Open Access Article
Open Access ArticleMulti-functionalized biologically active isatin-tagged dihydropyrimidine derivatives: green synthesis by the use of recyclable Fe-dopped Ce-oxide nanoparticles, computational studies and ex vivo cytotoxic activity against breast cancer cells
Keshav Kumar Saini ab,
Ravindra Kumar Upadhyayac,
Diksha Ranid,
Smaranjot Kaurd,
Ravi Kante,
Sanjay Kumar Dey*d and
Rakesh Kumar
ab,
Ravindra Kumar Upadhyayac,
Diksha Ranid,
Smaranjot Kaurd,
Ravi Kante,
Sanjay Kumar Dey*d and
Rakesh Kumar *a
*a
aBioorganic Laboratory, Department of Chemistry, University of Delhi, Delhi-110007, India. E-mail: rakeshkp@email.com; Tel: +91 9810348999
bDepartment of Chemistry, Dyal Singh College, University of Delhi, India
cDepartment of Chemistry, Sri Venkateswara College, University of Delhi, New Delhi 110021, India
dLaboratory for Membrane Proteins and Structural Biology, Dr B. R. Ambedkar Center for Biomedical Research (ACBR), University of Delhi, Delhi-110007, India. E-mail: sdey@acbr.du.ac.in; Tel: +91 9205337595
eDepartment of Chemistry, Government Post Graduate College, G.B. Nagar, Noida, UP 201301, India
First published on 12th December 2025
Abstract
A new class of compounds was designed and synthesised using a molecular hybridisation technique based on the dihydropyridine, isatin, triazole, thiazole, benzothiazole, and glucose scaffolds. The alkyne derivatives of dihydropyridine were synthesised by using reusable Fe-doped Ce-oxide nanoparticles via a three-component Biginelli reaction. The substances were then tested for their cytotoxic effects on two human breast tumour cell lines, MCF-7 and MDA-MB-231, as well as non-cancerous breast epithelial cell lines (i.e., HEK293). The primary objective was to synthesise the more efficient breast cancer cell inhibitory molecules, which contain multiple scaffolds, and to investigate their potential as anti-breast cancer cell inhibitory therapeutic agents. Notably, compound-6, 7 and 14a exhibited remarkable anti-cancer activity against two prominent breast-cancer cell-lines, MCF-7 and MDA-MB-231. The IC50 values of the compounds 6, 7, and 14a against MCF-7 and MDA-MB-231breast cancer cell lines were found to be the lowest. The successful synthesis of these dihyropyrimidines using an environmentally friendly approach emphasises the significance of sustainable and eco-friendly methodologies in pharmaceutical research.
Introduction
Today, breast cancer is the most common disease and the second most prevalent cause of cancer death among women.1 Many commercially available anti-breast cancer medications, such as alpelisib, anastrozole, cyclophosphamide, docetaxel, and tamoxifen citrate, are used to treat breast cancer (Fig. 1). Drug resistance to cancer chemotherapeutic agents is a profound medical issue. Moreover, many breast cancer patients are non-responsive or non-tolerant to most of the commercially available anti-breast cancer drugs.2 As a result, there is a pressing need to design hybrid chemotherapeutic medication with multiple modes of action in a single molecule, apart from exploiting novel targets. These merged pharmacophores may address the active sites of many targets and provide a way to overcome drug resistance. The design of procedures with the lowest environmental impact is another significant element of modern organic synthetic methodology.3 The use of one-time catalysts (metallic, acidic, and basic) and organic solvents is the primary source of environmental pollution.4 The development of a new efficient and environment friendly improved process in organic synthesis is a need of the hour. In the past few years, carrying out organic reactions using new recyclable and waste-derived catalysts in green solvents has gained popularity due to environmental concerns.5,6 One-pot cascade reactions are preferred over conventional methods since they are simple, quick, efficient, highly selective, give higher yields, maintain good atom economy, involve minimum steps, and eliminate waste production.7 Biginelli reaction is a three-component reaction and it has received a lot of attention in the last few years because a variety of catalysts have been found that enable the synthesis of the resulting dihydropyrimidines (DHPMs) with exceptional yield, in contrast to the inconsistent performance observed in the early studies.8 The Biginelli reaction is a one-pot condensation reaction with three components, which produces biologically active dihyropyrimidine by using β-keto esters, aldehydes, and urea/thiourea.9 Several one-pot strategies for synthesising DHPM derivatives have been documented over the past decade, and several modifications have also been established to enhance the yield and efficiency of the reaction. These modifications involved the use of Brønsted acids, Lewis acids, nanoparticles, mixed metal or metal oxide nanoparticles, ionic liquids, visible light photocatalysts, microwave assistance, clay, organic polymers, biocatalysts, ultrasound-irradiation, zeolites, and organic-inorganic mesoporous materials, etc.10Dihydropyrimidines (DHPMs) have a variety of pharmacological properties, mainly anti-HIV,11 calcium channel blocker,12 anticonvulsant,13 anti-fungal,14 anti-tubercular,15 antibacterial,16 anti-cancer,17 antihyperglycemic,18 antihypertensive,19 analgesic,20 and antioxidant.21 They are essential for the production of both RNA and DNA and are an essential component for regulating the cell lifecycle.
In synthetic organic chemistry, isatin plays an invaluable key role. It is one of the most important building block and a physiologically active pharmacophore that has been demonstrated to possess a variety of biological properties, including anti-cancer,22 anti-fungal,23 antibacterial,24 antimicrobial,25 anti-tubercular,26 and anti-HIV27 etc.
Single-drug, multi-target is a new emerging research field that has gained a lot of attention in recent years among the research community. This has been proven to be a powerful tool against complex diseases and drug resistance.28 Molecular hybridization is an important strategy for the synthesis of single drug with enhanced biological features. In which two or more biologically active moieties are linked to enhance the efficiency of the drug. Triazoles are important linkers to link more than one scaffold,29 which itself possesses a wide range of biological activities.30 The 1,6-dihydropyrimidine, isatin, thiazole/benzothiazole, and 1,2,3-triazole rings are the basis for many clinically approved and under-trial medications with strong antifungal properties, in addition to the aforementioned claims. This makes the 1,6-dihydropyrimidine, isatin, thiazole/benzothiazole, and 1,2,3-triazole core an intriguing and little-studied pharmacophore.31,32 During transcription, topoisomerase-II alpha, a naturally occurring enzyme, regulates and modifies the topologic properties of DNA.33 Chromosome condensation, chromatid separation, and the alleviation of torsional stress during DNA replication and transcription are among the functions of this nuclear enzyme. It catalyses the temporary separation and reunification of two duplex DNA strands, allowing each strand to flow past each other and changing the DNA's structure. There are two variations of this enzyme that are most likely the result of a gene duplication. Drug resistance has been linked to a number of gene alterations, and the gene encoding this enzyme is the target of multiple anticancer drugs.34 Targeting topoisomerase can cause damage to DNA, interfere with DNA replication, and eventually cause cellular death or stop the proliferation of cancer cells. Topoisomerases are appropriate targets for therapeutic interventions because of their increased activity in cancer cells.35
Although there are drugs available like tamoxifen,36 trastuzumab,37 and paclitaxel.38 There is still a need for new and improved drugs because (a) existing drugs develop resistance; (b) there are several subtypes of breast cancer, and all of them need to be targeted; and (c) various side effects have been observed for existing drugs.39
In this study, the new hybrids have been designed by combining thiazole/benzothiazole fused 1,6-dihydropyrimidine, isatin, and 1,2,3-triazole scaffolds into a single target molecule, continuing the strong interest in creating a single medication with multiple targets. One glucose derivative of each series was also prepared in order to check and enhance the solubility of the target compound. The utility of recyclable Fe-doped Ce-oxide nanoparticles as a catalyst in the Biginelli reaction was investigated at ambient temperature with the goal of developing an eco-friendlier procedure for the synthesis of dihydropyrimidine derivatives 6 and 7, which further react with corresponding isatin azide derivatives to give target compounds 14a–d, 15, 16a–d and 17. To the best of our knowledge, the use of recyclable Fe doped Ce-oxide nanoparticles in green solvents like ethanol for the Biginelli reaction has not been reported previously. HEK-293, MDA-MB-231 and MCF-7 were the three cell lines used to screen the synthesised compounds in order to determine their breast cancer cell inhibitory potential. Utilising both MCF-7 and MDA-MB-231 cell-lines enables us to evaluate the synthesised compound's efficacy against various forms of breast cancer. It provides a broader perspective on how the compounds might be efficacious in various breast cancer subtypes, enabling researchers to identify potential compounds with broader applicability. These compounds can be potential drug candidates for targeting breast cancer and may have better efficacy than the existing ones.
On the basis of literature survey the new hybrids have been designed by combining thiazole/benzothiazole fused 1,6-dihydropyrimidine, isatin, 1,2,3-triazole and glucose scaffolds by molecular hybridisation into a single target molecule, this continues our keen interest in designing a single drug containing moieties that individually show anticancer activity.
Experimental procedure for chemical synthesis
Method of preparation
The target compounds that were synthesised had three biologically active scaffolds, namely (i) thiazole/benzothiazole-fused 1,6-dihydropyrimidine, (ii) a substituted isatin moiety and (iii) triazole. Triazole-threaded thiazole/benzothiazole-fused dihydropyrimidine and isatin were synthesised in eight steps. In order to synthesise the target molecules 14a–d, 15, 16a–d, and 17, the following synthetic process was used:In step 1, Fe-doped nanoparticles have been prepared using the method described in the literature.40 15% FeCl3 in ethanol (w/w) was added to a solution of Ce(NO3)3·6H2O in ethanol and toluene (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The ethanol-based oxalic acid solution is then added to the reaction mixture. An orange-coloured solution was formed, which was dried in a muffle furnace to get red-coloured Fe doped Ce-oxide nanoparticles (Scheme 1). These nanoparticles were further used for the synthesis of compounds 6 and 7 (Scheme 4).
1). The ethanol-based oxalic acid solution is then added to the reaction mixture. An orange-coloured solution was formed, which was dried in a muffle furnace to get red-coloured Fe doped Ce-oxide nanoparticles (Scheme 1). These nanoparticles were further used for the synthesis of compounds 6 and 7 (Scheme 4).
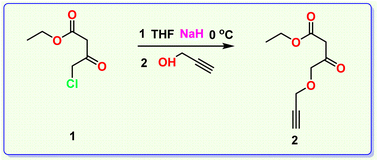
In step 2, ethyl 4-chloroacetoacetate was converted to its alkyne derivative, ethyl-3-oxo-4-(prop-2-yn-1-yloxy) butanoate (2) by treating ethyl-4-chloroacetoacetate (1) with propargyl alcohol in the presence of NaH and in THF (Scheme 2).
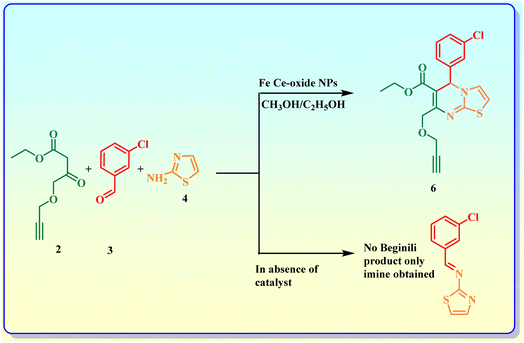
With the aim of developing a more efficient and eco-friendly procedure for the Beginili reaction, in step 3, recyclable Fe doped Ce-oxide nanoparticles have been used for the synthesis of reactants 6 and 7 (Scheme 4). First of all, reaction conditions were screened to determine the appropriate solvents and the mole ratio of reactants for the targeted Beginili multicomponent reaction. With the goal of maximising the product yield in a relatively brief time frame, the reaction's optimisation was investigated for the synthesis of compound 6 using catalyst-free conditions and with a catalyst (Scheme 3) in a variety of solvents. It has been observed that no reaction was observed in solvent-less conditions or in polar aprotic solvents (ACN and THF). Imine is formed from the reaction of an aldehyde and 2-aminobenzothiazole. However, compound 6 was formed when CH3OH and C2H5OH were used as solvents in the presence of catalyst, Fe doped Ce-Oxide nanoparticles on heating for 24 h at 30 °C (Scheme 3).
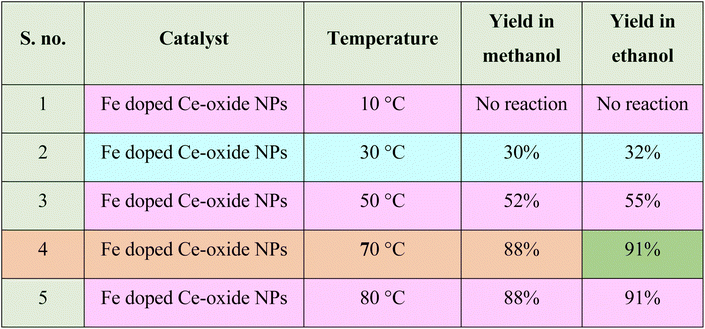
Further reaction conditions were optimised by carrying out reactions with Fe doped Ce-oxide nanoparticles at various temperatures to get maximum yield. It was observed that the maximum yield observed at 70 °C (Table 1). Heating the reaction mixture beyond 70 °C temperature gave side products along with the product.
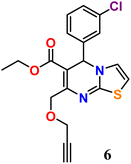
To the solution of propargylated ethyl 4-chloroacetoacetate (2), 3-chlorobenzaldehyde (3) and 2-aminothiazole (4), were dissolved in C2H5OH. Novel starting material 6 was obtained by adding 0.05 M% Fe-doped Ce oxide nanoparticle in ethanol and refluxing the reaction liquid for eight hours., via a one-pot cascade reaction as shown in (Scheme 4). Likewise, after refluxing compounds 2, 3, and 5 in ethanol for eight hours with catalyst, compound 7 was produced.
 | ||
| Scheme 4 Synthesis, of alkyne derivatives of thiazole and benzothiazole-based dihydropyrimidine 6 and 7, respectively, using Fe doped Ce-oxide nanoparticles in C2H5OH. | ||
Since synthesised compounds 6 and 7 are novel, to check the efficiency of nanoparticles in practice, compounds 6 and 7 were also synthesised by the reported method41 i.e., using conc. HCl in methanol by refluxing for 10 h. It was observed that the yield of Fe doped Ce-oxide nanoparticle-catalysed reactions was quite higher than that of acid-catalysed reactions as listed in Table 2.
In step 4, azide derivatives of acyl-protected D-glucose (13) and isatin (10a–d) and were prepared using the procedure described in the litrature42,43 as illustrated in Schemes 5 and 6.
In step 5, compound 6 was reacted with various substituted azides 10a–d and 13 to form 1,2,3-triazole derivatives 14a–d and 15, respectively via [2 + 3] cycloaddition reactions using DMF at 60 °C using a catalytic quantity of sodium ascorbate and CuSO4·5H2O (Scheme 7).
 | ||
| Scheme 7 Synthesis of novel 1, 2, 3- triazole derivatives of thiazole-fused dihydropyrimidines 14a–d and 15. | ||
Similarly, compounds 16a–d and 17 were synthesised by the click reaction of compound 7 with 10a–d and 13, respectively (Scheme 8).
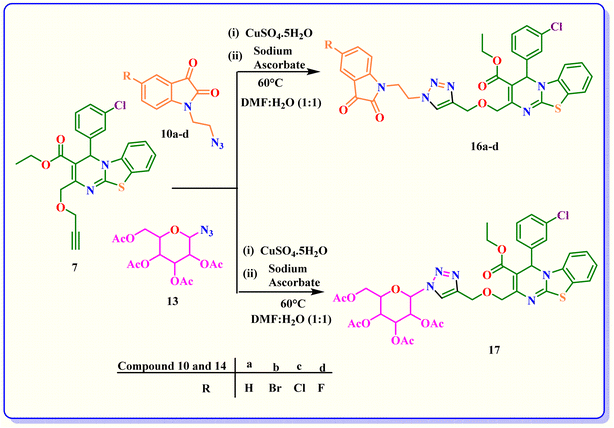 | ||
| Scheme 8 Synthesis of 1, 2, 3- triazole derivatives of benzothiazole-fused dihydropyrimidine 16a–d and 17. | ||
Results and discussions for chemical synthesis
The structures of all the synthesised compounds were validated using standard analytical techniques, including 1H and 13C nuclear magnetic resonance (NMR), infrared spectra (IR) and HRMS (high resolution mass spectrum) data. A triplet was detected at δ 1.21 ppm for three protons of the CH3 group of the ethyl ester in compound 7's 1H NMR. In the 1H NMR of compound 7, a triplet was observed at δ 1.21 ppm for three protons of the CH3 group of the ethyl ester. A triplet was observed at δ 3.43 ppm for 1 proton of the terminal alkyne. A singlet was observed at δ 6.56 ppm for 1 proton of the dihydropyrimidine ring. Compound 6's13C NMR spectra showed a peak at δ 164.77 ppm, which was attributed to the ester group's C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O carbon. The terminal alkyne carbon was identified as the source of a peak seen at δ 77.25 ppm. The peak that corresponds to the carbon of the ethyl ester's CH3 group is evident at δ 13.98 ppm. Further HRMS (high-resolution mass spectrum) peak for the m/z [M + H]+ ion peak appeared at 439.0942, which supports the synthesis of the compound 7 with the molecular formula C23H17ClN2O3S.
O carbon. The terminal alkyne carbon was identified as the source of a peak seen at δ 77.25 ppm. The peak that corresponds to the carbon of the ethyl ester's CH3 group is evident at δ 13.98 ppm. Further HRMS (high-resolution mass spectrum) peak for the m/z [M + H]+ ion peak appeared at 439.0942, which supports the synthesis of the compound 7 with the molecular formula C23H17ClN2O3S.
Compound 14a's 1H NMR revealed a triplet for the ester CH3 group's three protons at δ 1.08 ppm. A singlet for the dihydropyrimidine ring's single proton was detected at δ 6.37 ppm. A singlet for a single proton of the 1,2,3-triazole ring was identified at δ 8.19 ppm. The compound 14a's13C NMR spectra identified a peak at δ 182.93 ppm, which was attributed to the carbonyl carbon on the isatin ring's C-3 position. The peak that showed up at δ 165.26 ppm was attributed to the ester group's C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O carbon. The triazole ring's C-4 carbon is responsible for the peak seen at δ 130.93 ppm. Further HRMS (high-resolution mass spectrum) peak for the m/z [M + H]+ ion peak appeared at 605.1353, which supports the synthesis of the compound 14a with the molecular formula C29H25ClN6O5S.
O carbon. The triazole ring's C-4 carbon is responsible for the peak seen at δ 130.93 ppm. Further HRMS (high-resolution mass spectrum) peak for the m/z [M + H]+ ion peak appeared at 605.1353, which supports the synthesis of the compound 14a with the molecular formula C29H25ClN6O5S.
Methods for biological evaluation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded per well of 24 well flat bottom plates followed by incubation in CO2 incubator at 37 °C for 20–24 h. The cells were treated with 100 µM concentration of compounds dissolved in DMEM media in triplicates along with the positive control drug (tamoxifen), vehicle control (DMSO), and solvent control (PBS) for 48 h. Following incubation, cells were treated with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma-Merck, USA) and incubated for 2 h at 37 °C in dark, then formazan granules were dissolved in DMSO with gentle shaking. The cell viability was evaluated by taking absorbance at 570 nm using a Tecan Multiplate Reader (SpectraMax® iD3 multi-mode microplate reader). For titration of the best four compounds and calculation of IC50 against MCF-7 and MDA-MB-231 cells, further diluted stocks of 1 µM, 10 µM, and 100 µM were made in DMSO and treated in three replicates.
000 cells were seeded per well of 24 well flat bottom plates followed by incubation in CO2 incubator at 37 °C for 20–24 h. The cells were treated with 100 µM concentration of compounds dissolved in DMEM media in triplicates along with the positive control drug (tamoxifen), vehicle control (DMSO), and solvent control (PBS) for 48 h. Following incubation, cells were treated with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma-Merck, USA) and incubated for 2 h at 37 °C in dark, then formazan granules were dissolved in DMSO with gentle shaking. The cell viability was evaluated by taking absorbance at 570 nm using a Tecan Multiplate Reader (SpectraMax® iD3 multi-mode microplate reader). For titration of the best four compounds and calculation of IC50 against MCF-7 and MDA-MB-231 cells, further diluted stocks of 1 µM, 10 µM, and 100 µM were made in DMSO and treated in three replicates.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, hydrogen bond acceptors (HBA), hydrogen bond donors (HBD), rotatable bonds (RB), heavy atom count, polar surface area (PSA), and molar refractivity (MR). Comprehensive cheminformatics analysis, including the computation of several molecular descriptors, grouping, similarity evaluations, and data visualization, was made possible by the Canvas module in particular. This allowed for a thorough assessment of compound drug-likeness and structural variety. To find any parameter violations, all computed attributes were methodically evaluated in comparison to Lipinski's rule of five.44
P, hydrogen bond acceptors (HBA), hydrogen bond donors (HBD), rotatable bonds (RB), heavy atom count, polar surface area (PSA), and molar refractivity (MR). Comprehensive cheminformatics analysis, including the computation of several molecular descriptors, grouping, similarity evaluations, and data visualization, was made possible by the Canvas module in particular. This allowed for a thorough assessment of compound drug-likeness and structural variety. To find any parameter violations, all computed attributes were methodically evaluated in comparison to Lipinski's rule of five.44Results and discussions for biological evaluation of the synthesized compounds
Functional evaluation of the synthesised compounds
In order to determine the IC50 values, further investigation was conducted on four compounds (6, 14a, 7, and 16c) by treating MDA-MB-231 and MCF-7 cell-lines with varying doses of the compounds. Compounds 6 and 14a were tested from 10 nM to 100 µM, while compound 7 was tested in the dose range of 100 pM to 100 µM due to its potency even at low doses (Fig. 3A–C) However, the IC50 value of 16c was found to be higher than >100 µM, indicating it is not a viable candidate for further research, and therefore, it was excluded from the plot. Based on the results presented in Fig. 3A–C, these results indicate that Compound 7 is the most effective compound with both MCF-7 and MDA-MB-231 cell-lines.
 | ||
| Fig. 4 Cells before compound/drug treatment. (A) MCF-7, a breast cancer cell line; (B) HEK-29, a human embryonic kidney cell line that serves as a non-cancerous control. | ||
Docking glide scores were obtained for two compounds 6 and 14d indicating low efficacy as their binding score is around −4 kcal mol−1. However, further analysis revealed that out of all the 10 compounds, 14a, 14b, 14c, 15, 7, 16a, 16b, 16c, 16d, and 17 exhibited high efficacy, with binding scores ranging from approximately −5.02 to −8.28 kcal mol−1 (Fig. 8). Among these, 14c, 7, 16a and 16d showed the lowest binding energies (−5.77, −6.32, −5.67, and −8.28 kcal mol−1, respectively), indicating a strong interaction with topoisomerase II alpha (Fig. 9 and 10).
 | ||
| Fig. 8 Graph showing binding energies based on docking scores against topoisomerase II of 12 compounds (blue: less negative, green: more negative), and tamoxifen (red: negative control). | ||
 | ||
| Fig. 9 Three dimensional docked structures of the best four compounds (14c, 7, 16a and 16d) with topoisomerase (PDB ID: 5GWK) visualized using Maestro visualizer. | ||
 | ||
| Fig. 10 Two-dimensional docked structures of the best four compounds (14c, 7, 16a and 16d) with topoisomerase (PDB ID: 5GWK) visualised using Maestro visualizer. | ||
The docking conformations of the protein structures (PDB ID 5GWK) with the compounds revealed that specific amino acids, including GLN-1095, THR-825, PHE-828, LYS-287, GLN-1049, ALA-588, ILE-574, LYS-821, and LEU-826, played a vital role in the binding process. Topoisomerase II alpha consists of several domains such as TOPORIM, WHD, TOWER, ATPase, and CTD, which contribute to functions like metal binding, DNA cleavage, and dimerization. Notably, the WHD and CTD domains are primarily involved in binding, with functions related to DNA intercalation and DNA cleavage (Table S1 in SI). Protein-ligand interacting amino acid residue details of compounds 7, 14c, 16a, 16d and the standard drug tamoxifen with topoisomerase II alpha using UCSF Chimera software are shown in Table S2 (in SI)
Table S1 (in the SI) provides a comprehensive illustration of the molecular interactions between the ligand molecules 6, 7, 14a–d, 15, 16a–d, and 17 and different residues of various amino acids in the binding pocket of the enzyme topoisomerase II alpha.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (5.45), Table 3. It's interesting to note that tamoxifen, a clinically proven treatment for breast cancer, also surpasses the Lipinski-specified A
P (5.45), Table 3. It's interesting to note that tamoxifen, a clinically proven treatment for breast cancer, also surpasses the Lipinski-specified A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P threshold, which may have an impact on its pharmacokinetic profile. Similar to tamoxifen, compound 7 exhibits significantly greater cytotoxic efficacy and considerable biological inhibition at concentrations far lower than those reported for tamoxifen under comparable experimental settings, although it only slightly exceeds the optimum A
P threshold, which may have an impact on its pharmacokinetic profile. Similar to tamoxifen, compound 7 exhibits significantly greater cytotoxic efficacy and considerable biological inhibition at concentrations far lower than those reported for tamoxifen under comparable experimental settings, although it only slightly exceeds the optimum A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P limit. The idea was that molecular structure of compound 7 provides strong target engagement and cellular efficacy without significantly compromising drug-likeness is thus supported by the SAR study. All things considered, these findings identify compound 7 as a very promising lead that successfully strikes a balance between favourable physicochemical characteristics and strong nanomolar cytotoxicity (63.7 ± 2.17 nM and 55.0 ± 1.19 nM), providing benefits over well-known clinical standards like tamoxifen.
P limit. The idea was that molecular structure of compound 7 provides strong target engagement and cellular efficacy without significantly compromising drug-likeness is thus supported by the SAR study. All things considered, these findings identify compound 7 as a very promising lead that successfully strikes a balance between favourable physicochemical characteristics and strong nanomolar cytotoxicity (63.7 ± 2.17 nM and 55.0 ± 1.19 nM), providing benefits over well-known clinical standards like tamoxifen.
| S. no. | Compound | Docking score (kcal mol−1) | Predicted activity | MW (Da) | A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
HBA | HBD | RB | Heavy atom | PSA (Å2) | MR (cm3 mol−1) | Polar (%) | Lipinski violations |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
a MW: molecular weight, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P: partition coefficient (log P: partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, computationally predicted), HBA: hydrogen bond acceptors, HBD: hydrogen bond donors, RB: rotatable bonds, PSA: polar surface area, MR: molar refractivity. P, computationally predicted), HBA: hydrogen bond acceptors, HBD: hydrogen bond donors, RB: rotatable bonds, PSA: polar surface area, MR: molar refractivity. |
|||||||||||||
| 1 | 6 | −3.52 | −0.08 | 388.87 | 3.69 | 3 | 1 | 6 | 26 | 86.00 | 95.89 | 44.54 | None |
| 2 | 7 | −5.02 | −0.14 | 438.93 | 5.45 | 3 | 1 | 6 | 30 | 86.00 | 111.60 | 52.89 | A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 5 P > 5 |
| 3 | 14a | −6.32 | −0.17 | 605.06 | 3.12 | 7 | 0 | 10 | 42 | 154.09 | 154.54 | 70.25 | MW > 500 |
| 4 | 14b | −5.17 | −0.13 | 683.96 | 3.87 | 7 | 0 | 10 | 43 | 154.09 | 162.16 | 72.88 | MW > 500 |
| 5 | 14c | −3.58 | −0.12 | 639.51 | 3.78 | 7 | 0 | 10 | 43 | 154.09 | 159.34 | 72.22 | MW > 500 |
| 6 | 14d | −5.78 | −0.12 | 623.05 | 3.32 | 7 | 0 | 10 | 43 | 154.09 | 154.76 | 70.16 | MW > 500 |
| 7 | 15 | −4.03 | −0.12 | 762.18 | 1.66 | 14 | 0 | 17 | 52 | 231.14 | 177.26 | 77.68 | MW > 500, HBA > 10 |
| 8 | 16a | −5.68 | −0.09 | 655.12 | 4.88 | 7 | 0 | 10 | 46 | 154.09 | 170.25 | 78.61 | MW > 500 |
| 9 | 16b | −5.63 | −0.10 | 734.02 | 5.63 | 7 | 0 | 10 | 47 | 154.09 | 177.87 | 81.24 | MW > 500, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 5 P > 5 |
| 10 | 16c | −5.03 | −0.11 | 689.57 | 5.55 | 7 | 0 | 10 | 47 | 154.09 | 175.05 | 80.58 | MW > 500, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 5 P > 5 |
| 11 | 16d | −8.28 | −0.14 | 673.11 | 5.09 | 7 | 0 | 10 | 47 | 154.09 | 170.46 | 78.52 | MW > 500, A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 5 P > 5 |
| 12 | 17 | −5.14 | −0.09 | 812.24 | 3.42 | 14 | 0 | 17 | 56 | 231.14 | 192.97 | 86.04 | MW > 500, HBA > 10 |
| 13 | Tamoxifen | −5.28 | −0.12 | 371.51 | 6.32 | 1 | 0 | 8 | 28 | 12.47 | 119.26 | 56.02 | A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 5 P > 5 |
Conclusion
This work involved the synthesis of a number of dihydropyrimidine derivatives using a greener, environment-friendly method using recyclable Fe-doped Ce-oxide nanoparticles in ethanol. The nanoparticles are recyclable and give excellent yields up to 5 cycles. Further novel isatin-linked triazole derivatives were synthesized by starting with dihydropyrimidine derivatives. All synthesized compounds were subjected to screening against human breast-cancer cell-lines in vitro. The results were encouraging, as these derivatives exhibited remarkable anti-cancer activity against two prominent breast-cancer cell-lines, MCF-7 and MDA-MB-231. The IC50 values of the compounds 6, 14a, and 7 were 52.7 ± 0.73 µM, 42.3 ± 0.83 µM, and 63.7 ± 2.17 nM, respectively against MDA-MB-321 cancer cell lines. In the case of MCF-7 cells, 6, 14a, and 7 demonstrated IC50 values of 62.6 ± 0.86 µM, 81.9 ± 2.26 µM, and 55.0 ± 1.19 nM, respectively. These findings highlighted the significant potential of these dihydropyrimidine derivatives aseffective agents against breast cancer. This is especially true of compound 7, which showed an incredibly low IC50 value of 63.7 ± 2.17 nM against MDA-MB-231 cells and 55.0 ± 1.19 nM against MCF-7 cells. In molecular interaction studies, intriguing results were obtained, indicating that compounds 14a, 7, 16b, and 16d exhibited the lowest binding scores. This suggested that by preventing the topoisomerase enzyme from performing its intended function, these compounds could be able to prevent breast cancer. Inhibition of this enzyme can impede the survival of cancer cells, consequently providing a potentially effective therapeutic strategy for breast cancer. These findings highlighted the possibility of utilising these compounds as effective therapeutic agents, warranting further exploration and investigation into their precise mechanisms of action. This study contributes to the growing body of knowledge in the area of breast cancer treatments and emphasises the significance of additional research into the pharmacological characteristics and modes of action associated with these compounds. These results provided a solid foundation for future studies aimed at developing these derivatives into promising anti-breast cancer drugs.Author contributions
The notion was put out by KKS, who also created the manuscript portions. The final manuscript was written and reviewed by all of the writers. KKS, DR, and SJK conducted experiments, analysed data, and prepared the diagrams and tables. SKD and RS supervised the study and acquired funding for the study.Conflicts of interest
The authors declare that none of the information in this study has any conflicts of interest. This work has been submitted for a patent with an Application no. 202411007373.Data availability
Research data has been attached in the uploaded supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra07279d.Acknowledgements
KKS is grateful to the Institutions of Eminence (IoE) for providing funds to carry out the present research work. KKS is also grateful to the USIC (University Science Instrumentation Centre), University of Delhi, for providing the required instrumentation facilities. SKD acknowledges financial and infrastructural support from the University of Delhi (R&D and seed grants) and Dr B. R. Ambedkar Centre for Biomedical Research [ACBR] (start-up and research grants). SKD also acknowledges the Institution of Eminence grant (grant ID: IoE/2021/12/FRP) from the University of Delhi, India; SERB-DST ANRF grant (CRG/2023/007380) and ICMR extramural grant (grant ID: 5/4/1-11/CVD/2022-NCD-I) for providing financial support for the current work. RKU acknowledges the fellowship support from CSIR. DR acknowledges her Junior Research fellowship from the DBT (grant ID: DBT/2021-22/CBR/1702), and SKJ acknowledges her Project Assistant fellowship from the ICMR (grant ID: 5/4/1-11/CVD/2022-NCD-I) to complete this work. The authors also gratefully acknowledge CSIR for financial support for this work.References
- X. B. Hong R, Cancer Commun., 2022, 42, 913–936 CrossRef PubMed.
- G. P. Dailey, E. J. Crosby and Z. C. Hartman, Cancer Gene Ther., 2023, 30, 794–802 CrossRef CAS PubMed.
- S. Lakshmanan and N. Ramalakshmi, Synth. Commun., 2016, 46, 2045–2052 CrossRef CAS.
- E. S. Araújo, M. F. G. Pereira, G. M. G. da Silva, G. F. Tavares, C. Y. B. Oliveira and P. M. Faia, Toxics, 2023, 11(8), 658 CrossRef PubMed.
- S. E. Kim, H. J. Yang, S. Choi, E. Hwang, M. Kim, H. J. Paik, J. E. Jeong, Y. Il Park, J. C. Kim, B. S. Kim and S. H. Lee, Green Chem., 2022, 24, 251–258 RSC.
- B. Ramesh Naidu and K. Venkateswarlu, Green Chem., 2022, 24, 6215–6223 RSC.
- S. Maddila, O. A. Abafe, H. N. Bandaru, S. N. Maddila, P. Lavanya, N. Seshadri and S. B. Jonnalagadda, Arab. J. Chem., 2019, 12, 3814–3824 CrossRef CAS.
- X. H. Chen, X. Y. Xu, H. Liu, L. F. Cun and L. Z. Gong, J. Am. Chem. Soc., 2006, 128, 14802–14803 CrossRef CAS PubMed.
- Y. Li, T. Tan, Y. Zhao, Y. Wei, D. Wang, R. Chen and L. Tao, ACS Macro Lett., 2020, 9, 1249–1254 CrossRef CAS PubMed.
- M. Marinescu, Molecules, 2021, 26, 1–34 CrossRef PubMed.
- S. M. Rida, S. A. M. El-Hawash, H. T. Y. Fahmy, A. A. Hazzaa and M. M. M. El-Meligy, Arch. Pharm. Res., 2006, 29, 826–833 CrossRef CAS PubMed.
- M. H. El-Wakil, M. Teleb, M. M. Abu-Serie, S. Huang, G. W. Zamponi and H. Fahmy, Bioorg. Chem., 2021, 115, 105262 CrossRef CAS PubMed.
- A. Singadi, K. Venkateswarlu and I. J. Adv, Pharm. Biotechnol., 2020, 6, 4–9 Search PubMed.
- M. A. M. Abdel Reheim, I. S. Abdel Hafiz and H. S. E. Abdel Rady, Mol. Divers., 2022, 26, 741–755 CrossRef CAS PubMed.
- N. C. Desai, G. M. Kotadiya, K. A. Jadeja, K. N. Shah, A. H. Malani, V. Manga and T. Vani, Bioorg. Chem., 2021, 115, 105173 CrossRef CAS PubMed.
- N. Afradi, N. Foroughifar, H. Pasdar and M. Qomi, Res. Chem. Intermed., 2019, 45, 3251–3271 CrossRef CAS.
- V. Sharma, S. K. Gupta and M. Verma, Cancer Chemother. Pharmacol., 2019, 84, 1157–1166 CrossRef CAS PubMed.
- K. M. Bairagi, N. S. Younis, P. M. Emeka, K. N. Venugopala, O. I. Alwassil, H. E. Khalil, E. Sangtani, R. G. Gonnade, V. Mohanlall and S. K. Nayak, J. Mol. Struct., 2021, 1227, 129412 CrossRef CAS.
- O. Alam, S. A. Khan, N. Siddiqui, W. Ahsan, S. P. Verma and S. J. Gilani, Eur. J. Med. Chem., 2010, 45, 5113–5119 CrossRef CAS PubMed.
- M. Teleb, F. X. Zhang, J. Huang, V. M. Gadotti, A. M. Farghaly, O. M. AboulWafa, G. W. Zamponi and H. Fahmy, Bioorganic Med. Chem., 2017, 25, 1926–1938 CrossRef CAS PubMed.
- C. G. Neochoritis, T. Zarganes-Tzitzikas, C. A. Tsoleridis, J. Stephanidou-Stephanatou, C. A. Kontogiorgis, D. J. Hadjipavlou-Litina and T. Choli-Papadopoulou, Eur. J. Med. Chem., 2011, 46, 297–306 CrossRef CAS PubMed.
- R. E. Ferraz de Paiva, E. G. Vieira, D. Rodrigues da Silva, C. A. Wegermann and A. M. Costa Ferreira, Front. Mol. Biosci., 2021, 7, 1–24 Search PubMed.
- G. Singh, P. Kalra, A. Singh, G. Sharma, P. Sanchita, Mohit, C. Espinosa-Ruíz and M. A. Esteban, J. Organomet. Chem., 2021, 953, 122051 CrossRef CAS.
- S. Nain, G. Mathur, T. Anthwal, S. Sharma and S. Paliwal, Pharm. Chem. J., 2023, 57, 196–203 CrossRef CAS PubMed.
- V. K. R. Tangadanchu, Y. F. Sui and C. H. Zhou, Bioorganic Med. Chem. Lett., 2021, 41, 128030 CrossRef CAS PubMed.
- Y. Q. Hu, S. Zhang, F. Zhao, C. Gao, L. S. Feng, Z. S. Lv, Z. Xu and X. Wu, Eur. J. Med. Chem., 2017, 133, 255–267 CrossRef CAS PubMed.
- D. Sriram, P. Yogeeswari and K. Meena, Pharmazie, 2006, 61, 274–277 CAS.
- X. H. Makhoba, C. Viegas, R. A. Mosa, F. P. D. Viegas and O. J. Pooe, Drug Des. Devel. Ther., 2020, 14, 3235–3249 CrossRef CAS PubMed.
- A. Bastero, D. Font and M. A. Pericàs, J. Org. Chem., 2007, 72, 2460–2468 CrossRef CAS PubMed.
- R. Kharb, P. C. Sharma and M. S. Yar, J. Enzyme Inhib. Med. Chem., 2011, 26, 1–21 CrossRef CAS PubMed.
- T. A. Wani, N. Alsaif, M. M. Alanazi, A. H. Bakheit, S. Zargar and M. A. Bhat, Eur. J. Pharm. Sci., 2021, 158, 105686 CrossRef CAS PubMed.
- A. Mushtaq, R. Asif, W. A. Humayun and M. M. Naseer, RSC Adv., 2024, 14, 14051–14067 RSC.
- E. D. Dalvie, J. C. Stacy, K. C. Neuman and N. Osheroff, Biochemistry, 2022, 61, 2148–2158 CrossRef CAS PubMed.
- A. J. Lang, S. E. L. Mirski, H. J. Cummings, Q. Yu, J. H. Gerlach and S. P. C. Cole, Gene, 1998, 221, 255–266 CrossRef CAS PubMed.
- J. L. Delgado, C. M. Hsieh, N. L. Chan and H. Hiasa, Biochem. J., 2018, 475, 373–398 CrossRef CAS PubMed.
- A. Ismail, A. S. Doghish, B. E. M. Elsadek, S. A. Salama and A. D. Mariee, Steroids, 2020, 160, 108656 CrossRef CAS PubMed.
- J. Cortés, S.-B. Kim, W.-P. Chung, S.-A. Im, Y. H. Park, R. Hegg, M. H. Kim, L.-M. Tseng, V. Petry, C.-F. Chung, H. Iwata, E. Hamilton, G. Curigliano, B. Xu, C.-S. Huang, J. H. Kim, J. W. Y. Chiu, J. L. Pedrini, C. Lee, Y. Liu, J. Cathcart, E. Bako, S. Verma and S. A. Hurvitz, N. Engl. J. Med., 2022, 386, 1143–1154 CrossRef PubMed.
- H. A. Bashmail, A. A. Alamoudi, A. Noorwali, G. A. Hegazy, G. M. Ajabnoor and A. M. Al-Abd, Molecules, 2020, 25, 2–15 CrossRef PubMed.
- V. Jain, H. Kumar, H. V. Anod, P. Chand, N. V. Gupta, S. Dey and S. S. Kesharwani, J. Control. Release, 2020, 326, 628–647 CrossRef CAS PubMed.
- P. K. Mishra, P. Gahlyan, R. Kumar and P. K. Rai, ACS Sustain. Chem. Eng., 2018, 6, 10668–10678 CrossRef CAS.
- K. N. Venugopala, R. Govender, M. A. Khedr, R. Venugopala, B. E. Aldhubiab, S. Harsha and B. Odhav, Drug Des. Devel. Ther., 2015, 9, 911–921 CrossRef CAS PubMed.
- K. Bhagat, J. Bhagat, M. K. Gupta, J. V. Singh, H. K. Gulati, A. Singh, K. Kaur, G. Kaur, S. Sharma, A. Rana, H. Singh, S. Sharma and P. M. S. Bedi, ACS Omega, 2019, 4, 8720–8730 CrossRef CAS PubMed.
- V. Castro, H. Rodríguez and F. Albericio, ACS Comb. Sci., 2016, 18, 1–14 CrossRef CAS PubMed.
- S. Kaur, J. Kaur, B. A. Zarger, N. Islam and N. Mir, Heliyon, 2024, 10, e35897 CrossRef CAS PubMed.
- V. C. Jordan, Nat. Rev. Drug Discovery, 2003, 2, 205–213 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |