Open Access Article
Open Access ArticleA switchable magnetic resonance imaging nanoplatform for in situ microRNA imaging†
Yan
Tan
 a,
Junren
Wang
a,
Qian
Wan
a,
Junren
Wang
a,
Qian
Wan
 b,
Jinlong
Yang
b,
Jinlong
Yang
 a,
Jinkun
Huang
a,
Jinkun
Huang
 a,
Zijia
Zhou
a,
Haifeng
Dong
a,
Zijia
Zhou
a,
Haifeng
Dong
 *a and
Xueji
Zhang
*ac
*a and
Xueji
Zhang
*ac
aMarshall Laboratory of Biomedical Engineering, Precision Medicine and Health Research Institute, Research Center for Biosensor and Nanotheranostic, Guangdong Key Laboratory of Biomedical Measurements and Ultrasound Imaging, School of Biomedical Engineering, Shenzhen University Medical School, Shenzhen University, Shenzhen 518060, China. E-mail: hfdong@szu.edu.cn; zhangxueji@szu.edu.cn
bPaul C. Lauterbur Research Center for Biomedical Imaging, Institute of Biomedical and Health Engineering, Shenzhen Institutes of Advance Technology, Chinese Academy of Sciences, Shenzhen 518055, P. R. China
cGuangdong Laboratory of Artificial Intelligence and Digital Economy (SZ), Shenzhen, 518060, China
First published on 19th November 2024
Abstract
Aberrant microRNA (miRNA) expression is associated with various types of carcinogenesis, making miRNA a promising candidate for diagnostic and therapeutic biomarkers. However, in situ miRNA diagnostics remains a significant challenge owing to the various biological barriers. Herein, we report a novel miRNA imaging probe consisting of PEG-polylysine-PNIPAM polymer matrix-modified small Fe3O4 (PAA-Fe3O4-DNA@PPP) nanoparticles with an improved circulatory half-life, efficient tissue permeability, and enhanced tumor accumulation, for in situ miRNA magnetic resonance imaging (MRI). In this strategy, we employed large size PAA-Fe3O4-DNA@PPP to improve circulatory time and utilized PEG-polylysine-PNIPAM as a GSH-responsive moiety to dissociate PAA-Fe3O4-DNA@PPP and release small size PAA-Fe3O4-DNA for enhanced tumor permeability. Specifically, the target miRNA acts as a cross-linker for PAA-Fe3O4-DNA, forming larger assemblies that not only amplify the MRI signal for detection but also enhance retention for prolonged observation. Both the in vitro and in vivo results validate that the imaging probe exhibits an enhanced MRI signal with 3.69-fold amplification for tumor interior miRNA detection, allowing the dynamic changes in miRNA to be monitored by the probe. Given its long circulation, efficient penetration, and enhanced tumor accumulation, the PAA-Fe3O4-DNA@PPP probe holds great promise for in situ miRNA imaging and spatial genomics analysis in situ.
Introduction
In situ biomarker imaging that utilizes advanced imaging techniques such as microscopy, magnetic resonance imaging (MRI), or positron emission tomography (PET) to identify and monitor these biomarkers within their natural environment, holds great promise for personalized medicine in cancer diagnosis and advancing our understanding of the complex pathological process. However, several challenges and limitations persist in efficiently delivering imaging agents to tumor tissue to achieve optimal efficacy. As reported, nanoparticles (NPs) with a diameter of 100 nm can be stable in the circulation, but not efficiently pass through tumor vessels.1 In contrast, small-size NPs with an approximate size of 4–20 nm easily penetrate through deep tumor tissues, yet they are more inclined to clearance and lack retention at tumor sites.2 Therefore, rational designs of NPs with size-tunable performance under various biological conditions to achieve effective tumor accumulation and enhanced tumor permeability have been continuously developed.3–5As a significant small non-coding RNA molecular, microRNA (miRNA) is the post-transcriptional regulator in biological and pathological effects, especially serving as a crucial biomarker for tumor classification.6–8 In our previous studies, selective and sensitive detection of miRNA in living cells based on nanoparticle systems has been developed.9–11In situ miRNA detection is vital for biological function and mechanism research and monitoring of cancerous pathological conditions; however, it is still a huge challenge because of their low-abundant expression level and complex biological environments. Photoacoustic imaging (PAI) of miRNA at the tumor site was realized via miRNA-triggered self-assembly of DNA-coated nanostructures to form particle aggregation with enhanced PAI signals.11 A cancer cell membrane camouflaged nanoprobe with homotypic-targeting capability and remarkable amplification was also designed for PAI of miRNA in living mice.12 Smartly designed hierarchical nanoparticles with size-tunable properties in response to physiological conditions to satisfy long circulation, efficacy permeability and tumor site-specific miRNA activatable signal enhancement have never been reported.
