Open Access Article
Open Access ArticleData-driven discovery of near-infrared type I photosensitizers for RNA-targeted tumor photodynamic therapy†
Wen
Chen
 *a,
Xiao-Qiong
Mao
a,
Xiao-Zhi
Wang
b,
Ya-Cong
Liao
c,
Xiao-Yue
Yin
b,
Hai-Long
Wu
*a,
Xiao-Qiong
Mao
a,
Xiao-Zhi
Wang
b,
Ya-Cong
Liao
c,
Xiao-Yue
Yin
b,
Hai-Long
Wu
 b,
Tai-Yi
Chen
a,
Meng-Qing
Liu
a,
Tong
Wang
b,
Tai-Yi
Chen
a,
Meng-Qing
Liu
a,
Tong
Wang
 *b and
Ru-Qin
Yu
*b and
Ru-Qin
Yu
 b
b
aKey Laboratory of Functional Organometallic Materials of College of Hunan Province, College of Chemistry and Materials Science, Hengyang Normal University, Hengyang 421008, P. R. China. E-mail: wenchen@hnu.edu.cn
bState Key Laboratory of Chemo and Biosensing, College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China. E-mail: wangtong@hnu.edu.cn
cDepartment of Ultrasound Diagnosis, The Second Xiangya Hospital, Central South University, Changsha 410011, P. R. China
First published on 14th July 2025
Abstract
Type I photodynamic therapy (PDT) offers a promising solution for treating tumors with inherent hypoxia, but developing near-infrared (NIR) type I photosensitizers (PSs) remains a significant challenge. In this work, we propose a data-driven multi-stage screening workflow to rapidly discover novel type I PSs for RNA-targeted tumor PDT. Two custom-built databases were developed: one for training a type I PS identification model (1-PS-GCN) and the other for artificial intelligence (AI)-assisted multi-stage screening. The top 10 candidates with potential type I photosensitizing properties, high synthetic accessibility and RNA-targeting capability were identified. Experts and AI reached a consensus to select the most promising type I PS (PYD). Ultimately, PYD was synthesized and its properties were verified through spectral, cellular, and in vivo PDT experiments. It showed satisfactory phototoxicity and significant tumor inhibition. Our approach establishes an intelligent discovery paradigm to accelerate the identification of NIR type I PSs for advanced PDT.
Introduction
Photodynamic therapy (PDT) is an emerging cancer treatment method that selectively destroys abnormal cells using photosensitizers (PSs) under specific wavelength light irradiation. It has garnered attention for its high selectivity, low toxicity, controllability, and low invasiveness.1–3 During PDT, PSs can be categorized into type I and type II based on their interaction mechanisms with oxygen. However, the effectiveness of conventional type II PDT is significantly constrained by its reliance on oxygen, which is compromised by the tumor's hypoxic microenvironment. This limitation prevents it from reaching the desired therapeutic outcome.4,5 In contrast, type I PDT operates independently of oxygen. Thanks to disproportionation or Fenton reactions, type I PSs can generate reactive oxygen species, such as O2−˙, H2O2, or OH˙, even in the presence of severe hypoxia (2% O2), thus effectively eliminating tumor cells.6–9 In recent years, researchers have developed some targeted NIR type I PSs with excellent performance, such as the ENBS series of NIR type I PSs developed by Li et al. for tumor PDT research.10,11 A series of type I PSs based on aggregation-induced emission (AIE) luminescence modules were developed by the team of Liu et al.12 A class of type I BODIPY PSs that can exclusively generate superoxide anion radicals was developed by Teng et al.13 Our research team has engineered a series of acridine derivatives to develop a targeted G-quadruplex NIR type I PS that triggers anti-tumor immunity and effectively treats cold tumors.14 These studies provide new insights into tumor PDT in hypoxic microenvironments.Although type I PDT is a very promising treatment for tumors under hypoxic conditions, the development mechanism of type I PSs with NIR absorption properties still needs improvement. Currently, Chen et al. have proposed a strategy to successfully design type I PSs by enhancing the molecular electron-donating ability and the intersystem crossing (ISC) capability.15 Wan et al. have introduced energy level regulation into anionic-π+ type AIE-active luminogens (AIEgens) to successfully design type I PSs.16 Additionally, time-dependent density functional theory (TD-DFT) can be combined to increase the intramolecular charge transfer (ICT) ability, thereby reducing the energy gap (ΔEst) between singlet and triplet states, enhancing the ISC capability to ensure sufficient triplet state energy is produced, and creating a charge-rich environment to provide electrons to the excited PS, thus designing type I PSs.17–20 These principles are suitable for explaining the generation mechanism of type I PSs, but their design principles require further research. Moreover, the development of NIR type I PSs with specific properties/functions is still at a bottleneck stage, severely hindering the clinical application of PDT for tumors.
Over the past decade, artificial intelligence (AI), particularly deep learning (DL) technology, has demonstrated enormous application potential. In 2024, the Nobel Prizes in Chemistry and Physics were both awarded for work related to AI.21 These technologies can rapidly explore chemical space and accurately capture patterns and principles hidden in data, thereby accelerating molecular discovery,22,23 structure prediction,24,25 property prediction,26 and drug design.27,28 For example, Stokes et al. trained a deep neural network capable of predicting molecules with antibacterial activity, leading to the discovery of eight antibacterial compounds structurally distant from known antibiotics.29 Subsequently, Wong et al. used a graph-based interpretable DL model to identify a class of compounds with novel structures that exhibit antibacterial activity against methicillin-resistant S. aureus.30 Smer-Barreto et al. developed a cost-effective machine learning (ML) algorithm trained solely on published data, which identified three senolytic drugs with efficacy comparable to known anti-aging drugs, reducing the cost of drug screening by several hundredfold.31 These developments herald the Fourth Paradigm of science, i.e., data-intensive knowledge discovery, offering unprecedented opportunities for the efficient discovery of PSs in PDT.
In this study, a workflow for discovering type I PSs with AI assistance is reported for the first time. A type I PS molecule named PYD was identified, exhibiting excellent performance in in vivo tumor PDT. Two distinct data sets were constructed from scratch through literature mining: the first data set (data set 1) was used to train a novel DL model (1-PS-GCN) for predicting whether a molecule is a type I PS. Unlike traditional machine learning approaches that rely on single-modal features, 1-PS-GCN integrates molecular graph topologies, structural fingerprints, and physicochemical descriptors into a unified DL framework, enabling more comprehensive molecular representation and improved classification performance. The second data set (data set 2), containing 2768 NIR small molecules, was employed for multi-stage screening of compounds with the desirable properties, such as NIR excitation/emission wavelength, ease of synthesis, and RNA targeting. From this screening, the top 10 candidate compounds were identified, with molecule 10 (PYD) achieving expert-AI consensus as the best candidate. The type I PS properties of PYD were further explored through interpretability analysis, and its RNA-binding capability was confirmed via molecular docking experiments. Using an innovative strategy, PYD was synthesized and subjected to a series of wet-lab experiments, including absorption spectroscopy analysis, fluorescence spectroscopy analysis and electron paramagnetic resonance (EPR) spectroscopy analysis. Finally, PYD was successfully applied for RNA-targeted ablation of tumors in both cellular and in vivo tumor PDT.
Results and discussion
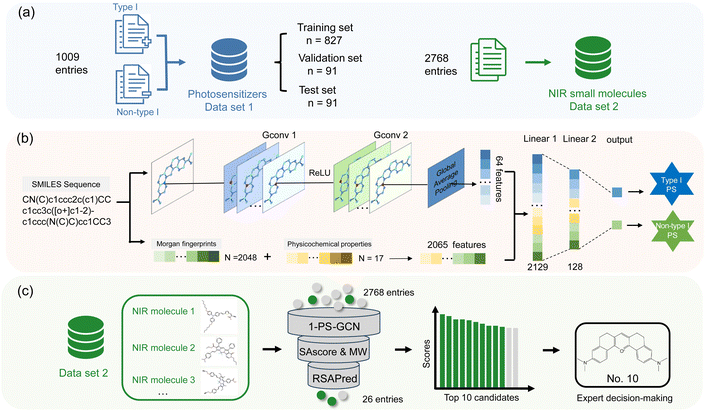
Data sets for PS discovery
Our goal is to develop a data-driven AI model capable of accurately identifying type I PSs and subsequently screening them from a large data set of NIR small molecules. To achieve this, two data sets were constructed from scratch: data set 1 for training the data-driven AI model to identify type I PSs, and data set 2 for high-throughput screening. As shown in Fig. 1a, in the PS data set (data set 1), we collected 1009 unique small-molecule PSs from the relevant literature published over the past two decades,4,8,32 containing 224 type I PSs (positive) and 785 non-type I PSs (negative). The latter were selected using the Nearest of Kin (NoK) concept and consist entirely of type II PSs. Moreover, we only considered small-molecule PSs, excluding complexes and polymers. The molecules in data set 1 were randomly split into training, validation, and test sets in an 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The training set was used for establishing the 1-PS-GCN model, the validation set for hyperparameter tuning, and the test set for evaluating model performance. Data set 2, an NIR small molecule data set (2768 compounds), was constructed by expanding existing databases and conducting supplementary literature reviews.33–41 Notably, most of them had not been reported as PSs. Detailed distributions of molecular weights and physicochemical properties for data set 2 are provided in Fig. S1.† The Morgan fingerprints, physicochemical descriptors, and molecular graph representations of all molecules were calculated before modeling (ESI and Table S1†).
1 ratio. The training set was used for establishing the 1-PS-GCN model, the validation set for hyperparameter tuning, and the test set for evaluating model performance. Data set 2, an NIR small molecule data set (2768 compounds), was constructed by expanding existing databases and conducting supplementary literature reviews.33–41 Notably, most of them had not been reported as PSs. Detailed distributions of molecular weights and physicochemical properties for data set 2 are provided in Fig. S1.† The Morgan fingerprints, physicochemical descriptors, and molecular graph representations of all molecules were calculated before modeling (ESI and Table S1†).
Machine learning models for screening type I PSs
To classify type I PSs and non-type I PSs, we developed six binary classification models, including three classical ML models: k-nearest neighbor (KNN), random forest (RF), and support vector machine (SVM), as well as three DL models: the fingerprint-enhanced graph convolutional network for type I PS recognition (1-PS-GCN), deep neural network (DNN), and convolutional neural network (CNN). Details of these models and hyperparameter optimization are summarized in the ESI and Tables S2–S4.† The 1-PS-GCN is a novel DL model we developed, which integrates molecular Morgan fingerprints with an advanced graph convolutional network. Its architecture, shown in Fig. 1b, processes molecular simplified molecular input line entry system (SMILES) sequences by utilizing the RDKit package42 to extract molecular graph information, Morgan fingerprints and 17 physicochemical properties. Two graph convolutional layers, combined with global average pooling, are used to extract molecular graph features. These features, together with Morgan fingerprints and physicochemical properties, are concatenated into a 2129-dimensional feature vector, which is subsequently processed through fully connected layers for classification.In summary, the 1-PS-GCN model enhances classification accuracy by integrating multimodal molecular information. The related code is publicly available as open-source (https://github.com/TWang-93/1-PS-GCN). Model performance metrics, including classification accuracy, precision, and the area under the receiver operating characteristic curve (ROC-AUC), were used to evaluate its performance. As shown in Fig. 2a–c, the 1-PS-GCN achieved the highest classification accuracy (0.8791), along with superior precision and ROC-AUC values (both exceeding 0.9). Accordingly, the 1-PS-GCN model was selected for the subsequent screening and discovery of type I PSs.
Procedure for the NIR type I PS discovery
Researchers often aim to design PSs with multiple desirable properties to meet practical application requirements. In this study, apart from the fundamental NIR characteristics, we also sought PSs that are easy to synthesize and capable of binding to RNA to enhance the PDT efficacy against tumors. Thus, we implemented a multi-stage screening process to discover PSs with multiple desired properties (Fig. 1c). The multi-stage screening process began with the 1-PS-GCN model, which identified 782 type I PSs from 2768 molecules in Data set 2, leaving the remaining 1986 classified as non-type I PSs. Fig. 3a illustrates the data volume changes across each screening stage, while Fig. 3e visualizes the chemical space of Data set 2 using t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction based on Morgan fingerprints. This visualization demonstrates the AI screening process refining a broad chemical space into a focused subset of interest. Type I and non-type I PSs exhibit distinct distributions in chemical space, as shown in Fig. 3e, reflecting structural differences that facilitate the construction of accurate classification models. In the second stage, molecules were screened for synthetic accessibility using thresholds of synthetic accessibility score43 (SAscore) < 3.5 and molecular weight (MW) < 400. Fig. 3b and c depict the SAscore and MW distributions of the 782 molecules, highlighting the selected ranges. The threshold of 3.5 was chosen based on the distribution of SAscore in our dataset, where it corresponds to a local maximum. After this stage, 60 easily synthesizable molecules remained. Recent studies have shown that certain RNAs (such as rRNA, mRNA, and G4s RNA) are specifically enriched in tumor cells. Photosensitizers (PS) capable of RNA-targeted activation can specifically generate reactive oxygen species (ROS) in RNA-rich regions, thereby enhancing precise and efficient killing of tumor cells.14,20,44 Recognizing the therapeutic potential of RNA-binding molecules for PDT therapy, we employed the RSAPred45 platform to predict the log-scale dissociation constant (pKd) values of the 60 molecules. 26 molecules with pKd > 8 were retained (Fig. 3d) and ranked based on their scores. As the ML models provide probabilistic predictions, expert evaluation was necessary for the final selection. Two experts specializing in PSs and small-molecule fluorescent probes assessed the top 10 candidates for resistance to photobleaching, excitation wavelength, and structural uniqueness (see the ESI and Fig. S2†). Based on this expert-AI collaboration, molecule No. 10 (PYD) was selected for its rigid, photobleaching-resistant structure and similarity to the commercial RNA detection reagent pyronine, suggesting that it has the greatest potential to become a high-performance PS.Model interpretability analysis
DL models are often regarded as “black-box” tools due to their impressive predictive performance but limited interpretability. Uncovering the reasoning behind their decisions remains a significant challenge. In this study, we attempted to explore how 1-PS-GCN identifies type I PSs using interpretability methods. The challenge was amplified by the model's hybrid architecture, which integrates molecular fingerprints and graph-based information.Using the selected PYD molecule as an example, we calculated the importance of individual features within its Morgan fingerprints (Fig. 2d). Features 10, 326, and 1376 exhibited higher importance scores, corresponding to substructures identified in Fig. 2e. All three features were linked to the pyran structure, likely due to the oxygen cation's strong electron-withdrawing effect, which enhances the molecule's intramolecular charge transfer (ICT) capability. This structural characteristic appears to promote the formation of type I PSs, aligning with findings in the literature.46
Additionally, graph interpretability methods were employed to evaluate the importance of each node (i.e., atom) in the molecular graph (Fig. 2f). The most critical graph-based features were associated with the molecule's conjugated structure, including properties such as if the atom. is in a ring (cyclic structure), sp2 (planar configuration with delocalized π electrons), total degree (3) (the atom with three chemical bonds), and If the atom is aromatic (aromatic system). These findings suggest that the graph component of the model recognizes the necessity of a basic conjugated structure for producing molecular fluorescence in type I PSs. This interpretability analysis provided preliminary insights into why the model classified the PYD molecule as a type I PS. From a data-driven perspective, it offers a promising approach to exploring the mechanisms underlying the formation of type I PSs.
To provide an intuitive demonstration of the model's discrimination mechanism and its decision boundaries in feature space, we extracted high-dimensional feature representations from the 128-dimensional embedding layer of the 1-PS-GCN model and performed two-dimensional visualization using the t-SNE algorithm, as shown in Fig. S3.† In this figure, four types of samples are distinctly color-coded: true positives (TP, red), true negatives (TN, blue), false positives (FP, yellow), and false negatives (FN, green), comprehensively presenting the classification decision distribution in the feature space.
The visualization reveals that the model forms relatively clear cluster structures in the t-SNE projection, with type I and non-type I samples demonstrating good overall separation. The majority of TP and TN samples form distinct clusters, indicating that the model can effectively learn and differentiate structural features of different categories in the high-dimensional feature space. To further strengthen the interpretability of the decision boundaries, we specifically selected several representative samples near the decision boundaries and visualized their molecular structures. The upper left section of the figure displays two molecular structures: one TP sample and one TN sample. Despite sharing certain structural similarities in their frameworks, the model successfully identified their critical differences and made correct classifications, demonstrating its capability to discern subtle structural features. Additionally, three molecules shown in the lower right corner are also located near the decision boundaries. These three structurally diverse molecules would be challenging to classify based solely on empirical knowledge. Nevertheless, our model effectively determined their photosensitization types, indicating its generalization ability in capturing potential structure–function relationships.
In vitro characterization
To verify that the PYD molecules selected by the AI-expert consensus possess RNA-binding and type I PS properties, we successfully synthesized the PYD molecule through condensation reaction under strong acid conditions (ESI and Scheme S1†). All intermediates and final products were characterized using 1H NMR, 13C NMR, and ESI-MS spectroscopy (Fig. S17–S23†). After obtaining the PYD molecule, we investigated whether it possesses properties similar to pyronine, a commercial nucleic acid reagent. First, we studied the photophysical properties of PYD. UV-vis absorption spectroscopy analysis revealed that PYD exhibits a strong absorption peak in the range of 500 to 700 nm. The absorption peak was red-shifted by the addition of RNA, with the maximum absorption peak around 640 nm (Fig. 4a), indicating that PYD interacted with RNA. Subsequently, we investigated the alterations in fluorescence intensity of PYD before and after its interaction with varying concentrations of RNA. As depicted in Fig. 4b and c, the fluorescence of PYD in PBS was initially very weak prior to the addition of RNA. However, upon the introduction of RNA, the fluorescence emission peak of PYD at 695 nm gradually intensified. Within the RNA concentration range of 0 to 0.25 mg mL−1, the fluorescence intensity of PYD exhibited a linear correlation with the RNA concentration, achieving a coefficient of determination (R2) value of 0.98. The fluorescence assay results further suggested that PYD may bind to the hydrophobic pocket (a low-polarity region) of RNA. Subsequently, we validated this hypothesis through spectroscopic experiments conducted in different solvents, with the results shown in Fig. S4.† In low-polarity solvents (e.g., dichloromethane, DCM), both UV absorption and fluorescence emission spectra exhibited varying degrees of red shift, with the fluorescence emission intensity reaching its maximum in DCM. This indicates that the mechanism by which RNA activates PYD fluorescence is likely related to its binding with the hydrophobic pocket. However, as the water content increases, the fluorescence gradually weakens, suggesting that PYD has poor dispersibility in aqueous solutions and is prone to aggregation-caused quenching (ACQ).44,47,48 Molecular docking experiments revealed that PYD was embedded within the nucleic acid structure, forming not only a salt bridge with G22 on the nucleic acid but also hydrogen bonding with G17. These interactions constitute the basis for their stable binding (Fig. 4d).To verify that PYD possesses the properties of type I PS, we conducted real-time illumination (660 nm, 10 mW cm−2) on both PYD and PYD + RNA systems using a general fluorescent indicator for ROS 2′,7′-dichlorodihydrofluorescein (DCFH). The experimental results indicated that the fluorescence emission peak intensity at 530 nm in PYD + RNA systems, which contained DCFH, gradually increased with the duration of illumination, whereas in the PYD system, the fluorescence intensity of DCFH only exhibits a slight change with the increase in illumination time (Fig. S5†). Next, we conducted a more detailed investigation using various specificity indicators. We employed the superoxide anion (O2−˙) indicator dihydrorhodamine 123 (DHR123), the specific hydroxyl radical (OH˙) indicator hydroxyphenyl fluorescein (HPF), the singlet oxygen (1O2) indicator singlet oxygen sensor green (SOSG) and 9, 10-anthracenediyl-bis (methylene) dimalonic acid (ABDA), to perform real-time illumination (660 nm, 10 mW cm−2) on both PYD and PYD + RNA systems. It was observed that the fluorescence intensity of the ROS probe in the PYD and PYD + RNA systems, when treated with DHR123 and HPF reagents, gradually increased with the duration of illumination. Moreover, the fluorescence intensity of various indicators in the RNA-containing PYD system was found to be stronger than that in the system with PYD alone (Fig. 4e, S6 and S7†). However, the UV and fluorescence intensity of the PYD and PYD + RNA systems with ABDA and SOSG reagents remained unchanged (Fig. S8 and S9†), indicating that ROS produced by illuminating the PYD and PYD + RNA systems were not the conventional PS-generated 1O2, and that RNA enhances the production of ROS by PYD. The in vitro indicator detection experiments revealed that PYD could efficiently generate ROS following exposure to light, suggesting its promise for type I PDT.
To confirm the generation of O2−˙ during the PDT process by PYD + RNA, we used 5,5-dimethyl-1-pyrroline N-oxide (DMPO) as a radical trap and further employed electron paramagnetic resonance (EPR) spectroscopy for testing. The experimental results indicated that after irradiation of PYD + RNA, a characteristic peak in the paramagnetic signal of the DMPO-O2−˙ complex could be observed, and this EPR signal was consistent with the well-known signal of O2−˙ generator KO2. In the control experiment EPR spectra of PYD + RNA under dark conditions, the signal peak of DMPO-O2−˙ adducts could not be detected, which proved the mechanism of type I PDT of PYD (Fig. 4f). In order to gain a deeper understanding of PYD's strong ROS generation capability upon light irradiation in PYD, we carried out TD-DFT calculations at the B3LYP/6311++G (d, p) level. The calculated energy gap between the lowest unoccupied molecular orbital (LUMO) and the highest occupied molecular orbital (HOMO) of the PYD in the S0 state is 2.55 eV, and the corresponding energy gap in the S1 state is 2.36 eV (Fig. 4g). According to the Kohn–Sham frontier orbital analysis, an energy gap below 0.3 eV favors the triggering of the ISC. The ΔEST (singlet–triplet energy level splitting) value of PYD is 0.16 eV, which is significantly less than 0.3 eV (Fig. 4h). These data combined suggest that PYD exhibits a strong ROS generation capability under light irradiation. Comprehensive results from multiple ROS detection experiments and EPR spectroscopy confirm that PYD is a type I photosensitizer.
An ideal PS should possess good photobleaching resistance and specific selectivity. Subsequently, we investigated the photobleaching properties, fluorescence quantum yield, and sensitivity of PYD both in the presence and absence of RNA. As depicted in Fig. S10,† the real-time illumination of PYD after 5400 s, both in the presence and absence of RNA, demonstrated ideal photobleaching resistance. In contrast, the fluorescence intensity of the control commercial dye cyanine 5 (Cy5) steadily declined, thereby further emphasizing PYD's superior photobleaching resistance. Subsequently, we investigated the fluorescence lifetimes of PYD, which were found to be 1.86 ns in the absence of RNA and 2.44 ns in its presence (Fig. S11†). Additionally, the fluorescence quantum yields were determined to be 0.02 and 0.15, respectively. These results indicate that after binding with RNA, the lifetime of the excited state of PYD is prolonged, the energy level remains stable, non-radiative decay is reduced, and the fluorescence quantum yield is enhanced, thereby improving the ability of PYD molecules to generate ROS. Subsequently, we conducted in vitro selective fluorescence activation tests on different structural nucleic acid chains (Table S5†), small molecules, and ions. As shown in Fig. 5a and b, the fluorescence activation of PYD by other nucleic acid chains, small molecules, and ions was almost negligible, but RNA can effectively activate the fluorescence of PYD. These excellent photophysical properties suggest that PYD is an ideal type I PS for RNA fluorescence activation.
Cellular assays
To verify that the fluorescence of PYD can be selectively activated by cellular RNA, we conducted dual fluorescence channel imaging of living cells using PYD and Hoechst. Confocal fluorescence images revealed that the breast cancer cell line (MCF-7) emitted strong red fluorescence in the cytoplasm and nucleolus, while the nucleus exhibited blue fluorescence. When MCF-7 cells were treated with DNase I, a significant loss of blue fluorescence in the nucleus was observed, whereas the red fluorescence in the cells remained. Following treatment with RNase A, the red fluorescence in MCF-7 cells almost completely disappeared, but the blue fluorescence in the nucleus remained largely unchanged (Fig. 5c–e). These results indicated that PYD can activate fluorescence upon binding to RNA within cells. Subsequently, we utilized MCF-7 cells to investigate the capacity of PYD to target RNA in living cells for PDT. Initially, we conducted imaging studies to examine the production of ROS by PYD in cells upon light irradiation. As depicted in Fig. 5f, MCF-7 cells exhibited strong green fluorescence from activated 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) in the light-irradiated group. In contrast, the non-irradiated group and MCF-7 cells pre-treated with N-acetylcysteine (NAC: a potent antioxidant) showed no green fluorescence and significantly reduced fluorescence, respectively. These results indicated that ROS are produced within cells upon light irradiation. Cells treated with the PYD and O2−˙ indicator hydroethidine (DHE) emitted strong red fluorescence after light exposure, suggesting that O2−˙ is produced within cells upon light irradiation. Additionally, we used the OH˙ indicator HPF in conjunction with PYD and observed specific green fluorescence in cells after light exposure, which was markedly attenuated by treatment with the superoxide dismutase (SOD) inhibitor 2-methoxyestradiol (2ME2). Cells treated with the 1O2 fluorescent probe SOSG showed no fluorescence before and after light exposure. These results suggest that light irradiation of PYD produces O2−˙ or that O2−˙ produced during light exposure is catalytically dismutated into OH˙ by intracellular SOD, which is consistent with previous reports. Collectively, these findings indicate that PYD can generate highly reactive O2−˙ and OH˙ within living cells.Subsequently, we investigated the ability of PYD to ablate tumor cells during PDT. Firstly, we explored the phototoxicity of different concentrations of PYD on MCF-7 and mouse breast cancer cells (4T1) under normoxic conditions. The experimental results indicated that the cells exhibited dose-dependent cytotoxicity following light exposure (660 nm, 100 mW cm−2, 10 min), whereas the unexposed control group demonstrated good biocompatibility. The IC50 values for MCF-7 and 4T1 cells were determined to be 0.75 and 0.83 μM, respectively (Fig. 6a, b and S12†). Simultaneously, we investigated the phototoxicity of PYD in cells cultured under hypoxic conditions (2% O2). The experimental results demonstrated that PYD also displays remarkable phototoxicity in hypoxic environments (Fig. 6a and b), with minor increases in the IC50 values for MCF-7 and 4T1 cells, which were 0.76 and 0.90 μM, respectively (Fig. S12†). These experimental results suggest that PYD can effectively ablate tumor cells upon light irradiation. Subsequently, we conducted live/dead cell staining analysis on cells post PDT using calcein AM and propidium iodide (PI), as depicted in Fig. 6c. Prior to irradiation, there was strong fluorescence in the calcein AM channel and no fluorescence in the PI channel. As irradiation time increased, the fluorescence in the calcein AM channel gradually weakened until it essentially disappeared, whereas the red fluorescence channel of PI became very bright after 5 min of irradiation. After 10 min of irradiation, the calcein AM channel exhibited almost no fluorescence, and the cell death process could be directly observed under bright field illumination. Compared with conventional PSs, PYD significantly induced cell death within a short period of time, suggesting that the AI-expert consensus RNA-targeted NIR type I PS PYD has an efficient PDT process.
To further illustrate the potent ablation effect of RNA-targeted type I PYD on tumor cells, we next examined the phototoxic mechanism of PYD on cells. We utilized Annexin V-FITC and PI to stain MCF-7 cells post-photodynamic therapy, and the flow cytometry analysis revealed that in the cell group treated with PYD and exposed to light for 10 min, the majority of cells succumbed through necrosis at 88.4% (PI+), while a smaller fraction died via apoptosis (6.7%, Annexin V+ and PI−). Moreover, the proportion of necrotic cells notably increased with extended irradiation time (Fig. 6d). In conclusion, these findings suggest that PYD possesses significant phototoxic properties and holds promise for tumor ablation in hypoxic microenvironments.
Tumor mouse model and PDT
To further investigate the potential of the RNA-targeted type I PS PYD for in vivo anti-tumor therapy, we established tumor-bearing mice by injecting 4T1 cells into the subcutaneous tissue of the abdominal cavity of BALB/c mice (Fig. 7a). The capability of PYD to image tumors in vivo was initially explored through intratumoral injection. As depicted in Fig. 7b, the fluorescence signal at the tumor site was quite intense, and the fluorescence intensity persisted at a high level 6 h post-injection, indicating that PYD can be activated by RNA within tumor tissues to emit fluorescence, exhibiting outstanding tumor imaging properties. To validate the activation of PYD by intratumoral RNA, fluorescence imaging analysis of enzymatically digested tumor tissues demonstrated a significant decrease in fluorescence after RNase A treatment (Fig. S13†), further indicating that intratumoral RNA can fluorescence-activate PYD. The pharmacokinetic profile of PYD in mice is crucial for tumor photodynamic therapy (PDT) experiments. We conducted an in-depth analysis of PYD's pharmacokinetic characteristics in mice, with experimental results showing that PYD is primarily cleared through the liver (blood half-life ≈ 3.6 min) (Fig. S14†). This finding aligns with most reported small-molecule drugs.49,50Subsequently, we evaluated the effect of PYD on PDT. BALB/c mice with 4T1 tumors were randomly assigned to four groups (n = 4). When the tumor size reached approximately 50 mm3, the mice were injected intra-tumorally with PBS (group II) and PYD (group IV). They were then subjected to light treatment experiments, which resulted in a significant reduction of tumor growth rate in group IV, treated with PYD in combination with light, as compared with the control group. The control group was injected with the same PBS (group I) and PYD (group III) without light treatment (Fig. 7c). However, the tumor growth rate in group II, which was injected with PBS and subjected to light treatment, remained unaffected, indicating that combining the RNA-targeted activated type I PS PYD with phototherapy exhibits a significant and highly efficient effect in inhibiting tumor growth, demonstrating substantial potential value in the field of tumor therapy. Ultimately, we dissected the tumors from mice after a 14-day treatment period, and the photographs of the dissected tumors further demonstrated that the PDT-treated group exhibited an anti-tumor effect (Fig. 7d). Meanwhile, the weight of mice in all groups remained within the normal range, indicating that the treatment has good biosafety (Fig. 7e). Furthermore, hematoxylin and eosin (H&E) staining of tumor sections revealed the highest levels of tumor cell death and cavity formation in tumor samples from group IV (Fig. 7f). Additional H&E staining of major organ sections, including heart, liver, spleen, lung, and kidney, did not exhibit abnormal cellular morphology or tissue damage, indicating that our treatment did not cause significant damage to these organs (Fig. S15†). In addition, the results of the blood biochemistry tests were within normal limits. All of this indicates that PYD has an excellent biosafety profile (Fig. 7g–j and S16†). In summary, these results suggested that PDT mediated by PYD shows effective anti-tumor treatment in vivo.
Conclusions
In this study, we proposed a data-driven multi-stage screening workflow to rapidly discover type I PSs with desirable properties. Two comprehensive data sets were constructed: one containing 1009 molecules to train a type I PS identification model, and the other containing 2768 NIR small molecules for screening using the newly developed 1-PS-GCN DL model. The 1-PS-GCN demonstrated superior classification accuracy (0.8791) on the test set, outperforming baseline models. Using this model, we successfully identified a set of candidate molecules with potential NIR type I PS properties. Through multi-stage screening considering synthesizability, molecular weight, and RNA-targeting properties, experts and AI reached a consensus, identifying PYD as the most promising type I PS. Model interpretability studies indicated that the type I PS properties of PYD might be related to its pyran structure and conjugated system, enhancing its ICT capability. Molecular docking experiments further validated the RNA-binding properties of PYD.We successfully synthesized PYD based on a novel approach, and validated its properties through a series of wet-lab experiments. In vitro experiments demonstrated changes in PYD's photophysical properties upon interaction with RNA, such as red-shifted absorption peaks and enhanced fluorescence intensity, which showed a linear relationship with RNA concentration, confirming its RNA-binding capability. ROS fluorescence probes and EPR spectroscopy confirmed that PYD could efficiently produce O2−˙ and OH˙ upon irradiation, demonstrating its potential for type I PDT. Moreover, PYD exhibited excellent photophysical properties, including photobleaching resistance, high fluorescence quantum yield, and remarkable selectivity towards RNA, making it an ideal RNA fluorescence-activatable type I PS. In cellular studies, PYD was capable of binding RNA in living cells to activate fluorescence and generating highly reactive O2−˙ and OH˙ species. It exhibited significant phototoxicity against tumor cells, effectively ablating them through necrosis and apoptosis mechanisms. Notably, PYD also showed strong tumor ablation potential under hypoxic conditions. In a tumor-bearing mouse model, PYD demonstrated excellent in vivo tumor imaging capability and significantly inhibited tumor growth through PDT. Furthermore, it exhibited good biocompatibility with no significant damage to major organs.
In summary, the AI-based method we proposed offers an efficient approach for discovering NIR type I PSs. As a newly discovered type I PS, PYD holds significant potential in the field of tumor PDT. It is expected to strongly support the further development of efficient photodynamic therapies and promote advancements in the field of tumor treatment.
Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Central South University and approved by the Animal Ethics Committee of Central South University (No. CSU-2024-0144).Data availability
The data supporting this article has been included as part of the ESI.† All data and custom codes used in this work are also open source and can be accessed without restriction at https://github.com/TWang-93/1-PS-GCN.Author contributions
W. C. and T. W. provided the research direction and contributed the basic framework and feasible technical route of the project. W. C., T. W., and X. Y. wrote the first draft. R. Y., H. W. and M. L. supervised the entire experiment and guided the research process. X. M., Y. L., T. C. and X. Y. were responsible for data collection and preprocessing. X. W. and T. W. conducted the AI modeling and algorithm development. All authors reviewed the manuscript and approved the submission.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 22304047 and 22204049), the Key Scientific Research Project of Hunan Provincial Department of Education (No. 23A0028) and Hengyang Science and Technology Innovation Programme Project (No. 202330046097).References
- S. S. Lucky, K. C. Soo and Y. Zhang, Nanoparticles in Photodynamic Therapy, Chem. Rev., 2015, 115, 1990–2042 CrossRef CAS PubMed.
- D. E. J. G. J. Dolmans, D. Fukumura and R. K. Jain, Photodynamic therapy for cancer, Nat. Rev. Cancer, 2003, 3, 380–387 CrossRef CAS PubMed.
- J. P. Celli, B. Q. Spring, I. Rizvi, C. L. Evans, K. S. Samkoe, S. Verma, B. W. Pogue and T. Hasan, Imaging and Photodynamic Therapy: Mechanisms, Monitoring, and Optimization, Chem. Rev., 2010, 110, 2795–2838 CrossRef CAS PubMed.
- T. C. Pham, V.-N. Nguyen, Y. Choi, S. Lee and J. Yoon, Recent Strategies to Develop Innovative Photosensitizers for Enhanced Photodynamic Therapy, Chem. Rev., 2021, 121, 13454–13619 CrossRef CAS PubMed.
- X. Zhao, J. Liu, J. Fan, H. Chao and X. Peng, Recent progress in photosensitizers for overcoming the challenges of photodynamic therapy: from molecular design to application, Chem. Soc. Rev., 2021, 50, 4185–4219 RSC.
- M. He, Z. Ma, L. Zhang, Z. Zhao, Z. Zhang, W. Liu, R. Wang, J. Fan, X. Peng and W. Sun, Sonoinduced Tumor Therapy and Metastasis Inhibition by a Ruthenium Complex with Dual Action: Superoxide Anion Sensitization and Ligand Fracture, J. Am. Chem. Soc., 2024, 146, 25764–25779 CrossRef CAS PubMed.
- R. Zhang, H. Xu, Y. Yao, G. Ran, W. Zhang, J. Zhang, J. L. Sessler, S. Gao and J.-L. Zhang, Nickel(II) Phototheranostics: A Case Study in Photoactivated H2O2-Enhanced Immunotherapy, J. Am. Chem. Soc., 2023, 145, 23257–23274 CrossRef CAS.
- M. Li, Y. Xu, X. Peng and J. S. Kim, From Low to No O2-Dependent Hypoxia Photodynamic Therapy (hPDT): A New Perspective, Accounts Chem. Res., 2022, 55, 3253–3264 CrossRef CAS PubMed.
- K.-X. Teng, L.-Y. Niu and Q.-Z. Yang, Supramolecular Photosensitizer Enables Oxygen-Independent Generation of Hydroxyl Radicals for Photodynamic Therapy, J. Am. Chem. Soc., 2023, 145, 4081–4087 CrossRef CAS.
- M. Li, J. Xia, R. Tian, J. Wang, J. Fan, J. Du, S. Long, X. Song, J. W. Foley and X. Peng, Near-Infrared Light-Initiated Molecular Superoxide Radical Generator: Rejuvenating Photodynamic Therapy against Hypoxic Tumors, J. Am. Chem. Soc., 2018, 140, 14851–14859 CrossRef CAS PubMed.
- M. Li, T. Xiong, J. Du, R. Tian, M. Xiao, L. Guo, S. Long, J. Fan, W. Sun, K. Shao, X. Song, J. W. Foley and X. Peng, Superoxide Radical Photogenerator with Amplification Effect: Surmounting the Achilles' Heels of Photodynamic Oncotherapy, J. Am. Chem. Soc., 2019, 141, 2695–2702 CrossRef CAS PubMed.
- L. Liu, C. Li, J. Gong, Y. Zhang, W. Ji, L. Feng, G. Jiang, J. Wang and B. Z. Tang, A Highly Water-Soluble Aggregation-Induced Emission Luminogen with Anion-π+ Interactions for Targeted NIR Imaging of Cancer Cells and Type I Photodynamic Therapy, Angew. Chem., Int. Ed., 2023, 62, e202307776 CrossRef CAS.
- K.-X. Teng, W.-K. Chen, L.-Y. Niu, W.-H. Fang, G. Cui and Q.-Z. Yang, BODIPY-Based Photodynamic Agents for Exclusively Generating Superoxide Radical over Singlet Oxygen, Angew. Chem., Int. Ed., 2021, 60, 19912–19920 CrossRef CAS PubMed.
- W. Chen, Y. Zhang, H.-B. Yi, F. Wang, X. Chu and J.-H. Jiang, Type I Photosensitizer Targeting G-Quadruplex RNA Elicits Augmented Immunity for Cancer Ablation, Angew. Chem., Int. Ed., 2023, 62, e202300162 CrossRef CAS PubMed.
- D. Chen, Z. Wang, H. Dai, X. Lv, Q. Ma, D.-P. Yang, J. Shao, Z. Xu and X. Dong, Boosting O2˙− Photogeneration via Promoting Intersystem-Crossing and Electron-Donating Efficiency of Aza-BODIPY-Based Nanoplatforms for Hypoxic-Tumor Photodynamic Therapy, Small Methods, 2020, 4, 2000013 CrossRef CAS.
- Q. Wan, R. Zhang, Z. Zhuang, Y. Li, Y. Huang, Z. Wang, W. Zhang, J. Hou and B. Z. Tang, Molecular Engineering to Boost AIE-Active Free Radical Photogenerators and Enable High-Performance Photodynamic Therapy under Hypoxia, Adv. Funct. Mater., 2020, 30, 2002057 CrossRef CAS.
- Z. Wang, C. Wu and W. Liu, NAC-TDDFT: Time-Dependent Density Functional Theory for Nonadiabatic Couplings, Accounts Chem. Res., 2021, 54, 3288–3297 CrossRef CAS PubMed.
- J. Yu, J. Wu, J. Huang, C. Xu, M. Xu, C. Z. H. Koh, K. Pu and Y. Zhang, Hypoxia-tolerant polymeric photosensitizer prodrug for cancer photo-immunotherapy, Nat. Commun., 2025, 16, 153 CrossRef PubMed.
- J. An, S. Tang, G. Hong, W. Chen, M. Chen, J. Song, Z. Li, X. Peng, F. Song and W.-H. Zheng, An unexpected strategy to alleviate hypoxia limitation of photodynamic therapy by biotinylation of photosensitizers, Nat. Commun., 2022, 13, 2225 CrossRef CAS PubMed.
- Y. Jiang, S. Huang, H. Ma, J. Weng, X. Du, Z. Lin, J. Kim, W. You, H. Zhang, D. Wang, J. S. Kim and H. Sun, RNA-Activatable Near-Infrared Photosensitizer for Cancer Therapy, J. Am. Chem. Soc., 2024, 146, 25270–25281 CrossRef CAS PubMed.
- L. A. Abriata, The Nobel Prize in Chemistry: past, present, and future of AI in biology, Commun. Biol., 2024, 7, 1409 CrossRef PubMed.
- P. Das, T. Sercu, K. Wadhawan, I. Padhi, S. Gehrmann, F. Cipcigan, V. Chenthamarakshan, H. Strobelt, C. dos Santos, P.-Y. Chen, Y. Y. Yang, J. P. K. Tan, J. Hedrick, J. Crain and A. Mojsilovic, Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations, Nat. Biomed. Eng., 2021, 5, 613–623 CrossRef CAS.
- W. Huang, S. Huang, Y. Fang, T. Zhu, F. Chu, Q. Liu, K. Yu, F. Chen, J. Dong and W. Zeng, AI-Powered Mining of Highly Customized and Superior ESIPT-Based Fluorescent Probes, Adv. Sci., 2024, 11, 2405596 CrossRef CAS PubMed.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritzel, O. Ronneberger, L. Willmore, A. J. Ballard, J. Bambrick, S. W. Bodenstein, D. A. Evans, C.-C. Hung, M. O'Neill, D. Reiman, K. Tunyasuvunakool, Z. Wu, A. Žemgulytė, E. Arvaniti, C. Beattie, O. Bertolli, A. Bridgland, A. Cherepanov, M. Congreve, A. I. Cowen-Rivers, A. Cowie, M. Figurnov, F. B. Fuchs, H. Gladman, R. Jain, Y. A. Khan, C. M. R. Low, K. Perlin, A. Potapenko, P. Savy, S. Singh, A. Stecula, A. Thillaisundaram, C. Tong, S. Yakneen, E. D. Zhong, M. Zielinski, A. Žídek, V. Bapst, P. Kohli, M. Jaderberg, D. Hassabis and J. M. Jumper, Addendum: Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature, 2024, 636, E4 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- S. Xu, X. Liu, P. Cai, J. Li, X. Wang and B. Liu, Machine-Learning-Assisted Accurate Prediction of Molecular Optical Properties upon Aggregation, Adv. Sci., 2022, 9, 2101074 CrossRef CAS.
- F. Ren, A. Aliper, J. Chen, H. Zhao, S. Rao, C. Kuppe, I. V. Ozerov, M. Zhang, K. Witte, C. Kruse, V. Aladinskiy, Y. Ivanenkov, D. Polykovskiy, Y. Fu, E. Babin, J. Qiao, X. Liang, Z. Mou, H. Wang, F. W. Pun, P. Torres-Ayuso, A. Veviorskiy, D. Song, S. Liu, B. Zhang, V. Naumov, X. Ding, A. Kukharenko, E. Izumchenko and A. Zhavoronkov, A small-molecule TNIK inhibitor targets fibrosis in preclinical and clinical models, Nat. Biotechnol., 2024, 43, 63–75 CrossRef.
- O. Zhang, H. Lin, H. Zhang, H. Zhao, Y. Huang, C.-Y. Hsieh, P. Pan and T. Hou, Deep Lead Optimization: Leveraging Generative AI for Structural Modification, J. Am. Chem. Soc., 2024, 146, 31357–31370 CrossRef CAS.
- J. M. Stokes, K. Yang, K. Swanson, W. Jin, A. Cubillos-Ruiz, N. M. Donghia, C. R. MacNair, S. French, L. A. Carfrae, Z. Bloom-Ackermann, V. M. Tran, A. Chiappino-Pepe, A. H. Badran, I. W. Andrews, E. J. Chory, G. M. Church, E. D. Brown, T. S. Jaakkola, R. Barzilay and J. J. Collins, A Deep Learning Approach to Antibiotic Discovery, Cell, 2020, 180, 688–702 CrossRef CAS PubMed.
- F. Wong, E. J. Zheng, J. A. Valeri, N. M. Donghia, M. N. Anahtar, S. Omori, A. Li, A. Cubillos-Ruiz, A. Krishnan, W. Jin, A. L. Manson, J. Friedrichs, R. Helbig, B. Hajian, D. K. Fiejtek, F. F. Wagner, H. H. Soutter, A. M. Earl, J. M. Stokes, L. D. Renner and J. J. Collins, Discovery of a structural class of antibiotics with explainable deep learning, Nature, 2024, 626, 177–185 CrossRef CAS PubMed.
- V. Smer-Barreto, A. Quintanilla, R. J. R. Elliott, J. C. Dawson, J. Sun, V. M. Campa, Á. Lorente-Macías, A. Unciti-Broceta, N. O. Carragher, J. C. Acosta and D. A. Oyarzún, Discovery of senolytics using machine learning, Nat. Commun., 2023, 14, 3445 CrossRef CAS PubMed.
- V.-N. Nguyen, Y. Yan, J. Zhao and J. Yoon, Heavy-Atom-Free Photosensitizers: From Molecular Design to Applications in the Photodynamic Therapy of Cancer, Accounts Chem. Res., 2021, 54, 207–220 CrossRef CAS.
- J. F. Joung, M. Han, J. Hwang, M. Jeong, D. H. Choi and S. Park, Deep Learning Optical Spectroscopy Based on Experimental Database: Potential Applications to Molecular Design, JACS Au, 2021, 1, 427–438 CrossRef CAS PubMed.
- C.-W. Ju, H. Bai, B. Li and R. Liu, Machine Learning Enables Highly Accurate Predictions of Photophysical Properties of Organic Fluorescent Materials: Emission Wavelengths and Quantum Yields, J. Chem. Inf. Model., 2021, 61, 1053–1065 CrossRef CAS PubMed.
- Q. Sun, D. He, L. Zhang, Z. Li, L. Qu and Y. Sun, Coumarin-hemicyanine-based far-red to near-infrared fluorescent probes: A new generation of fluorescent probe design platform, Trac. Trends Anal. Chem., 2023, 167, 117272 CrossRef CAS.
- L. Dai, Q. Zhang, Q. Ma and W. Lin, Emerging near infrared fluorophore: Dicyanoisophorone-based small-molecule fluorescent probes with large stokes shifts for bioimaging, Coord. Chem. Rev., 2023, 489, 215193 CrossRef CAS.
- G. Hong, A. L. Antaris and H. Dai, Near-infrared fluorophores for biomedical imaging, Nat. Biomed. Eng., 2017, 1, 0010 CrossRef CAS.
- Z. Zeng, S. S. Liew, X. Wei and K. Pu, Hemicyanine-Based Near-Infrared Activatable Probes for Imaging and Diagnosis of Diseases, Angew. Chem. Int. Ed., 2021, 60, 26454–26475 CrossRef CAS PubMed.
- M. Zhao, Y.-S. Guo, W.-N. Xu, Y.-F. Zhao, H.-Y. Xie, H.-J. Li, X.-F. Chen, R.-S. Zhao and D.-S. Guo, Far-red to near-infrared fluorescent probes based on silicon-substituted xanthene dyes for sensing and imaging, Trac. Trends Anal. Chem., 2020, 122, 115704 CrossRef CAS.
- V. J. Pansare, S. Hejazi, W. J. Faenza and R. K. Prud’homme, Review of Long-Wavelength Optical and NIR Imaging Materials: Contrast Agents, Fluorophores, and Multifunctional Nano Carriers, Chem. Mater., 2012, 24, 812–827 CrossRef CAS PubMed.
- X. Wang, H. Wu, T. Wang, Y. Chen, B. Jia, H. Fang, X. Yin, Y. Zhao and R. Yu, NIRFluor: A Deep Learning Platform for Rapid Screening of Small Molecule Near-Infrared Fluorophores with Desired Optical Properties, Anal. Chem., 2025, 97, 1992–2002 CrossRef CAS PubMed.
- G. Landrum, RDKit: a software suite for cheminformatics, computational chemistry, and predictive modeling. https://www.rdkit.org/RDKit_Overview.pdf.
- P. Ertl and A. Schuffenhauer, Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions, J. Cheminf., 2009, 1, 8 Search PubMed.
- Q. Yao, J. Fan, S. Long, X. Zhao, H. Li, J. Du, K. Shao and X. Peng, The concept and examples of type-III photosensitizers for cancer photodynamic therapy, Chem, 2022, 8, 197–209 CAS.
- S. R. Krishnan, A. Roy and M. M. Gromiha, Reliable method for predicting the binding affinity of RNA-small molecule interactions using machine learning, Brief. Bioinform., 2024, 25, bbae002 CrossRef.
- Y. Gu, H. Lai, Z.-Y. Chen, Y. Zhu, Z. Sun, X. Lai, H. Wang, Z. Wei, L. Chen, L. Huang, Y. Zhang, F. He and L. Tian, Chlorination-Mediated π–π Stacking Enhances the Photodynamic Properties of a NIR-II Emitting Photosensitizer with Extended Conjugation, Angew. Chem., Int. Ed., 2023, 62, e202303476 CrossRef CAS PubMed.
- J. Zhu and X. Jiang, How does aggregation-induced emission aggregate interdisciplinary research?, Aggregate, 2024, 5, e451 CrossRef CAS.
- J. Gong, W. Gong, B. Wu, H. Wang, W. He, Z. Dai, Y. Li, Y. Liu, Z. Wang, X. Tuo, J. W. Y. Lam, Z. Qiu, Z. Zhao and B. Z. Tang, ASBase: The universal database for aggregate science, Aggregate, 2023, 4, e263 CrossRef CAS.
- K. S. de Valk, H. J. Handgraaf, M. M. Deken, B. G. Sibinga Mulder, A. R. Valentijn, A. G. Terwisscha van Scheltinga, J. Kuil, M. J. van Esdonk, J. Vuijk, R. F. Bevers, K. C. Peeters, F. A. Holman, J. V. Frangioni, J. Burggraaf and A. L. Vahrmeijer, A zwitterionic near-infrared fluorophore for real-time ureter identification during laparoscopic abdominopelvic surgery, Nat. Commun., 2019, 10, 3118 CrossRef PubMed.
- P. Lu, X. Liu, X. Chu, F. Wang and J.-H. Jiang, Membrane-tethered activation design of a photosensitizer boosts systemic antitumor immunity via pyroptosis, Chem. Sci., 2023, 14, 2562–2571 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc03648h |
| This journal is © The Royal Society of Chemistry 2025 |