Open Access Article
Open Access ArticleTopMT-GAN: a 3D topology-driven generative model for efficient and diverse structure-based ligand design†
Shen
Wang
 a,
Tong
Lin
bc,
Tianyi
Peng
a,
Tong
Lin
bc,
Tianyi
Peng
 d,
Enming
Xing
d,
Enming
Xing
 a,
Sijie
Chen
a,
Levent Burak
Kara
a,
Sijie
Chen
a,
Levent Burak
Kara
 b and
Xiaolin
Cheng
b and
Xiaolin
Cheng
 *ae
*ae
aCollege of Pharmacy, The Ohio State University, Columbus, OH 43210, USA. E-mail: cheng.1302@osu.edu
bMechanical Engineering Department, Carnegie Mellon University, Pittsburgh, PA 15213, USA
cMachine Learning Department, Carnegie Mellon University, Pittsburgh, PA 15213, USA
dElectrical and Computer Engineering Department, Carnegie Mellon University, Pittsburgh, PA 15213, USA
eTranslational Data Analytics Institute (TDAI), The Ohio State University, Columbus, OH 43210, USA
First published on 8th January 2025
Abstract
Recent advancements in 3D structure-based molecular generative models have shown promise in expediting the hit discovery process in drug design. Despite their potential, efficiently generating a focused library of candidate molecules that exhibit both effective interactions and structural diversity at a large scale remains a significant challenge. Moreover, current studies often lack comprehensive comparisons to high-throughput virtual screening methods, resulting in insufficient evaluation of their effectiveness. In this study, we introduce Topology Molecular Type assignment (TopMT-GAN), a novel approach using Generative Adversarial Networks (GANs) for direct structure-based design. TopMT-GAN employs a two-step strategy: constructing 3D molecular topologies within a protein pocket with one GAN, followed by atom and bond type assignment with a second GAN. This integrated approach enables TopMT-GAN to efficiently generate diverse and potent ligands with precise 3D poses for specific protein pockets. When tested on five diverse protein pockets, TopMT-GAN exhibits promising and robust performance, demonstrating a potential enrichment of up to 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fold compared to traditional high-throughput virtual screening methods. This highlights its potential as a powerful tool in early-stage drug discovery, such as hit and lead generation.
000 fold compared to traditional high-throughput virtual screening methods. This highlights its potential as a powerful tool in early-stage drug discovery, such as hit and lead generation.
Introduction
The identification of optimal candidate ligands with both high affinity and specificity for protein targets remains a central challenge in drug discovery, often hampered by the immense size1 and inherent complexity of chemical space. Traditional computational methods such as high-throughput virtual screening,2,3 while valuable, are often resource-intensive and limited to existing chemical space. In contrast, structure-based generative models have emerged as transformative tools,4 demonstrating significant potential in effectively navigating the enormous unknown chemical space. Their ability to generate compounds that align with specific protein pockets makes them a targeted and streamlined approach in identifying promising ligand candidates in early-stage drug discovery.A crucial aspect of structure-based generative modeling lies in generating 3D molecular structures that accurately capture protein–ligand interactions. Molecular structure generation has been the subject of various studies, primarily categorized into three types based on molecular structure featurization:5 cubic grid-based, Euclidean distance matrix (EDM)-based, and Cartesian coordinate-based. Besides valid chemical connectivity and appropriate conformational geometry, it is essential for generated molecules to adopt binding poses that complement the target pocket and facilitate favorable interactions between ligands and the target. However, the integration of pocket information into molecule generation remains limited. For instance, LiGANN6 maps the pocket shape to ligand shape and decodes this into SMILES strings, representing an initial attempt in structure-based ligand design with generative models. Nevertheless, it does not generate 3D molecular structures.
Most structured-based models adopt autoregressive methods to sequentially build molecules atom by atom. Some models, such as SBDD,7 Pocket2Mol,8 GraphBP,9 SurfGen,10 ResGen,11 and PocketFlow12 use Graph Neural Networks (GNNs) to process pocket context and predict atom placements. In contrast, DeepLigBuilder13 utilizes Monte-Carlo Tree Search (MCTS) with smina14 docking scores to guide its L-Net generation process. Diverging from autoregressive approaches, TargetDiff,15 ShapeMol16 and PMDM17 employ diffusion models to generate entire molecules conditioned on the pocket environment. Additionally, fragment-based methods such as Lingo3DMol,18 ligandED,19 and DESERT20 assemble molecules by concatenating fragments within pockets. Although promising, methods like Pocket2Drug21 and SBMolGen,22 which rely on SMILES strings coupled with subsequent conformational sampling, are not 3D molecular generative models, and we there exclude them from comparison and discussion below.
Despite their promising potential, there are notable limitations that most current implementations have yet to overcome.
Limited scale
Most models, including SBDD, Pocket2Mol, GraphBP, DESERT, TargetDiff, SurfGen, ResGen, and Lingo3DMol, typically generate only around 100 molecules per target during evaluation. However, this falls short of the scale required for real-world drug discovery, where ultra-large high-throughput virtual screening against up to billions of compounds is now achievable.23,24 In such situations, these generative models offer no significant advantage in terms of chemical space exploration.Diversity challenges
Scaffold diversity is crucial in drug discovery, as a greater variety of scaffolds increases the likelihood of identifying active compounds and enhances the potential to discover novel structures distinct from those protected by patents. However, the limited scale of generation often raises concerns regarding the true diversity of the generated molecules. Relying on small pool of generated structures may lead to unreliable conclusions. Additionally, as the scale of molecule generation increases to tens of thousands, the risk of mode collapse—where the model repeatedly generates a narrow range of similar structures—significantly rises. This necessitates careful scaling and robust model validation to ensure a diverse output of molecular structures.Generation efficiency
While DeepLigBuilder reports generating approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules for the SARS-CoV-2 main protease and PocketFlow has produced 100
000 molecules for the SARS-CoV-2 main protease and PocketFlow has produced 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules for their case studies, generation speed remains a significant challenge across most models. For instance, DeepLigBuilder requires 3–5 hours to generate 100 molecules using a single RTX 3070 graphics card. Similarly, Pocket2Mol, TargetDiff, Lingo3DMol, and PMDM take about 1000 seconds to produce the same number on various hardware setups. DESERT, on the other hand, claims a rate of 20 good molecules per hour, though the specifics of their computing resources are not disclosed. PocketFlow demonstrates the ability to generate 50
000 molecules for their case studies, generation speed remains a significant challenge across most models. For instance, DeepLigBuilder requires 3–5 hours to generate 100 molecules using a single RTX 3070 graphics card. Similarly, Pocket2Mol, TargetDiff, Lingo3DMol, and PMDM take about 1000 seconds to produce the same number on various hardware setups. DESERT, on the other hand, claims a rate of 20 good molecules per hour, though the specifics of their computing resources are not disclosed. PocketFlow demonstrates the ability to generate 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules using 2 tesla P100s within two days; however, their analysis primarily focuses on the top few hundred molecules. Scaling up generation while maintaining efficiency will be crucial for practical application, as generating larger numbers of potentially duplicate molecules will further exacerbate speed concerns.
000 molecules using 2 tesla P100s within two days; however, their analysis primarily focuses on the top few hundred molecules. Scaling up generation while maintaining efficiency will be crucial for practical application, as generating larger numbers of potentially duplicate molecules will further exacerbate speed concerns.
In addition to the limitations of scale, diversity, and efficiency, another critical issue is the lack of a standardized benchmark for comparing generative models. SBDD, Pocket2Mol, DESERT, TargetDiff, Lingo3DMol, and PMDM utilized proteins from the DUD-E25 or CrossDock26 datasets as test cases, averaging docking results across 10–100 protein targets. However, this approach fails to capture the crucial diversity of protein pockets. Some pockets are large and deep, facilitating strong binding, while others are shallow and surface-exposed, leading to weaker interactions. Using a single, averaged score over such varied targets obscures a model's specific performance on individual pocket types, impeding meaningful comparison and optimization. In addition to internal comparisons among the generative models, SurfGen and ResGen also included a comparison with molecular docking of random compound libraries, but their docking evaluations were limited to just 200 randomly selected molecules. A comprehensive approach is needed to assess the true effectiveness of generative models, particularly in the context of large-scale high-throughput virtual screening (HTVS).
To overcome these prevailing limitations in scale, diversity, and efficiency, we have developed a novel two-step 3D structure-based generative model called Topology Molecular Type assignment (TopMT-GAN). The initial phase of our approach focuses on generating valid molecular topologies that align closely with the contours of the target pockets. This shape complementarity is critical in guiding the exploration of the discovery space.27 The generation is achieved through the use of a Graph Translation Generative Adversarial Network (GAN),28 which effectively models potential molecular configurations to match the spatial characteristics of the pocket. This phase also includes an advanced search strategy and a local topology filter to ensure the validity and diverse sampling of topologies. In the second step, we employ another GAN for molecular assignment that predicts atom and bond types for each topology to generate valid molecules. This process, combined with local minimization, allows for accurate positioning of generated molecules within the target pocket, facilitating the rapid scoring of these molecules.
In addition to the novel architecture of TopMT-GAN, we have implemented a rigorous benchmarking method for evaluating 3D molecular generative models that is different from those employed in previous studies.7–22 As shown in Table S6,† existing approaches vary significantly in their evaluation methods, from generating just 100 molecules per target to using different baseline comparisons, making it difficult to assess relative performance. To address these limitations, our benchmarking task is designed to reflect the complexities of real-world drug design scenarios and address the corresponding critical challenges. This comprehensive task involves selecting a diverse set of five protein targets with distinct pocket features, including enzymes, kinases, G-protein coupled receptors (GPCRs), and nuclear receptors. We then generated a substantial number of ligands—50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for each of the five protein pockets. For comparison, we also conducted moderate-scale high-throughput virtual screening (HTVS) of over 1 million compounds from the Enamine HTS collection43 against these targets. Our evaluation across the diverse targets revealed that TopMT-GAN could efficiently generate tens of thousands of molecules, which exhibit robust binding scores and strong scaffold diversity. Furthermore, the results indicated that molecules generated by TopMT-GAN could achieve up to a 46
000 for each of the five protein pockets. For comparison, we also conducted moderate-scale high-throughput virtual screening (HTVS) of over 1 million compounds from the Enamine HTS collection43 against these targets. Our evaluation across the diverse targets revealed that TopMT-GAN could efficiently generate tens of thousands of molecules, which exhibit robust binding scores and strong scaffold diversity. Furthermore, the results indicated that molecules generated by TopMT-GAN could achieve up to a 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fold enrichment compared to random HTVS, marking the first quantitative demonstration of a structure-based generative model's superior efficiency over traditional HTVS approaches. These findings showed not only the accuracy (i.e., potential binding in the pocket) but also the effectiveness (i.e., the speed and scaffold diversity) of our model, supporting its practical utility in real-world drug discovery.
000 fold enrichment compared to random HTVS, marking the first quantitative demonstration of a structure-based generative model's superior efficiency over traditional HTVS approaches. These findings showed not only the accuracy (i.e., potential binding in the pocket) but also the effectiveness (i.e., the speed and scaffold diversity) of our model, supporting its practical utility in real-world drug discovery.
Results
Overview of TopMT-GAN's framework
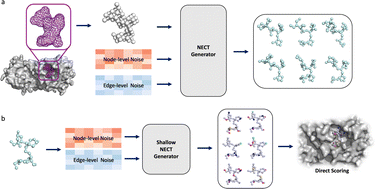
In the design of Topology Molecular Type assignment (TopMT-GAN), we have structured the framework into two distinct submodules: a topology generation module and a molecular type assignment module (Fig. 1). The topology generation module utilizes a graph translation GAN to create a diverse range of molecular topologies with 3D coordinates, specifically adapted to fit within the target protein pockets. Following this, the molecular type assignment module employs a second graph GAN to assign atom and bond types to each of the generated topologies. Both stages are highly efficient and parallelizable, enabling rapid sampling of a vast library of molecules within the spatial constraints of the target pockets. Additionally, fast scoring methods can be applied to prioritize and filter these molecules without the need for expensive conformational sampling.TopMT-GAN works in two distinct modes to generate ligands for specific targets: scaffold-hopping and pocket-mapping. When co-crystal structures for a target pocket bound with a known ligand are available, pocket binding molecules can be generated through the scaffold-hopping strategy. In this case, the initial pocket shape input to TopMT-GAN is derived from the structure of the bound ligand. On the other hand, when no ligand data for a target pocket are available, TopMT-GAN can map the pocket shape directly by using small fragment probes. The mapped pocket shape can then serve as the basis for ligand generation, allowing for the design of molecules that are tailored to fit the target site. Results from both strategies were evaluated and compared.
To rigorously evaluate the performance of TopMT-GAN in addressing the limitations of existing structure-based generative models, we conducted a benchmark comparison with high-throughput virtual screening against a library of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 327
327![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 116 molecules from the Enamine HTS library. For this purpose, we generated 50
116 molecules from the Enamine HTS library. For this purpose, we generated 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules for each protein target under investigation. Concurrently, we performed molecular docking of each compound in the library into the target pockets using AutoDock Vina.29,30 Additionally, we also gathered all known actives for each protein pocket from the Binding Database,31 and employed them as an additional benchmark to assess TopMT-GAN's performance. To facilitate performance comparisons, we report the hit rates for our 5 protein targets using Enamine HTS collections across different Vina score thresholds (Table S7†).
000 molecules for each protein target under investigation. Concurrently, we performed molecular docking of each compound in the library into the target pockets using AutoDock Vina.29,30 Additionally, we also gathered all known actives for each protein pocket from the Binding Database,31 and employed them as an additional benchmark to assess TopMT-GAN's performance. To facilitate performance comparisons, we report the hit rates for our 5 protein targets using Enamine HTS collections across different Vina score thresholds (Table S7†).
The performance of structure-based generative models can be influenced by the characteristics of target pockets. To demonstrate the robustness of TopMT-GAN, we selected five protein systems that represent a range of distinct pocket characteristics. These include 3C-like protease (PDB ID 7d3i)32 and androgen receptor (PDB ID 1e3g),33 both of which feature typical deep and buried protein pockets. In contrast, allosteric sites are often shallow and situated on protein surfaces, posing a greater challenge for ligand design. To facilitate a direct comparison between these different pocket types, we focused on two kinases with analogous overall structures yet distinct pocket types: c-Src kinase, which is bound with ponatinib at its orthosteric site (PDB ID 7wf5 (ref. 34)), and checkpoint kinase 1 (CHK1) that accommodates an allosteric ligand (PDB ID 3jvs35). Additionally, we selected the glucagon-like peptide-1 (GLP-1) receptor (PDB ID 5vew)36 to further evaluate TopMT-GAN's ability to design allosteric ligands for unusually large and shallow pockets. The five selected proteins are summarized in Table S1,† which includes their PDB ID, calculated solvent accessible surface area (SASA), pocket volume, ratio of SASA/volume and number of known actives.
Ligand design for orthosteric pockets
To evaluate ligand design for a typical substrate-binding pocket, we compare the Vina score distributions for molecules generated by TopMT-GAN with those obtained from HTVS and known actives. For comparison, 101 and 108 active compounds were collected for 3C-like protease and androgen receptor, respectively. As shown in Fig. 2 and S4,† the Vina score distributions show a notable shift towards lower Vina scores for our generated molecules compared to those from HTVS for both 3C-L protease and androgen receptor. These molecules demonstrated superior docking scores even when compared to known actives. Although these results do not necessarily indicate that the generated molecules will be more potent than the known actives, they highlight our model's capability to produce compounds with promising docking profiles, suggesting potential for favorable molecular interactions and binding potency. We then calculated enrichment factors (as defined in eqn (1)) across various Vina score thresholds, and these are detailed in Table 1. Notably, for 3C-like protease, at a Vina score threshold of −10 kcal mol−1, TopMT-GAN achieved impressive enrichment factors of 5751 and 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 272 for scaffold-hopping and pocket-mapping modes, respectively. These results underscore the remarkable effectiveness of TopMT-GAN in identifying potent ligands for protein orthosteric pockets.
272 for scaffold-hopping and pocket-mapping modes, respectively. These results underscore the remarkable effectiveness of TopMT-GAN in identifying potent ligands for protein orthosteric pockets.
 | ||
| Fig. 2 Distributions of TopMT-GAN generated molecules for 3C-L protease. (a1 and a2) Vina docking score distributions for scaffold-hopping and pocket-mapping modes. (b1 and b2) Scatter plots of QED versus docking scores for scaffold-hopping and pocket-mapping modes. (c1–c4) NPR space distributions. (c1) For scaffold-hopping, (c2) for pocket-mapping, (c3) for Enamine HTS collection, and (c4) for random PubChem compounds. (d) RDKit generic scaffold of the original ligand from PDB 7d3i (red box) alongside representative examples of generated scaffolds. Top row: scaffold-hopping mode, bottom row: pocket-mapping mode (e) T-map visualization of generated molecules for pocket-mapping mode. Salmon: generated molecules, light blue: drug-bank molecules, light green: known actives. | ||
| Target | Redock score kcal mol−1 | Sampling mode | EF < redock | EF < −8 | EF < −9 | EF < −10 | EF < −11 | EF < −12 |
|---|---|---|---|---|---|---|---|---|
| 3C-like protease PDB 7d3i | −8.3 | Scaffold-hopping | 69 | 32 | 440 | 5751 | N/A | N/A |
| Pocket-mapping | 115 | 43 | 1389 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 272 272
|
N/A | N/A | ||
| Androgen receptor PDB 1e3g | −11.7 | Scaffold-hopping | 770 | 9 | 121 | 1262 | 4725 | N/A |
| Pocket-mapping | 743 | 9 | 105 | 915 | 4247 | N/A | ||
| c-SRC kinase PDB 7wf5 | −13.2 | Scaffold-hopping |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 042 042
|
2 | 5 | 29 | 288 | 3300 |
| Pocket-mapping |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 228 228
|
2 | 5 | 27 | 261 | 2883 | ||
| CHK1 kinase PDB 3jvs | −8.1 | Scaffold-hopping | 52 | 43 | 330 | N/A | N/A | N/A |
| Pocket-mapping | 64 | 52 | 441 | N/A | N/A | N/A | ||
| GLP-1 receptor PDB 5vew | −7.2 | Scaffold-hopping | 39 | 272 | 1497 | N/A | N/A | N/A |
| Pocket-mapping | 40 | 329 | 2580 | N/A | N/A | N/A |
The diversity of the molecules generated by TopMT-GAN is quantified using an internal diversity metric based on Morgan fingerprints (eqn (2)). All generated sets exhibited a diversity score exceeding 0.8, indicating a broad spectrum of molecular structures (see Table 2 for details). Although TopMT-GAN is not prone to the issue of mode collapse or mere replication of training set outcomes, we sought to further validate its ability to generate novel scaffolds. This was achieved by comparing the similarity of the generated molecules to known actives, using a metric defined in eqn (3) which is also based on Morgan fingerprints. The distribution of maximum similarity score to known actives is depicted in Fig. S7,† the chart shows that the majority of generated molecules have low similarities to known actives, mostly ranging from 0 to 0.2. The average similarity scores to known actives were 0.1 or lower, indicating minimal overlap with the chemical space of the known ligands. This distribution suggests that the generated ligands exhibit minimal resemblance to known actives, emphasizing the model's ability to generate novel scaffolds and structures.
| Target | Mode | Internal diversity | Averaged similarity to actives | Max. similarity to actives | # unique scaffolds |
|---|---|---|---|---|---|
| 3C-like protease PDB 7d3i | Scaffold-hopping | 0.87 | 0.09 | 0.33 | 7574 |
| Pocket-mapping | 0.87 | 0.08 | 0.32 | 5961 | |
| Androgen receptor PDB 1e3g | Scaffold-hopping | 0.88 | 0.07 | 0.28 | 3527 |
| Pocket-mapping | 0.88 | 0.07 | 0.31 | 2824 | |
| c-SRC kinase PDB 7wf5 | Scaffold-hopping | 0.88 | 0.08 | 0.42 | 6558 |
| Pocket-mapping | 0.88 | 0.08 | 0.33 | 4802 | |
| CHK1 kinase PDB 3jvs | Scaffold-hopping | 0.88 | 0.09 | 0.38 | 9474 |
| Pocket-mapping | 0.88 | 0.09 | 0.39 | 5463 | |
| GLP-1 receptor PDB 5vew | Scaffold-hopping | 0.88 | 0.10 | 0.31 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 745 745 |
| Pocket-mapping | 0.87 | 0.10 | 0.32 | 6704 |
We also identified RDKit scaffolds within the generated sets and highlighted the most common scaffolds and their structures in Fig. 2d. Fig. 2d depicts the scaffold of the ligand bound in 3C-like protease (PDB ID 7d3i, in a red box), alongside representative generated scaffolds: those from the scaffold-hopping mode on the top row, and those from the pocket-mapping mode on the bottom row. These generated scaffolds showed very similar 3 hydrophobic interaction sites as the original ligand. For both modes, each 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-molecule set contained over 2500 unique scaffolds, as detailed in Table 2.
000-molecule set contained over 2500 unique scaffolds, as detailed in Table 2.
The spatial distribution of these molecules is illustrated in T-map37 space (Fig. 2e), with molecules color-coded as follows: salmon for generated molecules, light blue for DrugBank38 molecules, and light green for known actives. The T-map offers a tree-like visualization, where similar molecules cluster closely, often on the same branch, providing an intuitive view of the molecular diversity within the sets. The extensive branching observed in the T-map confirms TopMT-GAN's ability to explore diverse chemical spaces, encompassing the range of DrugBank compounds and known actives, and even extending beyond them. Collectively, these findings demonstrate that TopMT-GAN excels in generating molecules that are both potent and structurally diverse for orthosteric pockets.
Ligand design for allosteric pockets
Allosteric pockets present unique challenges for ligand design due to their shallow and surface-exposed characteristics. We characterized five protein pockets under study using the ratio of the solvent-accessible surface area (SASA) to the pocket volume. As shown in Table S1,† these ratios showed a clear distinction between orthosteric and allosteric pockets. Orthosteric pockets, which are deep and buried, exhibit higher ratios, indicating their potential for more extensive ligand interactions. In contrast, allosteric pockets have smaller ratios, suggesting a limited capacity for ligand interaction. This highlights the complexity involved in designing highly potent ligands for allosteric pockets, which necessitates a delicate balance between the buried portion of the molecules interacting with the protein and the exposed surface of the molecules interacting with the solvent.To further elucidate these differences, we selected two kinase targets for comparison: the c-Src kinase (PDB ID 7wf5), which has a ligand bound in the orthosteric pocket, and the serine/threonine–protein kinase CHK1 (PDB ID 3jvs), which is in complex with a ligand at an allosteric site. Fig. 3a provides a detailed comparison of these two pockets. As expected, the orthosteric pocket in the c-Src kinase, located in a cleft between two domains (pink mesh), is significantly deeper and more buried than the allosteric pocket in the CHK1 kinase (cyan mesh), which is much shallow and exposed on the surface. The SASA/volume ratios for the c-Src kinase and CHK1 are 1.48 and 1.12, respectively. Moreover, the Vina redock scores for their co-crystalized ligands are −13.2 kcal mol−1 and −8.2 kcal mol−1, respectively, supporting the notion that allosteric ligands do not bind as strongly as orthosteric ones.
 | ||
| Fig. 3 Comparison of two kinases and their ligand binding poses. (a) Structural alignment of c-Src kinase (PDB ID 7wf5, light pink) and CHK1 kinase (PDB ID 3jvs, pale cyan) with the orthosteric pocket of c-Src kinase and the allosteric pocket of CHK1 shown in a mesh surface representation. (b1) Ligand from the crystal structure of c-Src kinase. (b2 and b3) Two ligands generated for the orthosteric pocket of c-Src kinase, with generated poses in salmon and redocked poses in cyan. Vina scores are reported as redocked scores and RMSDs are calculated between generated and redocked poses of the ligand. (c1) Ligand from the crystal structure of CHK1 kinase. (c2 and c3) Two generated ligands for the allosteric pocket of CHK1 kinase, with their corresponding redocked scores and RMSDs. | ||
The ligands generated for c-Src kinase show similar results to those for 3C-like protease and androgen receptor, with the enrichment factor reaching impressive 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 042 and 46
042 and 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 228 at the redock score threshold of −13.2 kcal mol−1. This demonstrates TopMT-GAN's capability in designing exceptionally potent ligands. However, due to the inherent challenges of allosteric pockets, the generation of potent allosteric ligands is notably more difficult. Nonetheless, for CHK1 allosteric ligand generation, TopMT-GAN achieved significant enrichment factors of 330 and 441 at a threshold of −9 kcal mol−1 for the scaffold-hopping and pocket-mapping modes, respectively. Moreover, the Vina score distribution of the molecules generated for allosteric sites was better than or equal to that of known actives (Fig. S4†), underscoring TopMT-GAN's efficiency in generating ligands for allosteric pockets.
228 at the redock score threshold of −13.2 kcal mol−1. This demonstrates TopMT-GAN's capability in designing exceptionally potent ligands. However, due to the inherent challenges of allosteric pockets, the generation of potent allosteric ligands is notably more difficult. Nonetheless, for CHK1 allosteric ligand generation, TopMT-GAN achieved significant enrichment factors of 330 and 441 at a threshold of −9 kcal mol−1 for the scaffold-hopping and pocket-mapping modes, respectively. Moreover, the Vina score distribution of the molecules generated for allosteric sites was better than or equal to that of known actives (Fig. S4†), underscoring TopMT-GAN's efficiency in generating ligands for allosteric pockets.
To further evaluate the robustness of TopMT-GAN, we selected the GLP-1 receptor, a prominent protein in diabetes treatment. Its native ligand, GLP-1,39 comprises approximately 30 receptor-interacting amino acids,40 posing a significant challenge for the design of small molecule agonists that can produce similar biological effects as GLP-1. Consequently, the design of allosteric ligands has emerged as a more viable strategy. The allosteric pocket of the GLP-1 receptor is shown in Fig. S3b,† featuring a shallow and irregular shape, which, however, complicates the design process. This challenge is evidenced by the weak redocking score of −7.2 kcal mol−1 for the co-crystalized ligand at the allosteric site (PDB ID 5vew).
Despite these complexities, TopMT-GAN successfully generated allosteric ligands for the GLP-1 receptor. The results were promising; compared to HTVS, TopMT-GAN achieved enrichment factors of 1497 and 2580 (at a threshold of −9 kcal mol−1) for scaffold-hopping and pocket-mapping modes, respectively, Furthermore, the Vina score distributions of our generated molecules were superior even when compared to 796 known actives (Fig. 4). Fig. 4c shows the top 10 scaffolds for pocket-mapping mode, along with their molecular structures and occurrence counts. Despite their structural diversity, most of these scaffolds notably feature two branches and an anchoring site, allowing them to fully occupy the pocket and interact extensively with the receptor, facilitating similar binding as in the crystal structure.
Molecular properties
Following the examination of scaffold diversity, the internal diversity and similarity to the known actives of our generated molecules are summarized in Table 2. These metrics displayed consistent results across all five systems, with the lowest diversity score recorded at 0.87 for the 3C-like protease and the highest average similarity of 0.1 observed in GLP-1. The detailed profile of max. similarity distribution of generated molecules to actives are shown in Fig. S7.† Only a very small fraction of generated molecules exhibited similarities greater than 0.5, confirming the ability of our model to generate novel structures from known actives. Shape complementarity is a crucial aspect of structure-based generative design, and to this end, we have illustrated the shape distribution of our generated molecules within the Normalized Principal Moments of Inertia Ratio (NPR) descriptors41 space (Fig. 2c). In this NPR space, linear molecules are located in the top-left area, spherical molecules in the top-right, and planar molecules occupy the bottom region. This visualization reveals that our molecules are concentrated within a specific region of the chemical space, unlike the broader distribution observed for the Enamine HTS library (Fig. 2c3) or randomly selected molecules from PubChem (Fig. 2c4). Additionally, the distribution in NPR space reveals a preference for linear-shaped molecules in the pocket-mapping mode for the 3C-like protease, reflecting the extended length of this protein's pocket compared to its ligand in the crystal structure. Such a focused occupancy by our molecules highlights TopMT-GAN's efficiency in navigating the relevant chemical space, potentially leading to more effective interactions with the intended protein targets.We next examined the drug-likeness properties of our generated molecules by computing their Quantitative Estimate of Drug-likeness (QED). QED is a dimensionless score, whose value ranges between 0 and 1, with 1 being the most drug-like. The correlation between QED and Vina docking scores is depicted in Fig. 2b and 4b. The upper left quadrants represent potential high-quality hit ligands characterized by both high QED and low docking scores. Notably, a substantial fraction of the molecules generated by TopMT-GAN outperforms the co-crystalized ligands and known actives in terms of both QED and docking scores. This indicates a superior potential of TopMT-GAN for hit and lead identification. Other molecular properties, such as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and synthetic accessibility score are provided in Fig. S4.† The distribution of log
P and synthetic accessibility score are provided in Fig. S4.† The distribution of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P resembles the distribution of Enamine HTS collection and PubChem dataset. Regarding the SAS, our generated molecules tend to have higher scores, indicating more complex molecular structures, as seen in the comparison with the PubChem dataset. This is expected as we didn't set out to optimize this property during the initial version of our TopMT-GAN implementation.
P resembles the distribution of Enamine HTS collection and PubChem dataset. Regarding the SAS, our generated molecules tend to have higher scores, indicating more complex molecular structures, as seen in the comparison with the PubChem dataset. This is expected as we didn't set out to optimize this property during the initial version of our TopMT-GAN implementation.
Binding poses
A key strength of 3D structure-based generative models lies in their ability to directly produce binding poses. This feature facilitates rapid scoring and ranking of generated molecules. We examined a variety of poses from our generated ligands targeting the 3C-L protease, which were then compared with those captured in the crystal structures (Fig. 5). In the scaffold-hopping mode, the generated poses mirrored the shape as the original ligands, exhibiting binding patterns that were strikingly similar to the crystal poses (indicated by the salmon color in Fig. 5). On the other hand, in the pocket-mapping mode, the generated ligands could explore different regions within the pocket, leading to alternative binding poses that were not observed in the crystal structures. | ||
| Fig. 5 Selected ligand poses for 3C-like protease. (a) Ligand in the crystal structure of 3C-like protease (PDB ID 7d3i). (b1) Pose and shape of the original ligand in the crystal structure; (b2) detected pocket. (c1 and c2) Selected poses generated in the scaffold-hopping mode with generated poses in salmon and redocked poses in cyan, including Vina redocked scores and RMSD values between generated and redocked poses. (d1 and d2) Selected ligands and their poses generated in the pocket-mapping mode. | ||
In the subsequent analysis, the generated ligands were subjected to a redocking process, with the poses with the lowest redocking scores highlighted in cyan. The comparison revealed that while some generated poses align closely with the best redocked poses, there were instances where the redocked poses showed stronger binding. It is worth noting that our model was not explicitly trained to predict the best binding mode. Additionally, the docking algorithms don't guarantee to produce the most favorable binding poses either. Nevertheless, the generated poses set a baseline for potentially favorable binding interactions, with the redocked poses either meeting or surpassing this baseline. For all five systems, an average of 87.4% of redocked poses were either equally good or better than the generated poses (Table S4†). This capability is crucial for the initial ranking and selection of promising hit candidates for further detailed analysis.
To assess the quality of our generated binding poses, we performed a molecular geometry analysis using PoseCheck,42 focusing on two key metrics: steric clashes and strain energy. The performance of TopMT-GAN on these metrics was compared with other leading structure-based generative models (Table S3†). TopMT-GAN demonstrates encouraging performance in steric clashes, averaging 27 clashes per molecule, comparable to Pocket2Mol's 15 clashes. This relatively low clash count reflects the effectiveness of our two-stage geometric optimization strategy. In the first stage, only the ligand structure is optimized to establish a reasonable initial geometry. In the second stage, the structure is refined within the binding pocket, minimizing unfavorable steric interactions.
However, our analysis revealed a limitation in strain energy for TopMT-GAN. It generated poses with an average strain energy of 1170 kcal mol−1. While this is significantly lower than PMDM (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 822 kcal mol−1) and PocketFlow (6939 kcal mol−1), it remains higher than Pocket2Mol's remarkably low strain energy of 91 kcal mol−1. This discrepancy may stem from our use of AutoDock Vina for rapid pose scoring, which prioritizes computational efficiency over accurate energetic evaluation. Although Vina effectively captures protein–ligand interactions, it does not explicitly account for internal ligand strain, potentially resulting in less energetically favorable conformations. Future improvements could incorporate explicit strain energy terms into the scoring function to generate more energetically favorable conformations.
822 kcal mol−1) and PocketFlow (6939 kcal mol−1), it remains higher than Pocket2Mol's remarkably low strain energy of 91 kcal mol−1. This discrepancy may stem from our use of AutoDock Vina for rapid pose scoring, which prioritizes computational efficiency over accurate energetic evaluation. Although Vina effectively captures protein–ligand interactions, it does not explicitly account for internal ligand strain, potentially resulting in less energetically favorable conformations. Future improvements could incorporate explicit strain energy terms into the scoring function to generate more energetically favorable conformations.
Generation efficiency
The efficiency of molecule generation is essential for exploring relevant chemical spaces, especially when scaling up to produce tens of thousands of molecules. Given that TopMT-GAN operates through two sequential stages, topology generation and molecular assignment, it is imperative to evaluate the time efficiency of each step. We report the generation speed for 5 systems in Table S5.† Generation speed varies for different systems and generation modes. In general, sampling within a larger pocket demands more time. The most time-consuming step is the topology generation process, which takes approximately 2 seconds per topology on an Nvidia RTX A5000 GPU card. Once the molecular topologies are generated, the subsequent molecular assignment and Vina scoring steps can be executed in parallel. For these processes, we deployed 16 nodes, each equipped with Dual Intel Xeon 8268s Cascade Lake processors with 48 cores. The computational cost is quantified as CPU-core time per molecule for each of these tasks. Remarkably, both the molecular assignment and scoring phases are executed with high speed, completing within less than 5 hours for all systems.The generation speed varies depending on the size of the pocket to be explored and the size of the desired molecules. The pocket-mapping mode, which encompasses larger pocket volumes, typically results in longer sampling times. Notably, the slowest process—pocket-mapping mode for the c-Src kinase orthosteric pocket, the largest among the five pockets tested—generates molecules at a rate of 330 seconds per 100 molecules. Overall, TopMT-GAN requires between 0.25 to 1.89 days to generate 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 promising ligands. These results underscore TopMT-GAN's exceptional potential in scaling up and generating a modestly sized, focused library of potent and diverse molecules. To the best of our knowledge, this positions TopMT-GAN as the fastest among 3D structure-based generative models to date. The model's ability to rapidly produce a large number of potential ligands highlights its efficiency in exploring chemical spaces, significantly enhancing its potential utility in drug development processes.
000 promising ligands. These results underscore TopMT-GAN's exceptional potential in scaling up and generating a modestly sized, focused library of potent and diverse molecules. To the best of our knowledge, this positions TopMT-GAN as the fastest among 3D structure-based generative models to date. The model's ability to rapidly produce a large number of potential ligands highlights its efficiency in exploring chemical spaces, significantly enhancing its potential utility in drug development processes.
Comparison with existing models
To benchmark of TopMT-GAN against existing molecular generative models, we conducted a focused comparative study that balances computational feasibility with meaningful evaluation. We selected three 3D structure-based generative models – Pocket2Mol, PocketFlow, and PMDM – based on their computational efficiency and similar input requirements, specifically their ability to work with protein structures without requiring manual pocket definition. For this study, we used five representative protein targets from our benchmark dataset, generating 500 molecules per target with each model. While this scale is smaller than our full evaluation (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules), it allows for a robust and meaningful comparative analysis. For PMDM, all valid molecules generated within reasonable computational constraints were included, whereas for some models, fewer than 500 molecules were produced.
000 molecules), it allows for a robust and meaningful comparative analysis. For PMDM, all valid molecules generated within reasonable computational constraints were included, whereas for some models, fewer than 500 molecules were produced.
The generated molecules were docked to their respective crystal structure pockets using AutoDock Vina. The docking score distributions for all models are shown in Fig. S6.† Across all five targets, TopMT-GAN consistently outperformed other generative models. While other models produced molecules with good docking scores, they did not show significant improvements over the HTVS baseline. This finding underscores the importance of contextualizing docking scores within specific protein pockets rather than relying on docking scores alone. For instance, deep and buried pockets, such as kinase orthosteric sites, tend to yield high docking scores regardless of the generative method used. Thus, while generated molecules may achieve impressive absolute docking scores, the relative improvement over the HTVS baseline provides a more meaningful measure of generative model performance.
In addition to docking performance, we evaluated other model quality metrics across the generative models (Table S2†), focusing on general molecular properties such as drug-likeness (QED), synthetic accessibility (SAS), and internal diversity. Each model shows distinct strengths: Pocket2Mol achieves the highest QED (0.65 ± 0.17), while TopMT-GAN and PocketFlow show moderate QED values (0.45 ± 0.19 and 0.46 ± 0.17, respectively). For synthetic accessibility, TopMT-GAN and PMDM generated molecules with higher SAS scores (5.6 ± 0.7), indicating potentially more complex structures, whereas PocketFlow and Pocket2Mol produced molecules with lower synthetic complexity (3.4 ± 1.1 and 3.1 ± 1.3, respectively). All models demonstrated high internal diversity (0.88–0.91), suggesting effective exploration of diverse chemical spaces. These additional metrics provide valuable context for comparing generative models, though binding performance remains the primary evaluation criterion.
Discussion
The field of 3D deep molecular generative models is experiencing significant advancements. A variety of models have been developed by the integration of deep learning techniques with molecular design. Especially, recent advances in graph and diffusion generative models have opened opportunities for more accurate predictions of molecular conformations and interactions. However, the field faces several challenges. One of the primary issues is the generation efficiency. While there have been improvements, the discrete nature of chemical space and the vast number of possible drug-like compounds present a significant challenge for the published models to sample a meaningful number of potential hit molecules. This issue is compounded by the fact that only a small fraction of these compounds is therapeutically relevant, necessitating models that can effectively navigate this space to identify a diverse set of compounds. Additionally, there is a need for high-precision evaluation metrics to accurately assess the performance of these generative models, ensuring that the molecules generated have the desired properties and are viable for drug development.To address these challenges, we introduced TopMT-GAN, a novel structure-based 3D molecular generative model, for generating chemically valid 3D molecular structures within target binding sites. This model leverages deep learning techniques, such as generative adversarial network (GAN) models and A-star search, to predict high-affinity drug-like compounds with novel chemical structures. Comprehensive testing across five diverse protein targets demonstrated its robust performance in generating large-scale, highly diverse potent ligand sets targeting both orthosteric and allosteric pockets. Importantly, this work represents the first extensive comparison of a structure-based generative model with high-throughput virtual screening. Our results reveal TopMT-GAN's superior efficiency in exploring the relevant chemical space.
The current version of TopMT-GAN prioritizes the generation of potent and diverse ligands. This focus may result in some generated molecules with complex synthetic accessibility. However, the flexible two-step framework offers significant potential to address this limitation. By integrating synthetic accessibility score (SAS) with our generative model through tuning or constraining the generation space, TopMT-GAN will be able to generate molecules with enhanced synthetic accessibility.
In conclusion, we developed TopMT-GAN – a highly efficient 3D generative model for ligand design based on protein target pockets, which has demonstrated superior performance in efficiently generating diverse and potent ligands with precise 3D poses within protein pockets. With continued improvement and development, this model offers a promising tool for the efficient identification of viable therapeutic candidates, holding the promise of revolutionizing the process of drug discovery by significantly shortening the path from concept to clinic.
Methods
Dataset
The two GAN modules in TopMT-GAN utilize distinct training datasets tailored to their specific tasks. For training the graph translation GAN, we utilized a subset of the PubChem44 dataset, initially comprising 10 million molecules. After filtering to exclude molecules larger than 50 heavy atoms, the dataset was reduced down to approximately 7 million molecules, providing a diverse range for training. In contrast, the molecular type assignment module focuses on assigning valid atom and bond types, requiring only information about local environments, and connected functional groups rather than entire molecular topologies and structures. Therefore, a smaller, specialized dataset of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Enamine fragments45 was employed. These fragments provide the advantage of a stronger focus on readily synthesizable substructures relevant to drug development.
000 Enamine fragments45 was employed. These fragments provide the advantage of a stronger focus on readily synthesizable substructures relevant to drug development.
Topology generation as graph translation
We define the binding pocket shape using two complementary strategies: a scaffold-hopping mode that leverages known ligand structures and a probe-based pocket-mapping mode developed specifically for cases where reference ligands are unavailable. In the pocket-mapping mode, probes were selected from Enamine's curated mini-fragment library, which comprises 80 small fragments with heavy atom count ranging from 5 to 7. To ensure comprehensive pocket coverage, each fragment was docked into the pocket, retaining the top 10 docking poses. These poses were manually inspected to remove outliers positioned outside the primary binding region. The combined spatial volume occupied by the validated probe poses defines the binding pocket shape. We opted to use physical small molecule fragments as probes because accurately defining the interface volume is critical for our model's performance. Overestimating the pocket volume could lead to generated molecules with steric clashes, while underestimating it might result in missed opportunities for favorable interactions or overly constrained designs.
Spheres with a radius of 1.6 Å, resembling the van der Waals radius of carbon, are compactly arranged in a face-centered cubic (FCC) lattice to fill the pre-defined shape space. A detailed evaluation of this approach can be found in the ESI.† This explicit shape depiction constrains the generative space to relevant pockets. To account for the discrete nature of the sphere stacking and enhance the diversity in molecular generation, random orientations and minor perturbations are introduced during the stacking process. Once the spheres are stacked, a fully connected graph is constructed by connecting all the first and second nearest neighbors among the spheres. This graph effectively captures the spatial relationships within the chosen pocket.
For the topology generation GAN, we trained the model for 5 epochs, corresponding to approximately 360![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 training steps, with a batch size of 96. Both the generator and discriminator networks were optimized using the Adam optimizer with a learning rate of 0.0002. To ensure stable GAN dynamics, we employed a dynamic training strategy, selectively pausing discriminator updates when its performance became overly dominant. The model was trained using only adversarial loss; however, we monitored multiple metrics during training to identify the optimal model checkpoint. These metrics included generator loss, discriminator loss, and statistical properties of generated graphs compared to real molecular graphs, such as edge number distributions. At each checkpoint, we generated sample graphs for visual inspection to ensure the quality of the outputs. Based on these metrics and manual evaluation of the generated samples, we empirically selected the model checkpoint that produced the most suitable molecular graph topologies.
000 training steps, with a batch size of 96. Both the generator and discriminator networks were optimized using the Adam optimizer with a learning rate of 0.0002. To ensure stable GAN dynamics, we employed a dynamic training strategy, selectively pausing discriminator updates when its performance became overly dominant. The model was trained using only adversarial loss; however, we monitored multiple metrics during training to identify the optimal model checkpoint. These metrics included generator loss, discriminator loss, and statistical properties of generated graphs compared to real molecular graphs, such as edge number distributions. At each checkpoint, we generated sample graphs for visual inspection to ensure the quality of the outputs. Based on these metrics and manual evaluation of the generated samples, we empirically selected the model checkpoint that produced the most suitable molecular graph topologies.
Atom & bond assignment
For the type assignment GAN, we extended the training duration to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 epochs with a batch size of 128, while maintaining the same optimizer configuration and learning rate. To improve training stability, we employed Wasserstein GAN (wGAN) loss instead of the standard adversarial loss. Throughout training, generator and discriminator losses were continuously monitored, and generated type assignments were regularly inspected to ensure diversity and prevent mode collapse. Since mode collapse cannot be effectively assessed from loss values alone, we carefully examined the diversity of generated type assignments and then empirically selected the model checkpoint that produced sufficiently diverse molecular type assignments for inference.
000 epochs with a batch size of 128, while maintaining the same optimizer configuration and learning rate. To improve training stability, we employed Wasserstein GAN (wGAN) loss instead of the standard adversarial loss. Throughout training, generator and discriminator losses were continuously monitored, and generated type assignments were regularly inspected to ensure diversity and prevent mode collapse. Since mode collapse cannot be effectively assessed from loss values alone, we carefully examined the diversity of generated type assignments and then empirically selected the model checkpoint that produced sufficiently diverse molecular type assignments for inference.
For the training data, we applied a straightforward filtering criterion: molecules containing more than 50 heavy atoms were excluded to focus on drug-like molecules and maintain computational efficiency. Since GANs learn to generate data by matching distributions rather than memorizing specific examples, we did not use a traditional training/validation split. Instead, the evaluation focused on the quality and diversity of generated molecules, which serve as the primary metrics for assessing model performance.
The graphs generated by this GAN may encounter validity issues. However, most of these can be effectively resolved with minimal adjustments in bond types. Instances where validity issue cannot be resolved are simply discarded.
Sampling
The sampling process within TopMT-GAN consists of two distinct steps: topologies sampling and assignment sampling. By introducing randomness at each stage, we can generate molecules with a variety of scaffolds. In the topology sampling phase for a given molecular or pocket shape, we vary orientations and positions of the stacked spheres such that random noise is introduced to these stacked, fully connected graphs, enabling the generation of multiple graph variations from the same input. Additionally, the diversity also arises from the A-star search by introducing minor local structural perturbations. During the molecular assignment stage, a single topology can lead to hundreds of possible atom and bond combinations, further enhancing the diversity of generated molecules. Both the topology generation and molecular assignment steps are parallelizable, greatly enhancing the efficiency of the process. Despite this, the scoring phase remains most time-intensive, even without the need for conformational search. The detailed breakdown and analysis of the sampling times are discussed in the Results section.Evaluation metrics
To evaluate our structure-based generative model, the primary metric utilized is the binding affinity to the target pocket, as determined by the Vina score. To compare the effectiveness of our model with traditional high throughput screening, we report the enrichment factor at different threshold level t, the enrichment factor is defined as: | (1) |
In addition, our primary focus is on generating a wide variety of diverse molecules on a large scale. To assess this diversity, we applied four complementary metrics: internal diversity,51 average similarity to the known actives, maximum similarity distribution to the known actives, and the count of unique scaffolds.
While properties like the quantitative estimate of drug-likeness (QED)52 and synthetic accessibility score (SAS)53 are important, we placed less emphasis on them in this initial implementation.
Internal diversity is defined:
 | (2) |
While molecular similarity can have different definitions, in this study, we specifically employed the Tanimoto similarity of Morgan fingerprints54 with a radius of 2. The radius of 2 refers to the consideration of the molecule's environment up to two bonds away from each atom. This also applies to the calculation of similarity to the actives. The average similarity to the actives is quantitatively defined as the average Tanimoto similarity between the generated molecules and known active compounds:
 | (3) |
Using the same similarity definition, we calculated the maximum similarity between each generated molecule and the known actives and plotted their distributions. Beyond average similarity, these histograms facilitate a detailed comparison of similarity between active compounds and our generated molecules.
There are many ways to define the scaffold of a molecule, here, we use the definition from Bemis and Murcko scaffolds55 and remove the atom types. We count the number of unique scaffolds for each generated set of molecules as an indication of the diversity of generated molecules.
Data availability
The code for TopMT-GAN can be found at https://github.com/aeghnnsw/TopMT.git. This study was carried out using publicly available data from PubChem at https://pubchem.ncbi.nlm.nih.gov and Enamine at https://enamine.net.Author contributions
S. W. and X. C. designed research; S. W., T. L., T. P., E. X. and S. C. performed research and analyzed data. L. B. K. contributed to discussions and supervised T. L. and T. P. on machine learning methods. S. W. and X. C. led the writing of the manuscript with contributions from all co-authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Research in the Cheng lab was supported by The Ohio State University (OSU)’s Translational Data Analytics Institute (TDAI) Interdisciplinary Research Pilot Award and College of Pharmacy Start-up fund.References
- P. G. Polishchuk, T. I. Madzhidov and A. Varnek, Estimation of the size of drug-like chemical space based on GDB-17 data, J. Comput.-Aided Mol. Des., 2013, 27, 675–679 CrossRef CAS PubMed.
- L. G. Ferreira, R. N. Dos Santos, G. Oliva and A. D. Andricopulo, Molecular docking and structure-based drug design strategies, Molecules, 2015, 20(7), 13384–13421 CrossRef CAS.
- B. K. Shoichet, Virtual screening of chemical libraries, Nature, 2004, 432(7019), 862–865 CrossRef CAS.
- M. Thomas, A. Bender and C. de Graaf, Integrating structure-based approaches in generative molecular design, Curr. Opin. Struct. Biol., 2023, 79, 102559 CrossRef CAS PubMed.
- W. Xie, F. Wang, Y. Li, L. Lai and J. Pei, Advances and challenges in de novo drug design using three-dimensional deep generative models, J. Chem. Inf. Model., 2022, 62(10), 2269–2279 CrossRef CAS PubMed.
- M. Skalic, D. Sabbadin, B. Sattarov, S. Sciabola and G. De Fabritiis, From target to drug: generative modeling for the multimodal structure-based ligand design, Mol. Pharm., 2019, 16(10), 4282–4291 CrossRef CAS.
- S. Luo, J. Guan, J. Ma and J. Peng, A 3D generative model for structure-based drug design, Adv. Neural Inf. Process. Syst., 2021, 34, 6229–6239 Search PubMed.
- X. Peng, S. Luo, J. Guan, Q. Xie, J. Peng and J. Ma, Pocket2mol: efficient molecular sampling based on 3d protein pockets, in International Conference on Machine Learning, PMLR, 2022, pp. 17644–17655 Search PubMed.
- M. Liu, Y. Luo, K. Uchino, K. Maruhashi and S. Ji, Generating 3d molecules for target protein binding, arXiv, 2022, preprint, arXiv:2204.09410, DOI:10.48550/arXiv.2204.09410.
- O. Zhang, T. Wang, G. Weng, D. Jiang, N. Wang, X. Wang and T. Hou, Learning on topological surface and geometric structure for 3D molecular generation, Nat. Comput. Sci., 2023, 3(10), 849–859 CrossRef.
- O. Zhang, J. Zhang, J. Jin, X. Zhang, R. Hu, C. Shen and T. Hou, ResGen is a pocket-aware 3D molecular generation model based on parallel multiscale modelling, Nat. Mach. Intell., 2023, 5(9), 1020–1030 CrossRef.
- Y. Jiang, G. Zhang, J. You, H. Zhang, R. Yao, H. Xie and S. Yang, PocketFlow is a data-and-knowledge-driven structure-based molecular generative model, Nat. Mach. Intell., 2024, 1–12 Search PubMed.
- Y. Li, J. Pei and L. Lai, Structure-based de novo drug design using 3D deep generative models, Chem. Sci., 2021, 12(41), 13664–13675 RSC.
- D. R. Koes, M. P. Baumgartner and C. J. Camacho, Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise, J. Chem. Inf. Model., 2013, 53(8), 1893–1904 CrossRef CAS PubMed.
- J. Guan, W. W. Qian, X. Peng, Y. Su, J. Peng and J. Ma, 3d equivariant diffusion for target-aware molecule generation and affinity prediction, arXiv, 2023, preprint, arXiv:2303.03543, DOI:10.48550/arXiv.2303.03543.
- Z. Chen, B. Peng, S. Parthasarathy and X. Ning, Shape-conditioned 3D Molecule Generation via Equivariant Diffusion Models, arXiv, 2023, preprint, arXiv:2308.11890, DOI:10.48550/arXiv.2308.11890.
- L. Huang, T. Xu, Y. Yu, P. Zhao, X. Chen, J. Han and H. Zhang, A dual diffusion model enables 3D molecule generation and lead optimization based on target pockets, Nat. Commun., 2024, 15(1), 2657 CrossRef CAS.
- W. Feng, L. Wang, Z. Lin, Y. Zhu, H. Wang, J. Dong and W. Zhou, Generation of 3D molecules in pockets via a language model, Nat. Mach. Intell., 2024, 1–12 Search PubMed.
- L. Wang, R. Bai, X. Shi, W. Zhang, Y. Cui, X. Wang and B. Huang, A pocket-based 3D molecule generative model fueled by experimental electron density, Sci. Rep., 2022, 12(1), 15100 CrossRef CAS.
- S. Long, Y. Zhou, X. Dai and H. Zhou, Zero-shot 3d drug design by sketching and generating, Adv. Neural Inf. Process. Syst., 2022, 35, 23894–23907 Search PubMed.
- W. Shi, M. Singha, L. Pu and M. Brylinski, Pocket2Drug: an encoder-decoder deep neural network for the target-based drug design, Front. Pharmacol., 2022, 13, 837715 CrossRef CAS.
- B. Ma, K. Terayama, S. Matsumoto, Y. Isaka, Y. Sasakura, H. Iwata and Y. Okuno, Structure-based de novo molecular generator combined with artificial intelligence and docking simulations, J. Chem. Inf. Model., 2021, 61(7), 3304–3313 CrossRef CAS.
- A. A. Sadybekov, A. V. Sadybekov, Y. Liu, C. Iliopoulos-Tsoutsouvas, X. P. Huang, J. Pickett and V. Katritch, Synthon-based ligand discovery in virtual libraries of over 11 billion compounds, Nature, 2022, 601(7893), 452–459 CrossRef CAS.
- A. Luttens, H. Gullberg, E. Abdurakhmanov, D. D. Vo, D. Akaberi, V. O. Talibov and J. Carlsson, Ultralarge virtual screening identifies SARS-CoV-2 main protease inhibitors with broad-spectrum activity against coronaviruses, J. Am. Chem. Soc., 2022, 144(7), 2905–2920 CrossRef CAS PubMed.
- M. M. Mysinger, M. Carchia, J. J. Irwin and B. K. Shoichet, Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking, J. Med. Chem., 2012, 55(14), 6582–6594 CrossRef CAS.
- P. G. Francoeur, T. Masuda, J. Sunseri, A. Jia, R. B. Iovanisci, I. Snyder and D. R. Koes, Three-dimensional convolutional neural networks and a cross-docked data set for structure-based drug design, J. Chem. Inf. Model., 2020, 60(9), 4200–4215 CrossRef CAS.
- A. Fischer, M. Smiesko, M. Sellner and M. A. Lill, Decision making in structure-based drug discovery: visual inspection of docking results, J. Med. Chem., 2021, 64(5), 2489–2500 CrossRef CAS.
- I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair and Y. Bengio, Generative adversarial networks, Commun. ACM, 2020, 63(11), 139–144 CrossRef.
- J. Eberhardt, D. Santos-Martins, A. F. Tillack and S. Forli, AutoDock Vina 1.2. 0: new docking methods, expanded force field, and python bindings, J. Chem. Inf. Model., 2021, 61(8), 3891–3898 CrossRef CAS.
- O. Trott and A. J. Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31(2), 455–461 CrossRef CAS PubMed.
- M. K. Gilson, T. Liu, M. Baitaluk, G. Nicola, L. Hwang and J. Chong, BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology, Nucleic Acids Res., 2016, 44(D1), D1045–D1053 CrossRef CAS PubMed.
- J. Qiao, Y. S. Li, R. Zeng, F. L. Liu, R. H. Luo, C. Huang and S. Yang, SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model, Science, 2021, 371(6536), 1374–1378 CrossRef CAS.
- P. M. Matias, P. Donner, R. Coelho, M. Thomaz, C. Peixoto, S. Macedo and M. A. Carrondo, Structural evidence for ligand specificity in the binding domain of the human androgen receptor: implications for pathogenic gene mutations, J. Biol. Chem., 2000, 275(34), 26164–26171 CrossRef CAS.
- M. Guo, Y. Duan, S. Dai, J. Li, X. Chen, L. Qu and Y. Chen, Structural study of ponatinib in inhibiting SRC kinase, Biochem. Biophys. Res. Commun., 2022, 598, 15–19 CrossRef CAS PubMed.
- D. Vanderpool, T. O. Johnson, C. Ping, S. Bergqvist, G. Alton, S. Phonephaly and J. Ermolieff, Characterization of the CHK1 allosteric inhibitor binding site, Biochemistry, 2009, 48(41), 9823–9830 CrossRef CAS.
- G. Song, D. Yang, Y. Wang, C. De Graaf, Q. Zhou, S. Jiang and R. C. Stevens, Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators, Nature, 2017, 546(7657), 312–315 CrossRef CAS.
- TMap: a very fast visualization library for large, high-dimensional data sets, https://tmap.gdb.tools/ Search PubMed.
- D. S. Wishart, Y. D. Feunang, A. C. Guo, E. J. Lo, A. Marcu, J. R. Grant and M. Wilson, DrugBank 5.0: a major update to the DrugBank database for 2018, Nucleic Acids Res., 2018, 46(D1), D1074–D1082 CrossRef CAS.
- B. Kreymann, M. A. Ghatei, G. Williams and S. R. Bloom, Glucagon-like peptide-1 7-36: a physiological incretin in man, Lancet, 1987, 330(8571), 1300–1304 CrossRef.
- D. J. Drucker and M. A. Nauck, The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes, Lancet, 2006, 368(9548), 1696–1705 CrossRef CAS.
- W. H. Sauer and M. K. Schwarz, Molecular shape diversity of combinatorial libraries: a prerequisite for broad bioactivity, J. Chem. Inf. Comput. Sci., 2003, 43(3), 987–1003 CrossRef CAS.
- C. Harris, K. Didi, A. R. Jamasb, C. K. Joshi, S. V. Mathis, P. Lio and T. Blundell, Benchmarking Generated Poses: How Rational is Structure-based Drug Design with Generative Models?, arXiv, 2023, preprint, arXiv:2308.07413, DOI:10.48550/arXiv.2308.07413.
- Enamine HTS collection, https://enamine.net/compound-collections/screening-collection Search PubMed.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He and E. E. Bolton, PubChem 2023 update, Nucleic Acids Res., 2023, 51(D1), D1373–D1380 CrossRef PubMed.
- Enamine fragment libraries, https://enamine.net/compound-libraries/fragment-libraries Search PubMed.
- X. Guo, L. Wu and L. Zhao, Deep graph translation, IEEE Transact. Neural Networks Learn. Syst., 2022, 34(11), 8225–8234 Search PubMed.
- X. Guo, L. Zhao, C. Nowzari, S. Rafatirad, H. Homayoun and S. M. P. Dinakarrao, Deep multi-attributed graph translation with node-edge co-evolution, in 2019 IEEE International Conference on Data Mining (ICDM), IEEE, 2019, pp. 250–259 Search PubMed.
- P. E. Hart, N. J. Nilsson and B. Raphael, A formal basis for the heuristic determination of minimum cost paths, IEEE Trans. Syst. Sci. Cybern., 1968, 4(2), 100–107 Search PubMed.
- RDKit: open-source cheminformatics, https://www.rdkit.org Search PubMed.
- T. A. Halgren, Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94, J. Comput. Chem., 1996, 17(5–6), 490–519 CrossRef CAS.
- M. Benhenda, ChemGAN challenge for drug discovery: can AI reproduce natural chemical diversity?, arXiv, 2017, preprint, arXiv:1708.08227, DOI:10.48550/arXiv.1708.08227.
- G. R. Bickerton, G. V. Paolini, J. Besnard, S. Muresan and A. L. Hopkins, Quantifying the chemical beauty of drugs, Nat. Chem., 2012, 4(2), 90–98 CrossRef CAS.
- P. Ertl and A. Schuffenhauer, Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions, J. Cheminf., 2009, 1, 1–11 Search PubMed.
- H. L. Morgan, The generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service, J. Chem. Doc., 1965, 5(2), 107–113 CrossRef CAS.
- G. W. Bemis and M. A. Murcko, The properties of known drugs. 1. Molecular frameworks, J. Med. Chem., 1996, 39(15), 2887–2893 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc05211k |
| This journal is © The Royal Society of Chemistry 2025 |