Magnetic resonance imaging (MRI) has been proven as a mighty appliance for clinical diagnosis because of its resultful tissue contrast, high spatial resolution (about 50 μm), low ionizing radiation, and unstinted signal penetration depth.13–16 Thereinto, ultrasmall superparamagnetic iron oxide nanoparticles (IONPs) with appreciable biodistribution and metabolism have been widely used as T1-weighted MRI contrast agents.17,18 Interestingly, previous studies reported that the assemblies of IONP contrast agents dramatically affected the magnetic properties due to the aggregation-induced enhancement of the magnetic field resulting in T2 contrast imaging promotion.19–22 Such properties provided great opportunities to establish conditional activatable contrast agents that could further amplify the imaging contrast by abating the interference generated from nonspecific adsorption or endogenous iron-derived imaging contrast.23 A series of transformations of IONPs exhibited an excellent capacity for penetration and aggregation in the physiological environment and have been developed to evaluate deep tumor imaging in clinical applications.24 For example, “turn-on” MRI nanoprobes were fabricated that showed dark T2 contrast imaging in the vessels and transformed into bright T1 contrast imaging owing to GSH or pH-triggered cleavage in the tumor microenvironment (TME).25–28 Smart switchable MRI probes based on the self-assembly of IONPs were fabricated as a transformation from T2-weighted into T1-weighted contrast imaging at the tumor site under low pH-induced disassembly.29 However, multistep switchable MRI probes for in situ miRNA imaging have scarcely been explored.
Therefore, for the first time, we designed a “large-small-large” size-switchable PAA-Fe3O4-DNA@PPP nanoplatform, consisting of PEG-polylysine-PNIPAM polymer matrix-modified small Fe3O4, for in situ miRNA imaging. As shown in Scheme 1, PAA-Fe3O4-DNA@PPP maintains a large size for prolonged circulation time; it decomposes to a small size (4–5 nm) PAA-Fe3O4-DNA exposing to TME GSH conditions for deep penetration. After the internalization of PAA-Fe3O4-DNA, the small-size nanoparticles would be linked by the intracellular miRNA to self-assemble to form stable larger-size nanoparticles (more than 50 nm) that consequently enhance the T2-weighted MRI in the tumor for miRNA in situ detection. Meanwhile, fluorescent dye Cy5 was conjugated on the DNA strand to track the nanosystem accurately. The size switchable imaging system is anticipated to be applied to plenty of in situ biomolecular detections by rational detection probe design.
Results and discussion
“Small-to-large” PAA-Fe3O4-DNA@PPP assembly
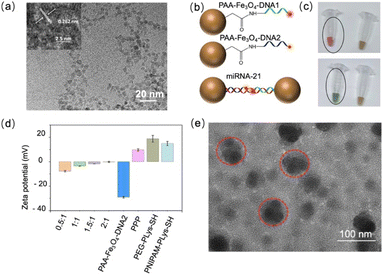
The construction procedure of PAA-Fe3O4-DNA@PPP is presented in Scheme 1. First, polyacrylic acid-modified Fe3O4 (PAA-Fe3O4) nanoparticles were synthesized according to the previous method with the hydrothermal decomposition in the existence of polyacrylic acid and ferric chloride raw materials.30 The average diameter of PAA-Fe3O4, as measured by transmission electron microscopy (TEM) is about 4.43 ± 0.6407 nm (Fig. 1a and S1†). The high solution TEM measurement of PAA-Fe3O4 shows that the lattice fringe spacing value is 0.262 nm, which is assigned to the (311) planes of Fe3O4 (inset of Fig. 1a). Fourier transform infrared spectrometry (FTIR) was chosen to confirm the carboxyl group of PAA conjugated on Fe3O4. The peak at 1639 cm−1 is due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond in the PAA.31 On the other hand, the vibration absorption peaks (3300 cm−1) for the O–H bond demonstrated that PAA-Fe3O4 was successfully synthesized (Fig. S2†). Additionally, the peak at around 580 cm−1 represented the vibration of FeO groups.32 Two fluorescent dye-labeled single-strand DNA sequences complementary to target miRNA were conjugated on PAA-Fe3O4via the amino-carboxyl group crosslinking reaction to form PAA-Fe3O4-DNA1 and PAA-Fe3O4-DNA2 (Fig. 1b to c). After the conjugation, the extra fluorescent DNA was removed with the help of a magnet (Fig. S3†). The successful modification of DNA molecules on PAA-Fe3O4 was determined via UV spectra at 260 nm (Fig. S4†). The amount of DNA1 and DNA2 on PAA-Fe3O4 was estimated to be 13.74 μmol g−1 Fe and 11.88 μmol g−1 Fe, respectively, according to the standard curve of fluorescence and concentration (Fig. S5†). These results suggested that the DNA was successfully conjugated on PAA-Fe3O4. To protect the PAA-Fe3O4-DNA nanoparticles, multiblock cationic polymer PEG-polylysine-PNIPAM (PPP) was synthesized subsequently. Polyethyleneglycol (PEG) modified polycationic polylysine (Plys) (PEG-PLys-SH) (Fig. S6†) and poly(N-isopropyl acrylamide) (PNIPAM) grafted Plys (PNIPAM-Plys-SH) (Fig. S7†) were prepared according to the previously reported method.33 The resulting polymers were characterized by 1H NMR, and the number of -SH was calculated to be 1.25 and 7 for PEG-PLys-SH (Fig. S8†) and PNIPAM-Plys-SH (Fig. S9†).
O bond in the PAA.31 On the other hand, the vibration absorption peaks (3300 cm−1) for the O–H bond demonstrated that PAA-Fe3O4 was successfully synthesized (Fig. S2†). Additionally, the peak at around 580 cm−1 represented the vibration of FeO groups.32 Two fluorescent dye-labeled single-strand DNA sequences complementary to target miRNA were conjugated on PAA-Fe3O4via the amino-carboxyl group crosslinking reaction to form PAA-Fe3O4-DNA1 and PAA-Fe3O4-DNA2 (Fig. 1b to c). After the conjugation, the extra fluorescent DNA was removed with the help of a magnet (Fig. S3†). The successful modification of DNA molecules on PAA-Fe3O4 was determined via UV spectra at 260 nm (Fig. S4†). The amount of DNA1 and DNA2 on PAA-Fe3O4 was estimated to be 13.74 μmol g−1 Fe and 11.88 μmol g−1 Fe, respectively, according to the standard curve of fluorescence and concentration (Fig. S5†). These results suggested that the DNA was successfully conjugated on PAA-Fe3O4. To protect the PAA-Fe3O4-DNA nanoparticles, multiblock cationic polymer PEG-polylysine-PNIPAM (PPP) was synthesized subsequently. Polyethyleneglycol (PEG) modified polycationic polylysine (Plys) (PEG-PLys-SH) (Fig. S6†) and poly(N-isopropyl acrylamide) (PNIPAM) grafted Plys (PNIPAM-Plys-SH) (Fig. S7†) were prepared according to the previously reported method.33 The resulting polymers were characterized by 1H NMR, and the number of -SH was calculated to be 1.25 and 7 for PEG-PLys-SH (Fig. S8†) and PNIPAM-Plys-SH (Fig. S9†).
The assembly of PAA-Fe3O4-DNA@PPP was optimized with different N/P ratios derived from PPP and PAA-Fe3O4-DNA. The zeta potential analysis confirmed that the PPP polymer was positive, and the negative charge of PAA-Fe3O4-DNA was neutralized as the N/P ratios increased (Fig. 1d). Agarose gel electrophoresis showed that the PAA-Fe3O4-DNA@PPP samples were gradually trapped in the beginning line of the gel as the N/P ratios increased, and no free PAA-Fe3O4-DNA was visualized at a ratio of 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. S10†). These results indicated that the negatively charged DNA has effectively condensed into the PPP polymer. As shown in Fig. 1e, the average diameter of PAA-Fe3O4-DNA@PPP is 82 ± 20 nm while the N/P ratio is 1.5. As reported, the PPP polymer would have a relatively smaller size when at a physiological temperature of 37 °C because of thermally responsive PNIPAM.34 When the temperature increases, PNIPAM could transform into the hydrophobic phase that provides an additional drive force to stimulate further condensation of PAA-Fe3O4-DNA@PPP and protect PAA-Fe3O4-DNA from enzymatic degradation in the physical environment. The high-resolution TEM image and the element mapping analysis indicated that PAA-Fe3O4-DNA was encapsulated into the PPP polymer structure (Fig. S11 & S12†).
1 (Fig. S10†). These results indicated that the negatively charged DNA has effectively condensed into the PPP polymer. As shown in Fig. 1e, the average diameter of PAA-Fe3O4-DNA@PPP is 82 ± 20 nm while the N/P ratio is 1.5. As reported, the PPP polymer would have a relatively smaller size when at a physiological temperature of 37 °C because of thermally responsive PNIPAM.34 When the temperature increases, PNIPAM could transform into the hydrophobic phase that provides an additional drive force to stimulate further condensation of PAA-Fe3O4-DNA@PPP and protect PAA-Fe3O4-DNA from enzymatic degradation in the physical environment. The high-resolution TEM image and the element mapping analysis indicated that PAA-Fe3O4-DNA was encapsulated into the PPP polymer structure (Fig. S11 & S12†).
“Large-to-small” and “small-to-large”transformation
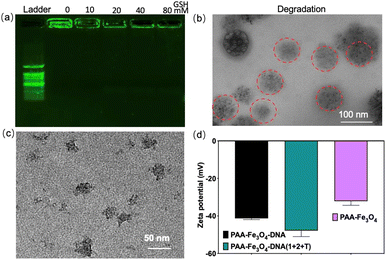
PPP was obtained by disulfide cross-linking formation of PEG-PLys-SH and PNIPAM-Plys-SH, and the resulting PPP could pack the small DNA-functionalized nanoparticles through electrostatic assemblage between cationic Plys-SH and anionic nucleic acid of PAA-Fe3O4-DNA to form PAA-Fe3O4-DNA@PPP. The disulfide cross-linking was readily cleaved under acidic and reductive conditions with glutathione (GSH). The existence of an ester bond and amide bond in PPP would accelerate the hydrolysis of PAA-Fe3O4-DNA@PPP nanocapsule in the acidic TME. The degradation of PAA-Fe3O4-DNA@PPP was evaluated by exposing it to GSH with different concentrations of 0 mM, 10 mM, 20 mM, 40 mM, and 80 mM (Fig. 2a). The gel electrophoresis results confirmed that PAA-Fe3O4-DNA@PPP gradually degraded with the increase in the GSH concentration. In addition, the TEM image showed the disassembly morphology of PAA-Fe3O4-DNA@PPP which was marked with red circles (Fig. 2b). These results confirmed the efficient degradation of PAA-Fe3O4-DNA@PPP to small PAA-Fe3O4-DNA, which was useful for enhanced tumor depth penetration. Using miRNA-21, a tumor-related up-regulated miRNA, as a model target miRNA, we investigated the feasibility of target miRNA-mediated PAA-Fe3O4-DNA assembly, leading to a conversion process from small to large size. As anticipated, the PAA-Fe3O4-DNA (1 + 2 + T) aggregates exhibited an average size of 50 nm when the concentration ratio of PAA-Fe3O4-DNA1, PAA-Fe3O4-DNA2, and miRNA-21 was maintained at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 2c). The resulting aggregates displayed more negative zeta potential (Fig. 2d). As the ratio increases from 1
1 (Fig. 2c). The resulting aggregates displayed more negative zeta potential (Fig. 2d). As the ratio increases from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, the size increases from about 50 nm to 80 nm (Fig. S13†). These results verified that PAA-Fe3O4-DNA1 and PAA-Fe3O4-DNA2 could be assembled through the bridge miRNA-21, implementing small-size to larger-size transformation.
10, the size increases from about 50 nm to 80 nm (Fig. S13†). These results verified that PAA-Fe3O4-DNA1 and PAA-Fe3O4-DNA2 could be assembled through the bridge miRNA-21, implementing small-size to larger-size transformation.
Penetration evaluation and in vitro magnetic resonance imaging
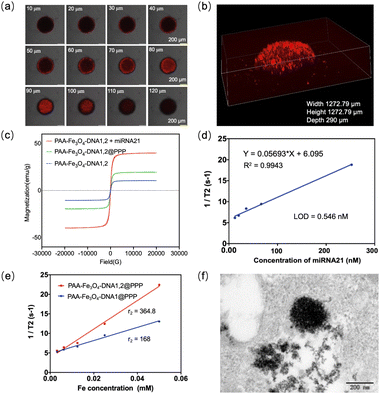
The penetration of PAA-Fe3O4-DNA@PPP was investigated by confocal imaging. MCF-7 multicellular tumor spheroids were formed in the agarose-based microwells and reached a diameter of more than 400 μm after culturing for several days according to previous methods.35,36 PAA-Fe3O4-DNA@PPP labeled with Cy5 in the DNA has efficiently penetrated the MCF-7 multicellular tumor spheroids characterized by confocal imaging through a layer scan model within 10 μm intervals (Fig. 3a). According to the confocal imaging, cross-sectional fluorescence can be observed when the scanning depth of the Z-axis is 100 μm. The three-dimensional fluorescence reconfiguration of multicellular tumor spheroids exposed to PAA-Fe3O4-DNA@PPP confirmed the deep penetration (Fig. 3b).This reported that ultrasmall Fe3O4 nanoparticles with a diameter of less than 5 nm have good T1-MRI contrast function. In contrast, the self-assembled ultrasmall Fe3O4 would enhance the magnetic size inducing a higher saturation magnetization, leading to an improved dark T2 MRI contrast.37–39 As seen in , the value of saturated magnetic intensity for PAA-Fe3O4-DNA1 increased from 10.5 to 39.8 meu g−1 when assembled with miRNA-21. Interestingly, the magnetic saturation value of PAA-Fe3O4-DNA@PPP was 19.5 meu g−1, indicating that PAA-Fe3O4-DNA1 and PAA-Fe3O4-DNA2 encapsulated into PPP. Furthermore, we measured the T1 and T2 relaxation times of PAA-Fe3O4-DNA1,2 with or without miRNA-21. The r1 and r2 relaxivity of PAA-Fe3O4-DNA1,2 in the presence of miRNA-21 was calculated to be 0.28 mM−1 s−1, 250.1 mM−1 s−1, while r1 and r2 relaxivity of the PAA-Fe3O4-DNA1,2 mixture without miRNA-21 was 0.83 mM−1 s−1 and 101.0 mM−1 s−1(Fig. S14 & S15†). These results suggested that the assembly PAA-Fe3O4-DNA1,2 triggered by miRNA-21 showed enhanced T2 suitable for T2 MRI contrast imaging. As the miRNA-21 concentration increased from 0 to 250 nM, the r2 increased from 6 to 20.4 s−1 (Fig. 3d), and the limit of detection was calculated to be 0.546 nM using T2-MRI. We further investigated its performance in living cells. PAA-Fe3O4-DNA1,2@PPP was incubated with the MCF-7 cells for 8 hours, and PAA-Fe3O4-DNA1@PPP was a negative control. Then the cells were all harvested and washed with PBS. After that, the cells were fixed using agarose and were detected under a 3.0 T MR (Fig. S16†). Fig. 3e shows a higher r2 relaxivity in the PAA-Fe3O4-DNA1,2@PPP treated cells compared with the PAA-Fe3O4-DNA1@PPP incubated cell group. Meanwhile, the r1 relaxivity was also measured (Fig. S17†), and the r2/r1 value was respectively calculated to be 46.0 and 112.7 for the PAA-Fe3O4-DNA1@PPP and PAA-Fe3O4-DNA1,2@PPP treated groups. As shown in Fig. 3f, the large aggregation structures were visualized in the bioTEM image of the PAA-Fe3O4-DNA1,2@PPP treated MCF-7 cells, suggesting that target miRNA-21 induced aggregation. The results illustrated that miRNA-21 in the cells would contribute to the aggregation of PAA-Fe3O4-DNA.
In vivo magnetic resonance imaging
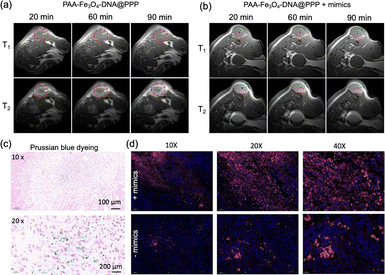
To investigate the in vivo MRI of PAA-Fe3O4-DNA@PPP. PAA-Fe3O4-DNA@PPP was tail intravenously injected into the MCF-7-bearing mice. The signal was observed after 20 min of injection (Fig. 4a). As indicated in the T2 weighted MRI, the assembled PAA-Fe3O4-DNA@PPP derived T2 signal reached the maximum at 90 minutes post-injection. Notably, the ratio of the T2/T1 signal was less than 1.0 at 20 min after injection (Fig. S18†), and the signal increased as time increased, verifying that PAA-Fe3O4-DNA@PPP was a good T2-weighted contrast reagent. Furthermore, to investigate the monitoring ability of miRNA change by PAA-Fe3O4-DNA@PPP, miRNA-21 mimics were injected intratumorally for 4 h before the imaging experiment. The mimic DNA sequence has the same function as miRNA-21. The T1 and T2 MRI signals were evaluated after being injected for 20 min, 40 min, 60 min, and 90 min. As shown in the T2 MRI images displayed in Fig. 4b, the hole in the green circle was the injection site. Many dark dots derived from the miRNA-21-induced PAA-Fe3O4-DNA assembly were observed (in red circles).The tumors were collected for sectioning and staining to verify whether PAA-Fe3O4-DNA@PPP entered into the tissues. Prussian blue dyeing was employed to observe iron. As exhibited in Fig. 4c, the blue granules were in the tumor cells, indicating that PAA-Fe3O4-DNA@PPP could reach the tumor site. Because PAA-Fe3O4-DNA@PPP was labeled with Cy5, the slices were also observed under a fluorescence microscope (Fig. 4d). The mimic miRNA-21-treated tissues displayed strong Cy5 fluorescence compared with that without treatment, which proved that PAA-Fe3O4-DNA was trapped in the tumor tissues with the assistance of the miRNA-induced assembly.
Conclusions
In this work, we report the construction of a switchable miRNA MRI imaging probe, PAA-Fe3O4-DNA@PPP, for in situ miRNA imaging. PAA-Fe3O4-DNA@PPP undergoes a “large-small-large” switchable process to cross various biological barriers, accumulate in tumor tissue, and achieve effective in situ miRNA MRI imaging. It maintains a large size for prolonged circulation time and then decomposes into small-sized (4–5 nm) PAA-Fe3O4-DNA in response to the TME GSH conditions, allowing for deep penetration. After cellular internalization, the small-sized nanoparticles are linked by intracellular miRNA, self-assembling into stable larger-sized nanoparticles (over 50 nm). This assembly enhances T2-weighted MRI signals in the tumor, enabling in situ miRNA detection and allowing in situ monitoring of the dynamic changes in miRNA. The long circulation time, efficient penetration, and enhanced tumor accumulation make the PAA-Fe3O4-DNA@PPP probe highly promising for in situ miRNA imaging and spatial genomics analysis.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization: YT, HFD, and XJZ; experiments and formal analysis: YT, JRW, and QW; methodology: JLY, JKH, and ZJZ; project administration: HFD and XJZ; writing the original draft: YT; writing – review & editing: YT, JRW and HFD. All authors have read and agreed to the final version of this manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was supported by the Special Foundation for State Major Research Program of China (grant 2022YFB3207202), the Shenzhen Medical Research Fund (grant 2301011), the Guangdong Province Pearl River Team (2021ZT09C289), and Shenzhen Key Laboratory for Nano-Biosensing Technology (ZDSYS20210112161400001). We thank the Instrumental Analysis Center of Shenzhen University for the assistance with the electron microscope, nuclear magnetic resonance spectroscope, mass spectrometer, confocal microscope, small animal diagnostics imaging, and material characterization technical support.References
- M. Kyu Shim, S. Yang, I.-C. Sun and K. Kim, Adv. Drug Delivery Rev., 2022, 183, 114177 CrossRef CAS PubMed.
- Z. Niu, L. Liu, L. Zhang, Q. Shao, W. Zhou, X. Chen and S. Xie, Adv. Mater., 2014, 26, 3681–3687 CrossRef CAS.
- F. Mo, K. Jiang, D. Zhao, Y. Wang, J. Song and W. Tan, Adv. Drug Delivery Rev., 2021, 168, 79–98 CrossRef CAS.
- B. Chen, L. Liu, R. Yue, Z. Dong, C. Lu, C. Zhang, G. Guan, H. Liu, Q. Zhang and G. Song, Nano Today, 2023, 51, 101931 CrossRef CAS.
- J.-M. Shen, T. Yin, X.-Z. Tian, F.-Y. Gao and S. Xu, ACS Appl. Mater. Interfaces, 2013, 5, 7014–7024 CrossRef CAS.
- P.-Y. Lui, D.-Y. Jin and N. J. Stevenson, Cell. Mol. Life Sci., 2015, 72, 3531–3542 CrossRef CAS PubMed.
- Y. Zhang, W. Chen, Y. Zhang, X. Zhang, Y. Liu and H. Ju, Angew. Chem., Int. Ed., 2020, 59, 21454–21459 CrossRef CAS PubMed.
- C. Xue, S.-X. Zhang, C.-H. Ouyang, D. Chang, B. J. Salena, Y. Li and Z.-S. Wu, Angew. Chem., Int. Ed., 2018, 57, 9739–9743 CrossRef CAS.
- Y. Zhang, F. Yang, W. Wei, Y. Wang, S. Yang, J. Li, Y. Xing, L. Zhou, W. Dai and H. Dong, ACS Nano, 2022, 16, 5587–5596 CrossRef CAS PubMed.
- F. Yang, H. Lu, X. Meng, H. Dong and X. Zhang, Small, 2022, 18, 2106281 CrossRef CAS.
- Y. Tan, J. Zhou, X. Xing, J. Wang, J. Huang, H. Liu, J. Chen, M. Dong, Q. Xiang, H. Dong and X. Zhang, Anal. Chem., 2023, 95, 11236–11242 CrossRef CAS.
- K. Zhang, X. Meng, Z. Yang, Y. Cao, Y. Cheng, D. Wang, H. Lu, Z. Shi, H. Dong and X. Zhang, Adv. Mater., 2019, 31, 1807888 CrossRef PubMed.
- Z. Liang, Q. Wang, H. Liao, M. Zhao, J. Lee, C. Yang, F. Li and D. Ling, Nat. Commun., 2021, 12, 3840 CrossRef CAS.
- H. Li, D. Luo, C. Yuan, X. Wang, J. Wang, J. P. Basilion and T. J. Meade, J. Am. Chem. Soc., 2021, 143, 17097–17108 CrossRef CAS PubMed.
- R. Qin, S. Li, Y. Qiu, Y. Feng, Y. Liu, D. Ding, L. Xu, X. Ma, W. Sun and H. Chen, Nat. Commun., 2022, 13, 1938 CrossRef CAS PubMed.
- S. Sheng, F. Liu, M. Meng, C. Xu, H. Tian and X. Chen, CCS Chem., 2022, 4, 2321–2332 CrossRef CAS.
- Z. Zhou, L. Yang, J. Gao and X. Chen, Adv. Mater., 2019, 31, 1804567 CrossRef PubMed.
- M. Jeon, M. V. Halbert, Z. R. Stephen and M. Zhang, Adv. Mater., 2021, 33, 1906539 CrossRef CAS PubMed.
- H. Zhou, M. Guo, J. Li, F. Qin, Y. Wang, T. Liu, J. Liu, Z. F. Sabet, Y. Wang, Y. Liu, Q. Huo and C. Chen, J. Am. Chem. Soc., 2021, 143, 1846–1853 CrossRef CAS.
- D. Ling, W. Park, S.-j. Park, Y. Lu, K. S. Kim, M. J. Hackett, B. H. Kim, H. Yim, Y. S. Jeon, K. Na and T. Hyeon, J. Am. Chem. Soc., 2014, 136, 5647–5655 CrossRef CAS PubMed.
- T. A. Meyer, C. Zhang, G. Bao and Y. Ke, Nano Lett., 2020, 20, 2799–2805 CrossRef CAS PubMed.
- T. Liu, M. Zhang, W. Liu, X. Zeng, X. Song, X. Yang, X. Zhang and J. Feng, ACS Nano, 2018, 12, 3917–3927 CrossRef CAS PubMed.
- J. Hou, H. Liu, Q. Ma, S. Xu and L. Wang, Anal. Chem., 2022, 94, 15578–15585 CrossRef CAS.
- R. Yue, C. Zhang, L. Xu, Y. Wang, G. Guan, L. Lei, X. Zhang and G. Song, Chem, 2022, 8, 1956–1981 CAS.
- Y. Cao, Z. Mao, Y. He, Y. Kuang, M. Liu, Y. Zhou, Y. Zhang and R. Pei, ACS Appl. Mater. Interfaces, 2020, 12, 26973–26981 CrossRef CAS.
- J. Lu, J. Sun, F. Li, J. Wang, J. Liu, D. Kim, C. Fan, T. Hyeon and D. Ling, J. Am. Chem. Soc., 2018, 140, 10071–10074 CrossRef CAS PubMed.
- J. Xu, G. Guan, Z. Ye, C. Zhang, Y. Guo, Y. Ma, C. Lu, L. Lei, X.-B. Zhang and G. Song, Sci. Bull., 2024, 69, 636–647 CrossRef CAS.
- M. Ma, H. Zhu, J. Ling, S. Gong, Y. Zhang, Y. Xia and Z. Tang, ACS Nano, 2020, 14, 4036–4044 CrossRef CAS PubMed.
- J. Hou, H. Liu, Q. Ma, S. Xu and L. Wang, Anal. Chem., 2022, 94, 15578–15585 CrossRef CAS PubMed.
- S.-H. Huang and D.-H. Chen, J. Hazard. Mater., 2009, 163, 174–179 CrossRef CAS.
- J. Liu, G. Liu and W. Liu, Chem. Eng. J., 2014, 257, 299–308 CrossRef CAS.
- J. L. Zhang, R. S. Srivastava and R. D. K. Misra, Langmuir, 2007, 23, 6342–6351 CrossRef CAS PubMed.
- O. Veiseh, F. M. Kievit, H. Mok, J. Ayesh, C. Clark, C. Fang, M. Leung, H. Arami, J. O. Park and M. Zhang, Biomaterials, 2011, 32, 5717–5725 CrossRef CAS PubMed.
- H. Ding, B. Li, Z. Liu, G. Liu, S. Pu, Y. Feng, D. Jia and Y. Zhou, Adv. Healthcare Mater., 2020, 9, 2000454 CrossRef CAS.
- L. Wang, J. Li, Z. Zhao, Y. Xia, Y. Xie, D. Hong, Y. Liu and W. Tan, Anal. Chem., 2024, 96, 154–162 CrossRef PubMed.
- F. Zhou, T. Fu, Q. Huang, H. Kuai, L. Mo, H. Liu, Q. Wang, Y. Peng, D. Han, Z. Zhao, X. Fang and W. Tan, J. Am. Chem. Soc., 2019, 141, 18421–18427 CrossRef CAS PubMed.
- W. Liu, S. Yin, Y. Hu, T. Deng and J. Li, Anal. Chem., 2021, 93, 14223–14230 CAS.
- W. Liu, S.-Y. Yin, Y. Hu, T. Deng and J. Li, ACS Appl. Mater. Interfaces, 2022, 14, 2629–2637 CrossRef CAS PubMed.
- C. Bai, P. Hu, N. Liu, G. Feng, D. Liu, Y. Chen, M. Ma, N. Gu and Y. Zhang, ACS Appl. Nano Mater., 2020, 3, 3585–3595 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc04675g |
| This journal is © The Royal Society of Chemistry 2025 |