Open Access Article
Open Access ArticleConstruction of DNA template sequences to generate fluorescent gold nanoclusters for the sensitive detection of DNA methyltransferase activity bioassay†
Fangyu
Zhou
 a,
Hui
Chen
a,
Tingting
Fan
a,
Zixia
Guo
a,
Xiangyan
Dong
a and
Feng
Liu
*ab
a,
Hui
Chen
a,
Tingting
Fan
a,
Zixia
Guo
a,
Xiangyan
Dong
a and
Feng
Liu
*ab
aState Key Laboratory of Chemical Oncogenomics, Tsinghua Shenzhen International Graduate School, Shenzhen, 518055, PR China. E-mail: liu.feng@sz.tsinghua.edu.cn
bNational & Local United Engineering Lab for Personalized Anti-tumor Drugs, Shenzhen Kivita Innovative Drug Discovery Institute, Tsinghua Shenzhen International Graduate School, Shenzhen, 518055, PR China
First published on 12th November 2024
Abstract
Abnormal DNA methylation mediated by DNA methyltransferases is one of the most common epigenetic modifications, and abnormal DNA methyltransferase activity is often responsible for serious diseases such as cancer. For rapid, sensitive and efficient detection of DNA methyltransferase activity, a bioassay system using gold nanoclusters (DNA-AuNCs) as output has been developed. In this study, dumbbell DNA substrate is recognized and methylated by methyltransferase followed by cleavage by endonuclease (GlaI). In the presence of terminal deoxynucleotidyl transferase (TdT), the poly-A DNA product eventually becomes the template for the reduction of gold nanoclusters and then generated with a strong fluorescence signal. The assay is highly sensitive and requires no amplification or complex material synthesis steps. The detection limit is 0.077 U mL−1. The bioassay showed good detection efficiency in both human serum, cell lysates and human-derived cells. Moreover, it can be used for screening and evaluation of M.SssI MTase inhibitors and hence has great potential use in disease diagnosis and drug discovery. The method was universal and allowed for other biological target detection by simply replacing the sequence of the substrate DNA recognition site; thus the proposed assay has a broad scope of application in both bioassay and drug screening.
1. Introduction
DNA methylation mediated by DNA methyltransferases is a common epigenetic modification.1,2 Methylation is the process in which a methyl group is transferred from S-adenosylmethionine (SAM) to adenine or cytosine remnants through the action of DNA methyltransferases.3,4 Methylated gene sequences affect the ability of transcription factor binding. Abnormal DNA methyltransferase activity is the main cause of abnormal DNA methylation.5,6 Methyltransferase expression is much higher in cancer cells than in normal cells. In the early stages of cancerous lesions, intracellular methyltransferases also showed abnormal activity. Several studies have shown that increased levels of methyltransferases accompany the progression of many major diseases, e.g. increased expression of DNA methyltransferase in lung cancer is associated with tumor cell growth and spread.7,8 In addition, DNA methyltransferases have been found to be overexpressed in many breast cancer cell lines such as MCF-7.8,9 As an important regulatory protein in epigenetics, methyltransferases provide new ideas for drug screening and disease treatment.10,11 Inhibitors of methyltransferases can affect the correct expression or silence of genes by affecting the activity of methyltransferases for therapeutic purposes.12,13 Therefore, the study of detecting methyltransferase activity and the screening and development of methyltransferase inhibitors is of great importance in the diagnosis and treatment of related diseases.14,15Common methods for detecting DNA methyltransferases include high-performance liquid chromatography (HPLC)16 and enzyme-linked immunosorbent assay (ELISA).17 Low sensitivity and the complexity of the experimental procedure are disadvantages of these methods. Therefore, DNA methyltransferase activity is now often detected through the construction of biosensors.18–21 To achieve lower detection limits, the lack of proper signal amplification also makes it difficult to detect low levels of DNA methyltransferase in real samples. To overcome this problem, different nucleic acid amplification strategies are rationally introduced to improve the performance of DNA methyltransferase assays. Nucleic acid amplification steps are often added to the assay: isothermic amplification of nucleic acids22,23 (RCA,24 HCR25), amplification using nicking endonucleases,26–29 amplification using the CRISPR/Cas system,30 the use of quantum dots (QDs) for increased sensitivity,31,32etc. The disadvantage of this is that the procedure is complex and the background signal tends to interfere with test results. It is therefore desirable to construct a bioassay system that can sensitively detect DNA methyltransferase activity without amplification of nucleic acid sequences. The construction of a biosensing strategy utilising metal nanoclusters as signal outputs is aligned with the objectives of the present study of methyltransferase activity. Metal nanoclusters are a class of nanomaterials composed of a few to several hundred metal atoms stacked together.33–35 The synthesis process involves the reduction of metal ions on biological ligands (proteins or DNA) in the presence of a reducing agent. Due to their small size, they have molecule-like properties and can emit fluorescence signals. Silver and copper nanoclusters are currently used for biosensing detection36,37 and bioimaging applications.38,39 Gold nanoclusters are of interest due to their simplicity of synthesis and low cytotoxicity compared to other metal nanoclusters. Gold nanoclusters using DNA as a template have high potential application value as signal outputs in biosensors because of the ready availability of raw materials and simple synthesis procedure.40 Based on previous studies,41 DNA methyltransferase activity was detected by quenching the fluorescence signal of gold nanoclusters. In this work, the method can more directly detect the activity of methyltransferase.
In this study, a bioassay system for the detection of DNA methyltransferase and other biological targets was successfully constructed using fluorescent gold nanoclusters as the signal output. In this assay strategy we generate the required template chains for gold nanoclusters by using endonuclease (GlaI)42 and terminal deoxynucleotidyl transferase (TdT).43 Following a simple reduction reaction, gold nanoclusters with fluorescence signals can be generated. This system is unable to detect the fluorescence signal from gold nanoclusters prior to detection and when the target is not present in the system. This method eliminates the need for an additional nucleic acid sequence amplification step to form new nucleic acid sequences to amplify the background signals due to the lower background baseline and fewer interfering signals received. Using this bioassay system, the activity of methyltransferase extracted from human cells was successfully detected. The bioassay system can also be used to screen inhibitors of DNA methyltransferases and assess the ability to inhibit the target's activity. Meanwhile, T4 PNK and EcoRI were used as targets to verify the generalisability of the bioassay system.
2. Experimental methods
2.1 Strategy for detecting M.SssI MTase
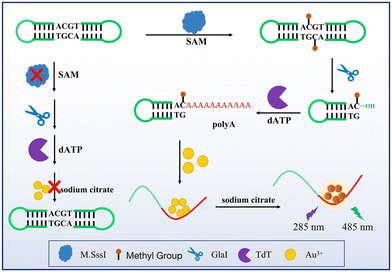
In this system (as shown in Scheme 1), a dumbbell DNA methylated by DNA methyltransferase is recognised by endonuclease (GlaI) and cleaved into two hairpin DNAs. dATP can be added to the hydroxyl terminus of hairpin DNA to form a poly-A sequence by terminal deoxynucleotidyl transferase (TdT), which becomes the template DNA strand for gold nanocluster reduction. The gold ions are then reduced to fluorescent gold nanoclusters in the presence of a reducing agent (sodium citrate). If there is no methyltransferase in the system, the dumbbell DNA cannot be cleaved by GlaI into hairpin DNA with hydroxyl terminations and TdT cannot act to close the dumbbell DNA, thereby preventing the formation of the DNA template strand. Gold nanoclusters with fluorescence could not be formed within the system.2.2 Reagents and materials
All the oligonucleotides used in this study were synthesized and HPLC-purified by Sangon Biotechnology Co., Ltd. (Shanghai, China), and the sequences are listed in Table S1.† T4 DNA ligase, ExoI exonuclease, ExoIII exonuclease, M.SssI MTase, and T4 PNK were obtained from New England Biolabs (Beijing, China). 5-Azacytidine (5-Aza) was obtained from Sigma-Aldrich and used as received. DNA endonuclease (GlaI) and 10 SEBuffer Y (330 mM Tris-acetate (Tris-Ac), 660 mM potassium acetate (KAc), and 100 mM magnesium acetate (Mg(Ac)2), 10 mM DTT, pH 7.9) were obtained from SibEnzyme Ltd. (Ak, Novosibirsk, Russia). All chemical reagents were of analytical grade and used without further purification. Fluorescence spectra were recorded by using a microplate reader (Tecan Infinite M1000 Pro, Männedorf, Switzerland). Transmission electron microscopy (TEM) measurements were obtained using a Tecnai G2 Spirit 120 kV instrument, and the samples used in TEM were prepared by dropping sample solutions onto carbon-coated copper grids followed by drying at room temperature. Native polyacrylamide gel electrophoresis (PAGE) was imaged using a Molecular Imager Pharos FXTM Plus system (BioRad, Hercules, CA, USA). Human blood samples were collected from the Shenzhen Second People's Hospital (Shenzhen, China) following the study protocol approved by the Institutional Review Board (IRB, No. 2020014). MCF-10A cell line and MCF-7 cell line were provided by the Chinese Academy of Sciences Cell Bank (Shanghai, China).2.3 Preparation of enclosed dumbbell DNA M.SssI MTase substrate
A total volume of 50 μL contains 10 μM of DNA dumbbell sequence, 1× T4 DNA ligase buffer, 400 U of T4 DNA ligase, 1 μL of ExoI (20 U μL−1) and 1 μL of ExoIII (100 U μL−1). The DNA dumbbell DNA sequence is now mixed with T4 DNA ligase buffer and incubated at 95 °C for 5 min. After the solution has cooled to room temperature, T4 DNA ligase is added to the reaction overnight. After this, 12 U of ExoI and 60 U of ExoIII were added to the reaction solution. The above solution was incubated at 37 °C for 1 h to ensure complete digestion of the leftover unreacted substrate. Finally, the mixture was heated at 80 °C for 20 min to stop this reaction. The solution obtained containing the closed dumbbell-shaped DNA probe can be used directly for subsequent experiments or stored at −20 °C.2.4 Detection of M.SssI MTase activity
2.5 Selectivity assay of M.SssI MTase activity
In order to investigate the selectivity of the assay system, we investigated whether the bioassay system is specific and sensitive for the detection of M.SssI MTase. BSA, Taq DNA ligase, T4 PNK, Dam MTase and M.SssI MTase were added to the system as targets for the assay. The assay process is consistent with that used to detect methyltransferase activity.2.6 Inhibition assay of M.SssI MTase activity
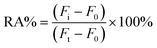 | (2-1) |
2.7 Detecting DNA methyltransferase activity in complex systems
2.8 M.SssI MTase recovery assay from human serum sample and cell lysates
To investigate the recovery of the established bioassay system in a complex assay environment 10% by volume of healthy human serum or 1% cell lysate was added to experimental groups containing different concentrations of DNA methyltransferase. This was followed by the steps described above for the detection of methyltransferase activity and the spiked recoveries were calculated from eqn (2-2) as follows:| Recovery (%) = (Cmean found/Cadded) × 100% | (2-2) |
2.9 Detection of human-derived intracellular methyltransferases
MCF-10A cell line and MCF-7 cell line were cultured in a sterile environment. Nuclear extracts were collected using a nuclear protein extraction kit. The resulting supernatant was stored at −80 °C until use. The cell lysate containing methyltransferase (Dnmt 1) was obtained after the above steps.2.10 Demonstrating the universality of the assay system
The solution was added to 10 mmol L−1 sodium citrate at pH 6 and 20 μL of 1 mmol L−1 HAuCl4 to a final volume of 300 μL and incubated at 90 °C for 30 min. The fluorescence signal was measured using an enzyme marker.
2.11 Native polyacrylamide gel electrophoresis (PAGE) analysis
Native polyacrylamide gel electrophoresis (PAGE) was used to characterize nucleotides based on their electrophoretic mobility and molecular weight. The reaction products were analyzed using 12% PAGE in 1× TBE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA, pH 8.3) at a 120 V constant voltage for 50 min.2.12 Characterisation of gold nanoclusters (DNA-AuNCs)
The prepared solution of DNA-AuNCs was uniformly dropped onto a copper grid and photographed under a transmission electron microscope (TEM) for measurement. The reduced solution was tested for fluorescence signal values on a microplate reader.2.13 Demonstrating the generality of the bioassay system
To demonstrate the generality of the bioassay system, T4 PNK and EcoRI were selected as targets for the assay. The exact methods are given in the ESI.†3. Results and discussion
3.1 Characterization strategies for the determination of M.SssI MTase activity
First, the feasibility of the test system was analyzed. The feasibility of the proposed bioassay system scheme was demonstrated using native polyacrylamide gel electrophoresis (PAGE) (Fig. 1A): lane 1 is the DNA probe formation of the dumbbell DNA in the presence of T4 DNA ligase and, using ExoI and ExoIII, removal of DNA that had not been joined into a closed dumbbell. Lane 2 shows where the dumbbell DNA was added to the methyl group in the presence of methyltransferase, which was then later recognized and cleaved by GlaI. In comparison, the migration rate of the DNA in lane 2 is faster than that in lane 1, showing that the dumbbell DNA is cleaved into a hairpin structure. This was followed by a TdT-catalyzed elongation reaction of the cleavage product with the addition of dATP at the hairpin tails sequentially to elongate a poly-A sequence, as can be seen from lane 3, where the rate of migration of the DNA band is altered because of the addition of tails at the ends. Hence this shows that the dumbbell DNA is eventually cleaved and a segment with a poly-A sequence is added in the presence of TdT.The feasibility of this scheme was verified by fluorescence signal spectroscopy: Fig. 1B shows that only when M.SssI MTase, dumbbell DNA probe, GlaI, TdT and dATP are present in the assay system. In the absence of any of M.SssI MTase, dumbbell DNA probe, GlaI, TdT and dATP, no significant fluorescence signal was detected at 485 nm, and no gold nanoclusters could be generated.
Gold nanoclusters with stable fluorescence were synthesized and characterized by heat-assisted reduction. The gold nanoclusters with stable fluorescence were characterized by transmission electron microscopy (TEM) and the results are shown in Fig. 1C. The gold nanoclusters were spherical, uniform in size, and well dispersed with an average particle size of about 3 nm. The maximum excitation wavelength of the gold nanoclusters formed by the reduction reaction was 285 nm and the maximum emission wavelength was 485 nm (Fig. 1D).
From the above validation and characterization results, it can be concluded that the proposed biosensing strategy can detect the activity of methyltransferase.
3.2 Selectivity of M.SssI MTase assay
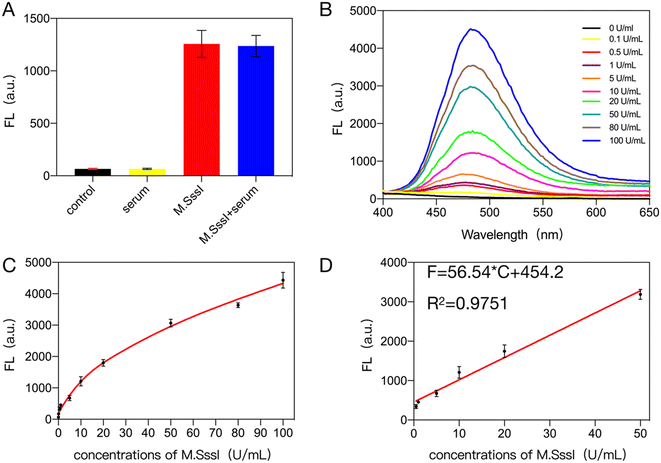
To verify the target specificity and selectivity of the proposed method, 0.1 g l−1 BSA, 10 U ml−1 T4 PNK, 10 U ml−1 Taq, 10 U ml−1 Dam MTase enzyme was added to the system instead of a target. Dam MTase is a common DNA adenine methyltransferase that adds methyl groups to adenine residues in the DNA sequence to modify methylation in the genome. The selectivity of the assay was further evaluated by adding 10 U mL−1 Dam MTase as an interfering agent to the assay system. The selectivity of the bioassay system was evaluated by recording the value of the fluorescence signal generated by the gold nanoclusters. According to the result in Fig. 2A, the final gold nanoclusters were generated and produced a stable and intense fluorescence signal only when M.SssI MTase was present in the system. Thus the proposed method has good specificity.3.3 Sensitivity of the M.SssI MTase assay
The best detection conditions for this bioassay were obtained through optimization experiments (Fig. S1†). After that, to verify the sensitivity of the bioassay system, different concentrations of M.SssI MTase were added to the system and the signal values of the fluorescence spectra of the generated gold nanoclusters were recorded. A linear relationship between the signal values and the targets can be seen by fitting a standard curve. The fluorescence signal spectrum as shown in Fig. 2B, the fluorescence signal of the generated gold nanoclusters increased with the increase of methyltransferase. Within this bioassay system (Fig. 2C and D), M.SssI MTase had a good linear relationship with the fluorescence signal value of the gold nanoclusters within 0.1–100 U mL−1. The equation is F = 57.91 × C + 467.8, R2 = 0.9834 (where F is the fluorescence signal value of the gold nanoclusters at 485 nm within the system and C is the concentration of the added methyltransferase), and then by evaluating the mean response of the blank control plus 3 times the standard deviation, the detection limit of the bioassay system was found to be 0.077 U ml−1. This was followed by a comparison of the sensitivity and linear detection range of biosensors that detect the same targets. As shown in Table S2,† the proposed bioassay system using fluorescent gold nanoclusters as the signal output has a wider linear range as well as a lower detection limit. Therefore, the method has great application value.3.4 Detection of M.SssI MTase activity in complex biological sample
To verify that the bioassay system detects the target in a complex biological environment, 10% human serum or 1% cell lysate was added to the system. As shown in Fig. 3A, the addition of 10% human serum to the system gave almost the same results as in the buffer solution. The sensitivity of the bioassay system after the addition of serum was also determined and as shown in Fig. 3B, the fluorescence signal generated in the system containing 10% human serum increased with the concentration of the target. Subsequently, by fitting the standard curves (Fig. 3C and D), it can be seen that there is a good linear relationship between the target concentration and the fluorescence value of the generated gold nanoclusters within 0.1–50 U ml−1 with the equation F = 56.54 × C + 454.2, R2 = 0.9751 (where F is the value of the fluorescence signal of the gold nanoclusters generated in the system with the addition of 10% human serum and C is the amount of methyltransferase added to the system) and its detection limit was 0.079 U ml−1, which was similar to that in the buffer solution. Recovery experiments were then performed. 10% human serum was added to the system along with various concentrations of methyltransferase and the recoveries were measured as shown in Table S3.† The target recovery in 10% serum ranged from 97.98% to 105.20%. The results of the study indicate that the bioassay system has good resistance to serum interference.The bioassay was further tested for interference resistance by adding 10%, 5% and 1% cell lysate to the system as shown in Fig. S2† and 4A. When 1% cell lysate was added to the system, the detection efficiency was also almost identical to that in the buffer. As can be seen in Fig. 4B and C, within the bioassay system containing 1% cell lysate, the fluorescence signal of the generated gold nanoclusters increased with increasing concentration of M.SssI MTase. As shown in Fig. 4D, there is a good linear relationship between the concentration of methyltransferase in the range of 1–50 U mL−1 and the fluorescence signal values of the gold nanoclusters. The linear equation was F = 24.53 × C + 770.8, R2 = 0.9354. The detection limit of the system was 0.109 U ml−1 in the presence of 1% cell lysate, which is almost identical to the detection efficiency in buffer solution. Subsequent recovery experiment results (Table S4†) showed that the bioassay system achieved recoveries between 99.48% and 114.60% in 1% cell lysate. All of the above experiments show that the proposed assay has good immunity to interference.
3.5 Detection of methyltransferase activity in human-derived cells (Dnmt 1) using this assay system
To further demonstrate the generality and wide application of the bioassay, the methyltransferase activity assay was performed on human-derived cell samples. MCF-7 cells (human breast cancer cells) and MCF-10A cells (normal human breast epithelial cells) were used for the assay. Based on the fluorescence signal of the test results (Fig. S3†), it can be seen that the methyltransferase activity extracted from MCF-7 cells was significantly higher than that extracted from MCF-10A cells, which served as a control group. This may be because the expression level of methyltransferase in cancer cells is usually higher than that in normal cells.2,44 The results of this experiment therefore also suggest that the bioassay protocol can be applied to the detection of methyltransferase activity in human-derived cells.3.6 Analysis of the inhibitory activity of M.SssI MTase inhibitors
The screening of inhibitors is of great importance for drug development and also plays a crucial role in the treatment of serious diseases. This part of the study therefore assesses the ability of the bioassay system to screen for target inhibitors. 5-Azacytidine (5-Aza) and 5-aza-2-deoxycytidine (5-Aza-dC) were selected as model inhibitors of the target. Before screening the inhibitors, it was first ruled out whether the inhibitors had any effect on enzymes other than methyltransferases. The following experiment was designed: the inhibitor was added after the system had completed the methylation process. As shown in Fig. S4A and B,† both 5-Aza and 5-Aza-dC had no effect on the experimental steps other than methylation, and their fluorescence signal values were similar to those generated in the other assay steps. This was followed by inhibitor screening experiments, using IC50 values as a basis for assessing whether the bioassay system had the capacity to screen for target inhibitors. Using this bioassay system, an IC50 value of 2.23 μM for 5-Aza was obtained as shown in Fig. 5A, consistent with the results reported in the literature (5.24 μM,45 3.33 μM,46 3.6 μM (ref. 47)), and an IC50 value of 1.09 μM was obtained for 5-Aza-dC (Fig. 5B), which is similar to results reported in the literature (1.15 μM,48 1.3 μM,47 1.58 μM (ref. 49)). The above experiments demonstrate the utility of this bioassay system for screening target inhibitors, which can be applied in drug screening and other applications. | ||
| Fig. 5 (A) Relationship between target activity and 5-Aza concentration. (B) Relationship between target activity and 5-Aza-dC concentration. | ||
3.7 Generality of the proposed sensing system
To further validate the generality of the assay, other common nucleases that play an important role in many biological processes were tested using the assay system: T4 PNK and EcoRI. By simply replacing the recognition site on the dumbbell loop with the recognition sequence of the new target, the target activity can be detected according to the previously proposed assay.3.8 Detection of T4 PNK with this assay
T4 PNK is an important DNA repair enzyme that repairs the phosphorylated 3′ end to the –OH end. The detection process for T4 PNK is shown in Fig. 6A. The DNA sequence with the recognition site is identified and cleaved by T4 PNK, releasing the DNA sequence with the –OH. The hydroxyl-terminated DNA sequence is identified by TdT, resulting in the attachment of dATP to the end of the DNA sequence. The extended sequence is the DNA template required for the reduction of gold nanoclusters. The gold nanoclusters are then reduced by sodium citrate to form gold nanoclusters with a fluorescence signal.The feasibility of the assay was first verified as shown in Fig. 6B. When T4 PNK is present in the system, gold nanoclusters are generated and a strong fluorescence signal is generated, and when T4 PNK is not present in the system, no significant fluorescence signal is available in the system. This demonstrates the feasibility of the assay.
Subsequently, it can be seen from the fluorescence spectra (Fig. 6C and D) that the fluorescence signal of gold nanoclusters increased with the concentration of T4 PNK in the system when T4 PNK was present in the system. The linear equation of F = 1552 × C + 607.5, R2 = 0.9036 was calculated to give a detection limit of 3.03 × 10−3 U mL−1 for the detection of T4 PNK enzyme activity.
3.9 Detection of EcoRI with this assay
The restriction endonuclease EcoRI is a common tool for molecular shearing and is involved in many important physiological processes. EcoRI recognizes specific sequences and upon shearing releases hydroxyl-terminated DNA sequences. The assay to detect EcoRI is shown in Fig. 7A. The bases in the complementary part of the dumbbell DNA are replaced by the EcoRI recognition sequence. When EcoRI is added to the system, the dumbbell DNA is recognized and cleaved into two hydroxyl-terminated hairpin DNAs. The hairpin DNA is recognized and processed by TdT, which adds a poly-A sequence to the hydroxyl terminus of the hairpin DNA, forming a stencil strand that is eventually reduced by sodium citrate to form gold nanoclusters with a fluorescence signal. The feasibility of the detection scheme was demonstrated by the fluorescence signal shown in Fig. 7B: when EcoRI was not present in the system there was almost no fluorescence signal in the system, while when EcoRI was present in the system there was a strong fluorescence signal at 485 nm, demonstrating the feasibility of the detection scheme.The fluorescence spectra (Fig. 7C) show that the fluorescence signal of the gold nanoclusters increased with the concentration of EcoRI. The linear equation (Fig. 7D) F = 160.1 × C + 968.1, R2 = 0.9504, was calculated to give a detection limit of 2.8 × 10−2 U mL−1 for the detection of EcoRI enzyme activity.
4. Conclusions
In this study, a bioassay system for the sensitive detection of methyltransferase and other target activities was successfully developed by constructing DNA template sequences to generate gold nanoclusters. The activity of the targets can be specifically and sensitively detected by fluorescence signals. The bioassay has a good linear range and a limit of detection of 0.077 U mL−1 for methyltransferases and is selective in that it does not fluoresce for enzymes or proteins other than the target. The bioassay also demonstrates high interference resistance and sensitive detection of target activity in complex biological environments. The proposed bioassay system has a wide range of applications including detection of methyltransferase (Dnmt 1) activity derived from human-derived cells and screening of methyltransferase inhibitors. The proposed assay has been shown to be well generalised and can be used for rapid and easy detection of various endonucleases. All materials used to construct the bioassay system are readily available. Thus, there are promising applications for screening and detection of various disease targets as well as for the development of appropriate drugs.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization, methodology, writing – original draft preparation: Fangyu Zhou. Formal analysis and investigation: Fangyu Zhou, Hui Chen, Tingting Fan; Zixia Guo, Xiangyan Dong, and Feng Liu. Resources: Hui Chen, Tingting Fan; and Zixia Guo. Writing – review & editing: Zixia Guo and Feng Liu. Funding acquisition, supervision: Feng Liu.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Development and Reform Commission of Shenzhen Municipality (No. 2019156).References
- P. A. Jones and S. B. Baylin, The fundamental role of epigenetic events in cancer, Nat. Rev. Genet., 2002, 3(6), 415–428, DOI:10.1038/nrg816.
- A. Eden, F. Gaudet, A. Waghmare and R. Jaenisch, Chromosomal instability and tumors promoted by DNA hypomethylation, Science, 2003, 300(5618), 455, DOI:10.1126/science.1083557.
- Z. D. Smith and A. Meissner, DNA methylation: roles in mammalian development, Nat. Rev. Genet., 2013, 14(3), 204–220, DOI:10.1038/nrg3354.
- X. D. Cheng and R. J. Roberts, AdoMet-dependent methylation, DNA methyltransferases and base flipping, Nucleic Acids Res., 2001, 29(18), 3784–3795, DOI:10.1093/nar/29.18.3784.
- J. M. C. Tubio, Y. Li, Y. S. Ju, I. Martincorena, S. L. Cooke and M. Tojo, et al. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes, Science, 2014, 345(6196), 531, DOI:10.1126/science.1251343.
- E. Lee, R. Iskow, L. Yang, O. Gokcumen, P. Haseley and L. J. Luquette III, et al. Landscape of Somatic Retrotransposition in Human Cancers, Science, 2012, 337(6097), 967–971, DOI:10.1126/science.1222077.
- R. Ghemrawi, A. Al Qassem, A. Ramadan, R. Aldulaymi, N. Sammani and W. K. Mousa, et al. DNA and protein methyltransferases inhibition by adenosine dialdehyde reduces the proliferation and migration of breast and lung cancer cells by downregulating autophagy, PLoS One, 2023, 18(7), e0288791, DOI:10.1371/journal.pone.0288791.
- J. Y. Zhang, C. Yang, C. F. Wu, W. Cui and L. H. Wang, DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy, Cancers, 2020, 12(8), 2123, DOI:10.3390/cancers12082123.
- J. D. Roll, A. G. Rivenbark, W. D. Jones and W. B. Coleman, DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines, Mol. Cancer, 2008, 7, 15, DOI:10.1186/1476-4598-7-15.
- S. B. Baylin and P. A. Jones, A decade of exploring the cancer epigenome - biological and translational implications, Nat. Rev. Cancer, 2011, 11(10), 726–734, DOI:10.1038/nrc3130.
- H. Heyn and M. Esteller, DNA methylation profiling in the clinic: applications and challenges, Nat. Rev. Genet., 2012, 13(10), 679–692, DOI:10.1038/nrg3270.
- M. J. Topper, M. Vaz, K. B. Chiappinelli, C. E. D. Shields, N. Niknafs and R.-W. C. Yen, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer, Cell, 2017, 171(6), 1284–1300.e21, DOI:10.1016/j.cell.2017.10.022.
- P. A. Jones, J.-P. J. Issa and S. Baylin, Targeting the cancer epigenome for therapy, Nat. Rev. Genet., 2016, 17(10), 630–641, DOI:10.1038/nrg.2016.93.
- A. Gnyszka, Z. Jastrzebski and S. Flis, DNA Methyltransferase Inhibitors and Their Emerging Role in Epigenetic Therapy of Cancer, Anticancer Res., 2013, 33(8), 2989–2996 CAS.
- C. Stresemann and F. Lyko, Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine, Int. J. Cancer, 2008, 123(1), 8–13, DOI:10.1002/ijc.23607.
- A. Lopez Torres, E. Yanez Barrientos, K. Wrobel and K. Wrobel, Selective Derivatization of Cytosine and Methylcytosine Moieties with 2-Bromoacetophenone for Submicrogram DNA Methylation Analysis by Reversed Phase HPLC with Spectrofluorimetric Detection, Anal. Chem., 2011, 83(20), 7999–8005, DOI:10.1021/ac2020799.
- F. Yu, Y.-m. Xiong, S.-c. Yu, L.-l. He, S.-s. Niu and Y.-m. Wu, et al. Magnetic immunoassay using CdSe/ZnS quantum dots as fluorescent probes to detect the level of DNA methyltransferase 1 in human serum sample, Int. J. Nanomed., 2018, 13, 429–437, DOI:10.2147/ijn.S152618.
- S. Y. Niu, C. Bi and W. L. Song, Detection of DNA methyltransferase activity using template-free DNA polymerization amplification based on aggregation-induced emission, Anal. Biochem., 2020, 590, 113532, DOI:10.1016/j.ab.2019.113532.
- Y. L. Liu, Y. B. Tu, H. P. Wu, H. Zhang, H. H. Chen and G. H. Zhou, et al. A renewable DNA biosensor for sensitive detection of DNA methyltransferase activity based on cascade signal amplification, Sens. Actuators, B, 2020, 313, 128029, DOI:10.1016/j.snb.2020.128029.
- X. Y. Zhou, M. Zhao, X. L. Duan, B. Guo, W. Cheng and S. J. Ding, et al. Collapse of DNA Tetrahedron Nanostructure for “Off On” Fluorescence Detection of DNA Methyltransferase Activity, ACS Appl. Mater. Interfaces, 2017, 9(46), 40087–40093, DOI:10.1021/acsami.7b13551.
- H. Zhang, Y. Yang, H. L. Dong and C. X. Cai, A superstructure-based electrochemical assay for signal-amplified detection of DNA methyltransferase activity, Biosens. Bioelectron., 2016, 86, 927–932, DOI:10.1016/j.bios.2016.07.103.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Rolling circle amplification: Applications in nanotechnology and biodetection with functional nucleic acids, Angew. Chem., Int. Ed., 2008, 47(34), 6330–6337, DOI:10.1002/anie.200705982.
- R. Asadi and H. Mollasalehi, The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance, Anal. Biochem., 2021, 631, 114260, DOI:10.1016/j.ab.2021.114260.
- Y. Wang, Y. Han, F. Zhou, T. Fan and F. Liu, Simple Detection of DNA Methyltransferase with an Integrated Padlock Probe, Biosensors, 2022, 12(8), 569, DOI:10.3390/bios12080569.
- X. Peng, J. Zhu, W. Wen, T. Bao, X. Zhang and H. He, et al. Silver nanoclusters-assisted triple-amplified biosensor for ultrasensitive methyltransferase activity detection based on AuNPs/ERGO hybrids and hybridization chain reaction, Biosens. Bioelectron., 2018, 118, 174–180, DOI:10.1016/j.bios.2018.07.048.
- A. R. Connolly and M. Trau, Isothermal Detection of DNA by Beacon-Assisted Detection Amplification, Angew. Chem., Int. Ed., 2010, 49(15), 2720–2723, DOI:10.1002/anie.200906992.
- Y.-C. Du, S.-Y. Wang, X.-Y. Li, Y.-X. Wang, A.-N. Tang and D.-M. Kong, Terminal deoxynucleotidyl transferase-activated nicking enzyme amplification reaction for specific and sensitive detection of DNA methyltransferase and polynucleotide kinase, Biosens. Bioelectron., 2019, 145, 111700, DOI:10.1016/j.bios.2019.111700.
- H. Yuan, S. Wang, L.-j. Wang and C.-y. Zhang, A CpG methylation-powered dynamic three-dimensional-DNAzyme walker for single-molecule monitoring of multiple cytosine-C5 methyltransferases, Sens. Actuators, B, 2023, 390, 133959, DOI:10.1016/j.snb.2023.133959.
- L.-j. Wang, H. Liu, X.-F. Li, Y. Meng, J.-G. Qiu and C. Y. Zhang, Methylation-powered engineering of a dual-color light-up RNA nanosensor for label-free and ultrasensitive sensing of multiple DNA methyltransferases, Sens. Actuators, B, 2022, 371, 132524, DOI:10.1016/j.snb.2022.132524.
- Y. Yu, H. Zeng, Q. Wu, X. Jiang, C. Duan and J. Long, et al. A sensing strategy combining T7 promoter-contained DNA probe with CRISPR/Cas13a for detection of bacteria and human methyltransferase, Anal. Chim. Acta, 2022, 1227, 340266, DOI:10.1016/j.aca.2022.340266.
- Q. Zhang, Y. Wu, Q. Xu, F. Ma and C.-y. Zhang, Recent advances in biosensors for in vitro detection and in vivo imaging of DNA methylation, Biosens. Bioelectron., 2021, 171, 112712, DOI:10.1016/j.bios.2020.112712.
- Q. Zhang, X. Zhang, F. Ma and C.-y. Zhang, Advances in quantum dot-based biosensors for DNA-modifying enzymes assay, Coord. Chem. Rev., 2022, 469, 214674, DOI:10.1016/j.ccr.2022.214674.
- Y. Tao, M. Q. Li, J. S. Ren and X. G. Qu, Metal nanoclusters: novel probes for diagnostic and therapeutic applications, Chem. Soc. Rev., 2015, 44(23), 8636–8663, 10.1039/c5cs00607d.
- X. Yuan, Z. Luo, Y. Yu, Q. Yao and J. Xie, Luminescent Noble Metal Nanoclusters as an Emerging Optical Probe for Sensor Development, Chem. – Asian J., 2013, 8(5), 858–871, DOI:10.1002/asia.201201236.
- R. Jin, Atomically precise metal nanoclusters: stable sizes and optical properties, Nanoscale, 2015, 7(5), 1549–1565, 10.1039/c4nr05794e.
- M. Shamsipur, F. Molaabasi, S. Hosseinkhani and F. Rahmati, Detection of Early Stage Apoptotic Cells Based on Label-Free Cytochrome c Assay Using Bioconjugated Metal Nanoclusters as Fluorescent Probes, Anal. Chem., 2016, 88(4), 2188–2197, DOI:10.1021/acs.analchem.5b03824.
- H.-B. Wang, H.-D. Zhang, Y. Chen, K.-J. Huang and Y.-M. Liu, A label-free and ultrasensitive fluorescent sensor for dopamine detection based on double-stranded DNA templated copper nanoparticles, Sens. Actuators, B, 2015, 220, 146–153, DOI:10.1016/j.snb.2015.05.055.
- X. Mu, L. Qi, P. Dong, J. Qiao, J. Hou and Z. Nie, et al. Facile one-pot synthesis of L-proline-stabilized fluorescent gold nanoclusters and its application as sensing probes for serum iron, Biosens. Bioelectron., 2013, 49, 249–255, DOI:10.1016/j.bios.2013.05.019.
- Y. Noh, E.-J. Jo, H. Mun, Y.-d. Ahn and M.-G. Kim, Homogeneous and selective detection of cadmium ions by forming fluorescent cadmium-protein nanoclusters, Chemosphere, 2017, 174, 524–530, DOI:10.1016/j.chemosphere.2017.02.025.
- H.-B. Wang, A.-L. Mao, B.-B. Tao, H.-D. Zhang and Y.-M. Liu, Fabrication of multiple molecular logic gates made of fluorescent DNA-templated Au nanoclusters, New J. Chem., 2021, 45(9), 4195–4201, 10.1039/d0nj06192a.
- F. Zhou, H. Chen, T. Fan, Z. Guo and F. Liu, Fluorescence turn-off strategy for sensitive detection of DNA methyltransferase activity based on DNA-templated gold nanoclusters, Heliyon, 2023, 9(7), e17724, DOI:10.1016/j.heliyon.2023.e17724.
- G. V. Tarasova, T. N. Nayakshina and S. K. H. Degtyarev, Substrate specificity of new methyl-directed DNA endonuclease GlaI, BMC Mol. Biol., 2008, 9, 7, DOI:10.1186/1471-2199-9-7.
- I. Sarac and M. Hollenstein, Terminal Deoxynucleotidyl Transferase in the Synthesis and Modification of Nucleic Acids, ChemBioChem, 2019, 20(7), 860–871, DOI:10.1002/cbic.201800658.
- J. G. Herman and S. B. Baylin, Mechanisms of disease: Gene silencing in cancer in association with promoter hypermethylation, N. Engl. J. Med., 2003, 349(21), 2042–2054, DOI:10.1056/NEJMra023075.
- Z. Li, T. Pi, K. Yang, Z. Xia, Y. Feng and X. Zheng, et al. Label-free fluorescence strategy for methyltransferase activity assay based on poly-thymine copper nanoclusters engineered by terminal deoxynucleotidyl transferase, Spectrochim. Acta, Part A, 2021, 260, 119924, DOI:10.1016/j.saa.2021.119924.
- X. Luo, T. Kang, J. Zhu, P. Wu and C. Cai, Sensitivity-Improved SERS Detection of Methyltransferase Assisted by Plasmonically Engineered Nanoholes Array and Hybridization Chain Reaction, ACS Sens., 2020, 5(11), 3639–3648, DOI:10.1021/acssensors.0c02016.
- Y. Chen, H. Yi, Y. Xiang and R. Yuan, Commercial glucometer as signal transducer for simple evaluation of DNA methyltransferase activity and inhibitors screening, Anal. Chim. Acta, 2018, 1001, 18–23, DOI:10.1016/j.aca.2017.11.045.
- J. Hu, Y. Liu and C.-y. Zhang, Construction of a single quantum dot nanosensor with the capability of sensing methylcytosine sites for sensitive quantification of methyltransferase, Nanoscale, 2020, 12(7), 4519–4526, 10.1039/c9nr10376g.
- Z.-y. Wang, P. Li, L. Cui, Q. Xu and C.-y. Zhang, Construction of a Universal and Label-Free Chemiluminescent Sensor for Accurate Quantification of Both Bacteria and Human Methyltransferases, Anal. Chem., 2020, 92(19), 13573–13580, DOI:10.1021/acs.analchem.0c03303.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00215f |
| This journal is © The Royal Society of Chemistry 2025 |