Open Access Article
Open Access ArticleA supramolecular FRET signal amplification nanoprobe for high contrast and synchronous in situ imaging of cell surface receptor homodimers/heterodimers†
Ya
Wang
a,
Feng
Yao
a,
Lulu
Song
a,
Mengpan
Zhang
a,
Zitong
Gong
a,
Yunli
Zhao
a,
Yamin
Xiong
*b and
Leiliang
He
 *a
*a
aCollege of Public Health, Zhengzhou University, No. 100 Science Avenue, Zhengzhou City, 450001, China. E-mail: llhe2015@zzu.edu.cn
bSchool of Life Sciences, Zhengzhou University, No. 100 Science Avenue, Zhengzhou City, 450001, China. E-mail: xiongym@zzu.edu.cn
First published on 17th February 2025
Abstract
Epidermal growth factor receptor (EGFR) homodimers and heterodimers play significant roles in a variety of tumors, but current imaging probes remain problematic due to restricted contrast and sensitivity. Thus, we have developed aptamer-mediated activated conformational transitions to target the EGFR and HER2. Furthermore, based on signal amplification techniques, especially the FRET fluorescence enhancement properties of poly-β-CD, supramolecular FRET signal amplification nanoprobes were constructed to improve imaging contrast and sensitivity. The results confirmed that the fluorescence intensity of the supramolecular FRET group probe is 1.2 to 1.3 times that of the multi-FRET group and 11.3 to 23.2 times that of the single-FRET group. The results further confirmed that the supramolecular nanoprobe could not only be activated by tumor cells and tissues to achieve high-contrast imaging of EGFR/EGFR and EGFR/HER2 dimers, but also successfully distinguish tumor cells and tissues from normal cells and tissues. The strategy provides a generalized platform for high-contrast imaging of other dimers intending to deepen the understanding of the central roles of multiple dimers in cancer development.
Introduction
Cell membrane receptors, as essential components of cells, play crucial roles in fundamental cellular activities such as proliferation, differentiation, signal transduction, and immune recognition by recognizing various ligands.1,2 Consequently, many human diseases are closely associated with the aberrant expression and activation of receptors.3,4 Specifically, the oligomerization of receptor proteins on the cell membrane surface is typically the initial step in intracellular signal transduction.5 This process induces phosphorylation and activates key downstream signaling pathways, ultimately leading to changes in cell morphology and behavior.6 For instance, the overexpression and abnormal activation of human-derived epidermal growth factor receptor (HER or ERBB) family members, including EGFR (HER1), HER2, HER3, and HER4, are closely linked to the development of various cancers, such as non-small cell lung cancer, breast cancer, and pancreatic cancer.7–11 Further studies have shown that the EGFR can undergo not only homodimerization but also heterodimerization with other HER family members, such as HER2, further contributing to the oncogenic signaling pathways of the HER family.12–15 This affects patient sensitivity to drugs, directly impacting the effectiveness of targeted therapy and the choice of treatment options. Therefore, developing an efficient and accurate method to detect the dimerization status of the EGFR on the cell membrane has significant clinical value for early auxiliary diagnosis, efficacy monitoring, and prognostic assessment of lung cancer.Immunohistochemistry (IHC) remains the most commonly used method for detecting cell membrane receptors in clinical pathology laboratories. Regrettably, this method fails to accurately detect their dimerization status in situ. Additionally, some studies have employed fluorescent protein genetic modifications to enable in situ visualization of protein–protein interactions by advanced fluorescence imaging techniques.16–18 Nevertheless, these methods involve genetic modifications that can affect receptor dimerization efficiency or interfere with other cellular biological processes.19 Furthermore, they are time-consuming and can yield unpredictable results.20,21 Recently, the proximity ligation assay (PLA) technique has been introduced as a non-genetic alternative to analyze the dimerization status of cell surface receptors.22–29 This technology also provides an option for the study of artificially regulated cell surface receptor interactions.30–33 In this context, in the choice of affinity ligands for neighboring probes, aptamers (called “chemical antibodies”) have obvious advantages over antibodies, such as small size, easy synthesis and modification, and low production costs.34–36 Importantly, as oligonucleotides, aptamers eliminate the tedious coupling and purification process in the preparation of neighboring probes. However, these non-genetically engineered strategies still have limitations. For instance, the detection sensitivity of current strategies is insufficient for effective in situ imaging of low-abundance homo/heterodimers. Additionally, the signaling pattern of the probes in these current strategies results in a high imaging background, and requires cleaning of the probes, which prolongs the detection time during imaging analysis.
To address the above issues, this study designed an “activatable” aptamer fluorescent probe based on the PLA technique, combined with in situ hybridization chain reaction (HCR)-mediated FRET signal amplification and the FRET fluorescence enhancement properties of poly-β-CD, to develop a supramolecular FRET signal amplification nanoprobe with high specificity and high imaging contrast. When the dimer is present, the hairpin-type aptamer probe of this strategy is activated, triggering HCR-mediated amplification of the FRET signal to generate a significant fluorescent signal with a low fluorescence quenching background. This study demonstrates the practicality of supramolecular FRET amplification strategies for high-contrast imaging of receptor dimerization states through different human cell lines as well as tumor tissues from the A549 tumor mouse model. Thus, the supramolecular FRET signal amplification nanoprobe is promising to provide a convenient and generalized platform to sensitively monitor the expression of multiple dimers in samples simultaneously without an intermediate washing step.
Results and discussion
The principle of a supramolecular FRET signal amplification nanoprobe
Fig. 1 illustrates the working mechanism of the dynamic assembly of a supramolecular FRET signal amplification nanoprobe on living cell membranes to visualize homodimers/heterodimers. The sequence of nanoprobes is shown in Table S1.† Initially, activatable aptamer fluorescent probes R1 and R2 containing EGFR aptamer sequences37 (labeled with FAM and Dabcy1) and R3 containing HER2 aptamer sequences38 (labeled with Cy3 and BHQ2) were incubated with the cells. Whereafter, the hairpin types H1, H2, H3 and H4 were added. Aptamer-receptor recognition induces conformational transitions of the probes (R1, R2, and R3) to restore fluorescence for the EGFR and HER2 monomer imaging and exposes the terminal single strands. When dimers are present, the terminal single strands partially hybridize to trigger the subsequent signal amplification reaction (H1/H2/H1/H2… and H3/H4/H3/H4…) mediated by the proximity effect (R1/R2 and R2/R3). In situ self-assembled FRET signal amplification nanoprobes (R1/R2/H1/H2… and R2/R3/H3/H4…) generate “activated” FRET amplified signals for EGFR/EGFR homodimer (FAM as the donor and Cy5 as the acceptor) and EGFR/HER2 heterodimer detection (Cy3 as the donor and Cy7 as the acceptor). Then, the detection signal was further improved by adding poly-β-CD as a FRET fluorescence enhancer. Finally, high-contrast simultaneous imaging of homo/heterodimers on the same living cell surface can be achieved by monitoring Cy5 and Cy7 channel acceptor fluorescence signals.Characterization of FRET signal amplification nanoprobes
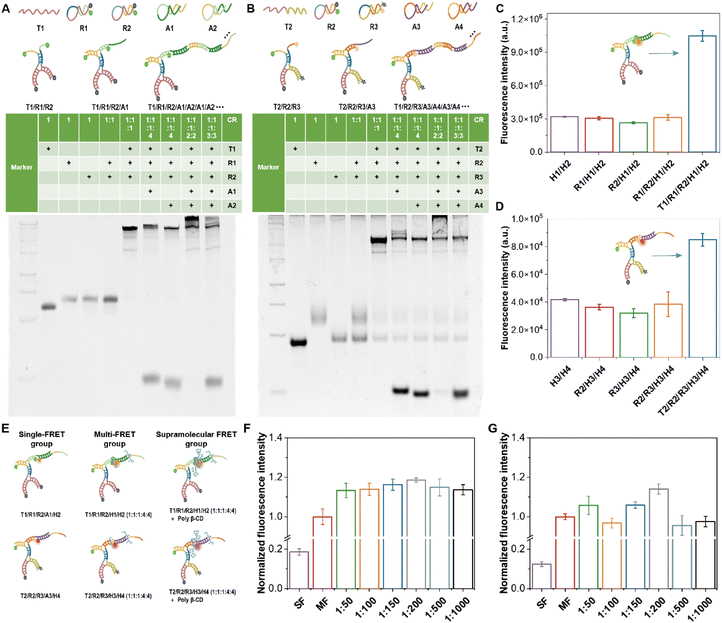
After that, whether the proximity ligation could activate HCR self-assembly amplification was further explored. Taking T1/R1/R2/A1/A2… as an example (Fig. 2A), the lanes of DNA single strand T1, R1 and R2 all showed only one clear band. Without dimer simulator strand T1, R1 and R2 did not hybridize. Adding T1 resulted in a clear delayed band, which indicated that T1 could open R1 and R2 to form a T1/R1/R2 double-stranded DNA complex. Based on the addition of the A1 chain, there was a more delayed band than T1/R1/R2, while with the addition of the A2 chain, the band was consistent with the position of T1/R1/R2, which indicated that T1/R1/R2 can only open A1. When all five strands were present, there were clear waterfall electrophoretic bands (T1/R1/R2/A1/A2/A1/A2…). The electrophoretic results of T2/R2/R3/A3/A4/A3/A4… were also similar (Fig. 2B). The above results indicated that the HCR self-assembled nanoprobe induced by proximity ligation has been successfully constructed.
Condition optimization of FRET signal amplification nanoprobes
The highly efficient assembly of FRET signal-amplifying nanoprobes is essential for obtaining sensitive and high-contrast receptor imaging effects. Therefore, the effects of different reaction times and reaction concentration ratios on the fluorescence signals of FRET signal amplification nanoprobes were explored. First, after mixing S1, H1 and H2, the fluorescence rapidly reached the highest intensity at 5 min, and then remained stable within 150 min, indicating that the amplification was rapid and stable (Fig. S3A†). As shown in the results of S2, H3 and H4 in Fig. S3B,† the fluorescence intensity also increased rapidly at 5 min, and then remained stable within 120 min, indicating rapid and relatively stable amplification. A too short reaction time may reduce the detection sensitivity, and a too long time may increase the cost. Therefore, 30 min was chosen as the HCR-mediated FRET signal amplification reaction time for the subsequent experiments. Then six different gradients were set to optimize the concentration ratios of FRET signal amplification nanoprobes (Fig. S4A and S4B†). Taking S1/H1/H2 as an example, the quantitative results (Fig. S4C†) showed that the fluorescence intensity of Cy5 increased rapidly in the early stage, and then the growth tendency slowed down after the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio. Similarly, the results of S2/H3/H4 showed that the fluorescence intensity of Cy7 was continuously enhanced (Fig. S4D†). Combining these results, 1
10 ratio. Similarly, the results of S2/H3/H4 showed that the fluorescence intensity of Cy7 was continuously enhanced (Fig. S4D†). Combining these results, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 was adopted as the optimal HCR-mediated FRET signal amplification concentration ratio for subsequent experiments.
10 was adopted as the optimal HCR-mediated FRET signal amplification concentration ratio for subsequent experiments.
Cyclodextrin (CD) enhances FRET signal amplification
Due to the unique structure of cyclodextrin (CD), which is hydrophilic on the outside and hydrophobic on the inside, when a dye forms an inclusion complex with it, the dye molecule is confined within the hydrophobic cavity of CD, reducing its freedom of movement. This encapsulation protects the excited state of the dye, prevents contact with water molecules, and maintains a suitable microenvironment, thereby enhancing the fluorescence signal.39,40 To construct a supramolecular FRET signal amplification nanoprobe and improve the sensitivity of the assay, the effects of four cyclodextrins (α-CD, β-CD, γ-CD, and poly-β-CD) on the fluorescence signals were then explored. For each cyclodextrin, one control group and four supramolecular groups were set up. Taking α-CD, T1/R1/R2/H1/H2 as an example, in which the control group was only T1/R1/R2/H1/H2 (the concentration ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), the concentration ratios of T1/R1/R2/H1/H2 to α-CD in the supramolecular group were 1
4), the concentration ratios of T1/R1/R2/H1/H2 to α-CD in the supramolecular group were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; 1
200; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000; 1
2000; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000; and 1
000; and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, respectively. The results (Fig. S5†) showed that poly-β-CD led to the best enhancement of receptor fluorescence at different ratios of concentration, reaching 1.21-fold at a concentration ratio of 1
000, respectively. The results (Fig. S5†) showed that poly-β-CD led to the best enhancement of receptor fluorescence at different ratios of concentration, reaching 1.21-fold at a concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. To further investigate the effect of poly-β-CD, the concentration gradient was re-created focusing on a concentration ratio of T1/R1/R2/H1/H2 to poly-β-CD of 1
200. To further investigate the effect of poly-β-CD, the concentration gradient was re-created focusing on a concentration ratio of T1/R1/R2/H1/H2 to poly-β-CD of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. Also, T1/R1/R2/A1/H2 was set up as the single-FERT group, T1/R1/R2/H1/H2 (1
200. Also, T1/R1/R2/A1/H2 was set up as the single-FERT group, T1/R1/R2/H1/H2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) as the multi-FRET group and T1/R1/R2/H1/H2/poly-β-CD as the supramolecular FRET group, respectively, and the schematic diagrams are shown in Fig. 2E. The quantitative results of T1/R1/R2/H1/H2 showed that the fluorescence enhancement of the supramolecular FRET group was 6.39-fold compared with that of the single-FRET group and about 1.19-fold compared with that of the multi-FRET group (Fig. 2F). The fluorescence enhancement of the supramolecular FRET group was the best when the ratio of the concentration of T1/R1/R2/H1/H2 and poly-β-CD was 1
4) as the multi-FRET group and T1/R1/R2/H1/H2/poly-β-CD as the supramolecular FRET group, respectively, and the schematic diagrams are shown in Fig. 2E. The quantitative results of T1/R1/R2/H1/H2 showed that the fluorescence enhancement of the supramolecular FRET group was 6.39-fold compared with that of the single-FRET group and about 1.19-fold compared with that of the multi-FRET group (Fig. 2F). The fluorescence enhancement of the supramolecular FRET group was the best when the ratio of the concentration of T1/R1/R2/H1/H2 and poly-β-CD was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. Similarly, when the ratio of T2/R2/R3/H3/H4 to poly-β-CD concentration was 1
200. Similarly, when the ratio of T2/R2/R3/H3/H4 to poly-β-CD concentration was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, which was 9.21-fold compared with that of single FRET and about 1.14-fold compared with that of the multi-FRET group (Fig. 2G).
200, which was 9.21-fold compared with that of single FRET and about 1.14-fold compared with that of the multi-FRET group (Fig. 2G).
Finally, it was adjusted to the optimal HCR-mediated FRET signal amplification concentration ratio (T/R/R/H/H concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) to explore the fluorescence enhancement of a poly-β-CD (T/R/R/H/H to poly-β-CD concentration ratio of 1
10) to explore the fluorescence enhancement of a poly-β-CD (T/R/R/H/H to poly-β-CD concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) for the nanoprobe (Fig. S6†). The results showed an 11.32-fold enhancement in the supramolecular FRET group (T1/R1/R2/H1/H2/poly-β-CD) compared with the single-FRET group (T1/R1/R2/A1/H2) and a 1.21-fold enhancement compared with the multi-FRET group. For T2/R2/R3/H3/H4, the fluorescence enhancement of the supramolecular FRET group was 23.13-fold higher than that of the single FRET group and 1.22-fold enhancement compared with the multi-FRET group. This fluorescence result confirmed the potential of poly-β-CD as a fluorescence enhancer involved in the construction of supramolecular FRET signal amplification nanoprobes.
200) for the nanoprobe (Fig. S6†). The results showed an 11.32-fold enhancement in the supramolecular FRET group (T1/R1/R2/H1/H2/poly-β-CD) compared with the single-FRET group (T1/R1/R2/A1/H2) and a 1.21-fold enhancement compared with the multi-FRET group. For T2/R2/R3/H3/H4, the fluorescence enhancement of the supramolecular FRET group was 23.13-fold higher than that of the single FRET group and 1.22-fold enhancement compared with the multi-FRET group. This fluorescence result confirmed the potential of poly-β-CD as a fluorescence enhancer involved in the construction of supramolecular FRET signal amplification nanoprobes.
Activatable aptamer fluorescent probe for cell membrane EGFR/HER2 monomer imaging
Then the ability of activatable probes (R1, R2 and R3) to recognize the receptor was further examined. First, the hairpin probe R1 (Fig. S7A†) was typically in a fluorescence-quenched state; fluorescence recovery occurred when incubated with A549 cells (human lung adenocarcinoma cells) and BEAS-2B cells (human normal bronchial epithelial cells). Also, the fluorescence intensity of A549 cells was significantly higher than that of BEAS-2B cells. The results of R2 and R3 probes were similar to those of R1 (Fig. S7B and C†). This phenomenon was consistent with the western blot results (Fig. S8†), that EGFR and HER2 were more expressed in A549 cells compared to BEAS-2B cells. Next, flow cytometry further validated the ability of R1, R2 and R3 to detect the receptor monomers. As shown in Fig. 3A and B, after incubating R1, R2 and R3 with A549 and BEAS-2B cells, respectively, the cells were seen to have obvious fluorescence enhancement. These results indicated that the constructed activatable probes can successfully target the corresponding receptors and produce activated fluorescence for detection.Additionally, R1, R2, and R3 were incubated with A549 and BEAS-2B cells respectively, and then subjected to confocal fluorescence imaging. As shown in Fig. 3C, the outline of the cell could be seen in A549 cells, whereas only a blurred cell boundary could be seen in BEAS-2B cells. The fluorescence quantification results (Fig. 3D) showed that the fluorescence intensity of the A549 cells was significantly higher than that of the BEAS-2B cells. Imaging results of the R2 and R3 probes were consistent with those of R1 (Fig. 3E–H). The above results demonstrated that the corresponding membrane receptors could be imaged with high contrast using the activatable probes to obtain the expression of the receptor profiles in different cell lines, and no obvious probe internalization was seen. This undoubtedly lays a solid foundation for further investigation of cell membrane EGFR homodimers as well as heterodimers in the subsequent steps.
Activatable FRET signal amplification nanoprobes for simultaneous in situ imaging of EGFR/EGFR and EGFR/HER2 dimers on cell membrane surfaces
Based on these results, we further validate the activatable probe and FRET signal amplification for cell membrane dimer imaging. First, flow cytometry was used to detect the activatable FRET signal amplification nanoprobe incubated with A549 cells and BEAS-2B cells, respectively. As shown in Fig. S9A and B,† EGFR/EGFR homodimers were detected by collecting the fluorescence of Cy5, and the fluorescence intensity of cells after incubation with the probe was significantly higher than that of the blank cell group. Moreover, the fluorescence intensity of A549 cells was significantly higher than that of BEAS-2B cells (Fig. S9C†), which indicated that the EFR/EGFR homodimer level was higher in A549 cells than BEAS-2B cells. Similarly, the EGFR/HER2 heterodimer level was also higher in A549 cells than in BEAS-2B cells (Fig. S10C†) by collecting fluorescence of Cy7 (Fig. S10A and B†). This correlated with the above results confirming the differences in EGFR and HER2 expression on the two types of cells. The above results indicated that the activatable FRET signal amplification nanoprobe could be activated by homo/heterodimers to generate the corresponding fluorescence amplification signals, thus realizing the detection of homo/heterodimers on different cell surfaces.Next, laser confocal microscopy was used to visualize EGFR homodimers/heterodimers on A549 and BEAS-2B cell membranes. The fluorescence of the donor FAM channel (first column) and the acceptor Cy5 channel (second column) of A549 cells was investigated under 488 nm and 638 nm excitation (Fig. S11A†). In addition, the fluorescence of acceptor Cy5 channels on untreated and EGF-treated A549 cells was investigated under 488 nm excitation, respectively (Fig. 4A). Fluorescence signals of EGFR/EGFR dimers were observed on the membrane of untreated A549 cells; the fluorescence signals were more than four-fold enhanced after EGF treatment, whereas no obvious fluorescence signals were seen in BEAS-2B cells (Fig. S12A†). This indicated that R1/R2/H1/H2 were activated to generate FRET amplification signals after recognizing EGFR/EGFR homodimers on the surface of A549 cell membranes. Additionally, under 552 nm and 638 nm excitation, fluorescence was investigated from the donor Cy3 channel and the acceptor Cy7 channel of A549 cells (Fig. S11B†). Then, the fluorescence of the Cy7 channel on untreated and EGF-treated A549 cells was investigated under 552 nm excitation (Fig. 4B). The results were consistent with EGFR/EGFR homodimers, and the fluorescent signal of EGFR/HER2 dimers could be observed on untreated A549 cells and more than two-fold enhancement of fluorescent signals could be observed on EGF-treated A549 cells, whereas no fluorescent signals were observed on BEAS-2B cells (Fig. S12B†). Furthermore, we also utilized probes to explore the expression of EGFR/EGFR and EGFR/HER2 dimers on human breast cancer cells (SKBR3) as well as human normal breast cells (MCF-10A). As shown in Fig. S13,† a small amount of dimer expression can be seen only on SKBR3 cells. Together, these results showed that the activatable FRET signal amplification nanoprobe could specifically recognize the corresponding dimer on the cell membrane and complete self-assembled FRET signal amplification to achieve high contrast imaging of homo/heterodimers in different cell lines by collecting the corresponding FRET fluorescence signals.
Finally, the activatable FRET signal amplification nanoprobe was also explored for simultaneous imaging of homo/heterodimers on cell membranes and effective differentiation of different cell lines. Live A549 cells were labeled using Calcein-AM, mixed with BEAS-2B cells and co-cultured in culture dishes. Then, the fluorescence signal of the Calcein-AM channel was collected. A549 cells showed bright green fluorescence, while no fluorescent signal was observed for BEAS-2B cells (Fig. S14†). This indicated that Calcein-AM does not crosstalk BEAS-2B cells and can be used to localize A549 cells. Next, mixed cells were imaged synchronously with two activatable FRET signal amplification nanoprobes. As shown in Fig. 4C, the fluorescence of the receptor Cy5 and Cy7 channels was only collected on the membrane surface of A549 cells pre-stained with Calcein-AM, while no obvious fluorescent signal of dimers was observed on BEAS-2B cells. This indicated that the constructed activatable FRET signal amplification nanoprobe was not only capable of simultaneous imaging of homo/heterodimers on the same cell surface, but also capable of visual differentiation of cells in mixed cells.
Supramolecular FRET signal amplification nanoprobes for imaging mouse lung adenocarcinoma tissue and normal lung tissue
To further validate the simultaneous visualization of EGFR/EGFR homodimers and EGFR/HER2 heterodimers in tissues by supramolecular FRET signal amplification nanoprobes, a tumor-bearing mouse model was constructed using A549 cells (Fig. S15A–C†). In the pathological results, HE staining (Fig. S15D†) showed that the tumor tissues showed dense and disordered growth; the results of IHC staining showed that the cells appeared to have brownish-yellow positive staining markers for the EGFR (Fig. S15E†) and HER2 (Fig. S15F†) in the tumor tissue sections, and the positive staining markers were higher than that of normal lung tissue. The above results indicated that the EGFR and HER2 were overexpressed in tumor tissues. Next, the activatable FRET signal amplification nanoprobe (FRET probe), as well as the supramolecular FRET signal amplification nanoprobe (FRET probe@poly-β-CD), were incubated with frozen sections of the two tissue types, respectively. The fluorescence of receptor Cy5 and receptor Cy7 was investigated. As shown in Fig. 5A, the tumor tissues showed bright fluorescence signals, and the fluorescence of the supramolecular FRET signal amplification probe group was stronger than that of the FRET probe. Normal lung tissues showed only weak fluorescence signals (Fig. 5B). The imaging results of supramolecular FRET signal amplification nanoprobes were compatible with the results of pathological HE staining and IHC staining. Additionally, we further explored the expression of dimers on the tumor-adjacent tissues. The results, as shown in Fig. S16,† were similar to those of normal lung tissues, and only weak fluorescence was seen in tumor-adjacent tissues. These results demonstrated that the supramolecular FRET signal amplification nanoprobe could not only distinguish lung cancer tissues from normal tissues and tumor-adjacent tissues, but also visualize the EGFR/EGFR and EGFR/HER2 dimers in lung cancer tissues with high contrast.Conclusions
In this study, we have developed a supramolecular FRET signal amplification nanoprobe for high contrast and synchronous in situ imaging of homodimer EGFR/EGFR and heterodimer EGFR/HER2 on the cell surface. Flow cytometry and confocal fluorescence imaging results revealed the receptor expression profiles of EGFR and HER2 monomers as well as EGFR/EGFR and EGFR/HER2 dimers in two cell types. This process was achieved using the activatable nanoprobe and without an intermediate washing step. In addition, based on HCR-mediated FRET signal amplification, especially FRET fluorescence enhancement properties of poly-β-CD, it was confirmed that the fluorescence intensity of the supramolecular FRET group is 1.2 to 1.3 times that of the multi-FRET group and 11.3 to 23.2 times that of the single-FRET group. This not only significantly enhances imaging contrast, but also facilitates improved sensitivity in the early detection of cancer. Finally, the A549 tumor-bearing mouse model was constructed, and the supramolecular FRET signal amplification nanoprobe was further validated on tumor tissues for high-contrast imaging of EGFR/EGFR and EGFR/HER2 dimers as well as for effectively distinguishing tumor tissues from normal lung tissues. In summary, the results demonstrated that the supramolecular FRET signal amplification nanoprobe is a convenient and high-contrast imaging strategy to simultaneously monitor multiple dimers in the samples.Data availability
The data supporting this article have been included in the ESI.† Additional data are available upon request from the corresponding author.Author contributions
Ya Wang: methodology, validation, formal analysis, writing – original draft, visualization. Feng Yao: validation, methodology, investigation. Lulu Song: investigation, validation. Mengpan Zhang: resources, formal analysis. Zitong Gong: formal analysis, validation. Yunli Zhao: resources. Yamin Xiong: conceptualization, funding acquisition, supervision. Leiliang He: conceptualization, supervision, writing – review & editing, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82373630, 82073606, and 82103893); the training grant of Henan Province for Young Teachers (No. 2023GGJS009). All protocols referring to animals in this study were approved by the Life Science Ethics Committee of Zhengzhou University (ZZUIRB2024-169).Notes and references
- J. Petschnigg, B. Groisman, M. Kotlyar, M. Taipale, Y. Zheng, C. F. Kurat, A. Sayad, J. R. Sierra, M. M. Usaj, J. Snider, A. Nachman, I. Krykbaeva, M. S. Tsao, J. Moffat, T. Pawson, S. Lindquist, I. Jurisica and I. Stagljar, Nat. Methods, 2014, 11, 585–592 CrossRef CAS PubMed.
- J. A. Ramilowski, T. Goldberg, J. Harshbarger, E. Kloppmann, M. Lizio, V. P. Satagopam, M. Itoh, H. Kawaji, P. Carninci, B. Rost and A. R. R. Forrest, Nat. Commun., 2015, 6, 7866 CrossRef CAS PubMed.
- D. Y. Li and M. H. Wu, Signal Transduction Targeted Ther., 2021, 6, 291 CrossRef CAS PubMed.
- C. J. Peach, L. E. Edgington-Mitchell, N. W. Bunnett and B. L. Schmidt, Physiol. Rev., 2023, 103, 717–785 CrossRef CAS PubMed.
- C. H. Heldin, Cell, 1995, 80, 213–223 CrossRef CAS PubMed.
- J. M. Yuan, X. D. Dong, J. J. Yap and J. C. Hu, J. Hematol. Oncol., 2020, 13, 113 CrossRef PubMed.
- S. Halder, S. Basu, S. P. Lall, A. K. Ganti, S. K. Batra and P. Seshacharyulu, Expert Opin. Ther. Targets, 2023, 27, 305–324 CrossRef CAS PubMed.
- A. Zaczek, M. Welnicka-Jaskiewicz, K. P. Bielawski, J. Jaskiewicz, A. Badzio, W. Olszewski, P. Rhone and J. Jassem, J. Cancer Res. Clin. Oncol., 2008, 134, 271–279 CrossRef CAS PubMed.
- A. Wang-Gillam, V. Rimkunas, A. Abu-Yousif, T. M. Nywening, F. Gao, D. G. DeNardo, D. Linehan, A. G. Czibere and A. Lugovskoy, J. Clin. Oncol., 2015, 33, 295 Search PubMed.
- N. V. Sergina and M. M. Moasser, Trends Mol. Med., 2007, 13, 527–534 CrossRef CAS PubMed.
- L. H. Huang and L. W. Fu, Acta Pharm. Sin. B, 2015, 5, 390–401 CrossRef PubMed.
- S. Q. Ma, L. Zhang, Y. Ren, W. Dai, T. Q. Chen, L. P. Luo, J. Zeng, K. Mi, J. Y. Lang and B. R. Cao, Oncogene, 2021, 40, 2596–2609 CrossRef CAS PubMed.
- M. Riudavets, I. Sullivan, P. Abdayem and D. Planchard, Esmo Open, 2021, 6, 100260 CrossRef CAS PubMed.
- A. E. G. Lenferink, R. Pinkas-Kramarski, M. L. M. van de Poll, M. J. H. van Vugt, L. N. Klapper, E. Tzahar, H. Waterman, M. Sela, E. J. J. van Zoelen and Y. Yarden, EMBO J., 1998, 17, 3385–3397 CrossRef CAS PubMed.
- X. Bai, P. Y. Sun, X. H. Wang, C. K. Long, S. Y. Liao, S. Dang, S. S. Zhuang, Y. T. Du, X. Y. Zhang, N. Li, K. M. He and Z. Zhang, Cell Discovery, 2023, 9, 18 CrossRef CAS PubMed.
- K. M. He, Y. N. Fu, W. Zhang, J. H. Yuan, Z. J. Li, Z. Z. Lv, Y. Y. Zhang and X. H. Fang, Biochem. Biophys. Res. Commun., 2011, 407, 313–317 CrossRef CAS PubMed.
- W. Zhang, Y. X. Jiang, Q. Wang, X. Y. Ma, Z. Y. Xiao, W. Zuo, X. H. Fang and G. Chen, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15679–15683 CrossRef CAS PubMed.
- D. Koschut, L. Richert, G. Pace, H. H. Niemann, Y. Mely and V. Orian-Rousseau, Biochim. Biophys. Acta, 2016, 1863, 1552–1558 CrossRef CAS PubMed.
- R. Ueki, S. Atsuta, A. Ueki and S. Sando, J. Am. Chem. Soc., 2017, 139, 6554–6557 CrossRef CAS PubMed.
- P. Liu, A. Calderon, G. Konstantinidis, J. Hou, S. Voss, X. Chen, F. Li, S. Banerjee, J. E. Hoffmann, C. Theiss, L. Dehmelt and Y. W. Wu, Angew. Chem., Int. Ed., 2014, 53, 10049–10055 CrossRef CAS PubMed.
- J. Y. Li, L. P. Wang, J. M. Tian, Z. L. Zhou, J. Li and H. H. Yang, Chem. Soc. Rev., 2020, 49, 1545–1568 RSC.
- D. Treppiedi, G. Marra, G. Di Muro, R. Catalano, F. Mangili, E. Esposito, D. Calebiro, M. Arosio, E. Peverelli and G. Mantovani, Front. Endocrinol., 2022, 13, 892668 CrossRef PubMed.
- C. Dong, X. Y. Fang, J. R. Xiong, J. J. Zhang, H. Y. Gan, C. Y. Song and L. H. Wang, ACS Nano, 2022, 16, 14055–14065 CrossRef CAS PubMed.
- H. Liang, S. Chen, P. P. Li, L. P. Wang, J. Y. Li, J. Li, H. H. Yang and W. H. Tan, J. Am. Chem. Soc., 2018, 140, 4186–4190 CrossRef CAS PubMed.
- Q. X. Mao, L. C. Wang, D. D. Xue, X. X. Lin, F. Sun, P. C. Xu, J. R. Chen, W. Y. Li, X. C. Li, F. Yan and C. Hu, Anal. Chem., 2024, 96, 2022–2031 CrossRef CAS PubMed.
- C. Dong, X. Y. Fang, X. C. Qin, Y. R. Wang, J. J. Zhang, C. Y. Song and L. H. Wang, Anal. Chem., 2023, 95, 6810–6817 CrossRef CAS PubMed.
- Y. H. Li, X. Zhang, W. Pan, N. Li and B. Tang, Anal. Chem., 2020, 92, 11921–11926 CrossRef CAS PubMed.
- L. L. Xu, Z. Zhou, X. L. Gou, W. C. Shi, Y. Gong, M. Yi, W. Cheng and F. Z. Song, Biosens. Bioelectron., 2021, 179, 113064 CrossRef CAS PubMed.
- S. Fredriksson, M. Gullberg, J. Jarvius, C. Olsson, K. Pietras, S. M. Gústafsdóttir, A. Östman and U. Landegren, Nat. Biotechnol., 2002, 20, 473–477 CrossRef CAS PubMed.
- L. P. Wang, H. Liang, J. Sun, Y. C. Liu, J. Y. Li, J. Y. Li, J. Li and H. H. Yang, J. Am. Chem. Soc., 2019, 141, 12673–12681 CrossRef CAS PubMed.
- H. Li, M. Wang, T. H. Shi, S. H. Yang, J. H. Zhang, H. H. Wang and Z. Nie, Angew. Chem., Int. Ed., 2018, 57, 10226–10230 CrossRef CAS PubMed.
- R. Ueki, A. Ueki, N. Kanda and S. Sando, Angew. Chem., Int. Ed., 2016, 55, 579–582 CrossRef CAS PubMed.
- B. Li, Y. N. Wang and B. H. Liu, ACS Nano, 2023, 17, 22571–22579 CrossRef CAS PubMed.
- C. L. Ji, J. Y. Wei, L. Zhang, X. R. Hou, J. Tan, Q. Yuan and W. H. Tan, Chem. Rev., 2023, 123, 12471–12506 CrossRef CAS PubMed.
- L. L. Wu, Y. D. Wang, X. Xu, Y. L. Liu, B. Q. Lin, M. X. Zhang, J. L. Zhang, S. Wan, C. Y. Yang and W. H. Tan, Chem. Rev., 2021, 121, 12035–12105 CrossRef CAS PubMed.
- L. Li, S. J. Xu, H. Yan, X. W. Li, H. S. Yazd, X. Li, T. Huang, C. Cui, J. H. Jiang and W. H. Tan, Angew. Chem., Int. Ed., 2021, 60, 2221–2231 CrossRef CAS PubMed.
- D. L. Wang, Y. L. Song, Z. Zhu, X. L. Li, Y. Zou, H. T. Yang, J. J. Wang, P. S. Yao, R. J. Pan, C. J. Yang and D. Z. Kang, Biochem. Biophys. Res. Commun., 2014, 453, 681–685 CrossRef CAS PubMed.
- A. Sett, B. B. Borthakur and U. Bora, Clin. Transl. Oncol., 2017, 19, 976–988 CrossRef CAS PubMed.
- L. L. He, K. Q. Shi, X. L. Liu, Y. Wang, L. L. Song, D. Wu, Y. J. Wu, Y. M. Xiong and P. L. Huang, Sens. Actuators, B, 2023, 393, 1341126 Search PubMed.
- W. R. Bergmark, A. Davis, C. York, A. Macintosh and G. Jones, J. Phys. Chem., 1990, 94, 5020–5022 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, methods and additional tables and figures. See DOI: https://doi.org/10.1039/d4sc08004a |
| This journal is © The Royal Society of Chemistry 2025 |