Open Access Article
Open Access ArticleDirect sequencing of DNA 5-methylcytosine by engineered dioxygenase NTET-assisted eNAPS†
Shan
Zhang‡
a,
Neng-Bin
Xie‡
a,
Li
Zeng
a,
Fang-Yin
Gang
a,
Yao-Hua
Gu
a,
Min
Wang
a,
Xia
Guo
a,
Tong-Tong
Ji
a,
Jun
Xiong
*a and
Bi-Feng
Yuan
 *ab
*ab
aDepartment of Occupational and Environmental Health, School of Public Health, Department of Radiation and Medical Oncology, Zhongnan Hospital of Wuhan University, State Key Laboratory of Metabolism and Regulation in Complex Organisms, Wuhan University, Wuhan 430071, China. E-mail: bfyuan@whu.edu.cn; jxiong@whu.edu.cn
bHubei Provincial Center for Disease Control and Prevention & NHC Specialty Laboratory of Food Safety Risk Assessment and Standard Development, Wuhan 430079, China
First published on 11th July 2025
Abstract
DNA methylation (5-methylcytosine, 5mC) represents the most prevalent modification in mammals, which is closely linked to disease pathogenesis and cancer development. Single-base resolution sequencing and quantitative analysis of 5mC are essential for elucidating its biological functions. However, current methods are still limited by resolution, sequencing bias, and false positives. In this study, we engineered a Naegleria TET-like protein (NTET), yielding a recombinant engineered NTET (eNTET), to improve both its oxidation activity and sequence compatibility for 5mC. Combined with pyridine borane reduction, we developed engineered NTET-assisted pyridine borane sequencing (eNAPS) to quantitatively detect 5mC in DNA at single-base resolution. In eNAPS, 5mC is oxidized by eNTET to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which are further reduced to dihydrouracil (DHU) by pyridine borane and read as thymine (T) in the subsequent sequencing. The direct conversion of 5mC-to-T allows for precise mapping of 5mC in DNA at single-base resolution. Compared with conventional bisulfite sequencing (BS-seq), eNAPS exhibits advantages such as non-destruction, enhanced sensitivity, improved accuracy, and greater efficiency. Using the eNAPS method, we achieved quantitative analysis of 5mC at single-base resolution in genomic DNA of lung tumor and tumor-adjacent normal tissues. Overall, eNAPS is a mild and bisulfite-free method with high accuracy, making it a valuable tool for investigating the dynamic interplay of 5mC in epigenetic regulation and disease pathogenesis.
Introduction
Eukaryotic DNA molecules contain a diverse range of modifications.1–5 DNA methylation (5-methylcytosine, 5mC) is a canonical epigenetic modification, prevalent in all species of living organisms.6,7 In mammalian cells, DNA methyltransferases (DNMTs) function by catalyzing the addition of a methyl group of S-adenosylmethionine (SAM) to the fifth position of cytosine (C), generating the genetically stable modification of 5mC.8,9 Despite the stability, 5mC can be reversibly converted to the unmodified state in several ways. Notably, ten-eleven translocation (TET) family proteins can mediate the iterative oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), and 5fC and 5caC can be excised by thymine DNA glycosylase (TDG) and restored to cytosine via base excision repair (BER).10–12 Alternatively, 5fC and 5caC can be directly reverted to cytosine through deformylation or decarboxylation.13–17 These mechanisms underscore that 5mC homeostasis is a dynamic and tightly regulated process in living systems.18Accumulating evidence has linked aberrant 5mC patterns to disease pathogenesis and cancer development.19 Consequently, single-base resolution sequencing of 5mC across the genome is necessary to elucidate its biological functions.20,21 Several immunoprecipitation-based methods, such as methylated DNA immunoprecipitation sequencing (MeDIP-seq) and methyl-CpG binding domain sequencing (MBD-Seq), were exploited to map DNA methylation. But these enrichment-based methods suffer from low resolution and semi-quantitative accuracy.22,23 Bisulfite sequencing (BS-seq) is considered the gold standard for single-base resolution profiling of 5mC in genomic DNA.24 However, the harsh bisulfite conversion conditions result in substantial DNA degradation, and BS-seq cannot distinguish between 5mC and 5hmC.25 Moreover, the C-to-U conversion mechanism reduces sequence complexity, leading to imbalance in base composition that compromises downstream analysis.26
The growing demands for high resolution and accuracy of 5mC detection have driven the advancement of bisulfite-free methods.21,27 Apolipoprotein B mRNA-editing enzyme catalytic subunit 3A (APOBEC3A) and its engineered variants have emerged as powerful tools for high-resolution sequencing of DNA cytosine modifications.28–31 Enzymatic methylation sequencing (EM-seq) utilizes the oxidation of 5mC to 5caC by TET, protecting it from deamination by A3A, thereby enabling precise 5mC sequencing.32 In addition to A3A, various other cytosine deaminases have been employed in DNA modification sequencing, including engineered apolipoprotein B mRNA-editing catalytic polypeptide-like 3C (eA3C) and double-stranded cytosine deaminases.33–36 However, these approaches still suffer from the challenge of indirect detection due to the conversion of unmodified C to T.37
Direct conversion methods targeting 5mC, such as methyltransferase-directed labeling combined with A3A deamination (MLAD-seq), direct methylation sequencing (DM-Seq), and carboxymethylation deamination sequencing (CMD-seq), utilize engineered methyltransferases for carboxymethylation-based protection of unmodified cytosine and integrate A3A deamination, enabling selective 5mC-to-T conversion for 5mC sequencing.38–40 However, these methods are limited to detecting 5mC in CG contexts due to the specificity of the methyltransferases. TET-assisted pyridine borane sequencing (TAPS) relies on TET enzymes to oxidize 5mC to 5fC and 5caC, which are then reduced by pyridine borane to dihydrouracil (DHU) and read as T during sequencing.41 However, mammalian TET enzymes (TET1, TET2, and TET3) require eukaryotic expression systems for their production and therefore are costly and unstable. Furthermore, these enzymes exhibit sequence bias at non-CG sites,42 resulting in incomplete methylation analysis and inaccurate quantification. Therefore, developing cost-effective, stable, and sequence-compatible recombinant TET enzymes is critical for advancing direct 5mC conversion-based sequencing technologies.
Here, we engineered the Naegleria TET-like protein (NTET), yielding a recombinant engineered NTET (eNTET) dioxygenase. eNTET can be cost-effectively expressed in Escherichia coli, with robust oxidation activity of 5mC to 5fC and 5caC in DNA across diverse sequence contexts. Using the properties of eNTET, we developed an engineered NTET-assisted pyridine borane sequencing (eNAPS) method for bisulfite-free and quantitative detection of 5mC in DNA at single-base resolution. The eNAPS method offers a direct method for selectively converting 5mC-to-T, while unmodified cytosine is retained. By applying eNAPS, we achieved quantification of 5mC in genomic DNA at single-base resolution from paired lung tumor and tumor-adjacent normal tissues, enabling direct comparison of 5mC dynamics at specific sites.
Experimental methods
Materials and reagents
2′-Deoxyguanosine (G), 2′-deoxyadenosine (A), 2′-deoxycytidine (C), thymidine (T), 2′-deoxynucleoside 5′-triphosphates (including dATP, dCTP, dGTP, and TTP), phosphodiesterase I, boracic acid, ethylene diamine tetraacetic acid (EDTA), tris(hydroxymethyl)aminomethane (Tris), and 2-morpholinoethanesulphonic acid (MES) were purchased from Sigma-Aldrich (St. Louis, MO, USA). 5-Methyl-2′-deoxycytidine (5mC), 5-hydroxymethyl-2′-deoxycytidine (5hmC), 5-formyl-2′-deoxycytidine (5fC), and 5-carboxyl-2′-deoxycytidine (5caC) were purchased from Berry & Associates (Dexter, MI). Pyridine borane was purchased from Energy Chemical (Shanghai, China). Dithiothreitol (DTT) was obtained from Beyotime Biotechnology (Wuhan, China). Recombinant T4 β-glucosyltransferase (β-GT), apurinic/apyrimidinic Endonuclease 1 (APE1), EpiMark Hot Start Taq polymerase, Taq DNA polymerase, and Q5 High-Fidelity polymerase were obtained from New England Biolabs (Ipswich, MA, USA). DNase I, S1 nuclease, and calf intestinal alkaline phosphatase (CIAP) were obtained from Takara Biotechnology Co. Ltd (Dalian, China).Preparation of DNA strands with various modifications
The 309-bp double-stranded DNA (dsDNA) substrates (Tables S1 and S2 in the ESI†) were synthesized by PCR amplification following a previous report.43 For the preparation of DNA-CG, DNA-CC, DNA-CT, and DNA-CA, 0.5 ng of synthetic DNA (TsingKe, Beijing, China) was used as the template for PCR amplification. The reaction was carried out in a 50 μL mixture containing 1 U of Taq DNA polymerase, 5 μL of 10× reaction buffer, 4 μL of dNTP mixture (2.5 mM each), and 1 μL each of forward and reverse primers (10 μM, Table S3 in the ESI†). For the synthesis of modified DNA strands (DNA-5mCG, DNA-5mCC, DNA-5mCT, DNA-5mCA, and DNA-5hmC, Table S2 in the ESI†), modified primers (Table S3 in the ESI†) were utilized for PCR amplification. All synthesized dsDNA substrates were separated via 2% agarose gel electrophoresis and purified using a Gel Extraction Kit (Omega Bio-Tek Inc., Norcross, GA, USA).Expression and purification of recombinant proteins
The genes encoding mCE7 (mutant colicin E7 DNase), wild-type NTET (wtNTET), and recombined human thymine DNA glycosylase (TDG, catalytic domain, amino acids 82–308) were synthesized by TsingKe Biological Technology (Beijing, China) using E. coli-optimized codons to ensure efficient expression in E. coli cells. The point mutation and fusion engineering of the wtNTET (eNTET) gene were achieved through overlapping PCR and confirmed by Sanger sequencing (TsingKe). The eNTET gene was subsequently subcloned into a pET-28a-MBP plasmid, encompassing an N-terminal 6× His and maltose binding protein (MBP) affinity tag. Similarly, the TDG and wtNTET genes were cloned into the pET-28a plasmid and the pET-28a-SUMO plasmid, respectively. The pET-28a-MBP-eNTET, pET-28a-SUMO-wtNTET, and pET-28a-TDG plasmids were then transformed into BL21 (DE3) E. coli cells. The expression of eNTET, wtNTET, and TDG was performed similarly to previous studies.44–47 Briefly, the expression of recombinant proteins was induced by the addition of 0.3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16 °C for 20 h under shaking at 200 rpm. For the purification of eNTET, cells were harvested and resuspended in a lysis buffer containing 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM Tris-HCl (pH 8.0) and 150
mM Tris-HCl (pH 8.0) and 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM NaCl, followed by lysis through sonication. After ultracentrifugation, the supernatant was applied to Ni-NTA affinity beads in a gravity-flow column for the selective enrichment of His-tagged recombinant proteins. The eluent was then concentrated and further purified by heparin–ion exchange chromatography (HiTrap Heparin HP column, GE Healthcare, Chicago, USA) and size-exclusion chromatography (Superdex G200 Increase10/300 column, GE Healthcare) using an AKTA pure system (GE Healthcare). The purification procedures for wtNTET and TDG proteins followed previously described methodologies.48,49 The purified proteins were validated by SDS-PAGE and stored at −80 °C. The amino acid sequences of eNTET, wtNTET, and TDG proteins are listed in Table S4 in the ESI.†
mM NaCl, followed by lysis through sonication. After ultracentrifugation, the supernatant was applied to Ni-NTA affinity beads in a gravity-flow column for the selective enrichment of His-tagged recombinant proteins. The eluent was then concentrated and further purified by heparin–ion exchange chromatography (HiTrap Heparin HP column, GE Healthcare, Chicago, USA) and size-exclusion chromatography (Superdex G200 Increase10/300 column, GE Healthcare) using an AKTA pure system (GE Healthcare). The purification procedures for wtNTET and TDG proteins followed previously described methodologies.48,49 The purified proteins were validated by SDS-PAGE and stored at −80 °C. The amino acid sequences of eNTET, wtNTET, and TDG proteins are listed in Table S4 in the ESI.†
Evaluation of the oxidation activity of eNTET by TDG/APE1 digestion
The enzymatic activity of eNTET was assessed by TDG/APE1 digestion using FAM-labeled dsDNA substrates containing 5mC (FAM-5mC annealing with 5mC-R, Table S5 in the ESI†). The eNTET (or wtNTET) oxidation and TDG cleavage reaction of FAM-5mC were combined and performed in a 20 μL solution containing 50 mM MES (pH 6.9), 150 mM NaCl, 10 mM α-KG, 200 μM (NH4)2FeSO4, 20 mM ascorbic acid, 1 mM DTT, 5 μM eNTET (or wtNTET), 500 nM TDG, 1U of APE1, and 2 pmol of FAM-labeled ds-DNA duplex. The reaction mixture was incubated at 34 °C for 12 h, followed by digestion with 2 μL of proteinase K at 55 °C for 30 min and 95 °C for 5 min. The sample was then mixed with an equal volume of deionized formamide, denatured at 95 °C for 10 min, and immediately cooled on an ice-water bath. The mixture was then analyzed by 20% denaturing polyacrylamide gel electrophoresis (PAGE) and visualized using a Tanon 4600SF gel imaging system (Shanghai, China).Evaluation of the oxidation activity of eNTET by liquid chromatography-mass spectrometry (LC-MS/MS) analysis
Four dsDNA containing 5mC sites at CG, CC, CT, and CA sequence contexts (DNA-5mCG, DNA-5mCC, DNA-5mCT, and DNA-5mCA, Table S2 in the ESI†) were used as substrates. The reaction was carried out in a 20 μL solution containing 100 nM dsDNA, 5 μM eNTET (or wtNTET), 50 mM MES (pH 6.9), 150 mM NaCl, 10 mM α-KG, 200 μM (NH4)2FeSO4, 20 mM ascorbic acid, and 1 mM DTT at 34 °C for 12 h. The reaction was terminated by proteinase K treatment. The resulting DNA was then enzymatically digested to nucleosides and analyzed by LC-MS/MS according to the established procedures.50,51 Details are described in the ESI.†Evaluation of the characteristics of eNAPS by sequencing
Five 309 bp dsDNA containing 5mC or 5hmC (DNA-5mCG, DNA-5mCC, DNA-5mCT, DNA-5mCA, and DNA-5hmC, Table S2 in the ESI†) were used as templates to evaluate the properties of eNAPS. The DNA template was first oxidized by eNTET as described previously. The products were then reduced in a 50 μl reaction solution containing 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM sodium acetate (pH 4.3) and 1
mM sodium acetate (pH 4.3) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M pyridine borane for 16
M pyridine borane for 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) h at 37 °C. The resulting DNA was purified using 1× KAPA Pure Beads (Roche). 5 ng of purified DNA was used as the template for extension and PCR amplification. The extension reaction was performed in a solution containing 1 U of Taq DNA polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, and 0.4 μM primer (Table S3 in the ESI†) at 60 °C for 30 min. The resulting DNA was purified with 1× KAPA Pure Beads. PCR amplification was performed with the purified DNA, 1 U of Q5 High-Fidelity DNA Polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, 0.4 μM forward primer, and 0.4 μM reverse primer. The PCR was carried out under the following gradient: 95 °C for 5 min, 30 cycles of 95 °C for 30 s, 68 °C for 1 min, 72 °C for 1 min, and 72 °C for 10 min. The PCR products were then subjected to Sanger sequencing or colony sequencing (TsingKe). Colony sequencing was performed according to previous procedures,35 involving random selection and sequencing of 50 positive colonies from each sample.
h at 37 °C. The resulting DNA was purified using 1× KAPA Pure Beads (Roche). 5 ng of purified DNA was used as the template for extension and PCR amplification. The extension reaction was performed in a solution containing 1 U of Taq DNA polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, and 0.4 μM primer (Table S3 in the ESI†) at 60 °C for 30 min. The resulting DNA was purified with 1× KAPA Pure Beads. PCR amplification was performed with the purified DNA, 1 U of Q5 High-Fidelity DNA Polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, 0.4 μM forward primer, and 0.4 μM reverse primer. The PCR was carried out under the following gradient: 95 °C for 5 min, 30 cycles of 95 °C for 30 s, 68 °C for 1 min, 72 °C for 1 min, and 72 °C for 10 min. The PCR products were then subjected to Sanger sequencing or colony sequencing (TsingKe). Colony sequencing was performed according to previous procedures,35 involving random selection and sequencing of 50 positive colonies from each sample.
Quantification of 5mC in synthetic DNA
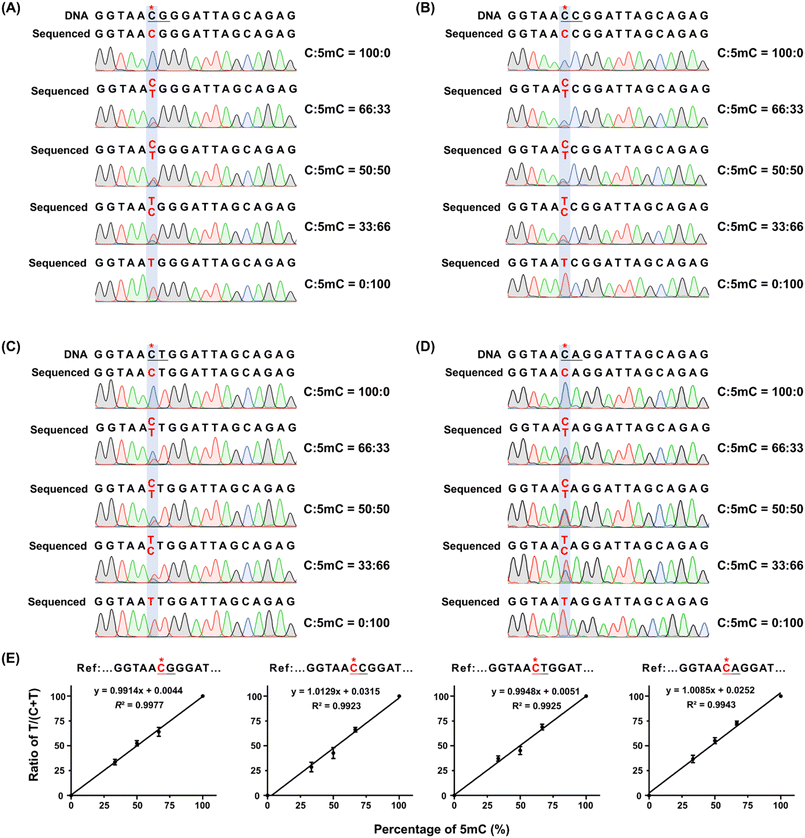
The 309 bp DNA template containing different sequence contexts (DNA-CG, DNA-CC, DNA-CT, and DNA-CA, Table S1 in the ESI†) was mixed with the corresponding 5mC-containing DNA (DNA-5mCG, DNA-5mCC, DNA-5mCT, and DNA-5mCA) at varying ratios (0%, 33%, 50%, 66%, and 100%). The mixture was first oxidized with eNTET, followed by pyridine borane reduction as previously described. The reduced DNA was amplified by PCR and analyzed by Sanger sequencing. The peak heights of C and T at CN sites (CG, CC, CT, and CA) were used to calculate the 5mC levels, enabling evaluation of the sensitivity and accuracy of eNAPS.Quantification of 5mC in specific sites of lung tissue DNA using eNAPS
The human non-small cell lung carcinoma (NSCLC) tissue and the matched tumor-adjacent normal tissue without preoperative target therapy/chemotherapy were obtained from the Zhongnan Hospital of Wuhan University. The study was approved by the Ethics Committee of Wuhan University and was conducted following the principles of the Declaration of Helsinki. All experiments were conducted in strict compliance with the guidelines and regulations set by the Ethics Committee of Wuhan University. Genomic DNA was extracted using a Tissue DNA Kit (Omega Bio-Tek Inc., Norcross, GA). The concentration of the purified genomic DNA was determined on a B-500 spectrophotometer (Metash Instruments Co., Ltd, Shanghai, China).The isolated DNA was fragmented to an average size of 300–500 bp under optimized conditions using an ultrasonic homogenizer (Scientz). The fragmented DNA was then analyzed by eNAPS. Briefly, 10 ng of fragmented DNA was incubated with 5 μM eNTET in a 20 μL solution of 50 mM MES (pH 6.9), 150 mM NaCl, 10 mM α-KG, 200 μM (NH4)2FeSO4, 20 mM ascorbic acid, and 1 mM DTT for 12 h at 34 °C. The products were then reacted in a 50 μL solution containing 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM sodium acetate (pH 4.3) and 1
mM sodium acetate (pH 4.3) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M pyridine borane for 16
M pyridine borane for 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) h at 37 °C. The resulting DNA was purified using 1× KAPA Pure Beads, and the purified DNA was used for extension and PCR amplification. The extension reaction was performed in a solution consisting of 1 U of Taq DNA polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, and 0.4 μM extension primer (Table S6 in the ESI†) for 30 min at 60 °C. The resulting product was purified using 1× KAPA Pure Beads. PCR amplification was carried out in a 50 μL solution containing purified DNA, 1 U of Q5 High-Fidelity DNA Polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, 0.4 μM forward primer, and 0.4 μM reverse primer. The sequences of the primers targeting the gene body of the tumor driver gene epidermal growth factor receptor (EGFR) are listed in Table S6 in the ESI.† PCR products were subjected to Sanger sequencing for locus-specific 5mC quantification. As for BS-seq, 400 ng of the fragmented DNA was processed using a Hieff DNA Bisulfite Conversion Kit (Yeasen, Shanghai, China). The sequences of the primers targeting the gene body of the EGFR for BS-seq are listed in Table S7 in the ESI.†
h at 37 °C. The resulting DNA was purified using 1× KAPA Pure Beads, and the purified DNA was used for extension and PCR amplification. The extension reaction was performed in a solution consisting of 1 U of Taq DNA polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, and 0.4 μM extension primer (Table S6 in the ESI†) for 30 min at 60 °C. The resulting product was purified using 1× KAPA Pure Beads. PCR amplification was carried out in a 50 μL solution containing purified DNA, 1 U of Q5 High-Fidelity DNA Polymerase, 5 μL of 10× reaction buffer, 0.2 mM dNTPs, 0.4 μM forward primer, and 0.4 μM reverse primer. The sequences of the primers targeting the gene body of the tumor driver gene epidermal growth factor receptor (EGFR) are listed in Table S6 in the ESI.† PCR products were subjected to Sanger sequencing for locus-specific 5mC quantification. As for BS-seq, 400 ng of the fragmented DNA was processed using a Hieff DNA Bisulfite Conversion Kit (Yeasen, Shanghai, China). The sequences of the primers targeting the gene body of the EGFR for BS-seq are listed in Table S7 in the ESI.†
Results and discussion
Principle of the engineered NTET-assisted pyridine borane sequencing (eNAPS)
TET proteins act as dioxygenases of 5mC and have the capacity to specifically oxidize 5mC sequentially to 5hmC, 5fC, and 5caC.32,41,48 Previous study have shown that mammalian TETs, including murine TET1, TET2, and TET3, exhibit a discernible sequence preference and exhibit constrained catalytic efficiency in oxidizing methylation at non-CG context sites, which affects the accuracy of methylation sequencing at non-CG sites.42 Additionally, these TETs possess a substantial molecular weight (typically exceeding 200 kDa), which frequently necessitates the presence of a chaperone or post-translational modifications to facilitate assisted folding and form a stable and active state. Therefore, these proteins are mainly expressed in eukaryotic cells, such as insects, and are difficult to express in prokaryotes such as E. coli (Fig. 1A), which invariably increases the cost of oxidizing enzymes in 5mC sequencing.32,41,52 The protozoan-derived NTET protein has better sequence compatibility for 5mC and a smaller molecular weight (36 kDa) and can be expressed exogenously in prokaryotic hosts such as E. coli, making it a potential enzyme for 5mC sequencing (Fig. 1A).53,54 However, it was shown that wtNTET only converts about 80% of 5mC to 5fC and 5caC, which seriously limits its application for DNA methylation sequencing methods.48,53,54In this study, we began with the engineering transformation of NTET through expanding the active site and enhancing the binding ability of DNA (Fig. 1B) and proposed an engineered NTET-assisted pyridine borane sequencing (eNAPS) method for the quantitative detection of site-specific 5mC at single-base resolution. In eNAPS, eNTET specifically and efficiently oxidizes 5mC to 5fC and 5caC and then converts it into DHU by pyridine borane, read as T in subsequent sequencing, while glycosylated 5hmC and C are not affected, they are interpreted as C in sequencing (Fig. 1C). Thus, only the original 5mC in DNA is converted to DHU and read as T (Fig. 1D). Differing from traditional deamination sequencing methods such as BS-seq, eNAPS is a nondestructive method and directly alters the readout of 5mC without affecting the original cytosines in DNA (Fig. 1E).
Characterization of the oxidation properties of eNTET
To cost-effectively produce TET enzymes with enhanced stability and broader sequence compatibility, we expressed and engineered NTET by targeted mutation (Fig. 2A). Previous study hypothesized the existence of a strictly conserved residue that may function as a gatekeeper to regulate 5mC oxidation.55 Moreover, the fusion expression of mCE7 has been shown to improve the binding ability of the DNA substrate.56 Here, we proposed an engineered NTET (eNTET) that mutates leucine (Leu) to alanine (Ala) at position 240 to expand the active space and fused mCE7 at the N-terminus of eNTET to enhance the binding ability of DNA (Fig. 2A–C). eNTET was expressed in E. coli. and purified for the development of the eNAPS method (Fig. 2C and S1 in the ESI†).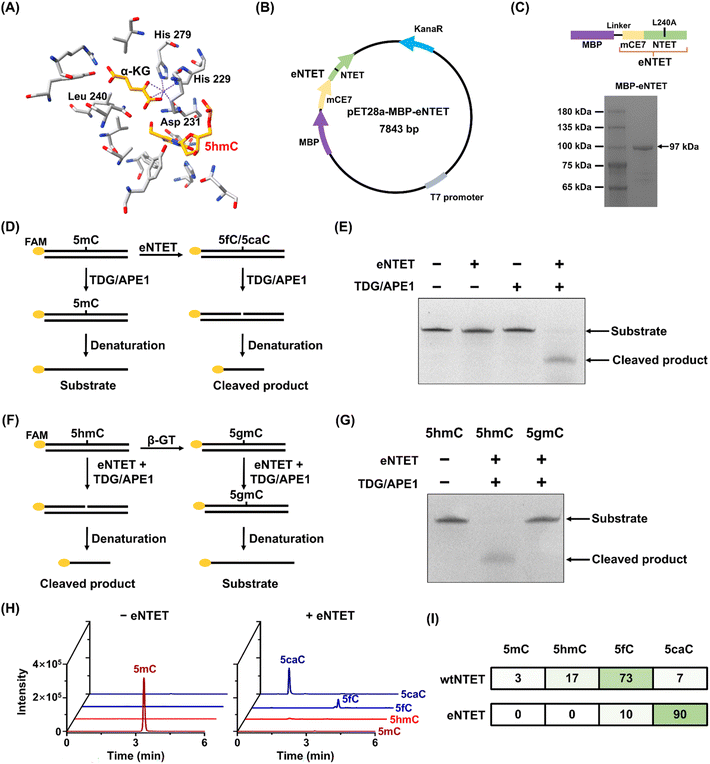 | ||
| Fig. 2 Evaluation of the oxidation activity of eNTET in dsDNA. (A) Structure of NTET bound with 5hmC-DNA (PDB:5CG8). (B) The plasmid for the expression of eNTET. (C) The analysis of purified eNTET by SDS-PAGE. (D) Workflow of the activity assay of the eNTET protein. (E) PAGE analysis of the oxidation of 5mC by eNTET. (F) Workflow of the oxidation activity assays of eNTET toward 5hmC and 5gmC. (G) PAGE analysis of the activities of eNTET toward 5hmC and 5gmC. (H) Extracted-ion chromatograms of 5mC, 5hmC, 5fC, and 5caC after eNTET treatment by LC-MS/MS analysis. (I) The percents of the oxidation products generated by eNTET or wtNTET. | ||
To assess the oxidation activity of eNTET, a 44 bp FAM-labeled dsDNA with a 5mC site (FAM-5mC, Table S5 in the ESI†) was utilized as the substrate and treated with eNTET. The oxidation products, 5fC and 5caC, could be recognized and excised by TDG to form an abasic (AP) site (Fig. S2 in the ESI†), which was subsequently excised by APE1 to produce truncated products (Fig. 2D). The results showed that 5mC was efficiently oxidized and cleaved after incubation with eNTET and TDG/APE1 (Fig. 2E), while wtNTET partially oxidized 5mC to 5fC and 5caC (Fig. S3 and S4 in the ESI†). Furthermore, eNTET also efficiently oxidized 5hmC but not glycosylated 5hmC (Fig. 2F and G).
Next, we further evaluated the oxidative activity and products of eNTET using LC-MS/MS. Four dsDNA strands with different 5mC contexts (CG, CC, CT, and CA sequence contexts, Table S2 in the ESI†) were utilized as the substrates. The results showed that eNTET possessed robust oxidation activity, with the 5mC substrate eliminated and both 5fC and 5caC produced after eNTET treatment (Fig. 2H), while the oxidation products of wtNTET retained a significant fraction of 5hmC (Fig. S5 in the ESI†). Quantitative analysis of the oxidation products indicated that 5mC was completely oxidized after eNTET treatment, yielding a 10% proportion of 5fC and 90% proportion of 5caC, while 97% of 5mC was oxidized after wtNTET treatment, with the products of 5hmC, 5fC and 5caC accounting for 17%, 73%, and 7%, respectively (Fig. 2I). Overall, we successfully constructed and expressed a recombined dioxygenase eNTET with excellent oxidation activity and sequence compatibility for 5mC to generate its two products, 5fC and 5caC, which were reported to be readily reduced by pyridine borane.
Development of eNAPS
After obtaining the eNTET protein, we proceeded to develop the eNAPS method for the sequencing and quantification of 5mC at single-base resolution (Fig. 3A). In eNAPS, 5mC is oxidized to 5fC and 5caC by eNTET treatment, and the following pyridine borane reduction induces 5fC-to-DHU and 5caC-to-DHU conversions, therefore enabling the differentiation of 5mC and C. We first evaluated the eNAPS method using a 309 bp dsDNA substrate containing a 5mC site (DNA-5mCG) by sequencing. The results demonstrated that 5mC in the CG context was read as T after eNTET and pyridine borane treatment, while unmodified C was still read as C in sequencing (Fig. 3B). Considering the sequence compatibility of eNTET, we hypothesized that eNAPS could also detect 5mC in non-CG contexts. Three 5mC-containing dsDNA substrates with 5mCC, 5mCT, and 5mCA contexts were used as substrates for eNAPS (Table S2 in the ESI†). The results showed that eNAPS could convert all these 5mC sites to T (Fig. 3B). To eliminate the interference of 5hmC, we utilized β-GT to selectively glycosylate 5hmC to form β-glucosyl-5-hydroxymethyl-cytosine (5gmC), which is resistant to eNTET oxidation (Fig. 3C). The sequencing results showed that 5hmC was read as T by eNAPS without β-GT treatment (Fig. S6 in the ESI†), but 5hmC was read as C after glycosylation protection prior to eNAPS (Fig. 3D). Collectively, these results demonstrated that eNAPS enables the detection of 5mC at single-base resolution by directly converting 5mC sites.We next quantitatively assessed the readouts of C and 5mC in eNAPS by colony sequencing (Fig. 4A). The results indicated that all 50 clones of the 5mC sites were read as T in eNAPS, while all the C sites were not affected and still read as C (Fig. 4B and S7 in the ESI†). In conclusion, these results highlight that the eNAPS approach enables the highly efficient 5mC-to-T conversion without impacting the normal cytosines in DNA.
 | ||
| Fig. 4 Evaluating the characteristics of eNAPS by colony sequencing. (A) Schematic illustration of eNAPS by colony sequencing. (B) The readouts of C and 5mC sites by eNAPS through colony sequencing. | ||
Quantitative performance of eNAPS in mapping 5mC
We subsequently examined the quantitative capability of eNAPS for the 5mC site. DNA substrates with C or 5mC in different CN contexts (N = G, C, T, and A) were mixed at various ratios, with the 5mC proportions being 0%, 33%, 50%, 66%, and 100%. The mixtures were subjected to eNAPS analysis, and the height ratios of T/(T + C) by Sanger sequencing were calculated. The results demonstrated that as the proportion of 5mC-containing DNA increased, the proportion of T increased, while the signal of the surrounding original C remained unchanged (Fig. 5A–D). Quantitative analysis revealed that the percentage of T/(T + C) exhibited a linear increase corresponding to the increase of 5mC at CG, CC, CT, and CA contexts, with R2 > 0.99 (Fig. 5E). The results demonstrated that the eNAPS approach could effectively quantify 5mC stoichiometry at single-base resolution in both CG and non-CG contexts.Determination of 5mC in genomic DNA by eNAPS
DNA methylation is an important regulatory mechanism controlling gene expression, and aberrant methylation at specific loci is closely associated with disease or tumorigenesis.57–61 For instance, the overexpression of the epidermal growth factor receptor (EGFR) has been shown to activate downstream signaling pathways, thereby facilitating tumor growth, invasion, and metastasis.62 Elevated expression of the EGFR has been observed in various cancer types, with lung cancer being particularly prevalent.63 Using the developed eNAPS method, we quantified 5mC levels in two specific sites within the gene body region of the EGFR in lung tumor and tumor-adjacent normal tissues (Fig. 6A and Table S8 in the ESI†).The results demonstrated that the two specific 5mC sites (chr7: 55109538 and chr7: 55109595) within the EGFR were read exclusively as T in the tumor tissue and partially converted to T in the adjacent normal tissue (Fig. 6B and C). The quantitative results showed that the 5mC levels at positions chr7: 55109538 and chr7: 55109595 were 96.5% and 100%, respectively, in tumor-adjacent normal tissue. The 5mC levels decreased by ∼50% on the two sites in tumor tissue, with methylation levels at positions chr7: 55109538 and chr7: 55109595 being 54.5% and 46.4%, respectively. To validate the accuracy of the quantitative results by eNAPS, we also performed BS-seq for the two specific sites (Table S7 in the ESI†). These results indicated that the quantitative results by eNAPS closely corresponded to those by BS-seq (Fig. 6B and C). In summary, these results demonstrated that the eNAPS approach is effective in the quantification of 5mC in genomic DNA at single-base resolution.
eNAPS offers several advantages over previous methods for quantitative detection of 5mC. First, compared to affinity enrichment sequencing, eNAPS enables single-base resolution sequencing and precise quantification of 5mC. Second, eNAPS employs mild conditions, as opposed to the traditional gold standard BS-seq method. Third, in contrast to deaminase-mediated sequencing methods, eNAPS directly induces 5mC-to-T conversion, rather than to the bulk unmodified C, thereby reducing background interference and false positives that may result from incomplete deamination. Fourth, previous direct sequencing methods, such as MLAD-seq, DM-seq, and CMD-seq, are limited to sequencing 5mC in CG contexts, whereas eNTET exhibits almost no sequence preference, enabling the detection of 5mC in non-CG contexts. Finally, compared with TAPS, eNAPS has the advantages of lower cost, more stability, and the ability to fully convert 5mC in CG and non-CG contexts, making it a more promising direct conversion sequencing method for DNA methylation. It is worth noting that a recent study reported a photoactivatable nano-demethylase for the selective oxidation of 5mC to 5fC.64 This presents an alternative chemical oxidation method for converting 5mC to 5fC, which could potentially be combined with pyridine borane in 5mC sequencing.
Conclusions
We developed a method termed eNAPS for bisulfite-free and single-base resolution quantification of 5mC in DNA. Specifically, we designed and constructed an engineered NTET that demonstrated superior sequence compatibility, cost-effectiveness, and enhanced activity for 5mC oxidation. eNAPS facilitates the direct conversion of 5mC-to-T and precise quantification of 5mC in genomic DNA by integrating eNTET oxidation with pyridine borane reduction, where C and 5mC are interpreted as C and T, respectively. In contrast to the conventional BS-seq method, eNAPS avoids base composition imbalance and harsh reaction conditions, minimizing false positives and mapping issues. Furthermore, 5hmC could be glycosylated by β-GT prior to eNTET treatment, allowing the efficient differentiation between 5mC and 5hmC in eNAPS. The eNAPS method was successfully applied to quantify 5mC at two specific sites within the EGFR gene body in lung tumor and tumor-adjacent normal tissues. The results revealed that the 5mC levels at these two sites were significantly decreased in tumor tissues. In conclusion, eNAPS represents a bisulfite-free, accurate, and cost-effective approach for DNA methylation sequencing at single-base resolution in both CG and non-CG contexts, making it a valuable tool for investigating the dynamic interplay of 5mC in epigenetic regulation and disease pathogenesis.Data availability
ESI available: Tables S1–S8 and Fig. S1–S7†.Author contributions
Bi-Feng Yuan: writing – review & editing, investigation, supervision, conceptualization, funding acquisition. Jun Xiong: writing – review & editing, investigation, supervision, conceptualization, funding acquisition. Shan Zhang: writing – original draft, methodology, formal analysis, data curation, conceptualization. Neng-Bin Xie: methodology, formal analysis, data curation, conceptualization. Li Zeng: methodology, data curation, conceptualization. Fang-Yin Gang: formal analysis, data curation, conceptualization. Yao-Hua Gu: data curation, conceptualization. Min Wang: methodology, data curation. Xia Guo: methodology, formal analysis. Tong–Tong Ji: methodology, formal analysis.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the Key Research and Development Project of Hubei Province (2023BCB094) and the National Natural Science Foundation of China (22277093 and 22207090).References
- W. Y. Lai, J. Z. Mo, J. F. Yin, C. Lyu and H. L. Wang, Trac. Trends Anal. Chem., 2019, 110, 173–182 CrossRef CAS.
- O. Deniz, J. M. Frost and M. R. Branco, Nat. Rev. Genet., 2019, 20, 417–431 CrossRef CAS PubMed.
- W. X. Shao, J. Wu, G. Li, Y. H. Min, Q. S. Hu, Y. L, W. Ci and B. F. Yuan, Chin. Chem. Lett., 2025, 36, 110747 Search PubMed.
- T. T. Ji, M. Wang, X. Guo, F. Y. Gang, S. Zhang, J. Xiong, Y. H. Gu, N. B. Xie and B. F. Yuan, Anal. Chem., 2025, 97, 8564–8573 CrossRef CAS PubMed.
- T. T. Ji, N. B. Xie, J. H. Ding, M. Wang, X. Guo, Y. Y. Chen, S. Y. Yu, Y. Q. Feng and B. F. Yuan, Anal. Chem., 2023, 95, 8384–8392 CrossRef CAS PubMed.
- A. Parry, S. Rulands and W. Reik, Nat. Rev. Genet., 2021, 22, 59–66 CrossRef CAS PubMed.
- C. Luo, P. Hajkova and J. R. Ecker, Science, 2018, 361, 1336–1340 CrossRef CAS PubMed.
- S. Adam, V. Klingel, N. E. Radde, P. Bashtrykov and A. Jeltsch, Nucleic Acids Res., 2023, 51, 6622–6633 CrossRef CAS PubMed.
- L. Zeng, F. Y. Gang, T. T. Ji, S. Zhang, X. Guo, Y. Hao, J. Xiong, Z. W. Wei, N. B. Xie and B. F. Yuan, Chin. J. Chem., 2025, 43, 1797–1805 CrossRef CAS.
- Y. F. He, B. Z. Li, Z. Li, P. Liu, Y. Wang, Q. Tang, J. Ding, Y. Jia, Z. Chen, L. Li, Y. Sun, X. Li, Q. Dai, C. X. Song, K. Zhang, C. He and G. L. Xu, Science, 2011, 333, 1303–1307 CrossRef CAS PubMed.
- S. Ito, L. Shen, Q. Dai, S. C. Wu, L. B. Collins, J. A. Swenberg, C. He and Y. Zhang, Science, 2011, 333, 1300–1303 CrossRef CAS PubMed.
- N. B. Xie, M. Wang, T. T. Ji, X. Guo, J. H. Ding, B. F. Yuan and Y. Q. Feng, Chem. Sci., 2022, 13, 7046–7056 RSC.
- K. Iwan, R. Rahimoff, A. Kirchner, F. Spada, A. S. Schroder, O. Kosmatchev, S. Ferizaj, J. Steinbacher, E. Parsa, M. Muller and T. Carell, Nat. Chem. Biol., 2018, 14, 72–78 CrossRef CAS PubMed.
- Y. Feng, N. B. Xie, W. B. Tao, J. H. Ding, X. J. You, C. J. Ma, X. Zhang, C. Yi, X. Zhou, B. F. Yuan and Y. Q. Feng, CCS Chem., 2021, 2, 994–1008 CrossRef.
- Y. Feng, J. J. Chen, N. B. Xie, J. H. Ding, X. J. You, W. B. Tao, X. Zhang, C. Yi, X. Zhou, B. F. Yuan and Y. Q. Feng, Chem. Sci., 2021, 12, 11322–11329 RSC.
- Y. Feng, Y. Q. Tian, Y. Q. Zhao, S. J. Chen and B. F. Yuan, Chin. Chem. Lett., 2024, 35, 109656 CrossRef CAS.
- Y. Feng, S. J. Chen and B. F. Yuan, Chin. J. Chem., 2024, 42, 645–651 CrossRef CAS.
- T. Liu, C. J. Ma, B. F. Yuan and Y. Q. Feng, Sci. China Chem., 2018, 61, 381–392 CrossRef CAS.
- K. D. Robertson, Nat. Rev. Genet., 2005, 6, 597–610 CrossRef CAS PubMed.
- F. Y. Gang, N. B. Xie, M. Wang, S. Zhang, T. T. Ji, W. Liu, X. Guo, S. Y. Gu and B. F. Yuan, Anal. Chem., 2024, 96, 20559–20567 CrossRef CAS PubMed.
- Y. Dai, B. F. Yuan and Y. Q. Feng, RSC Chem. Biol., 2021, 2, 1096–1114 RSC.
- R. A. Harris, T. Wang, C. Coarfa, R. P. Nagarajan, C. Hong, S. L. Downey, B. E. Johnson, S. D. Fouse, A. Delaney, Y. Zhao, A. Olshen, T. Ballinger, X. Zhou, K. J. Forsberg, J. Gu, L. Echipare, H. O'Geen, R. Lister, M. Pelizzola, Y. Xi, C. B. Epstein, B. E. Bernstein, R. D. Hawkins, B. Ren, W. Y. Chung, H. Gu, C. Bock, A. Gnirke, M. Q. Zhang, D. Haussler, J. R. Ecker, W. Li, P. J. Farnham, R. A. Waterland, A. Meissner, M. A. Marra, M. Hirst, A. Milosavljevic and J. F. Costello, Nat. Biotechnol., 2010, 28, 1097–1105 CrossRef CAS PubMed.
- D. Serre, B. H. Lee and A. H. Ting, Nucleic Acids Res., 2010, 38, 391–399 CrossRef CAS PubMed.
- M. Berney and J. F. McGouran, Nat. Rev. Chem., 2018, 2, 332–348 CrossRef CAS.
- K. Tanaka and A. Okamoto, Bioorg. Med. Chem. Lett., 2007, 17, 1912–1915 CrossRef CAS PubMed.
- Y. Kong, E. A. Mead and G. Fang, Nat. Rev. Genet., 2023, 24, 363–381 CrossRef CAS PubMed.
- Z. J. Liu, S. Martinez Cuesta, P. van Delft and S. Balasubramanian, Nat. Chem., 2019, 11, 629–637 CrossRef CAS PubMed.
- E. K. Schutsky, J. E. DeNizio, P. Hu, M. Y. Liu, C. S. Nabel, E. B. Fabyanic, Y. Hwang, F. D. Bushman, H. Wu and R. M. Kohli, Nat. Biotechnol., 2018, 36, 1083–1090 CrossRef CAS PubMed.
- J. H. Ding, G. Li, J. Xiong, F. L. Liu, N. B. Xie, T. T. Ji, M. Wang, X. Guo, Y. Q. Feng, W. Ci and B. F. Yuan, Anal. Chem., 2024, 96, 4726–4735 CrossRef CAS PubMed.
- N. B. Xie, M. Wang, W. Chen, T. T. Ji, X. Guo, F. Y. Gang, Y. F. Wang, Y. Q. Feng, Y. Liang, W. Ci and B. F. Yuan, ACS Cent. Sci., 2023, 9, 2315–2325 CrossRef CAS PubMed.
- Y. Hao, T. T. Ji, S. Y. Gu, S. Zhang, Y. H. Gu, X. Guo, L. Zeng, F. Y. Gang, J. Xiong, Y. Q. Feng, N. B. Xie and B. F. Yuan, Chem. Sci., 2025, 16, 8752–8763 RSC.
- R. Vaisvila, V. K. C. Ponnaluri, Z. Y. Sun, B. W. Langhorst, L. Saleh, S. X. Guan, N. Dai, M. A. Campbell, B. S. Sexton, K. Marks, M. Samaranayake, J. C. Samuelson, H. E. Church, E. Tamanaha, I. R. Correa, S. Pradhan, E. T. Dimalanta, T. C. Evans, L. Williams and T. B. Davis, Genome Res., 2021, 31, 1280–1289 CrossRef CAS PubMed.
- M. Wang, N. B. Xie, K. K. Chen, T. T. Ji, J. Xiong, X. Guo, S. Y. Yu, F. Tang, C. Xie, Y. Q. Feng and B. F. Yuan, Anal. Chem., 2023, 95, 1556–1565 CAS.
- R. Vaisvila, S. R. Johnson, B. Yan, N. Dai, B. M. Bourkia, M. Chen, I. R. Correa Jr, E. Yigit and Z. Sun, Mol. Cell, 2024, 84, 854 CrossRef CAS PubMed.
- S. Zhang, Y. Liang, T. Feng, X. Guo, M. Wang, T. T. Ji, J. Xiong, X. Xiao, Y. Liu, Y. B. Liu, W. M. Ci, N. B. Xie and B. F. Yuan, CCS Chem., 2025 DOI:10.31635/ccschem.025.202405023.
- N. B. Xie, M. Wang, T. T. Ji, X. Guo, F. Y. Gang, Y. Hao, L. Zeng, Y. F. Wang, Y. Q. Feng and B. F. Yuan, Chem. Sci., 2024, 15, 10073–10083 RSC.
- Y. Liu, Z. Hu, J. Cheng, P. Siejka-Zielinska, J. Chen, M. Inoue, A. A. Ahmed and C. X. Song, Nat. Commun., 2021, 12, 618 CrossRef PubMed.
- J. Xiong, K. K. Chen, N. B. Xie, T. T. Ji, S. Y. Yu, F. Tang, C. Xie, Y. Q. Feng and B. F. Yuan, Anal. Chem., 2022, 94, 15489–15498 CrossRef CAS PubMed.
- T. Wang, J. M. Fowler, L. Liu, C. E. Loo, M. Luo, E. K. Schutsky, K. N. Berrios, J. E. DeNizio, A. Dvorak, N. Downey, S. Montermoso, B. Y. Pingul, M. Nasrallah, W. S. Gosal, H. Wu and R. M. Kohli, Nat. Chem. Biol., 2023, 19, 1004–1012 CrossRef CAS PubMed.
- W. Liu, Z. C. Ma, S. Zhang, F. Y. Gang, T. T. Ji, Y. H. Gu, N. B. Xie, S. Y. Gu, X. Guo, T. Feng, Y. Liu, J. Xiong and B. F. Yuan, Chem. Sci., 2025, 16, 8788–8799 RSC.
- Y. Liu, P. Siejka-Zielinska, G. Velikova, Y. Bi, F. Yuan, M. Tomkova, C. Bai, L. Chen, B. Schuster-Bockler and C. X. Song, Nat. Biotechnol., 2019, 37, 424–429 CrossRef CAS PubMed.
- M. Ravichandran, D. Rafalski, C. I. Davies, O. Ortega-Recalde, X. Nan, C. R. Glanfield, A. Kotter, K. Misztal, A. H. Wang, M. Wojciechowski, M. Razew, I. M. Mayyas, O. Kardailsky, U. Schwartz, K. Zembrzycki, I. M. Morison, M. Helm, D. Weichenhan, R. Z. Jurkowska, F. Krueger, C. Plass, M. Zacharias, M. Bochtler, T. A. Hore and T. P. Jurkowski, Sci. Adv., 2022, 8, eabm2427 CrossRef CAS PubMed.
- X. Guo, J. Wu, T. T. Ji, M. Wang, S. Zhang, J. Xiong, F. Y. Gang, W. Liu, Y. H. Gu, Y. Liu, N. B. Xie and B. F. Yuan, Chem. Sci., 2025, 16, 3953–3963 RSC.
- W. X. Shao, Y. H. Min, W. Chen, J. Xiong, X. Guo, N. B. Xie, S. Zhang, S. Y. Yu, C. Xie, Y. Q. Feng and B. F. Yuan, Anal. Chem., 2023, 95, 10588–10594 CrossRef CAS PubMed.
- J. Xiong, K. K. Chen, N. B. Xie, W. Chen, W. X. Shao, T. T. Ji, S. Y. Yu, Y. Q. Feng and B. F. Yuan, Chin. Chem. Lett., 2024, 35, 108953 CrossRef CAS.
- Y. H. Min, W. X. Shao, Q. S. Hu, N. B. Xie, S. Zhang, Y. Q. Feng, X. W. Xing and B. F. Yuan, Anal. Chem., 2024, 96, 8730–8739 CrossRef CAS PubMed.
- X. Guo, N. B. Xie, W. Chen, T. T. Ji, J. Xiong, T. Feng, M. Wang, S. Zhang, S. Y. Gu, Y. Q. Feng and B. F. Yuan, Anal. Chem., 2024, 96, 847–855 CrossRef CAS PubMed.
- M. Wang, N. B. Xie, F. Y. Gang, S. Zhang, L. Zeng, T. T. Ji, J. Xiong, X. Guo, Y. Hao, Y. Liu and B. F. Yuan, Sci. China Life Sci., 2025 DOI:10.1007/s11427-024-2702-8.
- C. T. Coey, S. S. Malik, L. S. Pidugu, K. M. Varney, E. Pozharski and A. C. Drohat, Nucleic Acids Res., 2016, 44, 10248–10258 CAS.
- X. J. You, S. Zhang, J. J. Chen, F. Tang, J. He, J. Wang, C. B. Qi, Y. Q. Feng and B. F. Yuan, Nucleic Acids Res., 2022, 50, 9858–9872 CrossRef CAS PubMed.
- C. J. Ma, G. Li, W. X. Shao, Y. H. Min, P. Wang, J. H. Ding, N. B. Xie, M. Wang, F. Tang, Y. Q. Feng, W. Ci, Y. Wang and B. F. Yuan, ACS Cent. Sci., 2023, 9, 1799–1809 CrossRef CAS PubMed.
- M. Yu, G. C. Hon, K. E. Szulwach, C. X. Song, L. Zhang, A. Kim, X. Li, Q. Dai, Y. Shen, B. Park, J. H. Min, P. Jin, B. Ren and C. He, Cell, 2012, 149, 1368–1380 CrossRef CAS PubMed.
- H. Hashimoto, J. E. Pais, X. Zhang, L. Saleh, Z. Q. Fu, N. Dai, I. R. Correa Jr, Y. Zheng and X. Cheng, Nature, 2014, 506, 391–395 CrossRef CAS PubMed.
- H. Hashimoto, J. E. Pais, N. Dai, I. R. Correa Jr, X. Zhang, Y. Zheng and X. Cheng, Nucleic Acids Res., 2015, 43, 10713–10721 CrossRef CAS PubMed.
- S. Sappa, D. Dey, B. Sudhamalla and K. Islam, J. Am. Chem. Soc., 2021, 143, 11891–11896 CrossRef CAS PubMed.
- M. N. Vassylyeva, S. Klyuyev, A. D. Vassylyev, H. Wesson, Z. Zhang, M. B. Renfrow, H. Wang, N. P. Higgins, L. T. Chow and D. G. Vassylyev, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5138–E5147 CrossRef CAS PubMed.
- C. Seoighe, N. J. Tosh and J. M. Greally, Nat. Genet., 2018, 50, 1062–1063 CrossRef CAS PubMed.
- A. Nishiyama and M. Nakanishi, Trends Genet., 2021, 37, 1012–1027 CrossRef CAS PubMed.
- Z. D. Smith, S. Hetzel and A. Meissner, Nat. Rev. Genet., 2025, 26, 7–30 CrossRef CAS PubMed.
- S. Y. Gu, T. Feng, F. Y. Gang, S. Y. Yu, W. Chan, Z. C. Ma, Y. H. Gu and B. F. Yuan, Chin. Chem. Lett., 2025, 36, 110957 CrossRef.
- T. Feng, Y. L. Gao, D. Hu, K. Y. Yuan, S. Y. Gu, Y. H. Gu, S. Y. Yu, J. Xiong, Y. Q. Feng, J. Wang and B. F. Yuan, Chin. Chem. Lett., 2024, 35, 109259 CrossRef CAS.
- P. Wee and Z. Wang, Cancers, 2017, 9, 52 CrossRef PubMed.
- J. Jassem and R. Dziadziuszko, Lancet Respir. Med., 2020, 8, 528–529 CrossRef CAS PubMed.
- Y. D. Ma, H. D. Wang, R. Zhang, R. B. Li, H. Q. Zhang, X. J. Li and H. L. Wang, CCS Chem., 2025 DOI:10.31635/ccschem.025.202405046.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc03634h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |