Open Access Article
Open Access ArticleChemical proteomic profiling of lysine crotonylation using minimalist bioorthogonal probes in mammalian cells†
Yuan-Fei
Zhou‡
 a,
Shouli
Yuan‡
b,
Bin
Ma
a,
Jinjun
Gao
c and
Chu
Wang
a,
Shouli
Yuan‡
b,
Bin
Ma
a,
Jinjun
Gao
c and
Chu
Wang
 *ab
*ab
aSynthetic and Functional Biomolecules Center, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, China. E-mail: chuwang@pku.edu.cn
bPeking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China
cSchool of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen 518055, China
First published on 3rd January 2025
Abstract
Protein lysine crotonylation has been found to be closely related to the occurrence and development of various diseases. Currently, site identification of crotonylation is mainly dependent on antibody enrichment; however, due to the cost, heterogeneity, and specificity of antibodies, it is desired to develop an alternative chemical tool to detect crotonylation. Herein, we report an alkynyl-functionalized bioorthogonal chemical probe, Cr-alkyne, for the detection and identification of protein lysine crotonylation in mammalian cells. Our in-gel fluorescence and chemical proteomic analyses demonstrated that Cr-alkyne can be metabolically incorporated into lysine of histones and directly label known crotonylated proteins. We further applied Cr-alkyne to the proteome-wide profiling of crotonylation and revealed a large number of previously unreported modification sites, some of which could be validated by co-elution with synthetic peptides. Moreover, by integrating Cr-alkyne with quantitative chemical proteomics, we also explored the crotonylation sites regulated by HDACs, unveiling new HDAC regulated sites. Our study thus provides an enabling chemical tool for characterizing protein crotonylation and greatly expands our understanding of substrate proteins and functions of this important modification.
Introduction
Protein post-translational modifications (PTMs) are involved in regulating a variety of biological processes by affecting their substrate proteins' structure, activity, cellular localization and interactions with other biomolecules. Among PTMs, lysine acylation of proteins is widely distributed in organisms and known to play an indispensable role in regulating the chromatin structure, gene expression, enzyme activity, and protein–protein interactions.1 Currently, more than ten different types of lysine acylations have been discovered in mammalian cells, including lactylation,2 2-hydroxy(iso)butyrylation,3 crotonylation,4 malonylation,5 succinylation,6 glutarylation,7 β-hydroxybutyrylation,8 itaconylation,9etc.10,11Lysine crotonylation (Kcr), a covalent modification with a four carbon group containing a (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)-bond incorporated at the ε-amine of lysine's side chain, was identified as a novel PTM in 2011.4 More importantly, there has been an increase in the research between Kcr and various physiological and pathological processes, including differentiation,12 cancer development,13,14, kidney diseases,15 neurological diseases16 and metabolic disorders.17 Identification of crotonylated proteins and modification sites is an essential step to dissect the biological functions of crotonylation. Current studies are mainly dependent on pan anti-Kcr antibodies to detect modification sites. However, antibody-based approaches are limited by cost, heterogeneity, and specificity and, moreover, they are not suitable for monitoring dynamic turnover of crotonylation. Researchers have tried to identify crotonylated proteins by a TCEP-crotonyl reaction and evaluated their labeling efficiency. For this purpose, the method was only applied at the level of individual proteins.18 In order to overcome these limitations, an efficient, economic and accessible method is highly desired to detect and comprehensively analyze crotonylated proteins in mammalian cells.
C)-bond incorporated at the ε-amine of lysine's side chain, was identified as a novel PTM in 2011.4 More importantly, there has been an increase in the research between Kcr and various physiological and pathological processes, including differentiation,12 cancer development,13,14, kidney diseases,15 neurological diseases16 and metabolic disorders.17 Identification of crotonylated proteins and modification sites is an essential step to dissect the biological functions of crotonylation. Current studies are mainly dependent on pan anti-Kcr antibodies to detect modification sites. However, antibody-based approaches are limited by cost, heterogeneity, and specificity and, moreover, they are not suitable for monitoring dynamic turnover of crotonylation. Researchers have tried to identify crotonylated proteins by a TCEP-crotonyl reaction and evaluated their labeling efficiency. For this purpose, the method was only applied at the level of individual proteins.18 In order to overcome these limitations, an efficient, economic and accessible method is highly desired to detect and comprehensively analyze crotonylated proteins in mammalian cells.
Over the past few years, the strategy of using bioorthogonal chemical probes has emerged as a powerful alternative for studying PTMs.19–22 Incubation/administration of chemical probes with bioorthogonal handles (such as alkynes and azides) in cells or animals can selectively install these functional groups to target proteins at specific modification sites via metabolic labeling. Fluorescent dyes or/and biotin can be orthogonally conjugated at the modification sites by bioorthogonal chemistry reactions, such as the copper(I) catalyzed alkyne-azido cycloaddition (CuAAC) reaction,23,24 which enables selective visualization, enrichment and identification of metabolically labeled proteins.
Herein, we report the strategy of metabolic labelling using an alkyne-functionalized bioorthogonal probe, Cr-alkyne, for fluorescence detection and chemical proteomic profiling of protein crotonylation in mammalian cells (Fig. 1). Our in-gel fluorescence and chemoproteomic analyses demonstrated that Cr-alkyne can be metabolically incorporated into histones as well as other known crotonylated proteins. We further applied Cr-alkyne to the proteome-wide profiling of crotonylation and validated several newly discovered modification sites by co-elution with synthetic peptides. Furthermore, by using Cr-alkyne with quantitative chemoproteomics, we also explored the crotonylation sites regulated by HDACs.
 | ||
| Fig. 1 Schematic for the detection and identification of crotonylated proteins and site-specific quantification using a minimalist biorthogonal probe. | ||
Results and discussion
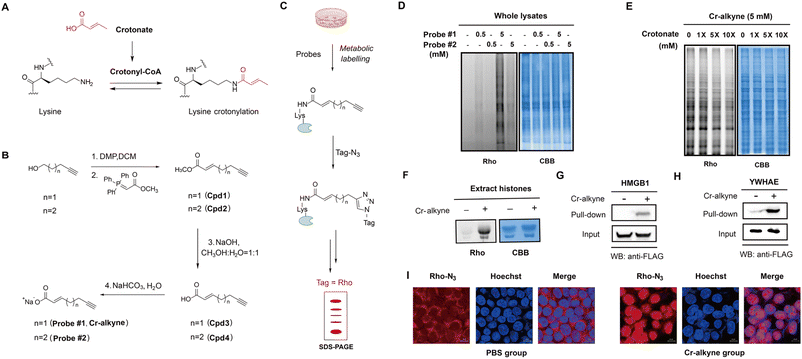
It was reported that exogenous treatment of sodium crotonate could increase Kcr levels in cells.25,26 Given that crotonate is the endogenous source of crotonyl-CoA (Fig. 2A), which is mediated by Acyl-CoA synthetase short chain family member 2,27 we hypothesized that an alkynyl analogue of crotonate should be metabolized and incorporated into crotonylated proteins in live mammalian cells. We thus designed and synthesized two crotonate derivative probes, probe #1 and #2 (Fig. 2B), both of which contain a terminal alkynyl group but with different linker lengths (n = 1 or 2). In comparison with the azide-containing probe, the alkyne-containing probe offers a more efficient synthetic route without safety risks and the probe is more stable under physiological conditions. The two probes were easily synthesized from 4-pentyn-1-ol and 5-hexyn-1-ol. First, 4-pentyn-1-ol and 5-hexyn-1-ol underwent Dess–Martin oxidation and the Wittig reaction to form compounds 1 and 2. Compounds 1 and 2 were used for methyl ester hydrolysis to obtain 3 and 4 and further reacted with NaHCO3 to get probe #1 and probe #2, respectively. With these two probes in hand, we first tested whether they could be metabolically incorporated into proteins in mammalian cells. HEK293T cells were incubated with each of the two probes and then lysed with denaturing buffer to maximize the extraction of proteins. The resulting lysates were then reacted with azido-rhodamine via the copper(I) catalyzed alkyne-azido cycloaddition (CuAAC) reaction, to conjugate probe-labelled proteins with rhodamine for fluorescence analysis (Fig. 2C). The results demonstrated that probe #1 that contains a shorter chain was more efficient for protein labeling than probe #2 in live cells at concentrations of 0.5 mM or 5 mM (Fig. 2D). Therefore, we chose probe #1 for subsequent studies and renamed it “Cr-alkyne” for clarity.Further experiments showed that Cr-alkyne could label cells in a dose- and time-dependent manner (Fig. S1A and S1B in the ESI†). More importantly, the labeling with Cr-alkyne could be effectively competed by co-administration of excessive crotonate (Fig. 2E), supporting that Cr-alkyne is metabolically incorporated into crotonylated proteins in a similar manner to crotonate. We also used the shorter acetic acid and longer palmitic acid as competitors, and no significant competition with the probe was observed. This selective competition pattern suggests that our probe maintains specificity for crotonylation (Fig. S1C†). Furthermore, robust protein labelling with Cr-alkyne was observed for all other cell types tested (Fig. S1D†), highlighting the general utility of Cr-alkyne.
We next investigated whether Cr-alkyne could metabolically label known crotonylated proteins. Since histone proteins have been well established to be crotonylated in the literature,4 we separated the nuclear fractions from Cr-alkyne-labelled HEK293T cells and extracted the core histones for conjugation with azido-rhodamine by click chemistry. Notably, in-gel fluorescence analysis clearly showed that core histones were labelled with Cr-alkyne (Fig. 2F). As HMGB1 (ref. 28) and YWHAE29 were reported as two non-histone proteins with crotonylation, we expressed them with FLAG tags in HEK293T cells and labeled with Cr-alkyne. After protein extraction, conjugation with azide-biotin and enrichment with streptavidin beads, western blot analysis of the immunoprecipitated target proteins showed that Cr-alkyne could metabolically label HMGB1 and YWHAE as expected (Fig. 2G and H).
We further performed fluorescence imaging of crotonylated proteins in cells using Cr-alkyne. In contrast to the highly diffused signals in PBS-treated cells, intense rhodamine signals were observed in the Cr-alkyne-treated cells, especially in the nucleus (Fig. 2I), suggesting that Cr-alkyne could be used for fluorescence imaging of crotonylated proteins in cells.
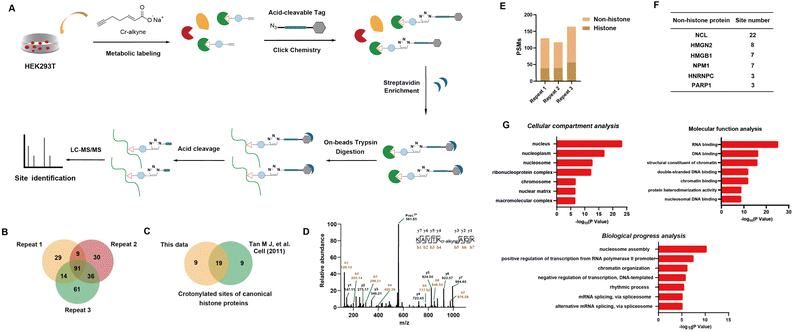
After verifying Cr-alkyne's effectiveness in in-gel and in-cell fluorescence, we identified its modification sites at the proteome level. Lysates of Cr-alkyne labelled HEK293T cells were prepared, conjugated with the acid-cleavable azido-DADPS-biotin tag (Fig. S2A†), and subjected to affinity purification, on-bead digestion and acid-mediated cleavage. The enriched peptides with Cr-alkyne modification were eluted for LC-MS/MS analysis (Fig. 3A). The acquired MS/MS spectra were analyzed by setting the fragment on Cr-alkyne-labelled lysine residues (K +249.1477 Da) as a variable modification (Fig. S2B†). To enhance the reliability and coverage of the site identification, only the sites that were identified in at least two out of the three replicates were considered for further analysis (Fig. 3B and Table S1†). The LC-MS/MS analysis demonstrated that we identified 28 crotonylated sites in canonical histone proteins, of which 19 sites were reported previously by Tan et al.4 (Fig. 3C). For example, a peptide containing the Cr-alkyne modification at K20 on H2B, KAVTKCr-alkyneAQK, was identified by LC-MS/MS analysis (Fig. 3D). Among the remaining 9 sites, 7 were also reported previously using the CHIMA strategy30 and the other two are potential novel histone crotonylation sites, including H1-K177 and H2B-K24 (Fig. S14 and S15†). In addition to these canonical histone crotonylation marks, we also identified five potential novel non-canonical histone crotonylation marks, including H2AX-K5, H2AX-K9, H2AZ1-K7, H2AZ1-K115 and H2AZ1-K120 (Fig. S16–S20†). We performed a detailed comparison of our identified crotonylation sites with known functional sites, including acetylation and lactylation sites on histones and found both common and differentiated sites (Fig. S3A†), which indicate potential crosstalks between these marks. Moreover, since crotonyl-CoA can be metabolized into beta-hydroxybutyryl-CoA and further acetoacetyl-CoA, we specifically searched for potential Kbhb-alkyne and Kacac-alkyne conjugations as variable modifications. Notably, we identified several peptides with potential Kacac modifications, including a modification site at H2B-K5 (Fig. S3B†), suggesting that crotonyl-alkyne may be metabolically converted to Kacac-alkyne in cells.
Aside from histone crotonylation marks, our profiling also revealed a large number of crotonylation sites in non-histone proteins as supported by the high peptide-spectra-match (PSM) counts (Fig. 3E). Some representative non-histone proteins, such as NCL, HMGN2, HMGB1, NPM1, HNRNPC and PARP1, were identified with multiple crotonylation sites (Fig. 3F). For some known crotonylated proteins, new modification sites were found with Cr-alkyne, such as EEF1A1-K457 and NUCKS1-K188 (Fig. S25 and 26†), and more importantly, several potential novel crotonylated proteins were discovered including P53, SETLP, WIZ, PI2R, UBE2T and ZN735 (Fig. S12–S17†).
We next performed a series of bioinformatics analyses of the identified crotonylated proteins. Cellular component analysis using the Gene Ontology (GO) database revealed that these Cr-alkyne labeled proteins are more represented in the nucleus, nucleoplasm, and nucleosome (Fig. 3G). In addition, GO analysis on biological processes indicated a significant enrichment in processes such as nucleosome assembly, positive regulation of transcription from the polymerase II promoter and chromatin organization. Molecular function analysis demonstrated that many of these proteins possess RNA, DNA, and chromatin binding activities. Collectively, these results suggest that crotonylation mainly occurs on proteins that are associated with diverse nuclear biological processes such as chromosome organization, RNA processing/metabolism and DNA repair.
In order to quantify the percentage of the metabolic crotonylation, we conducted a quantitative analysis using dimethyl labeling to determine the percentage of metabolic crotonylation. Specifically, we compared Cr-alkyne-treated and untreated cells by extracting histones from both groups and labeling them with light and heavy formaldehyde, respectively. Through this dimethyl quantification approach, we identified peptides whose unmodified forms were reduced due to probe modification. By analyzing the peak area ratios of light/heavy labeled co-eluting peptides, we were able to calculate the modification rate of metabolic crotonylation. We have demonstrated this with three representative sites (Fig. S4†), which show approximately 12.4% modification at H2AZ1-K115 and modification rates of Cr-alkyne at H2B-K85 and NCL-K124 for about 7.8% and 12.4%, respectively.
To ultimately validate the new modification sites, we performed the co-elution experiment with synthetic peptides, which is widely used as the golden standard to verify the structures of new crotonylated sites. We selected two potential novel crotonylated sites for validation, including H2AZ1-K115 (ATIAGGGVIPHIHKcrSLIGK) and NUCKS1-K188 (GKcrVGRPTASK), both of which were detected with Cr-alkyne labelling and could be consistently identified in our chemoproteomic experiments (Fig. S23 and S26†). By solid phase peptide synthesis (SPPS), we synthesized these two crotonylated peptides with high purity, in which the C-terminal lysine was replaced with an isotopically labeled lysine (+8 Da) (Fig. S5A in the ESI†). The histone of crotonate-stimulated HEK293T cells was extracted, digested and immunoprecipitated by the anti-Kcr antibody for endogenous H2AZ1-K115. For the endogenous NUCKS1-K188 peptide, FLAG-tagged NUCKS1 was immunoprecipitated from crotonate-stimulated HEK293T cells and digested by trypsin for further analysis (Fig. S5B in the ESI†). These two endogenous samples were spiked with the heavy peptide standards and analyzed for coelution and co-fragmentation by LC-MS/MS, respectively. The results showed that the synthetic standard has the exact same retention time and MS1 isotopic envelope as the endogenously crotonylated peptides (Fig. 4A–D). In addition, the MS/MS spectrum of the synthetic standard shows the identical fragmentation pattern as the endogenous one, and a mass shift of +8 Da could be clearly observed for the y fragment ions that cover the crotonylated site (Fig. 4C and F). Collectively, these results provided unambiguous evidence to confirm the existence and structure of these two novel crotonylated sites, which highlights the power of Cr-alkyne in identifying new crotonylation substrates with site-specific resolution on a proteome-wide scale.
Regulation of Kcr is a dynamic balance between the enzyme activity of writers and erasers, which add and remove Kcr, respectively. As class I HDACs have been reported31 as histone decrotonaylases in mammalian cells, we next investigated how HDAC1/3 affects the metabolic labeling of Cr-alkyne on histones, aiming to survey crotonylation sites that are dynamically regulated by these two enzymes. HDAC1/3 were knocked down or over-expressed in HEK293T cells and after the cells were incubated with Cr-alkyne, histones were extracted and reacted with azido-rhodamine for in-gel fluorescence analysis. The results showed that Cr-alkyne had a slightly stronger labeling in HDAC1 knockdown cells than in the untreated cells (Fig. S6 in the ESI†) and consistently, Cr-alkyne's labeling in HDAC1 overexpression cells was also slightly weaker (Fig. S6B in the ESI†).
We also conducted an in vitro kinetic analysis to characterize the decrotonylation activity of HDAC1. Specifically, we synthesized two fluorogenic substrates: Ac-Lys(cr)-AMC and Ac-Lys(cr-alkyne)-AMC. Using purified HDAC1, we performed a detailed study of enzyme kinetics with both substrates. Our results demonstrated that HDAC1 exhibited a significant decrotonylation activity towards both substrates. The kinetic parameters (Km and Kcat) determined for these substrates are similar to each other (Fig. S7†), suggesting HDAC1's decrotonylation to process both native and probe-modified crotonyl substrates.
To obtain more quantitative results, we next performed rdTOP-ABPP experiments using the Cr-alkyne probe to profile specific lysines that were perturbed by HDAC1/3. Briefly, proteomes from HDAC1/3 knockdown, over-expression and untreated cells were treated with the Cr-alkyne and then conjugated with acid-cleavable azido-biotin tags by CuAAC. After enrichment and on-bead digestion, the probe-adducted peptides from the knock-down, over-expression and untreated cells were isotopically labeled with light, medium and heavy dimethylation regents according to the rdTOP-ABPP protocol,32 respectively (Fig. 5A and S8†). The RH/L (“untreated/knock-down”) ratio reflects how much Cr-alkyne's labeling was increased upon HDAC knock-down while the RH/M (“untreated/over-expression”) ratio reflects how much Cr-alkyne's labeling was lost upon HDAC over-expression. Only the RH/L negatively regulated and RH/L positively regulated sites are considered HDAC regulated sites.
For example, H3-K23, H4-K8, H2AZ1-K8, H2B-K11 and H2B-K20 were negatively regulated while those sites were positively regulated (Fig. 5B). Among these crotonylation sites, H3-K23 was previously reported as a de-crotonylation site using HDAC1 and H4-K8 was considered to be regulated by HDAC1 and HDAC3 (ref. 31) (Fig. S8†), both of which are consistent with our data. In order to test whether the other identified Kcr sites can be removed by HDAC1/3 natively, we choose Kcr at H2B-K20 for further verification. In HDAC1/3 knock down cells, the H2B-K20cr level was up-regulated, indicating H2B-K20cr accumulation in knock down cells while Pan-Kcr and H4-K8cr levels increased as well (Fig. 5C). We also expressed FLAG-tagged HDAC1/3 in HEK293T cells, showing that H2B-K20cr levels were down-regulated. These results suggested that HDAC1/3 are erasers of H2B-K20cr. In general, Cr-alkyne probe could be used to unveil crotonylation sites regulated by HDACs and might be used for dynamic site-specific turnover profiling upon perturbation by other erasers or writers.
Conclusion
In summary, we have developed a new alkyne-functionalized bioorthogonal probe, Cr-alkyne, for effective metabolic labelling and profiling of lysine crotonylation in mammalian cells. Cr-alkyne has been characterized for metabolic labelling and efficient fluorescence detection of known crotonylated proteins. In combination with chemical proteomics, Cr-alkyne can be used to discover new crotonylated sites. It is also worth noting that the Cr-alkyne probe could be used to unveil crotonylation sites dynamically regulated by HDACs and related enzymes. Overall, our study not only provides a powerful and readily accessible chemical tool for the general detection and identification of protein crotonylation in different biological contexts, but also suggests novel substrate proteins and regulatory roles of crotonylation. We envision that the analytical and functional study of protein crotonylation will be greatly accelerated with this enabling chemical biology tool.Data availability
Data for the synthesis of Cr-alkyne, in-gel fluorescence characterization, proteomic analysis, and bioinformatics analysis are provided in the ESI.† The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD053632.Author contributions
Y.-F. Zhou and S. Yuan contributed equally to this work. Y.-F. Zhou performed the synthesis, in-gel fluorescence, proteomics, co-elution experiments, and biological experiments and wrote the manuscript. S. Yuan performed proteomics, kinetic, competition and biological experiments. B. Ma assisted in the co-elution analysis. J. Gao provided advice. C. Wang conceived and supervised the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Computing Platform of the Center for Life Science for supporting the proteomics data analysis. This work was supported by the National Key R&D Program of China (2022YFA1304700 to C.W.) and the National Natural Science Foundation of China (92153301, 21925701 and 22321005 to C.W.). Y.-F. Zhou acknowledged the support from the Boya Postdoctoral Fellowship and National Postdoctoral Program for Postdoctoral Researchers (GZB20230015). S. Yuan acknowledged support from the Postdoctoral Fellowship of the Peking-Tsinghua Center for Life Sciences.Notes and references
- B. R. Sabari, D. Zhang, C. D. Allis and Y. Zhao, Metabolic regulation of gene expression through histone acylations, Nat. Rev. Mol. Cell Biol., 2017, 18, 90–101 CrossRef CAS.
- D. Zhang, Z. Tang, H. Huang, G. Zhou, C. Cui, Y. Weng, W. Liu, S. Kim, S. Lee, M. Perez-Neut, J. Ding, D. Czyz, R. Hu, Z. Ye, M. He, Y. G. Zheng, H. A. Shuman, L. Dai, B. Ren, R. G. Roeder, L. Becker and Y. Zhao, Metabolic regulation of gene expression by histone lactylation, Nature, 2019, 574, 575–580 CrossRef CAS.
- L. Dai, C. Peng, E. Montellier, Z. Lu, Y. Chen, H. Ishii, A. Debernardi, T. Buchou, S. Rousseaux, F. Jin, B. R. Sabari, Z. Deng, C. D. Allis, B. Ren, S. Khochbin and Y. Zhao, Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark, Nat. Chem. Biol., 2014, 10, 365–370 CrossRef CAS PubMed.
- M. Tan, H. Luo, S. Lee, F. Jin, J. S. Yang, E. Montellier, T. Buchou, Z. Cheng, S. Rousseaux, N. Rajagopal, Z. Lu, Z. Ye, Q. Zhu, J. Wysocka, Y. Ye, S. Khochbin, B. Ren and Y. Zhao, Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification, Cell, 2011, 146, 1015–1027 CrossRef.
- C. Peng, Z. Lu, Z. Xie, Z. Cheng, Y. Chen, M. Tan, H. Luo, Y. Zhang, W. He, K. Yang, B. M. M. Zwaans, D. Tishkoff, L. Ho, D. Lombard, T.-C. He, J. Dai, E. Verdin, Y. Ye and Y. Zhao, The First Identification of Lysine Malonylation Substrates and Its Regulatory Enzyme, Mol. Cell. Proteomics, 2011, 10, M111.012658 CrossRef.
- Z. Zhang, M. Tan, Z. Xie, L. Dai, Y. Chen and Y. Zhao, Identification of lysine succinylation as a new post-translational modification, Nat. Chem. Biol., 2011, 7, 58–63 CrossRef CAS PubMed.
- M. Tan, C. Peng, K. A. Anderson, P. Chhoy, Z. Xie, L. Dai, J. Park, Y. Chen, H. Huang, Y. Zhang, J. Ro, G. R. Wagner, M. F. Green, A. S. Madsen, J. Schmiesing, B. S. Peterson, G. Xu, O. R. Ilkayeva, M. J. Muehlbauer, T. Braulke, C. Muehlhausen, D. S. Backos, C. A. Olsen, P. J. McGuire, S. D. Pletcher, D. B. Lombard, M. D. Hirschey and Y. Zhao, Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5, Cell Metab., 2014, 19, 605–617 CrossRef CAS.
- Z. Xie, D. Zhang, D. Chung, Z. Tang, H. Huang, L. Dai, S. Qi, J. Li, G. Colak, Y. Chen, C. Xia, C. Peng, H. Ruan, M. Kirkey, D. Wang, L. M. Jensen, O. K. Kwon, S. Lee, S. D. Pletcher, M. Tan, D. B. Lombard, K. P. White, H. Zhao, J. Li, R. G. Roeder, X. Yang and Y. Zhao, Metabolic Regulation of Gene Expression by Histone Lysine β-Hydroxybutyrylation, Mol. Cell, 2016, 62, 194–206 CrossRef CAS.
- D. Liu, W. Xiao, H. Li, Y. Zhang, S. Yuan, C. Li, S. Dong and C. Wang, Discovery of Itaconate-Mediated Lysine Acylation, J. Am. Chem. Soc., 2023, 145, 12673–12681 CrossRef CAS.
- K. Delaney, M. Tan, Z. Zhu, J. Gao, L. Dai, S. Kim, J. Ding, M. He, L. Halabelian, L. Yang, P. Nagarajan, M. R. Parthun, S. Lee, S. Khochbin, Y. G. Zheng and Y. Zhao, Histone lysine methacrylation is a dynamic post-translational modification regulated by HAT1 and SIRT2, Cell Discov., 2021, 7, 122 CrossRef CAS.
- Y. Gao, X. Sheng, D. Tan, S. Kim, S. Choi, S. Paudel, T. Lee, C. Yan, M. Tan, K. M. Kim, S. S. Cho, S. H. Ki, H. Huang, Y. Zhao and S. Lee, Identification of Histone Lysine Acetoacetylation as a Dynamic Post-Translational Modification Regulated by HBO1, Adv. Sci., 2023, 10, 2300032 CrossRef CAS.
- J. Zhang, G. Shi, J. Pang, X. Zhu, Q. Feng, J. Na, W. Ma, D. Liu and S. Zhou, Crotonylation of GAPDH regulates human embryonic stem cell endodermal lineage differentiation and metabolic switch, Stem Cell Res. Ther., 2023, 14, 63 CrossRef CAS PubMed.
- N. Mu, Y. Wang, X. Li, Z. Du, Y. Wu, M. Su, Y. Wang, X. Sun, L. Su and X. Liu, Crotonylated BEX2 interacts with NDP52 and enhances mitophagy to modulate chemotherapeutic agent-induced apoptosis in non-small-cell lung cancer cells, Cell Death Dis., 2023, 14, 645 CrossRef CAS.
- Y. Zheng, L. Zhu, Z.-Y. Qin, Y. Guo, S. Wang, M. Xue, K.-Y. Shen, B.-Y. Hu, X.-F. Wang, C.-Q. Wang, L.-X. Qin and Q.-Z. Dong, Modulation of cellular metabolism by protein crotonylation regulates pancreatic cancer progression, Cell Rep., 2023, 42, 112666 CrossRef CAS.
- L. Dang, X. Y. Cao, T. Y. Zhang, Y. Z. Sun, S. S. Tian, T. Y. Gong, H. Xiong, P. P. Cao, Y. H. Li, S. Q. Yu, L. Yang, L. R. Zhang, T. Liu, K. Zhang, J. Liang and Y. P. Chen, Nuclear Condensation of CDYL Links Histone Crotonylation and Cystogenesis in Autosomal Dominant Polycystic Kidney Disease, J. Am. Soc. Nephrol., 2022, 33, 1708–1725 CrossRef CAS.
- S. K. Dai, P. P. Liu, X. Li, L. F. Jiao, Z. Q. Teng and C. M. Liu, Dynamic profiling and functional interpretation of histone lysine crotonylation and lactylation during neural development, Development, 2022, 149, dev200049 CrossRef CAS PubMed.
- Y. Zhang, Y. L. Chen, Z. Zhang, X. Tao, S. Xu, X. Y. Zhang, T. Zurashvili, Z. P. Lu, J. R. Bayascas, L. P. Jin, J. Y. Zhao and X. Y. Zhou, Acox2 is a regulator of lysine crotonylation that mediates hepatic metabolic homeostasis in mice, Cell Death Dis., 2022, 13, 279 CrossRef CAS.
- J. Bos and T. W. Muir, A Chemical Probe for Protein Crotonylation, J. Am. Chem. Soc., 2018, 140, 4757–4760 CrossRef CAS.
- W. Qin, Y. Zhang, H. Tang, D. Liu, Y. Chen, Y. Liu and C. Wang, Chemoproteomic Profiling of Itaconation by Bioorthogonal Probes in Inflammatory Macrophages, J. Am. Chem. Soc., 2020, 142, 10894–10898 CrossRef CAS.
- N. Chen, Y. Liu, Y. Li and C. Wang, Chemical Proteomic Profiling of Protein 4′-Phosphopantetheinylation in Mammalian Cells, Angew. Chem., Int. Ed., 2020, 59, 16069–16075 CrossRef CAS PubMed.
- X. Bao, Y. Xiong, X. Li and X. D. Li, A chemical reporter facilitates the detection and identification of lysine HMGylation on histones, Chem. Sci., 2018, 9, 7797–7801 RSC.
- Y. Sun, Y. Chen and T. Peng, A bioorthogonal chemical reporter for the detection and identification of protein lactylation, Chem. Sci., 2022, 13, 6019–6027 RSC.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS.
- C. W. Tornoe, C. Christensen and M. Meldal, Peptidotriazoles on solid phase: 1,2,3 -triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides, J. Org. Chem., 2002, 67, 3057–3064 CrossRef CAS PubMed.
- Y. Fang, X. Xu, J. Ding, L. Yang, M. T. Doan, P. W. F. Karmaus, N. W. Snyder, Y. Zhao, J.-L. Li and X. Li, Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells, Cell Stem Cell, 2021, 28, 748–763 CrossRef CAS PubMed.
- W. Cai, D. Xu, C. Zeng, F. Liao, R. Li, Y. Lin, Y. Zhao, W. Dong, Q. Wang, H. Yang, D. Wen, J. Gu, W. Shentu, H. Yu, X. Zhang, J. Wei and J. Duan, Modulating Lysine Crotonylation in Cardiomyocytes Improves Myocardial Outcomes, Circ. Res., 2022, 131, 456–472 CrossRef CAS PubMed.
- B. R. Sabari, Z. Tang, H. Huang, V. Yong-Gonzalez, H. Molina, H. E. Kong, L. Dai, M. Shimada, J. R. Cross, Y. Zhao, R. G. Roeder and C. D. Allis, Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation, Mol. Cell, 2015, 58, 203–215 CrossRef CAS PubMed.
- H. Yu, C. Bu, Y. Liu, T. Gong, X. Liu, S. Liu, X. Peng, W. Zhang, Y. Peng, J. Yang, L. He, Y. Zhang, X. Yi, X. Yang, L. Sun, Y. Shang, Z. Cheng and J. Liang, Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair, Sci. Adv., 2020, 6, eaay4697 CrossRef CAS PubMed.
- Z. Zheng, Q. Zhong and X. Yan, YWHAE/14-3-3ε crotonylation regulates leucine deprivation-induced autophagy, Autophagy, 2023, 19, 2401–2402 CrossRef CAS.
- J. Gao, X. Sheng, J. Du, D. Zhang, C. Han, Y. Chen, C. Wang and Y. Zhao, Identification of 113 new histone marks by CHiMA, a tailored database search strategy, Sci. Adv., 2023, 9, eadf1416 CrossRef CAS.
- W. Wei, X. Liu, J. Chen, S. Gao, L. Lu, H. Zhang, G. Ding, Z. Wang, Z. Chen, T. Shi, J. Li, J. Yu and J. Wong, Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription, Cell Res., 2017, 27, 898–915 CrossRef CAS.
- F. Yang, J. Gao, J. Che, G. Jia and C. Wang, A Dimethyl-Labeling-Based Strategy for Site-Specifically Quantitative Chemical Proteomics, Anal. Chem., 2018, 90, 9576–9582 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc06745b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |