. Liu, A. J. Politza, Md. A. Ahamed, A. Kshirsagar, Y. Zhu and W. Guan, Biosens. Bioelectron., 2024, 116997 Search PubMed.
- X. Li, X. Li, D. Li, M. Zhao, H. Wu, B. Shen, P. Liu and S. Ding, Biosens. Bioelectron., 2020, 168, 112554 Search PubMed.
- C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef PubMed.
- J. Chen and N. C. Seeman, Nature, 1991, 350, 631–633 Search PubMed.
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2017, 3, 17068 Search PubMed.
- J. O'Brien, H. Hayder, Y. Zayed and C. Peng, Front. Endocrinol., 2018, 9, 402 Search PubMed.
- B. Hrdlickova, R. C. De Almeida, Z. Borek and S. Withoff, Biochim. Biophys. Acta, Mol. Basis Dis., 2014, 1842, 1910–1922 CrossRef CAS PubMed.
- J. Krol, I. Loedige and W. Filipowicz, Nat. Rev. Genet., 2010, 11, 597–610 CrossRef CAS PubMed.
- M. A. Kumar, S. K. Baba, H. Q. Sadida, S. Al. Marzooqi, J. Jerobin, F. H. Altemani, N. Algehainy, M. A. Alanazi, A.-B. Abou-Samra, R. Kumar, A. S. Al-Shabeeb Akil, M. A. Macha, R. Mir and A. A. Bhat, Signal Transduction Targeted Ther., 2024, 9, 27 Search PubMed.
- A. Gallo, M. Tandon, I. Alevizos and G. G. Illei, PLoS One, 2012, 7, e30679 Search PubMed.
- A. Turchinovich, L. Weiz, A. Langheinz and B. Burwinkel, Nucleic Acids Res., 2011, 39, 7223–7233 CrossRef CAS PubMed.
- J. Zhang, S. Li, L. Li, M. Li, C. Guo, J. Yao and S. Mi, Genomics, Proteomics Bioinf., 2015, 13, 17–24 Search PubMed.
- S. Mathivanan, H. Ji and R. J. Simpson, J. Proteomics, 2010, 73, 1907–1920 Search PubMed.
- D. Li, L. Xia, Q. Zhou, L. Wang, D. Chen, X. Gao and Y. Li, Anal. Chem., 2020, 92, 12769–12773 Search PubMed.
- Y. Huang, T. Sun, L. Liu, N. Xia, Y. Zhao and X. Yi, Talanta, 2021, 234, 122622 Search PubMed.
- S. Modani, D. Tomar, S. Tangirala, A. Sriram, N. K. Mehra, R. Kumar, D. K. Khatri and P. K. Singh, J. Drug Targeting, 2021, 29, 925–940 CrossRef CAS PubMed.
- Q. Guo, H. Wang, Y. Yan, Y. Liu, C. Su, H. Chen, Y. Yan, R. Adhikari, Q. Wu and J. Zhang, Front. Oncol., 2020, 10, 472 Search PubMed.
- B. Yue, H. Yang, J. Wang, W. Ru, J. Wu, Y. Huang, X. Lan, C. Lei and H. Chen, Cell Proliferation, 2020, 53, e12857 Search PubMed.
- Y. Chen, Y. Zhao, Y. Yin, X. Jia and L. Mao, Bioengineered, 2021, 12, 8186–8201 CrossRef CAS PubMed.
- F. Fabbiano, J. Corsi, E. Gurrieri, C. Trevisan, M. Notarangelo and V. G. D'Agostino, J. Extracell. Vesicles, 2020, 10, e12043 CrossRef CAS PubMed.
- X. Tang, M. Leng, W. Tang, Z. Cai, L. Yang, L. Wang, Y. Zhang and J. Guo, Molecules, 2024, 29, 1199 CrossRef CAS PubMed.
- B. Hannafon and W.-Q. Ding, Int. J. Mol. Sci., 2013, 14, 14240–14269 Search PubMed.
- Y. Wang, T. Xiao, C. Zhao and G. Li, Int. J. Mol. Sci., 2023, 25, 255 Search PubMed.
- B. Février and G. Raposo, Curr. Opin. Cell Biol., 2004, 16, 415–421 CrossRef PubMed.
- P. Perrin, L. Janssen, H. Janssen, B. Van Den Broek, L. M. Voortman, D. Van Elsland, I. Berlin and J. Neefjes, Curr. Biol., 2021, 31, 3884–3893.e4 Search PubMed.
- N. W. Lin, C. Liu, I. V. Yang, L. A. Maier, D. L. DeMeo, C. Wood, S. Ye, M. H. Cruse, V. L. Smith, C. A. Vyhlidal, K. Kechris and S. Sharma, Front. Genet., 2022, 13, 762834 CrossRef CAS PubMed.
- P. Kanlikilicer, R. Bayraktar, M. Denizli, M. H. Rashed, C. Ivan, B. Aslan, R. Mitra, K. Karagoz, E. Bayraktar, X. Zhang, C. Rodriguez-Aguayo, A. A. El-Arabey, N. Kahraman, S. Baydogan, O. Ozkayar, M. L. Gatza, B. Ozpolat, G. A. Calin, A. K. Sood and G. Lopez-Berestein, EBioMedicine, 2018, 38, 100–112 CrossRef CAS PubMed.
- G. K. Patel, M. A. Khan, A. Bhardwaj, S. K. Srivastava, H. Zubair, M. C. Patton, S. Singh, M. Khushman and A. P. Singh, Br. J. Cancer, 2017, 116, 609–619 CrossRef CAS PubMed.
- C. Li, T. Zhou, J. Chen, R. Li, H. Chen, S. Luo, D. Chen, C. Cai and W. Li, J. Transl. Med., 2022, 20, 6 CrossRef CAS PubMed.
- H. T. Nguyen, S. E. O. Kacimi, T. L. Nguyen, K. H. Suman, R. Lemus-Martin, H. Saleem and D. N. Do, Biology, 2021, 10, 417 CrossRef CAS PubMed.
- S. U. Khan, M. U. Khan, M. Azhar Ud Din, I. M. Khan, M. I. Khan, S. Bungau and S. S. U. Hassan, Front. Immunol., 2023, 14, 1166487 CrossRef CAS PubMed.
- E. Paccosi and L. Proietti-De-Santis, Int. J. Mol. Sci., 2023, 24, 9547 CrossRef CAS PubMed.
- C. Iaconetti, S. Sorrentino, S. De Rosa and C. Indolfi, Physiology, 2016, 31, 16–24 CrossRef CAS PubMed.
- H. Li, J. Zhan, C. Chen and D. Wang, Med. Rev., 2022, 2, 140–168 CrossRef PubMed.
- R. Xue, W. Tan, Y. Wu, B. Dong, Z. Xie, P. Huang, J. He, Y. Dong and C. Liu, Front. Cardiovasc. Med., 2020, 7, 592412 CrossRef CAS PubMed.
- S. Gurunathan, M. H. Kang and J.-H. Kim, Front. Immunol., 2021, 12, 716407 CrossRef CAS PubMed.
- Z. Zhang, T. Xing, Y. Chen and J. Xiao, Biomed. Pharmacother., 2018, 106, 1135–1143 Search PubMed.
- K. Liu, X. Gao, B. Kang, Y. Liu, D. Wang and Y. Wang, Front. Oncol., 2022, 12, 836548 CrossRef CAS PubMed.
- J. C. Santos, N. D. S. Lima, L. O. Sarian, A. Matheu, M. L. Ribeiro and S. F. M. Derchain, Sci. Rep., 2018, 8, 829 CrossRef PubMed.
- J. M. Cozar, I. Robles-Fernandez, A. Rodriguez-Martinez, I. Puche-Sanz, F. Vazquez-Alonso, J. A. Lorente, L. J. Martinez-Gonzalez and M. J. Alvarez-Cubero, Mutat. Res., Rev. Mutat. Res., 2019, 781, 165–174 CrossRef CAS PubMed.
- J. A. Deiuliis, Int. J. Obes., 2016, 40, 88–101 CrossRef CAS PubMed.
- J. Wang, B.-L. Yue, Y.-Z. Huang, X.-Y. Lan, W.-J. Liu and H. Chen, Int. J. Mol. Sci., 2022, 23, 2461 Search PubMed.
- Z. Zhang, T. Liu, M. Dong, Md. A. Ahamed and W. Guan, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2024, 16, e1969 Search PubMed.
- N. Al Rawi, N. Elmabrouk, R. Abu Kou, S. Mkadmi, Z. Rizvi and Z. Hamdoon, Arch. Oral Biol., 2021, 125, 105108 CrossRef CAS PubMed.
- M. Song, H. Bai, P. Zhang, X. Zhou and B. Ying, Int. J. Oral Sci., 2023, 15, 2 CrossRef CAS PubMed.
- Z. Xie, G. Chen, X. Zhang, D. Li, J. Huang, C. Yang, P. Zhang, Y. Qin, Y. Duan, B. Gong and Z. Li, PLoS One, 2013, 8, e57502 Search PubMed.
- J. Du and L. Zhang, Oncol. Lett., 2017, 14, 1387–1394 CrossRef PubMed.
- F. Li, J. M. Yoshizawa, K.-M. Kim, J. Kanjanapangka, T. R. Grogan, X. Wang, D. E. Elashoff, S. Ishikawa, D. Chia, W. Liao, D. Akin, X. Yan, M.-S. Lee, R. Choi, S.-M. Kim, S.-Y. Kang, J.-M. Bae, T.-S. Sohn, J.-H. Lee, M.-G. Choi, B.-H. Min, J.-H. Lee, J. J. Kim, Y. Kim, S. Kim and D. T. W. Wong, Clin. Chem., 2018, 64, 1513–1521 Search PubMed.
- J. Yang, R. Xu, C. Wang, J. Qiu, B. Ren and L. You, Cancer Commun., 2021, 41, 1257–1274 CrossRef PubMed.
- M. Koopaie, F. Abedinejad, S. Manifar, R. Mousavi, S. Kolahdooz and A. Shamshiri, Gene Rep., 2021, 25, 101317 Search PubMed.
- T. P. N. Nguyen, M. Kumar, E. Fedele, G. Bonanno and T. Bonifacino, Int. J. Mol. Sci., 2022, 23, 4718 CrossRef CAS PubMed.
- H. Li, L. Yu, M. Li, X. Chen, Q. Tian, Y. Jiang and N. Li, Mol. Genet. Genomic Med., 2020, 8, e1189 Search PubMed.
- I. Kawikova and P. W. Askenase, Brain Res., 2015, 1617, 63–71 CrossRef CAS PubMed.
- X. Xia, Y. Wang, Y. Huang, H. Zhang, H. Lu and J. C. Zheng, Prog. Neurobiol., 2019, 183, 101694 Search PubMed.
- Y. W. Chen, H. V. Lee and S. B. Abd Hamid, Carbohydr. Polym., 2017, 178, 57–68 Search PubMed.
- R. Shi, P.-Y. Wang, X.-Y. Li, J.-X. Chen, Y. Li, X.-Z. Zhang, C.-G. Zhang, T. Jiang, W.-B. Li, W. Ding and S.-J. Cheng, Oncotarget, 2015, 6, 26971–26981 CrossRef PubMed.
- E. V. Filatova, A. Kh. Alieva, M. I. Shadrina and P. A. Slominsky, Biochemistry, 2012, 77, 813–819 CAS.
- D. Zheng and J. Chen, Neuroscience, 2025, 568, 298–313 Search PubMed.
- L. Leggio, S. Vivarelli, F. L'Episcopo, C. Tirolo, S. Caniglia, N. Testa, B. Marchetti and N. Iraci, Int. J. Mol. Sci., 2017, 18, 2698 Search PubMed.
- Q. Shi, W. Kang, Z. Liu and X. Zhu, Heliyon, 2023, 9, e20595 CrossRef CAS PubMed.
- I. Manna, S. De Benedittis, A. Quattrone, D. Maisano, E. Iaccino and A. Quattrone, Pharmaceuticals, 2020, 13, 243 CrossRef CAS PubMed.
- X. Wu, T. Zheng and B. Zhang, Neurosci. Bull., 2017, 33, 331–338 Search PubMed.
- L. Yuan and J.-Y. Li, ACS Chem. Neurosci., 2019, 10, 964–972 Search PubMed.
- I. Manna, A. Quattrone, S. De Benedittis, B. Vescio, E. Iaccino and A. Quattrone, Parkinsonism Relat. Disord., 2021, 93, 77–84 CrossRef CAS PubMed.
- D. Chandran, S. Krishnan, M. Urulangodi and S. Gopala, Neurol. Sci., 2024, 45, 3625–3639 Search PubMed.
- J. R. Pinnell, M. Cui and K. Tieu, J. Neurochem., 2021, 157, 413–428 Search PubMed.
- H. Zhang, X. Liu, J. Shi, X. Su, J. Xie, Q. Meng and H. Dong, Front. Cell. Infect. Microbiol., 2024, 14, 1418168 CrossRef PubMed.
- R. E. Drury, D. O'Connor and A. J. Pollard, Front. Immunol., 2017, 8, 1182 Search PubMed.
- S. Zhou and J. Xu, Exp. Ther. Med., 2019, 18, 253–259 CAS.
- S. D. Hicks, D. Zhu, R. Sullivan, N. Kannikeswaran, K. Meert, W. Chen, S. Suresh and U. Sethuraman, Int. J. Mol. Sci., 2023, 24, 8175 CrossRef CAS PubMed.
- I. Saulle, M. Garziano, G. Cappelletti, F. Limanaqi, S. Strizzi, C. Vanetti, S. Lo Caputo, M. Poliseno, T. A. Santantonio, M. Clerici and M. Biasin, Int. J. Mol. Sci., 2023, 24, 10992 Search PubMed.
- R. Mishra and A. C. Banerjea, Front. Immunol., 2021, 12, 656700 CrossRef CAS PubMed.
- J. Zhang, X. Li, J. Hu, P. Cao, Q. Yan, S. Zhang, W. Dang and J. Lu, Virol. J., 2020, 17, 51 CrossRef CAS PubMed.
- R. D. Estep, A. N. Govindan, M. Manoharan, H. Li, S. S. Fei, B. S. Park, M. K. Axthelm and S. W. Wong, PLoS One, 2020, 15, e0228484 Search PubMed.
- M. A. Ahamed and W. Guan, Biophys. J., 2024, 123, 145a CrossRef.
- J. T. Granados-Riveron and G. Aquino-Jarquin, Cells, 2021, 10, 1655 CrossRef CAS PubMed.
- Md. A. Ahamed, M. A. U. Khalid, M. Dong, A. J. Politza, Z. Zhang, A. Kshirsagar, T. Liu and W. Guan, Biosens. Bioelectron., 2024, 246, 115866 Search PubMed.
- R. Ojha, R. Nandani, R. K. Pandey, A. Mishra and V. K. Prajapati, J. Cell. Physiol., 2019, 234, 1030–1043 CrossRef CAS PubMed.
- V. Martini, F. D’Avanzo, P. M. Maggiora, F. M. Varughese, A. Sica and A. Gennari, Ecancermedicalscience, 2020, 14, 1149 Search PubMed.
- R. Kaur and K. Kumar, Eur. J. Med. Chem., 2021, 215, 113220 CrossRef CAS PubMed.
- P. Zhou, M. Dong, J. Wang, F. Li, J. Zhang and J. Gu, Exp. Ther. Med., 2018, 16, 3805–3812 Search PubMed.
- A. Khanam, J. V. Chua and S. Kottilil, Int. J. Mol. Sci., 2021, 22, 5497 Search PubMed.
- Y. Yin, Y. Zhao, Q. Chen, Y. Chen and L. Mao, Front. Microbiol., 2022, 13, 1044832 Search PubMed.
- T.-L. Liao, Y.-M. Chen, S.-L. Hsieh, K.-T. Tang, D.-Y. Chen, Y.-Y. Yang, H.-J. Liu and S.-S. Yang, mBio, 2021, 12, e0276421 Search PubMed.
- S. Li, S. Li, S. Wu and L. Chen, BioMed Res. Int., 2019, 2019, 1–9 Search PubMed.
- Y. Hu, F. Cui, S. Wang, C. Liu, S. Zhang, R. Wang, J. Song and Y. Zhang, Front. Cell. Infect. Microbiol., 2023, 13, 1217984 Search PubMed.
- J. S. Nahand, M. Mahjoubin-Tehran, M. Moghoofei, M. H. Pourhanifeh, H. R. Mirzaei, Z. Asemi, A. Khatami, F. Bokharaei-Salim, H. Mirzaei and M. R. Hamblin, Epigenomics, 2020, 12, 353–370 Search PubMed.
- J. Li, Y. Zhang and B. Luo, Cancers, 2022, 14, 3552 CrossRef CAS PubMed.
- M. Mohammadinasr, S. Montazersaheb, V. Hosseini, H. Kahroba, M. Talebi, O. Molavi, H. Ayromlou and M. S. Hejazi, Cytokine, 2024, 179, 156624 CrossRef CAS PubMed.
- H. Dong, Q. Gao, X. Peng, Y. Sun, T. Han, B. Zhao, Y. Liu, C. Wang, X. Song, J. Wu and L. Yang, Front. Vet. Sci., 2017, 4, 186 CrossRef PubMed.
- A. Bayer, E. Delorme-Axford, C. Sleigher, T. K. Frey, D. W. Trobaugh, W. B. Klimstra, L. A. Emert-Sedlak, T. E. Smithgall, P. R. Kinchington, S. Vadia, S. Seveau, J. P. Boyle, C. B. Coyne and Y. Sadovsky, Am. J. Obstet. Gynecol., 2015, 212, 71.e1–71.e8 Search PubMed.
- M. V. Liberti and J. W. Locasale, Trends Biochem. Sci., 2016, 41, 211–218 Search PubMed.
- R. Mirzaei, S. Babakhani, P. Ajorloo, R. H. Ahmadi, S. R. Hosseini-Fard, H. Keyvani, Y. Ahmadyousefi, A. Teimoori, F. Zamani, S. Karampoor and R. Yousefimashouf, Mol. Med., 2021, 27, 34 CAS.
- D. De Gonzalo-Calvo, R. W. Van Der Meer, L. J. Rijzewijk, J. W. A. Smit, E. Revuelta-Lopez, L. Nasarre, J. C. Escola-Gil, H. J. Lamb and V. Llorente-Cortes, Sci. Rep., 2017, 7, 47 CrossRef CAS PubMed.
- D. Zheng, M. Huo, B. Li, W. Wang, H. Piao, Y. Wang, Z. Zhu, D. Li, T. Wang and K. Liu, Front. Cell Dev. Biol., 2021, 8, 616161 CrossRef PubMed.
- N. Yu-Qing, L. Xiao, Z. Jun-Kun and L. You-Shuo, Aging Dis., 2020, 11, 164 Search PubMed.
- M. Lu, S. Yuan, S. Li, L. Li, M. Liu and S. Wan, J. Cardiovasc. Transl. Res., 2019, 12, 68–74 CrossRef PubMed.
- J. Heo and H. Kang, Int. J. Mol. Sci., 2022, 23, 1002 Search PubMed.
- F. Sessa, M. Salerno, M. Esposito, G. Cocimano and C. Pomara, Int. J. Mol. Sci., 2023, 24, 5192 CrossRef CAS PubMed.
- H. Ling, Z. Guo, Y. Shi, L. Zhang and C. Song, Front. Physiol., 2020, 11, 654 CrossRef PubMed.
- L. Wang, J. Liu, B. Xu, Y. Liu and Z. Liu, Kaohsiung J. Med. Sci., 2018, 34, 626–633 CrossRef PubMed.
- F. Vacante, L. Denby, J. C. Sluimer and A. H. Baker, Vasc. Pharmacol., 2019, 112, 24–30 CrossRef CAS PubMed.
- H. Jia, L. Kang, Z. Ma, S. Lu, B. Huang, C. Wang, Y. Zou and Y. Sun, J. Cardiothorac. Surg., 2021, 16, 230 CrossRef PubMed.
- W. Pan, J. Liang, H. Tang, X. Fang, F. Wang, Y. Ding, H. Huang and H. Zhang, Int. J. Biochem. Cell Biol., 2020, 118, 105645 CrossRef CAS PubMed.
- Y. Fu and H. Sun, J. Ovarian Res., 2022, 15, 66 CrossRef CAS PubMed.
- C. Li, Y.-Q. Ni, H. Xu, Q.-Y. Xiang, Y. Zhao, J.-K. Zhan, J.-Y. He, S. Li and Y.-S. Liu, Signal Transduction Targeted Ther., 2021, 6, 383 Search PubMed.
- J. H. Lee, J. A. Kim, M. H. Kwon, J. Y. Kang and W. J. Rhee, Biomaterials, 2015, 54, 116–125 CrossRef CAS PubMed.
- Y. Wu, Y. Zhang, X. Zhang, S. Luo, X. Yan, Y. Qiu, L. Zheng and L. Li, Biosens. Bioelectron., 2021, 177, 112962 Search PubMed.
- A. Kögel, A. Keidel, M.-J. Loukeri, C. C. Kuhn, L. M. Langer, I. B. Schäfer and E. Conti, Nature, 2024, 635, 237–242 CrossRef PubMed.
- Y. Dong, C. Yao, Y. Zhu, L. Yang, D. Luo and D. Yang, Chem. Rev., 2020, 120, 9420–9481 CrossRef CAS PubMed.
- C. Yao, Y. Xu, P. Hu, J. Ou and D. Yang, Acc. Mater. Res., 2022, 3, 42–53 Search PubMed.
- W. Ma, Y. Zhan, Y. Zhang, C. Mao, X. Xie and Y. Lin, Signal Transduction Targeted Ther., 2021, 6, 351 CrossRef CAS PubMed.
- D.-X. Wang, J. Wang, Y.-X. Wang, Y.-C. Du, Y. Huang, A.-N. Tang, Y.-X. Cui and D.-M. Kong, Chem. Sci., 2021, 12, 7602–7622 RSC.
- T. Zhang, T. Tian, R. Zhou, S. Li, W. Ma, Y. Zhang, N. Liu, S. Shi, Q. Li, X. Xie, Y. Ge, M. Liu, Q. Zhang, S. Lin, X. Cai and Y. Lin, Nat. Protoc., 2020, 15, 2728–2757 CrossRef CAS PubMed.
- R. P. Goodman, R. M. Berry and A. J. Turberfield, Chem. Commun., 2004, 1372 Search PubMed.
- J. A. Adaskaveg and B. Blanco-Ulate, Curr. Opin. Biotechnol., 2023, 79, 102872 Search PubMed.
- D. Mao, M. Zheng, W. Li, Y. Xu, C. Wang, Q. Qian, S. Li, G. Chen, X. Zhu and X. Mi, Biosens. Bioelectron., 2022, 204, 114077 CrossRef CAS PubMed.
- H. Li, M. Han, X. Weng, Y. Zhang and J. Li, ACS Nano, 2021, 15, 1710–1717 CrossRef CAS PubMed.
- N. Xie, S. Liu, X. Yang, X. He, J. Huang and K. Wang, Analyst, 2017, 142, 3322–3332 RSC.
- L. Yu, S. Yang, Z. Liu, X. Qiu, X. Tang, S. Zhao, H. Xu, M. Gao, J. Bao, L. Zhang, D. Luo, K. Chang and M. Chen, Mater. Today Bio, 2022, 15, 100276 CrossRef CAS PubMed.
- H. Pei, N. Lu, Y. Wen, S. Song, Y. Liu, H. Yan and C. Fan, Adv. Mater., 2010, 22, 4754–4758 Search PubMed.
- H. Ding, J. Li, N. Chen, X. Hu, X. Yang, L. Guo, Q. Li, X. Zuo, L. Wang, Y. Ma and C. Fan, ACS Cent. Sci., 2018, 4, 1344–1351 CrossRef CAS PubMed.
- L. Shan, Y. Chen, X. Tan, S. Ge, L. Zhang, L. Li, J. Yu and L. Li, Anal. Chem., 2023, 95, 4760–4767 CrossRef CAS PubMed.
- L. Wang, Y. Ji, Y. Chen, S. Zheng, F. Wang and C. Li, TrAC, Trends Anal. Chem., 2024, 180, 117962 Search PubMed.
- J. Wu, H. Liu, W. Chen, B. Ma and H. Ju, Nat. Rev. Bioeng., 2023, 1, 346–360 CrossRef CAS PubMed.
- N. Liu, H. Lu, L. Liu, W. Ni, Q. Yao, G.-J. Zhang and F. Yang, Anal. Chem., 2021, 93, 5917–5923 CrossRef CAS PubMed.
- J. Zhang, J. Zhu, F. Guo, J. Jiang, M. Xie, L. Hao and J. Chao, Chem. Commun., 2023, 59, 6869–6872 RSC.
- H. Chai, Y. Tang and P. Miao, Anal. Chem., 2022, 94, 9975–9980 CrossRef CAS PubMed.
- J. Gao, H. Zhang and Z. Wang, Analyst, 2020, 145, 3535–3542 RSC.
- S. Wang, J. Guang, Y. Gao, B. Fan, Y. Liang, J. Pan, L. Li, W. Meng and F. Hu, Microchim. Acta, 2024, 191, 462 Search PubMed.
- E. S. Lee, J. Woo, J. Shin, B. S. Cha, S. Kim and K. S. Park, Biosens. Bioelectron., 2024, 250, 116055 CrossRef CAS PubMed.
- H. Wang, D. He, K. Wan, X. Sheng, H. Cheng, J. Huang, X. Zhou, X. He and K. Wang, Analyst, 2020, 145, 3289–3296 RSC.
- L. Luo, L. Wang, L. Zeng, Y. Wang, Y. Weng, Y. Liao, T. Chen, Y. Xia, J. Zhang and J. Chen, Talanta, 2020, 207, 120298 CrossRef CAS PubMed.
- Y. Li, X. Tang, R. Deng, L. Feng, S. Xie, M. Chen, J. Zheng and K. Chang, ACS Omega, 2024, 9, 19723–19731 CrossRef CAS PubMed.
- Y. Xu, X. Li, C. Niu, H. Wu, Y. Yong, C. Qi, W. Gong, H. Bai, Y. Chen, S. Ding and P. Liao, Biosens. Bioelectron., 2022, 212, 114405 CrossRef CAS PubMed.
- Z. Yu, Y. Zheng, H. Cai, S. Li, G. Liu, W. Kou, C. Yang, S. Cao, L. Chen, X. Liu, Z. Wan, N. Zhang, X. Li, G. Cui, Y. Chang, Y. Huang, H. Lv and T. Feng, Sci. Adv., 2024, 10, eadl6442 CrossRef CAS PubMed.
- L.-Y. Zhai, M.-X. Li, W.-L. Pan, Y. Chen, M.-M. Li, J.-X. Pang, L. Zheng, J.-X. Chen and W.-J. Duan, ACS Appl. Mater. Interfaces, 2018, 10, 39478–39486 Search PubMed.
- S. Jiang, Q. Li, C. Wang, Y. Pang, Z. Sun and R. Xiao, ACS Sens., 2021, 6, 852–862 CrossRef CAS PubMed.
- D. He, H. Wang, S.-L. Ho, H.-N. Chan, L. Hai, X. He, K. Wang and H.-W. Li, Theranostics, 2019, 9, 4494–4507 CrossRef CAS PubMed.
- R.-Y. Zhang, S.-H. Luo, X.-M. Lin, X.-M. Hu, Y. Zhang, X.-H. Zhang, C.-M. Wu, L. Zheng and Q. Wang, Anal. Chim. Acta, 2021, 1157, 338396 CrossRef CAS PubMed.
- Md. A. Ahamed, G. Kim, Z. Li and S.-J. Kim, Anal. Chim. Acta, 2022, 1236, 340587 CrossRef CAS PubMed.
- X. Wang and N. C. Seeman, J. Am. Chem. Soc., 2007, 129, 8169–8176 CrossRef CAS PubMed.
- Y. Wang, J. E. Mueller, B. Kemper and N. C. Seeman, Biochemistry, 1991, 30, 5667–5674 CrossRef CAS PubMed.
- N. R. Kallenbach, R.-I. Ma and N. C. Seeman, Nature, 1983, 305, 829–831 CrossRef CAS.
- A. Idili, A. Bonini, C. Parolo, R. Alvarez-Diduk, F. Di Francesco and A. Merkoçi, Adv. Funct. Mater., 2022, 32, 2201881 CrossRef CAS.
- Y. Li, Y. D. Tseng, S. Y. Kwon, L. d'Espaux, J. S. Bunch, P. L. McEuen and D. Luo, Nat. Mater., 2004, 3, 38–42 CrossRef CAS PubMed.
- S. H. Um, J. B. Lee, S. Y. Kwon, Y. Li and D. Luo, Nat. Protoc., 2006, 1, 995–1000 CrossRef CAS PubMed.
- S. W. Shin, B. S. Lee, K. Yang, L. Amornkitbamrung, M. S. Jang, B. M. Ku, S.-W. Cho, J. H. Lee, H. Bae, B.-K. Oh, M.-J. Ahn, Y. T. Lim and S. H. Um, Sci. Rep., 2017, 7, 13499 CrossRef PubMed.
- L. Yang, C. Yao, F. Li, Y. Dong, Z. Zhang and D. Yang, Small, 2018, 14, 1800185 CrossRef PubMed.
- Z. Zhang, Y. Liu, P. Liu, L. Yang, X. Jiang, D. Luo and D. Yang, Nanoscale, 2017, 9, 19367–19373 RSC.
- M. R. Hartman, D. Yang, T. N. N. Tran, K. Lee, J. S. Kahn, P. Kiatwuthinon, K. G. Yancey, O. Trotsenko, S. Minko and D. Luo, Angew. Chem., Int. Ed., 2013, 52, 8699–8702 CrossRef CAS PubMed.
- F. Li, Y. Dong, Z. Zhang, M. Lv, Z. Wang, X. Ruan and D. Yang, Biosens. Bioelectron., 2018, 117, 562–566 CrossRef CAS PubMed.
- L. Cheng, Z. Zhang, D. Zuo, W. Zhu, J. Zhang, Q. Zeng, D. Yang, M. Li and Y. Zhao, ACS Appl. Mater. Interfaces, 2018, 10, 34869–34877 CrossRef CAS PubMed.
- M. Lin, J. Wang, G. Zhou, J. Wang, N. Wu, J. Lu, J. Gao, X. Chen, J. Shi, X. Zuo and C. Fan, Angew. Chem., Int. Ed., 2015, 54, 2151–2155 CrossRef CAS PubMed.
- L. Mo, Y. Hong, M. Mo, D. Liang, R. Yuan, C. Yang and W. Lin, Sens. Actuators, B, 2024, 401, 134973 CrossRef CAS.
- J. Deng, J. Xu, M. Ouyang, Z. Zou, Y. Lei, J. Li, Z. Qing and R. Yang, Chin. Chem. Lett., 2022, 33, 773–777 CrossRef CAS.
- X. Sun, Y. Chen, H. Li, W. Xing, M. Chen, J. Wang and L. Ye, Chem. Commun., 2024, 60, 4777–4780 RSC.
- J. Chen, M. Xie, M. Shi, K. Yuan, Y. Wu, H.-M. Meng, L. Qu and Z. Li, Anal. Chem., 2022, 94, 2227–2235 CrossRef CAS PubMed.
- S. Zhao, S. Zhang, H. Hu, Y. Cheng, K. Zou, J. Song, J. Deng, L. Li, X. Zhang, G. Ke and J. Sun, Angew. Chem., Int. Ed., 2023, 62, e202303121 CrossRef CAS PubMed.
- W. Wang, C. Li, S. Luo and Z.-S. Wu, Anal. Chem., 2024, 96, 7091–7100 CrossRef CAS PubMed.
- J. Zhao, C. Liu, Y. Li, Y. Ma, J. Deng, L. Li and J. Sun, J. Am. Chem. Soc., 2020, 142, 4996–5001 CrossRef CAS PubMed.
- J. H. Lee, J. A. Kim, S. Jeong and W. J. Rhee, Biosen. Bioelectron., 2016, 86, 202–210 CrossRef CAS PubMed.
- H. Yang, J. Zhao, J. Dong, L. Wen, Z. Hu, C. He, F. Xu, D. Huo and C. Hou, Chem. Eng. J., 2022, 438, 135594 CrossRef CAS.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- C. A. Mirkin and S. H. Petrosko, ACS Nano, 2023, 17, 16291–16307 CrossRef CAS PubMed.
- L. Liu, H. Lu, R. Shi, X.-X. Peng, Q. Xiang, B. Wang, Q.-Q. Wan, Y. Sun, F. Yang and G.-J. Zhang, Anal. Chem., 2019, 91, 13198–13205 CrossRef CAS PubMed.
- Y. Lei, X. Fei, Y. Ding, J. Zhang, G. Zhang, L. Dong, J. Song, Y. Zhuo, W. Xue, P. Zhang and C. Yang, Sci. Adv., 2023, 9, eadi1556 CrossRef CAS PubMed.
- Q.-Q. Wu, Y.-Y. Wu, Y.-W. Liang, H.-Y. Yang, J. Xie, M.-M. Li, B.-P. Xie, J. Chen and W.-J. Duan, Sens. Actuators, B, 2024, 418, 136298 CrossRef CAS.
- R. Abolhasan, A. Mehdizadeh, M. R. Rashidi, L. Aghebati-Maleki and M. Yousefi, Biosens. Bioelectron., 2019, 129, 164–174 CrossRef CAS PubMed.
- J. Huang, X. Yang, X. He, K. Wang, J. Liu, H. Shi, Q. Wang, Q. Guo and D. He, TrAC, Trends Anal. Chem., 2014, 53, 11–20 CrossRef CAS.
- X. Liu, M. Zou, D. Li, R. Yuan and Y. Xiang, Anal. Chim. Acta, 2019, 1076, 138–143 CrossRef CAS PubMed.
- S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS PubMed.
- M. Xie, F. Wang, J. Yang, Y. Guo, F. Ding, X. Lu, Y. Huang, Y. Li, X. Zhu and C. Zhang, Anal. Chem., 2022, 94, 13043–13051 CrossRef CAS PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- Y. Zhang, J. Chen, H. Yang, W. Yin, C. Li, Y. Xu, S.-Y. Liu, Z. Dai and X. Zou, Anal. Chem., 2022, 94, 9665–9673 CrossRef CAS PubMed.
- D. He, L. Hai, H. Wang, R. Wu and H.-W. Li, Analyst, 2018, 143, 813–816 RSC.
- J. Kim, J. S. Shim, B. H. Han, H. J. Kim, J. Park, I.-J. Cho, S. G. Kang, J. Y. Kang, K. W. Bong and N. Choi, Biosens. Bioelectron., 2021, 192, 113504 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |



