Open Access Article
Open Access ArticleBottom-up design of peptide shapes in water using oligomers of N-methyl-L/D-alanine†
Jumpei
Morimoto‡
 *a,
Marin
Yokomine‡
*a,
Marin
Yokomine‡
 a,
Yota
Shiratori‡
b,
Takumi
Ueda
a,
Yota
Shiratori‡
b,
Takumi
Ueda
 cd,
Takayuki
Nakamuro
cd,
Takayuki
Nakamuro
 e,
Kiyofumi
Takaba
f,
Saori
Maki-Yonekura
e,
Kiyofumi
Takaba
f,
Saori
Maki-Yonekura
 f,
Koji
Umezawa
gh,
Koichiro
Miyanishi
i,
Yasuhiro
Fukuda
f,
Koji
Umezawa
gh,
Koichiro
Miyanishi
i,
Yasuhiro
Fukuda
 a,
Takumu
Watanabe
a,
Mayuko
Suga
a,
Ayumi
Inayoshi
a,
Takuya
Yoshida
c,
Wataru
Mizukami
i,
Koh
Takeuchi
a,
Takumu
Watanabe
a,
Mayuko
Suga
a,
Ayumi
Inayoshi
a,
Takuya
Yoshida
c,
Wataru
Mizukami
i,
Koh
Takeuchi
 d,
Koji
Yonekura
d,
Koji
Yonekura
 fjk,
Eiichi
Nakamura
fjk,
Eiichi
Nakamura
 e and
Shinsuke
Sando
e and
Shinsuke
Sando
 *ab
*ab
aDepartment of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo, Tokyo 113-8656, Japan. E-mail: jmorimoto@chembio.t.u-tokyo.ac.jp; ssando@chembio.t.u-tokyo.ac.jp
bDepartment of Bioengineering, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo, Tokyo 113-8656, Japan. E-mail: ssando@chembio.t.u-tokyo.ac.jp
cGraduate School of Pharmaceutical Sciences, Osaka University, 1-6, Yamadaoka, Suita, Osaka, Japan
dGraduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo, Tokyo 113-0033, Japan
eDepartment of Chemistry, The University of Tokyo, 7-3-1 Hongo, Bunkyo, Tokyo 113-0033, Japan
fRIKEN SPring-8 Center, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan
gDepartment of Biomedical Engineering, Graduate School of Science and Technology, Shinshu University, 8304 Minami-minowa, Kami-ina, Nagano 399-4598, Japan
hInstitute for Biomedical Sciences, Interdisciplinary Cluster for Cutting Edge Research, Shinshu University, Matsumoto, Nagano 390-8621, Japan
iCenter for Quantum Information and Quantum Biology, Osaka University, 1-2 Machikaneyama, Toyonaka, Osaka 560-0043, Japan
jAdvanced Electron Microscope Development Unit, RIKEN-JEOL Collaboration Center, RIKEN Baton Zone Program, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan
kInstitute of Multidisciplinary Research for Advanced Materials, Tohoku University, 2-1-1 Katahira, Aoba, Sendai, Miyagi 980-8577, Japan
First published on 2nd May 2025
Abstract
De novo design of peptide shapes is of great interest in biomolecular science since the local peptide shapes formed by a short peptide chain in the proteins are often key to biological activities. Here, we show that the de novo design of peptide shapes with sub-nanometer conformational control can be realized using peptides consisting of N-methyl-L-alanine and N-methyl-D-alanine residues. The conformation of N-methyl-L/D-alanine residue is largely fixed because of the restricted bond rotation and hence can serve as a scaffold on which we can build a peptide into a designed shape. The local shape control by per-residue conformational restriction by torsional strains starkly contrasts with the global shape stabilization of proteins based on many remote interactions. The oligomers allow the bottom-up design of diverse peptide shapes with a small number of amino acid residues and would offer unique opportunities to realize the de novo design of biofunctional molecules.
Introduction
Molecular functions depend on the three-dimensional structures of the molecules. Therefore, the establishment of the design principles of three-dimensional molecular shapes in water is essential in molecular science for producing bio-functional molecules. The biological system utilizes proteins composed of nature's choice of suitable sequences of amino acids. Their diverse functions originate from local three-dimensional peptide shapes formed by short peptide sequences within the global protein structure. For example, an antibody is composed of 600–700 amino acid residues, but its essential function, i.e., antigen recognition, is majorly achieved by one of the local loop structures called complementarity determining region H3 consisting of 2–26 amino acid residues.1 Each amino acid residue that constitutes a protein is intrinsically conformationally flexible, but the conformational freedom is restricted by remote interactions among amino acid residues, e.g., hydrogen bonds, van der Waals contacts, hydrophobic interactions, and electrostatic interactions, resulting in the construction of the tertiary structure of proteins (Fig. 1a). Many interactions among amino acid residues are required to realize a hierarchically stabilized global protein structure and peptide shapes therein,2–4 and previous studies suggest that approximately 50 amino acids are necessary for proteins to adopt their folds.5 | ||
| Fig. 1 The comparison of peptide shape in the globally folded protein structures and the bottom-up formation of peptide shape consisting of conformationally fixed amino acid residues. (a) The conformation of each amino acid residue in proteins changes depending on the amino acid sequence. The Ramachandran plot was generated using PyRAMA (https://github.com/gerdos/PyRAMA). The three-dimensional structure of chignolin (PDB ID: 1UAO) is shown as an example of miniature proteins. The peptide shapes in the representative miniature proteins are plotted on the PMI plot. Plotted are the peptide shapes of consecutive six amino acid residues (AAs) in the proteins. (b) The peptides consisting of the previously reported amino acid residues with fixed conformations. Oligoproline (brown), oligooxopiperazine (purple), and oligo-NMA (green) are plotted on the PMI plot. (Oligoproline and oligo-NMA are overlapped on the plot.) The structures of oligoproline and oligo-L-NMA were generated using CCDC-1014542 (P. Wilhelm, et al., J. Am. Chem. Soc., 2014, 136, 15829–15832) and CCDC-278108 (S. Zhang, et al., Chem. Commun., 2006, 497–499), respectively. The oligooxopiperazine structure was constructed using 3D molecular modelling software (Avogadro) based on the previous report (P. Tošovská and P. S. Arora, Org. Lett., 2010, 12, 1588–1591). (The modelled structure is included as a ESI† file.) (c) The peptides consisting of L-NMA and D-NMA residues that have fixed conformations. The peptide shapes realized by oligo-NMA of 6-AAs and 10-AAs are plotted on the PMI plot. | ||
There are rare exceptions called miniature proteins, e.g., Trpzip,6 Trp-cage,7 chignolin,8,9 HP7,10 and a small protein containing a WSXWS motif,11 that achieve global stabilization of a three-dimensional structure with less than 20 amino acid residues (Fig. 1a). The interplay of extensive inter-residual interactions can realize such a small protein. For example, the β-sheet structure of CLN025, a chignolin analog, is stabilized by hydrogen bonds and an aromatic–aromatic interaction.9 The helix–loop–strand structure of the miniature protein containing a WSXWS motif is stabilized by an extensive cation–π interaction network.11 However, ten or more amino acid residues are still required to achieve a stable global structure and local peptide shapes.
Bottom-up peptide design using conformationally fixed amino acid residues is a promising alternative approach to creating peptide shapes with a minimal number of amino acid residues. Having a fixed local conformation, some amino acid residues strongly stabilize specific structural motifs per amino acid residue and hence can be utilized for the bottom-up design of peptide shapes (Fig. 1b). Proline residue is a typical example. The ring constraint of proline allows its oligomer to stably adopt a rod-like shape called PPII helix.12 An oxopiperazine residue is another example of a conformationally-constrained building block. Oligomers of oxopiperazine are conformationally constrained and have been utilized as molecular scaffolds.13–15N-Methylalanine (NMA) residue has also been recently reported to be an amino acid residue with a fixed conformation. The conformation of the NMA residue in its oligomers is rigidified by pseudo-1,3-allylic strain, and the oligomer forms a stable extended shape.16–18 These amino acid residues have hence served as attractive building blocks for the bottom-up design of peptide shapes.19,20 The bottom-up approach enables the direct design of peptide shapes with a minimal number of amino acid residues. However, the bottom-up design of stable peptide shapes using a single building block has so far realized repetitive linear sequences with limited structural diversity.
The principal moment of inertia (PMI) analysis21 is a method for quantitatively comparing the three-dimensional structures of different classes of molecules. The PMI plots in the right panels of Fig. 1a and b illustrate the large gap in peptide shape diversity between proteins and bottom-up designed peptide. The analysis extracts the three principal moments of inertia from the molecules and plots the information on a triangular diagram indicating the rod-, disc-, and sphere-likeness of the three-dimensional molecular structure. Here, for a fair comparison, the PMI analysis was conducted using the main chain atoms (N, Cα, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of six consecutive amino acid residues in each miniature protein and peptide. As represented by the PMI plot, a wide range of three-dimensional structures including rod-like, disc-like, and sphere-like are realized by the peptide shapes within the hierarchically folded miniature proteins (Fig. 1a). In contrast, the bottom-up peptide shape designs have realized only rod-like shapes using conformationally-fixed amino acid residues (Fig. 1b). Since the molecular shape visualized by the PMI plot is intimately linked to the biological activity of the molecules,21 expansion of the peptide shape diversity realized by the bottom-up approach is important.
O) of six consecutive amino acid residues in each miniature protein and peptide. As represented by the PMI plot, a wide range of three-dimensional structures including rod-like, disc-like, and sphere-like are realized by the peptide shapes within the hierarchically folded miniature proteins (Fig. 1a). In contrast, the bottom-up peptide shape designs have realized only rod-like shapes using conformationally-fixed amino acid residues (Fig. 1b). Since the molecular shape visualized by the PMI plot is intimately linked to the biological activity of the molecules,21 expansion of the peptide shape diversity realized by the bottom-up approach is important.
Here, we report a non-hierarchical bottom-up design of diverse peptide shapes that self-stand in water using a combination of N-methyl-L-alanine (L-NMA) and N-methyl-D-alanine (D-NMA) residues as building blocks (Fig. 1c). We reasoned that the combinatorial usage of multiple building blocks with fixed conformations would realize diverse three-dimensional molecular shapes. While the incorporation of D-amino acids is a simple approach to expanding the conformational diversity of peptides,22–24 standard non-methylated D-amino acids are not conformationally fixed. We assumed combining L-NMA and D-NMA would be a promising strategy, as D-NMA should serve as a building block with a fixed conformation.
The concept of using L/D-NMA in combination was inspired by the seminal work of Kodadek and coworkers, where a library of oligomers composed of oligo-L/D-NMA backbones with functional groups at the amide nitrogens was utilized as a source of protein ligands.25–27 Notably, the stereoinversion of one residue in a protein ligand was shown to change the protein affinity of the oligomer,25 suggesting that different sequences of oligo-L/D-NMA form distinct shapes. Their work indicated that the oligomers consisting of L/D-NMA backbone realize diverse shapes, but oligomers only up to three consecutive L/D-NMA residues have been explored and the oligomer shapes have not been investigated in their studies.
In this study, we describe sub-nanometer conformational control of the NMA oligomers through a rational combination of multiple L-NMA and D-NMA residues, as studied by NMR, crystallographic, and computational analyses, as well as by direct imaging of the dynamics of the peptide's shape using a cinematographic electron microscopic technique.28,29 The diversity of the peptide shapes achieved by the bottom-up design using oligo-L/D-NMA approaches the level realized by miniature proteins (Fig. 1c, right). We also demonstrated the utility of oligo-L/D-NMAs as scaffolds by showing that the introduction of functional groups on an oligo-L/D-NMA does not significantly alter the backbone shape.
Results and discussion
Two conformationally constrained monomers as building blocks for the de novo design of the three-dimensional peptide shapes
Previously, density-functional theory (DFT) calculations suggested that a minimal model of L-NMA oligomer stably forms conformations that avoid pseudo-1,3-allylic strains.17,18 To further confirm that the building block maintains the same stable conformation in water, more detailed calculations, and experimental validations were conducted. The allowed conformational space of an amino acid residue in a peptide chain is commonly visualized using the Ramachandran plot.30 Here, we recruited a similar plot on which the free energy of an amino acid residue is plotted against the two conformational determinants: dihedral angles φ and ψ. We first reproduced the Ramachandran-type plot of acetyl-N-methyl-L-alanine dimethylamide (1), a minimal model of L-NMA oligomers, by DFT calculations with minor modifications on the procedures from the previous report.18 In these calculations, φ and ψ values of 1, which are the two determinants of the conformation of the molecule, were combinatorically rotated from −180° to 180° by 15° and the 625 conformers were geometry optimized by DFT calculations at the B3LYP/6-31G(d) level of theory with implicit water model. The energy of each conformer was calculated at the same level and plotted. The generated Ramachandran-type plot of 1 was shown to have a narrow low-energy region around the lowest energy conformer with (φ, ψ) = (−135°, 75°), which is consistent with the previous report18 (Fig. 2a). This result suggests that the minimal model structure forms a fixed conformation in water. | ||
| Fig. 2 Ramachandran-type plots of minimal model structures of oligo-N-methylalanine. (a) Ramachandran-type plot of acetyl-N-methyl-L-alanine dimethylamide (a minimal model of L-NMA oligomers). (b) The DFT-optimized structure of L-NMA. The lowest-energy conformer in the Ramachandran-type plot in Fig. 2a was geometry optimized. Hydrogen pairs on non-neighboring carbons showing medium to strong NOE signals are connected using red arrows. The hydrogens with NOE signals are shown in yellow. (c) The crystal structure of acetyl-N-methyl-L-alanine with a C-terminal Boc-piperazine. Hydrogen atoms are omitted for clarity. For (b and c), atoms and bonds other than the L-NMA residue are shown in gray. (d) Ramachandran-type plot of acetyl-N-methyl-D-alanine dimethylamide (a minimal model of D-NMA oligomers). | ||
To experimentally validate that the minimal L-NMA model stably forms the lowest-energy conformer from the DFT calculations, we conducted NMR measurements in water and obtained nuclear Overhauser effect (NOE) signals that reflect the distances between the pairs of protons (Fig. S1–S4 and Table S1, S2†). The consistency of the observed NOE signals with the proton proximities on the stable conformers was examined. On the stable conformers from the DFT calculations, the two proton pairs were found to come close to each other: intra-residual N-methyl protons and β-protons; the α-proton and N-methyl protons of the C-terminal structure. This is reasonable considering the conformational restrictions by pseudo-1,3-allylic strains. Consistent with the proton proximities in the DFT model (Fig. 2b), medium to strong NOE signals were observed for the two proton pairs (Fig. S4 and Table S1†). The NOE signals strongly suggest that the minimal L-NMA model stably forms the lowest energy conformer from the DFT calculations.
X-ray crystallographic studies were also conducted to obtain an atomic-resolution snapshot of the monomer structure. To generate single crystals, derivatives of 1 with different N/C-terminal groups were synthesized and subjected to crystallization. From several tested compounds, acetyl-N-methyl-L-alanine with a C-terminal 1-(tert-butyloxycarbonyl)piperazine (Boc-piperazine) (2) provided single crystals with good X-ray diffraction. The solved structure was close to the lowest-energy conformer of the minimal monomer model 1 from the DFT calculations. The φ and ψ angles of the solved structure were −134.6° and 77.7°, respectively (Fig. 2c), which is close to those of the most stable conformation predicted by the DFT calculation. These computational and experimental studies strongly suggest that the building block (L-NMA residue) forms a fixed conformation in water.
The Ramachandran-type plot of acetyl-N-methyl-D-alanine dimethylamide (3), which is a D-NMA minimal model, was also generated using the same procedures. As expected, the Ramachandran-type plot is point symmetric to the plot of the L-NMA minimal model 1 and has a narrow low-energy region around (φ, ψ) = (135°, −75°) (Fig. 2d). These results indicate that L-NMA residue and D-NMA residue can be used as building blocks with a fixed conformation for a bottom-up peptide shape design.
A minimal model study of the bottom-up peptide shape design using dimers consisting of L-NMA and D-NMA residues
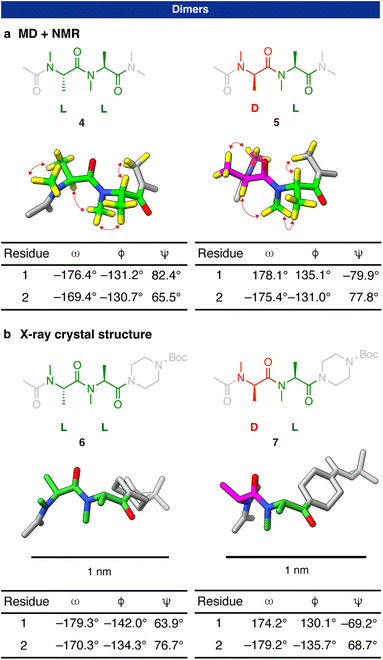
We hypothesized that combinatorial usages of the two building blocks can realize the rational control of diverse peptide shapes with sub-nanometer conformational control. Because the conformations of the building blocks are largely fixed, an oligomer is expected to form a predictable shape that is a simple connection of the stable conformations of the building blocks.To examine the hypothesis, we first analyzed the three-dimensional structures of dimers, which are the shortest oligomers. LL dimer (4) and DL dimer (5) were studied (Fig. 3). (L and D in the oligomer sequence denote L-NMA residue and D-NMA residue, respectively.) DD dimer and LD dimer were not examined since they are mirror images of the two dimers.
The stable structures of the two dimers were assessed using a combined computational and experimental approach. We first conducted multicanonical molecular dynamics (McMD) simulations31 to comprehensively explore the allowed conformational space of the two dimers. The generated conformers at 300 K were grouped into 10 distinct clusters (Table S3†). As a result, the representative conformation from the top cluster of both dimers was found to have dihedral angles (φ, ψ) within 10° of the lowest energy point on the Ramachandran-type plot, specifically (φ, ψ) = (−135°, 75°) for L-NMA residue and (135°, −75°) for D-NMA residue. In the conformations, the two amide bonds were found to be both in the trans geometry. These results suggest that the dimers stably form predictable three-dimensional structures in which the stable conformations of the two monomers are connected with a trans amide bond.
We conducted NMR measurements to experimentally validate the conformations of the dimers predicted by the computations (Fig. S5–S12 and Table S4–S7†). On the 1H-NMR spectrum of the LL dimer 4, there was a dominant set of peaks with two sets of minor peaks. According to the NOE signals, the dominant set of peaks corresponds to a conformational state with which the amide connecting the acetyl group and the 1st residue and the amide connecting the 1st and 2nd residues are both in the trans geometry (78% of the total observed population). The two minor sets of peaks correspond to conformational states in which one of the two amides is in trans, and the other is in cis. Approximately 90% of each amide bond in 4 was in the trans geometry (Fig. S6†), which is evidenced by the existence of the inter-residual NOE between N-methyl protons and β-protons and the absence of the inter-residual NOE between α-protons (Fig. S8 and Table S4†). This is consistent with the McMD result that the two amides in the dominant conformer are both in the trans geometry. As with the monomer structure, two kinds of hydrogen pairs are in close proximities on the stable conformer: (1) intra-residual N-methyl protons and β-protons; (2) the α-proton and N-methyl protons of the following NMA residue or C-terminal structure (Fig. 3a, left). Consistently, medium to strong NOE signals were observed for these hydrogen pairs (Table S4†). A similar result was obtained for the DL dimer 5. 82% of the conformational ensemble was in a conformational state in which all the amides were in the trans geometry. In addition, the NOE signals expected from the proton proximities on the calculated stable conformer were observed (Fig. 3a, right and Table S6†). These results have shown that the dimers stably form the three-dimensional structures in which the most stable conformation of L-NMA residue and D-NMA residue are simply connected with a trans amide bond.
It is worth noting that the amide bond connecting the two NMA monomers in the LL and DL dimers adopts a trans geometry in 88% and 93% of the population, respectively. This suggests that the trans geometry is preferred for both homochiral and heterochiral connections. On the other hand, in a previous study, the trans![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cis ratio of an N-methylamide differed significantly between a homochiral LL dimer and a heterochiral DL dimer.32 A key difference from our study is that, in the previous report, only one amide bond in the peptide sequence was N-methylated, while the other amides were not. These non-N-methylated amides are likely involved in intramolecular hydrogen bonding, which can influence the trans
cis ratio of an N-methylamide differed significantly between a homochiral LL dimer and a heterochiral DL dimer.32 A key difference from our study is that, in the previous report, only one amide bond in the peptide sequence was N-methylated, while the other amides were not. These non-N-methylated amides are likely involved in intramolecular hydrogen bonding, which can influence the trans![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cis equilibrium. In contrast, in our system, all amide bonds are N-meth-ylated, eliminating the possibility of intramolecular hydrogen bonding. Our results therefore suggest that N-methylamides generally prefer the trans geometry regardless of the stereochemistry when all amide bonds in the sequence are N-methylated.
cis equilibrium. In contrast, in our system, all amide bonds are N-meth-ylated, eliminating the possibility of intramolecular hydrogen bonding. Our results therefore suggest that N-methylamides generally prefer the trans geometry regardless of the stereochemistry when all amide bonds in the sequence are N-methylated.
X-ray crystallographic study of the dimers further confirmed the dimer structures. Dimers with the N-terminal acetamide and C-terminal Boc-piperazine amide (6 and 7) gave X-ray quality single crystals. The conformers found in the crystals are close to the most stable conformers predicted by the calculations. All the dihedral angles (φ, ψ) in the crystal structures (Fig. 3b) were within ±15° of the values corresponding to the lowest energy point on the Ramachandran-type plot in Fig. 2.
Bottom-up design of peptide shapes in water using the oligo-L/D-NMAs
To showcase the utility of L/D-NMA oligomers for the de novo design of diverse three-dimensional peptide shapes, we examined hexameric oligomers and generated a list of the predicted stable conformations using DFT calculations (Fig. 4a). The most stable conformations of L/D-NMA residues were connected, and the geometry of the generated conformer was optimized by DFT calculations at the B3LYP/6-311G(d) level. The collection of the predicted structures is shown in Fig. 4a. The shapes were also plotted on the PMI plot to evaluate the shape diversity (Fig. 1c, right, blue circles). The plot demonstrates that combining two amino acid residues, i.e., L- and D-NMA residues, realizes a large shape diversity. The peptide shape diversity approaches the level that is achieved by the same chain length (six amino acid residues) of peptides in miniature proteins (Fig. 1a, right). | ||
| Fig. 4 Diverse peptide shapes realized by the oligo-L/D-NMAs. (a) A list of peptide shapes realized by hexameric L/D-NMA oligomers. The stable conformations of L-NMA and D-NMA residues were connected with trans amides, and the resulting structures were optimized by DFT calculations to generate the model structures. Among the possible 64 diastereomers, six stereoisomers that have intramolecular steric crashes are omitted (their possible conformations are shown in Fig. S13†). Considering that the remained 58 stereoisomers are 29 enantiomer pairs, only 29 diastereomers that are not enantiomers with each other are shown. The hexamers that were experimentally examined are highlighted by black squares. (b) Solution structure of a hexameric oligomer LLDDLL. A representative conformation in the highest population cluster from the McMD simulations is shown. (c) A crystal structure of LLDDLL obtained by XFEL crystallography. (d) Solution structure of LLLLDL. A representative conformation in the highest population cluster from the McMD simulations is shown. For (b and d), the non-neighboring hydrogen pairs with significant NOE signals are indicated by red arrows. For (b–d), hydrogens are omitted for clarity, and the dihedral angle (φ, ψ) pair of each NMA residue is plotted on the Ramachandran-type plot of L-NMA residue on the right using a magenta circle. To discuss L- and D-NMA residues on the same plot, the sign of each dihedral angle value of the D-NMA residues was reserved, and then the values were plotted. (e) The PMI plot of the predicted shape of LLDDLLDDLL (red circle) and LLLLLLLLLL (green circle). (f–g) A SMART-EM analysis taken at 50 fps and an electron dose rate of 3.8–4.9 × 106 e− nm−2 s−1 of the LLDDLLDDLL decamer (f) and the LLLLLLLLLL decamer (g). A stable conformation predicted from the shapes of the L/D-NMA residue is shown (left). A simulated TEM image (second from left). The TEM image of the oligomer on an amino-CNT (second from right). A molecular model (right). (h–i) The conformations of LLDDLLDDLL (h) and LLLLLLLLLL (i) from MD simulations that match the observed TEM images. The time from the beginning of the observation is shown below the conformation. Dihedral angles (φ, ψ) of each L/D-NMA residue of the observed conformers are plotted on the Ramachandran-type plot of L-NMA as magenta circles. To discuss L- and D-NMA residues on the same plot, the sign of each dihedral angle value of the D-NMA residues was reserved, and then the values were plotted. | ||
To validate the predicted shapes, stable conformations of one of the oligomers (LLDDLL) were evaluated by a combination of computation and experiment. We first conducted McMD simulations of the oligomer (8) to systematically explore the allowed conformational space. The generated conformers at 300 K were grouped into 10 distinct clusters (Table S8†). To identify the valid conformations from the clustered structures, we next conducted NMR measurements in water (Fig. S14–S16 and Table S9, S10†). The dominant set of peaks corresponds to a conformational state with which all the amides are in the trans geometry according to the NOE signals (Fig. S16 and Table S9†). While minor peaks were observed in the NMR spectra corresponding to conformers with one or more cis amide bonds, the spectra are dominated by a set of peaks corresponding to the all-trans conformer, which accounts for 66% of the total population observed. This result indicates that the all-trans conformer is preferred in solution, although a minor population of alternative conformers is present due to the intrinsic cis–trans isomerization of tertiary amide bonds. The value is consistent with the NMR data for the LL dimer, in which approximately 10% of each amide bond adopts a cis geometry. As with the monomer and dimers, two kinds of hydrogen pairs are in close proximities on the stable conformer: (1) intra-residual N-methyl protons and β-protons; (2) α-proton and N-methyl protons of the following NMA residue or C-terminal structure. The representative conformer of the top cluster from McMD simulations was consistent with all the NMR signals (Fig. 4b and Table S9†); therefore, the conformer was determined to be the representative solution structure of oligo-NMA 8. The dihedral angles (φ, ψ) of all the NMA residues in the solution structure were within ±20° of the values corresponding to the lowest energy point on the Ramachandran-type plot of the L-NMA and D-NMA monomers (Fig. 4b). This result suggests that the NMA hexamer majorly forms the conformation determined by the low-energy conformations of the building blocks. We also conducted X-ray crystallographic studies to obtain atomic resolution snapshots of the oligomer structures. The LLDDLL oligomers with an N-terminal p-nitrobenzoyl group (9) yielded only tiny crystals that were too small for conventional single-crystal X-ray diffraction. Instead, still diffraction patterns were collected using high-intensity X-ray free electron lasers (XFEL), and the crystal structure was determined from the diffraction data by a serial XFEL crystallographic technique for smaller molecules recently introduced33 (Fig. 4c and Table S11†). The overall shape of the oligomer is close to the shape in the list and the solution structure. The dihedral angles (φ, ψ) of the 1st, 3rd, and 5th NMA residues are within ±15° of the values corresponding to the lowest energy point on the Ramachandran-type plot of the monomers, and the 2nd, 4th, and 6th NMA residues are slightly off from the lowest energy point. However, the dihedral angles of the 2nd, 4th, and 6th NMA residues are still within 2.5 kcal mol−1 from the lowest energy point on the Ramachandran-type plot of L/D-NMA residues. This deviation can be explained by the fact that the solution structure represents an ensemble-averaged conformation in dynamic equilibrium, whereas the X-ray crystal structure captures a single, static conformer stabilized under crystallization conditions. Such methodological differences are expected to result in small discrepancies in dihedral angles. This result, combined with the McMD simulations and NMR measurements, implies that the oligomer majorly forms the conformational state consisting of the lowest-energy conformations of the building blocks, but it has a small conformational freedom around the low-energy area on the Ramachandran-type plot.
To demonstrate the shape diversity realized by the oligo-L/D-NMA, another diastereomer, LLLLDL (10), was synthesized and analyzed by McMD simulations and NMR (Fig. 4d). The molecule is predicted to form a linear structure with a bent structure at the C-terminus (Fig. 4a). The oligomer was first analyzed by McMD simulations to obtain the allowed conformational space of the oligomer. The generated conformers at 300 K were grouped into 10 distinct clusters (Table S8†). We also conducted NMR measurements (Fig. S17–S19 and Table S12, S13†) to identify a valid conformation from the clustered conformations from McMD simulations. The representative conformer of the second cluster from McMD simulations was consistent with all the NMR signals (Fig. 4d and Table S12†); therefore, the conformer was determined to be the representative solution structure of oligo-NMA 10. The solution structure was found to have the dihedral angles (φ, ψ) of around (−135°, 75°) for L monomer and (135°, −75°) for D monomer. Similar to hexamer 8, the conformer in which all the amide bonds are in trans geometry accounted for the majority (74%) of the total population of hexamer 10, as indicated by the NMR spectra. These results further confirmed the conformational rigidity of the oligo-L/D-NMA.
As an additional structural analysis, we measured circular dichroism (CD) spectra of oligomers 8 and 10, as well as homochiral oligomers LLLLLL (S5) and DDDDDD (S6) (Fig. S20†). The homochiral oligomer S5 exhibited a β-strand-like CD spectrum, consistent with previous reports.17,25,34 The enantiomeric oligomer S6 showed a mirror-image spectrum, as expected. Oligomers 8 (LLDDLL) and 10 (LLLLDL) displayed similar spectral shapes to that of S5, but with lower intensity. This attenuation is consistent with the fact that the conformations of L- and D-NMA residues are defined on a per-residue basis, and that they produce CD spectra that are mirror images of each other. As a result, heterochiral oligomers exhibit similar spectral profiles to their homochiral analogues, but with reduced overall intensity.
These oligomer studies strongly support that the three-dimensional shapes of the oligopeptides can be designed with sub-nanometer conformational control by simply connecting the stable conformations of the building blocks.
Cinematographic microscopic imaging of structural dynamics of decamers
The foregoing experiments on oligopeptides consisting of two and six L/D-NMA residues provided strong enough evidence of the validity of our de novo shape-design strategy. However, none of the NMR, crystallographic, or computational methods can provide solid structural evidence for larger, flexible, and non-crystalline oligomers with huge conformational possibilities. Microscopic methods capable of sub-angstrom and sub-millisecond imaging over a long period of time should serve the purpose of probing this challenging issue. Thus, we examined the potential of the single-molecule atomic-resolution time-resolved electron microscopy (SMART-EM), which recently elucidated the dynamic structures of the oligomers of a lipopeptide antibiotic, daptomycin.35 The key feature of the method includes the chemical ligation of a peptide onto aminated conical carbon nanotubes (amino-CNTs) and cinematographic recording of the dynamic motions of peptide chains over minutes—the timeframe unavailable to molecular dynamics simulation.We examined the structures of oligomers consisting of ten L- and D-NMA residues, which is equal to the minimal amino acid residues for miniature proteins. The PMI plot of the oligomers indicates the shape diversity is further expanded and covers a wider area (Fig. 1c, right, red circles). We chose two oligomers, LLDDLLDDLL and LLLLLLLLLL, that locate at different regions on the PMI plot. The model structure of LLDDLLDDLL locates around the center of the PMI plot (Fig. 4e, red circle), while the model structure of LLLLLLLLLL is a rod-like shape and located at the top-left of the plot (Fig. 4e, green circle). DFT calculations suggest a shape of the alphabet “C” as the most stable conformation of the LLDDLLDDLL decamer, and a linear shape for the LLLLLLLLLL decamer (Fig. 4f, “Predicted shape”). The oligomer was synthesized with a C-terminal amide and conjugated to amino-CNTs with the aid of potassium acyltrifluoroborate (KAT) chemistry, as previously described.35 Since the TEM image contrast strongly depends on the atomic number of the atoms being imaged,36 we can selectively visualize the main chain of the peptide oligomers, especially by using a slightly larger defocus value (see ESI†). As shown in Fig. 4f “TEM image”, we indeed found that the LLDDLLDDLL decamer persistently took the shape of “C”, which matches a simulated image based on DFT calculation (Fig. 4f, “Simulation” and “Molecular model”). In sharp contrast, the LLLLLLLLLL decamer persistently took a linear shape, which matches a simulated image based on DFT calculation (Fig. 4g, “Simulation” and “Molecular model”).
Cinematographic atomic resolution imaging provides further structural information unavailable from the still pictures shown in Fig. 4f and g. During the SMART-EM observation for 10 s at 298 K at a frame rate of 50 frames per second (fps, 20.0 ms per frame), the “C” or the linear shape of the oligomers was maintained most of the time, but the shape fluctuated to some degree (Fig. S21†) indicating that minor conformations are populated occasionally. To assess whether the observed conformational freedom of the oligomer can be explained by the conformational freedom of the L/D-NMA residues on the Ramachandran-type plots, we conducted MD simulations in vacuo at 200 and 300 K for 500 ns starting from the DFT-derived stable conformation as the initial conformation. As found by the EM imaging, the MD data also indicated that alphabet “C”-like structures for the LLDDLLDDLL decamer and the linear structures for the LLLLLLLLLL decamer are the predominant conformers. The minor conformer results from the conformational fluctuation of L/D-NMA residues reflected in the low-energy region on the Ramachandran-type plots. We found a variety of conformers of the two decamers interchanging at the beginning of the SMART-EM observation and could assign the structure to most of them through comparison with the conformers found in the MD trajectories (Fig. 4h–i and Fig. S21, S22†). This agreement suggests that the EM observed conformational fluctuation falls into the range expected from the conformational fluctuation of the monomeric NMA residue at the low-energy region around (φ, ψ) = (−135°, 75°) on the Ramachandran-type plots. The dihedral angles (φ, ψ) of the observed conformers were restricted in a small region on the Ramachandran-type plot of the L/D-NMA monomer that is within 2.0 kcal mol−1 from the lowest energy point (Fig. 4h–i, right). The result is also consistent with the NMR analysis of the solution structure of the hexamers that the overall shape of an oligo-L/D-NMA is maintained in solution with a small degree of rotational freedom as assessed from the low-energy area on the Ramachandran-type plots. The EM observation has also given evidence that the combinatorial usage of the L- and D-NMA realizes the expansion of the peptide shape diversity that is produced from the bottom-up designs using conformationally fixed amino acid residues (Fig. 1c and 4e).
Utility of the peptide shapes as scaffolds for presenting functional groups
The defined peptide shapes are potentially useful for designing biofunctional molecules. They function as scaffolds to present multiple functional groups with specific spatial orientations.As demonstrated in our previous studies, as well as those of others, various functional groups can be readily introduced into oligo-L-NMAs at either the amide nitrogens or the α-carbons. Amide nitrogens can be functionalized via reductive amination17,37 or Fukuyama–Mitsunobu reactions,38 whereas α-carbons can be functionalized using amino acids other than alanine.18 The same strategies can be applied to functionalize oligo-L/D-NMAs.
We investigated whether the peptide shape of an oligo-L/D-NMA would be preserved upon the introduction of functional groups. To this end, we synthesized several oligo-NMA sequences bearing functional groups at both the amide nitrogens and the α-carbons. In designing these functionalized peptoids, we considered that introducing different N- and Cα-substituents would alter the electronic environments experienced by the backbone protons (i.e., α-, β-, and Nα-protons), thereby improving the chemical shift dispersion across residues. Specifically, we selected combinations such as benzyl and alkyl groups, as their differences in electronic characteristics were expected to create distinct local environments and help resolve overlapping signals in the NMR spectra. One of these sequences, designated as oligo-NMA 11 (Fig. 5a), showed well-separated 1H-NMR signals, enabling detailed conformational analysis of the peptide in solution using NMR spectroscopy and McMD simulations. We conducted McMD simulations and found that the representative conformation of the top cluster among the 10 clusters is consistent with the NMR spectroscopic data (Fig. S23–S25, and Table S14, S15†). Thus, the conformation shown in Fig. 5b was determined to be the representative solution structure (Table S16†). Notably, the oligo-NMA backbone conformation is maintained upon the introduction of substituents. More specifically, in the solution structure of functionalized oligo-NMA 11, all amide bonds adopt the trans geometry, and all φ and ψ angles are within 2 kcal mol−1 of the lowest-energy point on the Ramachandran-type plot. Consequently, the backbone shape of the functionalized oligo-NMA 11 overlapped well with that of its non-functionalized oligo-NMA counterpart (8) (root mean square deviation (RMSD) of the backbone atoms = 0.469 Å) (Fig. 5c). It is also noteworthy that no NOE signals were observed between the two aromatic groups at the 2nd and 6th residues, suggesting that the stabilized backbone conformation prevents their association. This finding supports the idea that heterochiral oligo-NMAs are useful as scaffolds for presenting functional groups.
Conclusions
In this study, we have shown that a rational combination of L-NMA residues and D-NMA residues allows us to de novo design a wide range of three-dimensional peptide shapes with sub-nanometer conformational control. We found that a single NMA residue predominantly exists as a single stable conformer in water and tends to maintain this conformation even when incorporated into an oligomeric sequence. This observation led us to propose a per-residue bottom-up design of peptide shapes by choosing appropriate combinations of L- and D-NMA residues incorporated into an oligomeric sequence.The combinatorial usage of the two NMA residues with fixed conformations, i.e., L-NMA and D-NMA residues, has enabled the bottom-up design of diverse peptide shapes in water (Fig. 4). The n-mer oligopeptides consisting of L/D-NMAs can potentially realize 2n different shapes. As demonstrated by the PMI plot, the bottom-up design realizes a large shape diversity (Fig. 1c), which approaches the diversity of the peptide shapes realized by miniature proteins. The bottom-up approach for the peptide shape design is more direct and straightforward compared to the design approach in proteins and foldamers,39–41 which are globally stabilized by a network of inter-residual interactions. The introduction of additional building blocks, such as a proline residue would further increase the peptide shape diversity in the future. It would realize the peptide shape diversity equal to or even larger than that realized by proteins.
Aside from the peptides that composed of α-amino acids discussed in this study, there are conceptually similar precedents in which oligomer shapes are designed using conformationally constrained building blocks. Among these, Spiroladder oligomers developed by Schafmeister's group are pioneering examples. The oligomers are conformationally constrained, and various shapes can be designed through the combinatorial use of multiple distinct bis-amino acid monomers.42–44
Diverse shape design realized by oligo-L/D-NMAs has a large advantage over the non-peptidic shape-programmable molecules in their minimal structures and high-density functionalizable sites. As discussed in our previous study on oligo-L-NMA,18 oligo-L/D-NMA does not require a hydrogen-bonding network or ring constraints for conformational constraints and, thus, requires a minimal number of atoms for achieving the fixed conformations. Furthermore, various functional groups can be easily installed both at the amide nitrogen and the α-carbon of each building block of the oligomers using a peptide-like solid-phase synthetic method.17,25,38,45 The facile design of functionalized peptide shapes with sub-nanometer precision in water would be useful for the de novo design of biofunctional molecules, such as protein ligands and organic catalysts. Despite these advantages, it should be noted that a fraction of the population of an oligo-NMA adopts cis-amide conformations due to the inherent cis–trans isomerization of tertiary amide bonds. While the all-trans conformer is dominant, the presence of minor cis conformers may influence molecular recognition or function in applications. Therefore, careful consideration of amide bond geometry may be important in the future design of functionalized oligo-NMAs.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. Authors Jumpei Morimoto, Marin Yokomine and Yota Shiratori contributed equally.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Takuma Kato for his help in solving the crystal structures, and Drs Ichiro Inoue and Kensuke Tono for setting up the XFEL data collection system. We also thank Prof. Koji Harano for the helpful discussion in the early stages of this work. The authors acknowledge the One-stop Sharing Facility Center for Future Drug Discoveries at the University of Tokyo for the use of micrOTOF II. A part of the computation was performed using Research Center for Computational Science, Okazaki, Japan (Project: 23-IMS-C071). Another part of the computation was carried out using the facilities of the Super-computer Center at the Institute for Solid State Physics, University of Tokyo. The XFEL experiments were performed at the BL2 of SACLA with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal No. 2022A8025). This work was supported by PRESTO, Japan Science and Technology Agency, Grant Number JPMJPR21AF (to J. M.), JSPS KAKENHI (JP23K26771 to J. M.), CREST, Japan Science and Technology Agency, Grant Numbers JPMJCR21N5 (to S. S.), JST-Mirai Program (Grant Number JPMJMI20G5 to K. Y.), Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP22ama121006 (to K. Y., S. M.-Y. and K. T.), JSPS KAKENHI (JP19H05459 to E. N., JP22K14704 to T. N.), a Grant-in-Aid for Scientific Research on Innovative Areas “Materials Science of Meso-Hierarchy” from Japan Society for the Promotion of Science (JSPS) JP23H04874, JSPS KAKENHI (JP20K21494, JP20H03378, and JP22K18374) (to K. T.), and MEXT/JSPS KAKENHI (JP23H02618, JP21H05509, and JP20H03375) (to T. U.). The NMR experiments were performed at NMR Platform of The University of Tokyo supported by MEXT, Japan.Notes and references
- T. T. Wu, G. Johnson and E. A. Kabat, Proteins: Struct. Funct. Bioinf., 1993, 16, 1–7 CrossRef CAS PubMed.
- W. F. DeGrado and I. V Korendovych, Q. Rev. Biophys., 2020, 53, e3 CrossRef.
- D. N. Woolfson, J. Mol. Biol., 2021, 433, 167160 CrossRef CAS.
- P. S. Huang, S. E. Boyken and D. Baker, Nature, 2016, 537, 320–327 CrossRef CAS.
- P. L. Privalov, Annu. Rev. Biophys. Biophys. Chem., 1989, 18, 47–69 CrossRef CAS PubMed.
- A. G. Cochran, N. J. Skelton and M. A. Starovasnik, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5578–5583 CrossRef CAS PubMed.
- J. W. Neidigh, R. M. Fesinmeyer and N. H. Andersen, Nat. Struct. Biol., 2002, 9, 425–430 CrossRef CAS.
- S. Honda, K. Yamasaki, Y. Sawada and H. Morii, Structure, 2004, 12, 1507–1518 CrossRef CAS.
- S. Honda, T. Akiba, Y. S. Kato, Y. Sawada, M. Sekijima, M. Ishimura, A. Ooishi, H. Watanabe, T. Odahara and K. Harata, J. Am. Chem. Soc., 2008, 130, 15327–15331 CrossRef CAS.
- N. H. Andersen, K. A. Olsen, R. M. Fesinmeyer, X. Tan, F. M. Hudson, L. A. Eidenschink and S. R. Farazi, J. Am. Chem. Soc., 2006, 128, 6101–6110 CrossRef CAS.
- T. W. Craven, M.-K. Cho, N. J. Traaseth, R. Bonneau and K. Kirshenbaum, J. Am. Chem. Soc., 2016, 138, 1543–1550 CrossRef CAS PubMed.
- P. Wilhelm, B. Lewandowski, N. Trapp and H. Wennemers, J. Am. Chem. Soc., 2014, 136, 15829–15832 CrossRef CAS PubMed.
- P. Tošovská and P. S. Arora, Org. Lett., 2010, 12, 1588–1591 CrossRef PubMed.
- B. B. Lao, K. Drew, D. a Guarracino, T. F. Brewer, D. W. Heindel, R. Bonneau and P. S. Arora, J. Am. Chem. Soc., 2014, 136, 7877–7888 CrossRef CAS.
- B. B. Lao, I. Grishagin, H. Mesallati, T. F. Brewer, B. Z. Olenyuk and P. S. Arora, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7531–7536 CrossRef CAS.
- S. Zhang, S. Prabpai, P. Kongsaeree and P. I. Arvidsson, Chem. Commun., 2006, 497–499 RSC.
- J. Morimoto, Y. Fukuda, D. Kuroda, T. Watanabe, F. Yoshida, M. Asada, T. Nakamura, A. Senoo, S. Nagatoishi, K. Tsumoto and S. Sando, J. Am. Chem. Soc., 2019, 141, 14612–14623 CrossRef CAS PubMed.
- M. Yokomine, J. Morimoto, Y. Fukuda, Y. Shiratori, D. Kuroda, T. Ueda, K. Takeuchi, K. Tsumoto and S. Sando, Angew. Chem., Int. Ed., 2022, 61, e202200119 CrossRef CAS PubMed.
- S. Dobitz, M. R. Aronoff and H. Wennemers, Acc. Chem. Res., 2017, 50, 2420–2428 CrossRef CAS PubMed.
- J. Morimoto and S. Sando, J. Synth. Org. Chem., Jpn., 2020, 78, 1076–1084 CrossRef CAS.
- W. H. B. Sauer and M. K. Schwarz, J. Chem. Inf. Comput. Sci., 2003, 43, 987–1003 CrossRef CAS PubMed.
- V. Nanda and W. F. DeGrado, J. Am. Chem. Soc., 2004, 126, 14459–14467 CrossRef CAS.
- V. Nanda and W. F. DeGrado, J. Am. Chem. Soc., 2006, 128, 809–816 CrossRef CAS.
- J. D. Kim, D. H. Pike, A. M. Tyryshkin, G. V. T. Swapna, H. Raanan, G. T. Montelione, V. Nanda and P. G. Falkowski, J. Am. Chem. Soc., 2018, 140, 11210–11213 CrossRef CAS PubMed.
- Y. Gao and T. Kodadek, Chem. Biol., 2013, 20, 360–369 CrossRef CAS PubMed.
- Y. Gao, S. Amar, S. Pahwa, G. Fields and T. Kodadek, ACS Comb. Sci., 2015, 17, 49–59 CrossRef CAS PubMed.
- T. M. Doran, Y. Gao, S. Simanski, P. McEnaney and T. Kodadek, Bioorg. Med. Chem. Lett., 2015, 25, 4910–4917 CrossRef CAS.
- M. Koshino, T. Tanaka, N. Solin, K. Suenaga, H. Isobe and E. Nakamura, Science, 2007, 316, 853 CrossRef CAS PubMed.
- E. Nakamura, Acc. Chem. Res., 2017, 50, 1281–1292 CrossRef CAS PubMed.
- G. N. Ramachandran, C. Ramakrishnan and V. Sasisekharan, J. Mol. Biol., 1963, 7, 95–99 CrossRef CAS PubMed.
- J. Ikebe, K. Umezawa, N. Kamiya, T. Sugihara, Y. Yonezawa, Y. Takano, H. Nakamura and H. Higo, J. Comput. Chem., 2011, 32, 1286–1297 CrossRef CAS PubMed.
- P. Lahiri, H. Verma, A. Ravikumar and J. Chatterjee, Chem. Sci., 2018, 9, 4600–4609 RSC.
- K. Takaba, S. Maki-Yonekura, I. Inoue, K. Tono, T. Hamaguchi, K. Kawakami, H. Naitow, T. Ishikawa, M. Yabashi and K. Yonekura, Nat. Chem., 2023, 15, 491–497 CrossRef CAS.
- S. Zhang, S. Prabpai, P. Kongsaeree and P. I. Arvidsson, Chem. Commun., 2006, 497–499 RSC.
- T. Nakamuro, K. Kamei, K. Sun, J. W. Bode, K. Harano and E. Nakamura, J. Am. Chem. Soc., 2022, 144, 13612–13622 CrossRef CAS.
- J. Xing, K. Takeuchi, K. Kamei, T. Nakamuro, K. Harano and E. Nakamura, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2114432119 CrossRef CAS.
- K. Pels and T. Kodadek, ACS Comb. Sci., 2015, 17, 152–155 CrossRef CAS PubMed.
- Y. Fukuda, M. Yokomine, D. Kuroda, K. Tsumoto, J. Morimoto and S. Sando, Chem. Sci., 2021, 12, 13292–13300 RSC.
- S. H. Gellman, Acc. Chem. Res., 1998, 31, 173–180 CrossRef CAS.
- W. S. Horne and S. H. Gellman, Acc. Chem. Res., 2008, 41, 1399–1408 CrossRef CAS PubMed.
- B. A. F. Le Bailly and J. Clayden, Chem. Commun., 2016, 52, 4852–4863 RSC.
- C. E. Schafmeister, Z. Z. Brown and S. Gupta, Acc. Chem. Res., 2008, 41, 1387–1398 CrossRef CAS.
- Z. Z. Brown, J. Alleva and C. E. Schafmeister, Biopolymers, 2011, 96, 578–585 CrossRef CAS.
- M. F. L. Parker, S. Osuna, G. Bollot, S. Vaddypally, M. J. Zdilla, K. N. Houk and C. E. Schafmeister, J. Am. Chem. Soc., 2014, 136, 3817–3827 CrossRef CAS PubMed.
- M. Yokomine, J. Morimoto, Y. Fukuda, T. Ueda, K. Takeuchi, K. Umezawa, H. Ago, H. Matsuura, G. Ueno, A. Senoo, S. Nagatoishi, K. Tsumoto and S. Sando, Chem. Sci., 2024, 15, 7051–7060 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 2251714 and 2258775–2258777. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d5sc01483b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |