Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Facile preparation of sulfonium peptide and protein probes for selective crosslinking of methyllysine readers†
Kun
Zou‡
ab,
Jinyu
Yang‡
ab,
Yingxiao
Gao
b,
Feng
Feng
c and
Mingxuan
Wu
 *bcd
*bcd
aDepartment of Chemistry, Zhejiang University, 310027, Hangzhou, Zhejiang Province, China. E-mail: wumingxuan@westlake.edu.cn
bDepartment of Chemistry, School of Science, Westlake University, 310030, Hangzhou, Zhejiang Province, China
cWestlake Laboratory of Life Sciences and Biomedicine, 310024, Hangzhou, Zhejiang Province, China
dInstitute of Natural Sciences, Westlake Institute for Advanced Study, 310024, Hangzhou, Zhejiang Province, China
First published on 12th December 2024
Abstract
Sulfonium is an electrophilic and biocompatible group that is widely applied in synthetic chemistry on small molecules. However, there have been few developments of peptide or protein-based sulfonium tools. We recently reported sulfonium-mediated tryptophan crosslinking and developed NleS+me2 (norleucine-dimethylsulfonium) peptides as dimethyllysine mimics that crosslink site-specific methyllysine readers. Therefore, sulfonium probes show great potential for investigating methyllysine readers and other aromatic cage-containing proteins. However, the current synthesis is not very efficient and is limited to peptide probes that, in many cases, cannot mimic protein–protein interactions. In addition to peptidyl conjugates that are valuable for reader identification, there are unavoidable methyl conjugates as side products. As a result, a robust method to prepare peptide and protein sulfonium tools with great crosslinking reactivity and selectivity is highly desirable. Here, we report a cysteine alkylation method to introduce site-specific sulfonium at protein level with excellent yield. In addition to dimethylsulfonium, we also developed cyclic sulfonium warheads that enhanced peptidyl conjugate selectivity. The method thus made it possible to prepare nucleosome probes in which LEDGF and NSD2, as H3K36 methylation readers were readily crosslinked. We thus believe this method will accelerate the development of sulfonium peptide and protein tool sets for broad applications in chemical biology studies.
Introduction
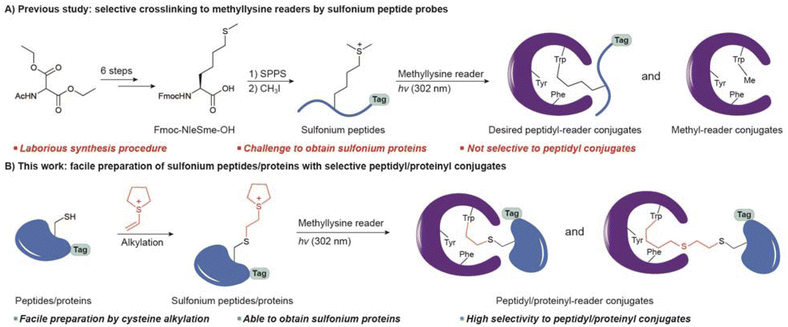
Lysine methylation is an important posttranslational modification. Histone lysine methylation regulates the chromatin structure and epigenetics.1,2 In addition, nonhistone lysine methylation controls diverse cellular processes such as MAP3K2 methylation3,4 and p53 methylation5–7 that drive cancer. Reader proteins, which bind to site-specifically modified protein motifs, are significant for downstream regulation upon lysine methylation.8,9 For example, CBX1 (a chromobox protein homolog, also known as heterochromatin protein HP1β) binds to histone H3 with K9 methylation and promotes the formation of heterochromatin and gene silencing.10,11 BPTF (bromodomain PHD finger transcription factor) contains a reader domain of H3K4 methylation that promotes gene transcription.12,13 Therefore, the identification and investigation of new methyllysine readers are essential to understanding the biological roles of lysine methylation at the molecular level.Covalent probes are powerful for identifying proteins of interest (POI) because covalent linkage allows efficient enrichment of POI from complicated cellular samples.14–16 Probes that contain photoreactive groups are widely used,17,18 but laborious synthesis is required to optimize the placement of diazirine or BPA (p-benzoyl-L-phenylalanine) for efficient crosslinking. In addition, it is more synthetically challenging to prepare protein probes that target reader proteins.19 We recently reported a sulfonium-mediated crosslinking strategy so that NleS+me2 probes bind to readers as Kme2 peptide mimics and selectively crosslink to tryptophan inside the binding pocket (Fig. 1A).20 Because NleS+me2 plays roles in both binding and crosslinking, the design and synthesis are more straightforward.
However, there are some limitations to the current sulfonium method. First, sulfonium probes are limited to peptides with low synthesis yield. To introduce the unnatural amino acid residue, 6 steps are needed to prepare Fmoc-L-NleSme-OH as a building block for solid-phase peptide synthesis (SPPS). After peptide synthesis, extra S-methylation by iodomethane is necessary to yield NleS+me2 peptides.21 Second, sulfonium peptides may not mimic proteins in cellular environments. For example, histone H3K36 is proximate to DNA in the nucleosome structure,22,23 so an H3K36 methylation reader cannot bind a peptide well. Third, the crosslinking selectivity is not optimal. Peptidyl conjugates are desirable products for enrichment via affinity tags on peptides. However, irradiation also leads to a methyl-conjugate that reduces the efficiency of downstream applications (Fig. 1A).
Chemoselective modification of proteins is a major strategy for preparing site-specifically modified proteins.24–26 Due to its unique reactivity, cysteine is the most targeted residue for reactions, including it being a methyllysine mimic,27 a methylarginine mimic,28 and an acetyllysine mimic.29–31 Given the electrophilic nature of sulfonium, we envision that such a warhead may be incorporated into peptides and proteins by the nucleophilic thiol of cysteine. We found some clues in the reported methods of sulfonium synthesis and bioconjugation.32–37 Vinyl sulfonium has shown alkylation capability to thiol in previous studies.32 A very recent study reported vinyl aryl sulfonium for selective cysteine alkylation on proteins, but the sulfonium intermediate is designed to leave from proteins. The resulting episulfonium on the protein is functionalized by additional nucleophiles.38 Here we report a cysteine alkylation method to introduce sulfonium as a dimethyllysine and NleS+me2 mimic at specific sites (Fig. 1B). Such a method makes sulfonium peptides and proteins much more synthetically accessible and could be widely used to develop sulfonium probes for tryptophan site-specific crosslinking and the study of methyllysine readers.
Results and discussion
Development of a robust alkylation method to prepare NleCS+me2 peptides
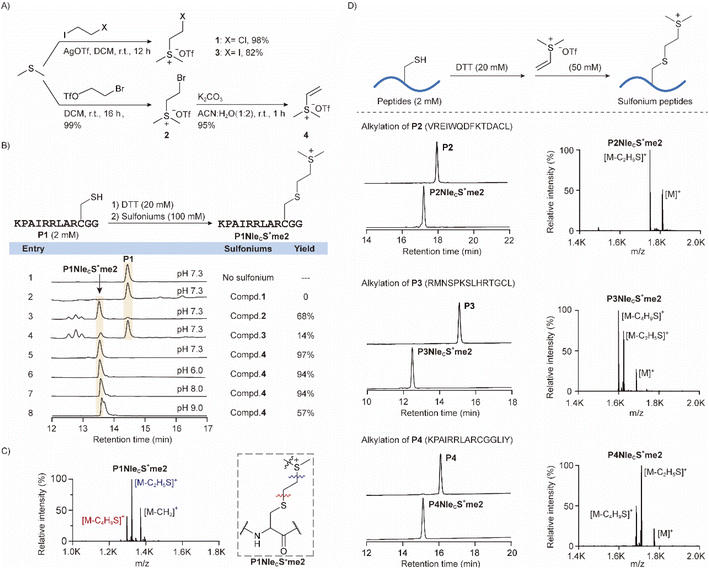
As we aim to introduce sulfonium to cysteine on free peptides and proteins, the alkylating agent should meet a few requirements. First, the agent should be electrophilic enough for alkylation by nucleophilic thiol on cysteine. Second, the reactivity should be highly selective to cysteine as peptides and proteins contain diverse functional groups. Third, the reaction should proceed under physiological conditions so that the protein products will still be well-folded. Fourth, the alkylation products should be structurally close to NleS+me2 for selective crosslinking to tryptophan inside a methyllysine-binding pocket in the readers. As a result, we designed several sulfonium compounds 1–4 for initial screening (Fig. 2A). Sulfonium compounds 1 and 3 were prepared from dimethylsulfide and haloethyl iodide reagents. Alternatively, bromide compound 2 was prepared with bromoethyltriflate. A subsequent elimination reaction of 2 under basic conditions yielded vinyl sulfonium 4.32,39With the agents in hand, we started to characterize reactivity and selectivity using a cysteine-containing peptide P1 (Fig. 2B). P1 was pre-treated with DTT as a general procedure for cysteine alkylation to break the possible disulfide bonds of cysteine-containing samples formed during storage (Fig. S5†)27,40 and was subsequently treated with sulfonium alkylation agents. By HPLC analysis, compound 2 yielded a significant amount of desired product, as confirmed by mass spectrometry (Fig. 2C), but there were several additional peaks from side products. Compounds 1 and 3, with different halogens, however, were much less active. Compound 4 exhibited the best performance with excellent yield. Therefore, we further characterized the reactivity of sulfonium 4 at different pH values (Fig. 2B, entries 5–8), times (Fig. S3A†), and temperatures (Fig. S3B†). Another product peak was observed at pH 9 and was later indicated to be a dialkylated product by mass spectrum analysis (Fig. S3C†). We suggest that the extra alkylation occurs on the N-amine or lysine side chain due to the enhanced nucleophilic substitution reactivity of amine at pH 9. Therefore, we explored potential amine reactivity using Boc-Lys-OH with free ε-amine. A clear alkylation product from sulfonium 4 was identified at pH 9, but no such reaction happened at pH 7.3 (Fig. S4†). The reaction rate was generally fast as 100% conversion could be achieved within 15 min at 27 °C. We thus later selected another three peptides that contain all functional groups of natural amino acids, and all the alkylation reactions of compound 4 were robust and clean (Fig. 2D).
Michael addition is a widely used method to selectively modify cysteine on peptides and proteins, but there is an instability issue due to the retro-Michael reaction.41,42 To characterize the stability of the sulfonium from cysteine alkylation, we prepared a glutathione conjugate 6 (Fig. S12†) to measure stability using 1H NMR and HPLC. It exhibited excellent stability after 48 h in both pH 7.3 buffer and pH 9.5 buffer (Fig. S21–23†).
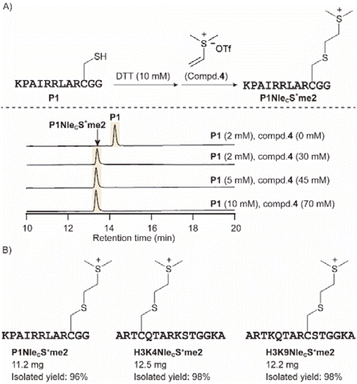
As the analytical yield with the vinyl sulfonium was promising, we next optimized the peptide concentration and alkylating agent 4 (Fig. 3A). 10 mM peptide was also readily converted to sulfonium product, and thus we can scale up the synthesis in a more concentrated mixture. We next applied peptide P1 at 10 mg scale and obtained the product P1NleCS+me2 with 96% isolated yield. Similarly, H3K4NleCS+me2 and H3K9NleCS+me2 peptides were obtained with excellent isolated yield, that will be studied for methyllysine reader crosslinking (Fig. 3B).
NleCS+me2 is a good mimic of NleS+me2 for tryptophan crosslinking
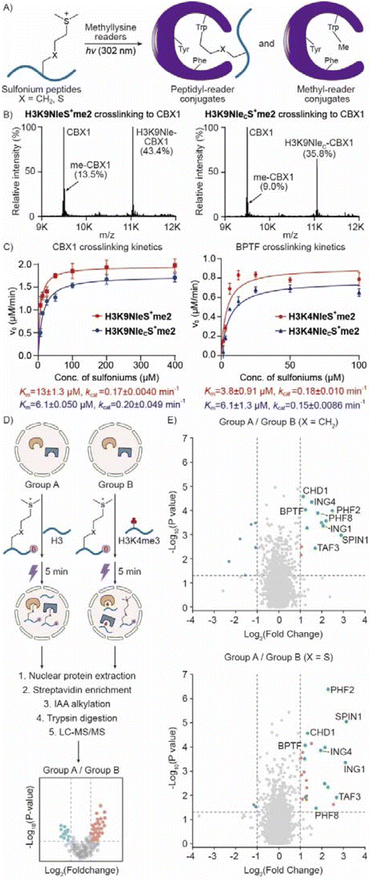
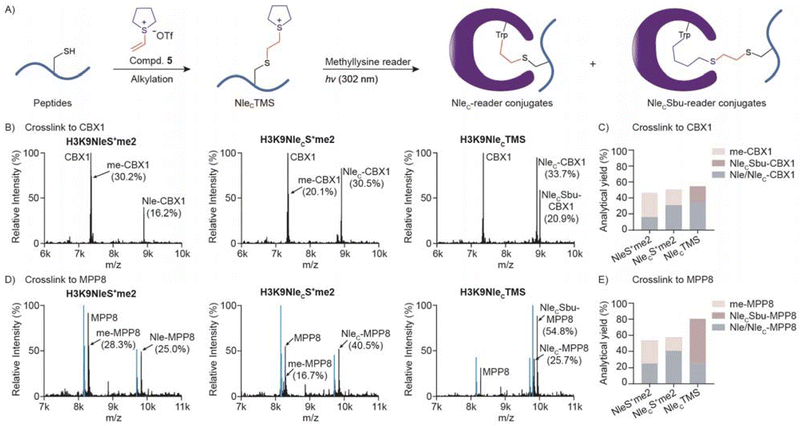
With the establishment of a method for sulfonium peptide preparation, we next wondered whether NleCS+me2 is a good mimic of NleS+me2. We thus set up photo-crosslinking reactions using several methyllysine readers (Fig. 4A). Generally, the reaction yields of peptidyl conjugate and methyl conjugate between NleCS+me2 and NleS+me2 peptides were very close (Fig. 4B and S15†). To compare the two sulfonium compounds in more detail, we carried out a kinetic analysis of crosslinking to CBX1 and BPTF. The correlation between reaction rate and sulfonium peptide concentration was quite similar (Fig. 4C). Regression analysis with the Michaelis–Menten equation indicated very close Km and kcat values. Furthermore, we applied H3K4NleS+me2 and H3K4NleCS+me2 to extracted cell nuclei and volcano plots demonstrated that similar groups of protein hits were selectively crosslinked and enriched by the two sulfonium peptides with biotin tags (Fig. 4D and E). Thus, we concluded that NleCS+me2 is a good mimic of NleS+me2 for reader crosslinking from individual recombinant proteins to the proteome.Cyclic sulfonium improved peptidyl-conjugation
From our previous studies, we found that photo-crosslinking between sulfonium peptide probes and readers led to two conjugates (Fig. 4A).20 Once sulfonium accepts an excited single electron from indole, we suggest that homolysis of the C–S bond yields a sulfide and an alkyl radical to crosslink Trp*+. Therefore, peptidyl and methyl conjugates were both obtained (Fig. 4B and S15†). Because only the peptidyl conjugate could be enriched by an affinity tag on the peptide, methyl conjugation reduces the efficiency for following protein ID.We thus designed cyclic sulfonium to overcome this limitation because any possible C–S homolytic cleavage would only result in a peptidyl conjugate. In addition, there were cyclic amine structures in methyllysine reader inhibitors, which indicate tolerance of such a group in the reader binding pocket.43,44 We thus made efforts to prepare a series of vinyl cyclic sulfonium compounds. Probably due to high strain, three- or four-member ring products could not be obtained. Instead, vinyl cyclic sulfonium with five- or six-member rings were readily synthesized and applied to cysteine alkylation to yield NleCTMS (tetramethylene sulfide) and NleCPMS (pentamethylene sulfide) (Fig. 5A, S8 and 9†). We also prepared a glutathione conjugate 7 that demonstrated the same stability as NleCS+me2 in buffers (Fig. S13 and S24–26†).
We next compared dimethylsulfonium and cyclic sulfonium for crosslinking reactivity and selectivity. H3K9NleCTMS resulted in two expected crosslinking products, NleC-CBX1 conjugate and NleCSbu-CBX1 conjugate, with a combined analytical yield of peptidyl conjugate that is significantly higher than the yield using H3K9NleS+me2 and H3K9NleCS+me2 (Fig. 5B and C). In the case of MPP8 (M-phase phosphoprotein 8), the peptidyl conjugate yield with H3K9NleCTMS exhibited over a 3-fold increase compared to H3K9NleS+me2 (Fig. 5D and E). We next compared NleCTMS and NleCPMS crosslinking activities to CBX1 and MPP8. Both cyclic sulfonium compounds exhibited a close overall analytical yield of peptidyl conjugates, and the NleC-reader was a preferred product over a NleCSbu-reader or NleCSpe-reader (Fig. S16†). Further kinetic analysis revealed faster NleC-reader conjugation but slower NleCSbu conjugation by NleCTMS (Fig. S17†). Overall, cyclic sulfonium-mediated crosslinking avoids a methyl conjugate with much superior peptidyl conjugation to dimethylsulfonium for downstream protein ID applications.
Development of sulfonium nucleosome probes for reader crosslinking
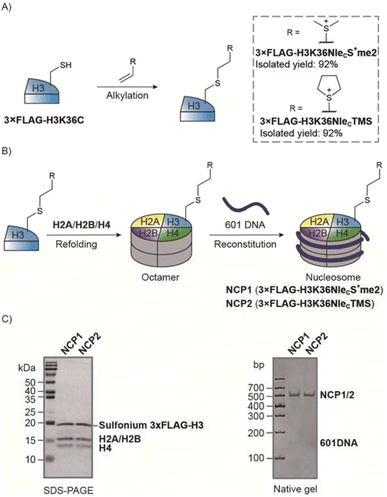
Peptides are frequently used to study posttranslational modifications that mimic the partial structure of proteins such as histone tails.45–47 However, for histone sites in a globular domain that have interactions with other core histones and DNA, peptide-based studies are not ideal.40,48 As the vinyl sulfonium exhibited high reactivity and selectivity on peptides, we think the reaction would also be robust on proteins to prepare protein sulfonium probes.We prepared a recombinant X. laevis histone 3xFLAG-H3K36C mutant for vinyl sulfonium alkylation. Both sulfonium 4 and 5 yielded the desired products with 92% isolated yield at multiple mg scale (Fig. 6A). We next used the sulfonium-containing histone H3 to prepare nucleosome core particles (NCP) NCP1 and NCP2 (Fig. 6B). The yield of octamer refolding and nucleosome reconstitution were the same as for histone 3xFLAG-H3Kc36me3. In addition, the mobile shifts of the nucleosomes were the same as for H3Kc36me3 nucleosome NCP3 on a native PAGE gel (Fig. 6C and S20†). Thus, the sulfonium group is well tolerated through the nucleosome preparation. To confirm the stability of sulfonium, we monitored NCP2 using mass spectrometry and the sulfonium group was intact over 45 days in storage buffer at 4 °C (Fig. S27†).
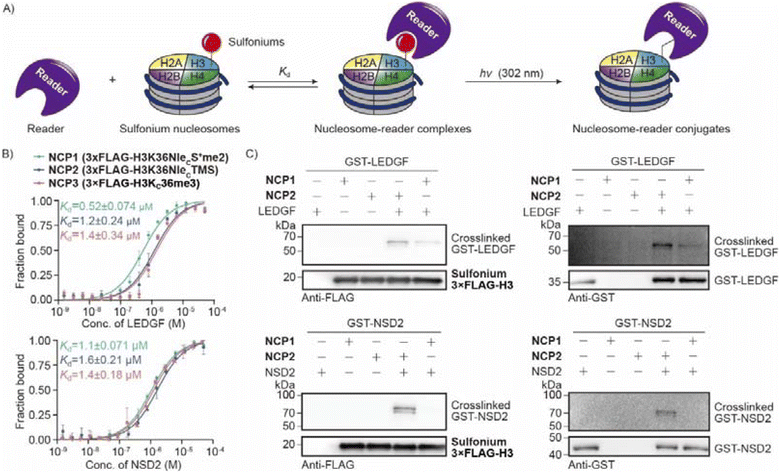
H3K36 methylation is an important PTM and there are several reported readers that interact with H3K36me3 by aromatic cages.49,50 In addition, the readers interact closely with DNA that is proximate to H3K36. As histone H3 peptides cannot mimic the local chemical environment in chromatin, we aimed to prepare nucleosome probes with sulfonium at H3K36 for reader crosslinking. We expressed recombinant PWWP domains of LEDGF and NSD2 that are well studied as H3K36 methylation readers and later measured binding affinities to the nucleosomes. The data demonstrated that sulfonium NCP1 and NCP2 are good mimics of H3Kc36me3 NCP3 with close Kd values (Fig. 7A and B). Next, the mixtures of NCP and readers were irradiated using UV-B and analyzed using western blot (Fig. 7A and C). The data demonstrated crosslinking activity between sulfonium NCP and the readers. Moreover, NCP2 which contains cyclic sulfonium, exhibited much better peptidyl conjugation than NCP1 (Fig. 7C). This result further confirms the improved efficiency of reader crosslinking by employment of cyclic sulfonium. Overall, the sulfonium-containing proteins are readily available by cysteine alkylation and nucleosome probes are active for crosslinking the site-specific readers.
Conclusions
Here we have developed a facile method to prepare sulfonium-containing peptides and proteins by cysteine alkylation. Vinyl sulfoniums are simple to prepare and exhibit robust reactivity and selectivity to cysteine. Therefore, a milligram scale of peptides and proteins with diverse sulfonium is easily achievable. Moreover, the resulting NleCS+me2 and derivatives are good mimics of methyllysine for site-specific tryptophan crosslinking inside the binding pocket of readers. In addition to sulfonium peptide probes, we demonstrated that the method enables sulfonium-containing nucleosome probes that mimic chromatin to crosslink readers that interact with globular domains. In addition, cyclic sulfonium enhanced crosslinking to readers for higher efficiency for protein ID. Consequently, we believe that sulfonium probes formed with vinylsulfonium could be widely used for tryptophan crosslinking and aromatic cage investigations.Data availability
The data that support the findings of the study are available in the ESI† of this article.Author contributions
M. W. conceived the idea and secured funding acquisition. K. Z. established the alkylation method, prepared sulfonium peptides and proteins, and performed kinetic experiments. J. Y. characterized crosslinking using recombinant readers and cell nuclei and prepared nucleosomes and readers with investigation of the binding and crosslinking. Y. G. prepared some reader proteins and F. F. synthesized Fmoc-NleSme-OH. K. Z. and J. Y. drew the figures and wrote the manuscript with the guidance of M. W., and all authors contributed to the editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the support from National Natural Science Foundation of China (no. 22161132006) and Westlake University startup. We acknowledge the Instrumentation and Service Center for Molecular Sciences for the instrument support. We also thank the Mass Spectrometry & Metabolomics Core Facility of the Biomedical Research Core Facilities at Westlake University.Notes and references
- J. C. Black, C. Van Rechem and J. R. Whetstine, Mol. Cell, 2012, 48, 491–507 CrossRef CAS.
- C. D. Allis and T. Jenuwein, Nat. Rev. Genet., 2016, 17, 487–500 CrossRef CAS PubMed.
- P. K. Mazur, N. Reynoird, P. Khatri, P. W. T. C. Jansen, A. W. Wilkinson, S. Liu, O. Barbash, G. S. Van Aller, M. Huddleston, D. Dhanak, P. J. Tummino, R. G. Kruger, B. A. Garcia, A. J. Butte, M. Vermeulen, J. Sage and O. Gozani, Nature, 2014, 510, 283–287 CrossRef CAS.
- S. Ikram, A. Rege, M. Y. Negesse, A. G. Casanova, N. Reynoird and E. M. Green, Sci. Adv., 2023, 9, eadi5921 CrossRef CAS.
- X. Shi, L. Kachirskaia, H. Yamaguchi, L. E. West, H. Wen, E. W. Wang, S. Dutta, E. Appella and O. Gozani, Mol. Cell, 2007, 27, 636–646 CrossRef CAS.
- S. Chuikov, J. K. Kurash, J. R. Wilson, B. Xiao, N. Justin, G. S. Ivanov, K. McKinney, P. Tempst, C. Prives, S. J. Gamblin, N. A. Barlev and D. Reinberg, Nature, 2004, 432, 353–360 CrossRef CAS PubMed.
- J. Huang, L. Perez-Burgos, B. J. Placek, R. Sengupta, M. Richter, J. A. Dorsey, S. Kubicek, S. Opravil, T. Jenuwein and S. L. Berger, Nature, 2006, 444, 629–632 CrossRef CAS.
- S. D. Taverna, H. Li, A. J. Ruthenburg, C. D. Allis and D. J. Patel, Nat. Struct. Mol. Biol., 2007, 14, 1025–1040 CrossRef CAS PubMed.
- C. A. Musselman, M. E. Lalonde, J. Côté and T. G. Kutateladze, Nat. Struct. Mol. Biol., 2012, 19, 1218–1227 CrossRef CAS PubMed.
- M. Lachner, N. O'Carroll, S. Rea, K. Mechtler and T. Jenuwein, Nature, 2001, 410, 116–120 CrossRef CAS PubMed.
- A. J. Bannister, P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire and T. Kouzarides, Nature, 2001, 410, 120–124 CrossRef CAS.
- J. Wysocka, T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu and C. D. Allis, Nature, 2006, 442, 86–90 CrossRef CAS PubMed.
- X. Shi, T. Hong, K. L. Walter, M. Ewalt, E. Michishita, T. Hung, D. Carney, P. Peña, F. Lan, M. R. Kaadige, N. Lacoste, C. Cayrou, F. Davrazou, A. Saha, B. R. Cairns, D. E. Ayer, T. G. Kutateladze, Y. Shi, J. Côté, K. Chua and O. Gozani, Nature, 2006, 442, 96–99 CrossRef CAS PubMed.
- X. Li and X. D. Li, Acc. Chem. Res., 2021, 54, 3734–3747 CrossRef CAS PubMed.
- J. Lin and X. D. Li, Chin. Chem. Lett., 2018, 29, 1051–1057 CrossRef CAS.
- N. D. Pham, R. B. Parker and J. J. Kohler, Curr. Opin. Chem. Biol., 2013, 17, 90–101 CrossRef CAS.
- X. Li and T. M. Kapoor, J. Am. Chem. Soc., 2010, 132, 2504–2505 CrossRef CAS.
- T. Yang, X. Li, X. Bao, Y. M. E. Fung and X. D. Li, Nat. Chem. Biol., 2016, 12, 70–72 CrossRef CAS.
- J. Lin, Y. Wu, G. Tian, D. Yu, E. J. Yang, W. Lam, Z. Liu, Y. Jing, S. Dang, X. Bao, J. W. H. Wong, Y. Zhai and X. D. Li, Science, 2023, 379, 717–723 CrossRef CAS PubMed.
- F. Feng, Y. Gao, Q. Zhao, T. Luo, Q. Yang, N. Zhao, Y. Xiao, Y. Han, J. Pan, S. Feng, L. Zhang and M. Wu, Nat. Chem., 2024, 16, 1267–1277 CrossRef CAS.
- Y. Gao, F. Feng and M. Wu, Tetrahedron Lett., 2024, 151, 155332 CrossRef CAS.
- R. van Nuland, F. M. A. van Schaik, M. Simonis, S. van Heesch, E. Cuppen, R. Boelens, H. T. M. Timmers and H. van Ingen, Epigenet. Chromatin, 2013, 6, 12 CrossRef CAS.
- J. O. Eidahl, B. L. Crowe, J. A. North, C. J. McKee, N. Shkriabai, L. Feng, M. Plumb, R. L. Graham, R. J. Gorelick, S. Hess, M. G. Poirier, M. P. Foster and M. Kvaratskhelia, Nucleic Acids Res., 2013, 41, 3924–3936 CrossRef CAS PubMed.
- X. Fu, Y. Yuan, A. Jha, N. Levin, J. Ren, D. Mamalis, S. Mohammed, B. G. Davis and A. M. Giltrap, ACS Cent. Sci., 2023, 9, 405–416 CrossRef CAS PubMed.
- M. M. Müller and T. W. Muir, Chem. Rev., 2015, 115, 2296–2349 CrossRef.
- S. R. G. Galan, J. R. Wickens, J. Dadova, W. L. Ng, X. Zhang, R. A. Simion, R. Quinlan, E. Pires, R. S. Paton, S. Caddick, V. Chudasama and B. G. Davis, Nat. Chem. Biol., 2018, 14, 955–963 CrossRef CAS.
- M. D. Simon, F. Chu, L. R. Racki, C. C. de la Cruz, A. L. Burlingame, B. Panning, G. J. Narlikar and K. M. Shokat, Cell, 2007, 128, 1003–1012 CrossRef CAS PubMed.
- D. D. Le, A. T. Cortesi, S. A. Myers, A. L. Burlingame and D. G. Fujimori, J. Am. Chem. Soc., 2013, 135, 2879–2882 CrossRef CAS.
- F. Li, A. Allahverdi, R. Yang, G. B. J. Lua, X. Zhang, Y. Cao, N. Korolev, L. Nordenskiöld and C. Liu, Angew. Chem., Int. Ed., 2011, 50, 9611–9614 CrossRef CAS PubMed.
- R. Huang, M. A. Holbert, M. K. Tarrant, S. Curtet, D. R. Colquhoun, B. M. Dancy, B. C. Dancy, Y. S. Hwang, Y. Tang, K. Meeth, R. Marmorstein, R. N. Cole, S. Khochbin and P. A. Cole, J. Am. Chem. Soc., 2010, 132, 9986–9987 CrossRef CAS PubMed.
- B. Josephson, C. Fehl, P. G. Isenegger, S. Nadal, T. H. Wright, A. W. J. Poh, B. J. Bower, A. M. Giltrap, L. Chen, C. Batchelor-McAuley, G. Roper, O. Arisa, J. B. I. Sap, A. Kawamura, A. J. Baldwin, S. Mohammed, R. G. Compton, V. Gouverneur and B. G. Davis, Nature, 2020, 585, 530–537 CrossRef CAS PubMed.
- H. Xu, X. Qin, Y. Zhang, C. Wan, R. Wang, Z. Hou, X. Ding, H. Chen, Z. Zhou, Y. Li, C. Lian, F. Yin and Z. Li, Chin. Chem. Lett., 2022, 33, 2001–2004 CrossRef CAS.
- Z. Hou, D. Wang, Y. Li, R. Zhao, C. Wan, Y. Ma, C. Lian, F. Yin and Z. Li, J. Org. Chem., 2020, 85, 1698–1705 CrossRef CAS PubMed.
- C. Wan, Y. Feng, Z. Hou, C. Lian, L. Zhang, Y. An, J. Sun, D. Yang, C. Jiang, F. Yin, R. Wang and Z. Li, Org. Lett., 2022, 24, 581–586 CrossRef CAS.
- N. Liu, Y. Zhang, Y. Lei, R. Wang, M. Zhan, J. Liu, Y. An, Y. Zhou, J. Zhan, F. Yin and Z. Li, J. Med. Chem., 2022, 65, 876–884 CrossRef CAS.
- G. Bao, X. Song, Y. Li, Z. He, Q. Zuo, R. E, T. Yu, K. Li, J. Xie, W. Sun and R. Wang, Nat. Commun., 2024, 15, 6909 CrossRef PubMed.
- D. Wang, M. Yu, N. Liu, C. Lian, Z. Hou, R. Wang, R. Zhao, W. Li, Y. Jiang, X. Shi, S. Li, F. Yin and Z. Li, Chem. Sci., 2019, 10, 4966–4972 RSC.
- P. Hartmann, K. Bohdan, M. Hommrich, F. Juliá, L. Vogelsang, J. Eirich, R. Zangl, C. Farès, J. B. Jacobs, D. Mukhopadhyay, J. M. Mengeler, A. Vetere, M. S. Sterling, H. Hinrichs, S. Becker, N. Morgner, W. Schrader, I. Finkemeier, K. J. Dietz, C. Griesinger and T. Ritter, Nat. Chem., 2024, 16, 380–388 CrossRef CAS.
- M. Yar, E. M. McGarrigle and V. K. Aggarwal, Angew. Chem., Int. Ed., 2008, 47, 3784–3786 CrossRef CAS PubMed.
- Y. Xiao, K. Zou, J. Yang and M. Wu, Cell Rep. Phys. Sci., 2023, 4, 101638 CrossRef CAS.
- A. D. Baldwin and K. L. Kiick, Bioconjugate Chem., 2011, 22, 1946–1953 CrossRef CAS PubMed.
- B. Bernardim, M. J. Matos, X. Ferhati, I. Compañón, A. Guerreiro, P. Akkapeddi, A. C. B. Burtoloso, G. Jiménez-Osés, F. Corzana and G. J. L. Bernardes, Nat. Protoc., 2019, 14, 86–99 CrossRef CAS PubMed.
- N. Milosevich and F. Hof, Biochemistry, 2016, 55, 1570–1583 CrossRef CAS.
- L. I. James, D. Barsyte-Lovejoy, N. Zhong, L. Krichevsky, V. K. Korboukh, J. M. Herold, C. J. MacNevin, J. L. Norris, C. A. Sagum, W. Tempel, E. Marcon, H. Guo, C. Gao, X. Huang, S. Duan, A. Emili, J. F. Greenblatt, D. B. Kireev, J. Jin, W. P. Janzen, P. J. Brown, M. T. Bedford, C. H. Arrowsmith and S. V. Frye, Nat. Chem. Biol., 2013, 9, 184–191 CrossRef CAS PubMed.
- T. Yang, Z. Liu and X. Li, Chem. Sci., 2015, 6, 1011–1017 RSC.
- X. Guo, Y. Wang, Y. An, Z. Liu, J. Liu, J. Chen, M. Zhan, M. Liang, Z. Hou, C. Wan, F. Yin, R. Wang and Z. Li, Chem.–Eur. J., 2023, 29, e20230162 Search PubMed.
- X. Li, E. A. Foley, K. R. Molloy, Y. Li, B. T. Chait and T. M. Kapoor, J. Am. Chem. Soc., 2012, 134, 1982–1985 CrossRef CAS.
- E. J. Worden, N. A. Hoffmann, C. W. Hicks and C. Wolberger, Cell, 2019, 176, 1490–1501 CrossRef CAS PubMed.
- U. T. F. Lam, B. K. Y. Tan, J. J. X. Poh and E. S. Chen, Epigenet. Chromatin, 2022, 15, 17 CrossRef CAS.
- H. Wang, L. Farnung, C. Dienemann and P. Cramer, Nat. Struct. Mol. Biol., 2020, 27, 8–13 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc05886k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |