Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel ene-reductase from Halomonas elongata for flow biocatalytic synthesis of 3-phenylpropionaldehyde and sustainable indigo-carmine dyeing
Lauriane Pilleta,
Cristina Lía Fernández Reguerio b,
Markus Richard Buscha,
David Roura Padrosa
b,
Markus Richard Buscha,
David Roura Padrosa b and
Francesca Paradisi
b and
Francesca Paradisi *a
*a
aDepartment of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Freiestrasse 3, CH-3012 Bern, Switzerland. E-mail: francesca.paradisi@unibe.ch
bInSEIT AG, Freiestrasse 3, CH-3012 Bern, Switzerland
First published on 12th November 2025
Abstract
In order to broaden the toolbox of enzymes available for biocatalytic reductions of carbon-carbon double bonds, we investigated four promising ene-reductases (ERs) stemming from extremophilic organisms or showing homology with thermophilic ERs. The novel ene-reductase from the halophilic organism Halomonas elongata showed consistently high activity across a range of tested substrates. Upon immobilisation of the ERs, the flow biocatalytic ene-reduction of cinnamaldhehyde into 3-phenylpropionaldehyde was successfully achieved with an intensification of the process of 62-fold with respect to batch (2173.9 mg L−1 h−1 and 34.7 mg L−1 h−1, respectively). Additionally, to expand the scope of ERs applications, we describe a proof-of concept of a novel enzymatic cascade to convert indole to indigo using an unspecific peroxygenase, and its subsequent reduction to the water-soluble leuco-indigo. In addition, the co-production of the valuable pharmaceuticals precursor 2-oxindole was demonstrated.
Introduction
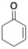
The increasing demand for enantiopure molecules in the fine-chemical, pharmaceutical and fragrance industries triggered the development of well-characterised, efficient and stable catalysts for their production. One of the most widely used industrial reactions to access chiral compounds is the asymmetric reduction of alkenes.1,2 Approaches for saturation of carbon–carbon double bonds rely on asymmetric hydrogenation technology, or on organocatalysis.1,2 However, these approaches require special catalysis expertise and/or specialised equipment, expensive hydrogen source and/or high catalyst loadings, which dramatically increases process costs and reduce their industrial viability. Moreover, the metals used for catalytic hydrogenation are becoming less readily available due to their high geological rarity, high demand, and extraction challenges, which has stimulated the search for more sustainable ways to reduce carbon-carbon double bonds.3 Enzymatic approaches, especially ene-reductases (ERs)/old yellow enzymes (OYEs), have emerged as a viable method for regio- and chemoselective alkene reduction, leading to shorter synthetic routes towards not only high-added value but also bulk chemicals.1–4 These flavin-mononucleotide (FMN)-dependant enzymes can catalyse the stereoselective reduction of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds activated with electron-withdrawing groups (EWG) at the expense of a nicotinamide cofactor (Fig. 1).
C bonds activated with electron-withdrawing groups (EWG) at the expense of a nicotinamide cofactor (Fig. 1).
 | ||
Fig. 1 Ene-reductases FMN-dependent reduction of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds activated with EWG at the expense of NAD(P)H cofactor.2 C bonds activated with EWG at the expense of NAD(P)H cofactor.2 | ||
In the past few years, ERs have become a hot research topic, with many innovative example showcasing their versatility.4,5 They can reduce not only α,β-unsaturated compounds activated in Cα by EWG such as carboxylic acids, esters, aldehydes, ketones, nitro or nitrile groups, but have also been shown to reduce oximes into imines, to perform reductive cyclisation of α,β-unsaturated compounds containing an additional electrophilic group to form chiral cyclopropanes,4,5 or even alkynes to alkenes followed by subsequent reduction to alkanes.6
In addition, they have been shown to work in the oxidative direction using O2 as an oxidant at 70 °C,5 or even be coupled to photoexcitation to form C–C bonds via stereoselective formal intramolecular hydroalkylation of non-activated alkenes.7 OYEs have also often been paired with dehydrogenases to form redox-neutral cascades without the requirement of an external electron donor,1–4 to transaminases,4 or to squalene hopene cyclases (SHC).4,8 Interestingly, alternative NAD(P)H regeneration systems relying on electrochemical reductions, photocatalysts, cheap hydride sources, cofactor analogues or even cofactor-free reductions with ERs have been reported.1–4 ERs have been used in several chemo-enzymatic cascades,1,4,5 including the preparative scale synthesis of the antiepileptic pregabalin with high enantiomeric excess (>99%) and good yields (69%),9 or the production of the powerful odorant decanal from 2E-decenal with 10 g L−1 of substrate.10 In addition, new OYE/ER homologues are becoming available through sequence similarity searches,2,11 site-directed mutagenesis or directed evolution towards desired substrates.4,12,13 However, despite the numerous applications of ERs at the lab scale, they are still underrepresented in industrial/preparative-scale reductions.4 While ERs have been immobilised in some studies with co-factor recycling enzymes partners (for example, glucose dehydrogenases (GDH)),5,14–16 applications of ERs in flow biocatalysis are even more scarce.4–6
Herein, to bring ERs closer to industrial applications requirements, we selected four promising ERs for rational immobilisation and explored the continuous biocatalytic transformation of cinnamaldehyde into 3-phenylpropionaldehyde. Then, to broaden the scope of potential applications of ERs, we propose a novel approach for sustainable dyeing at neutral pH with an enzymatic cascade to convert indole into leuco-indigo, the reduced water-soluble form of indigo that can intercalate into the fibres before being air oxidised back to the everlasting indigo.17,18
Experimental section
Reagents
All chemical reagents were acquired from ThermoFisher Scientific, Fluka (Honeywell), Sigma Aldrich (Merck) or VWR (Avantor) unless otherwise specified. Cofactors involved in biotransformations (NADH, NAD+, NADPH and NADP+) were acquired from Apollo Scientific. The HisTrapFF crude® column used for protein purification was purchased from Cytiva. All commercial chemicals were used without further purification. All enzymes were produced in house, except the rAaeUPO peroxidase that was generously donated by the lab of Prof. Dr Frank Hollmann.Ene-reductases expression and purification
The pET28b(+) plasmids harboring the genes of the different ERs (synthetised by Microsynth) or of BmGDH19 (Table S1 and Fig. S1) were transformed into chemically competent E. coli BL21 (DE3) cells for McOYE, HeOYE, PhENR and BmGDH or into E. coli BL21 Lemo21 (DE3) for TtENR. Then, 10 mL of LB media (yeast extract (5 g L−1), tryptone or N-Z amine (10 g L−1), NaCl (10 g L−1)) supplemented with the appropriate antibiotic(s) (Table S2) was inoculated with a single colony and incubated overnight at 37 °C and 150 rpm. 1 mL of this preculture were then added to 300 mL of LB or TB supplemented with the appropriate antibiotics and with 500 µM L-rhamnose for TtENR (Table S2) and cells were then grown at 37 °C and 150 rpm until an OD600 of approximately 0.6–0.8 was reached. After that, the appropriate concentration of isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce protein expression, and the flasks were incubated overnight at 25–37 °C (Table S2). Cells were then harvested by centrifugation (4500 rpm, 20 minutes, 4 °C), resuspended in 10 mL of loading buffer (Table S3) on ice and lysed by sonication at 40% amplitude for 8 min, with pulses of 5 s ON, 10 s OFF. After centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 4 °C, 45 min), the supernatant was filtered with a 0.45 µm filter, and the different enzymes were then purified by metal affinity chromatography (IMAC) using an AKTA™ Start. Pure fractions were pooled and dialysed for 20 h at 4 °C with dialysis buffer (Table S3) with one buffer exchange after 2 h. Purification of the enzyme was checked by analysing the different fractions with a 12% SDS PAGE (Fig. S2) and concentration of the purified enzymes was estimated measuring the absorbance at 280 nm in the EPOCH2 (nanodrop Take3plate) using extinction coefficients (ε) and molecular weights of predicted using the ExPASy ProtParam tool (Table S1).20 Low protein concentrations (<1 mg mL−1) were determined by Bradford assay by measuring the absorbance at 595 nm of a solution containing 5 µL of protein sample with 250 µL of Bradford reagent.
000 g, 4 °C, 45 min), the supernatant was filtered with a 0.45 µm filter, and the different enzymes were then purified by metal affinity chromatography (IMAC) using an AKTA™ Start. Pure fractions were pooled and dialysed for 20 h at 4 °C with dialysis buffer (Table S3) with one buffer exchange after 2 h. Purification of the enzyme was checked by analysing the different fractions with a 12% SDS PAGE (Fig. S2) and concentration of the purified enzymes was estimated measuring the absorbance at 280 nm in the EPOCH2 (nanodrop Take3plate) using extinction coefficients (ε) and molecular weights of predicted using the ExPASy ProtParam tool (Table S1).20 Low protein concentrations (<1 mg mL−1) were determined by Bradford assay by measuring the absorbance at 595 nm of a solution containing 5 µL of protein sample with 250 µL of Bradford reagent.
Activity assays
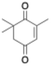
The activity of ERs was determined by monitoring the decrease in absorbance of NAD(P)H at 340 nm (ε = 6220 mol−1 L cm−1) in a 1 mL cuvette at 25, 30 or 75 °C for 10 minutes. A typical reaction mixture contained 100 µL of NADPH (2 mM), 50 µL of maleimide (400 mM), 800 µL of phosphate buffer (100 mM, pH 7.4) containing 20% DMSO and 50 µL of enzyme with appropriate dilution. Cofactor was incubated for 10 minutes with the enzyme at optimal assay's temperature before the assay was triggered by the addition of substrate. During the investigation of the optimal pH for activity assays, citrate buffer (100 mM, pH 5.4), phosphate buffer (100 mM, pH 7.4), or bicarbonate buffer (100 mM, pH 9.4) containing 20% of DMSO were used.The activity of BmGDH19 was determined by monitoring the increase in NADPH absorbance at 340 nm (ε = 6220 mol−1 L cm−1) in a 1 mL cuvette at 37 °C for 10 minutes. A typical reaction mixture contained 100 µL of NADP+ (2 mM), 50 µL of enzyme with appropriate dilution and 850 µL of glucose (40 mM) in phosphate buffer (100 mM, pH 7.4). Cofactor was incubated for 10 minutes at 37 °C with the enzyme before the assay was triggered by the addition of glucose.
The activity of the rAaeUPO peroxidase was determined by monitoring the increase in absorbance of ABTS (2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid)) at 420 nm (ε = 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mol−1 L cm−1) in a 96-well plate at 25 °C for 10 minutes. A typical reaction mixture contained 20 µL of ABTS (5 mM), 20 µL of H2O2 (2 mM), 155 µL of sodium citrate buffer (100 mM, pH 4.4), and 5 µL of the enzyme at the appropriate dilution (0.01 mg mL−1). The assay was triggered by the addition of hydrogen peroxide.
000 mol−1 L cm−1) in a 96-well plate at 25 °C for 10 minutes. A typical reaction mixture contained 20 µL of ABTS (5 mM), 20 µL of H2O2 (2 mM), 155 µL of sodium citrate buffer (100 mM, pH 4.4), and 5 µL of the enzyme at the appropriate dilution (0.01 mg mL−1). The assay was triggered by the addition of hydrogen peroxide.
All activity tests were performed in triplicates and one unit [U] is defined as the amount of enzyme which catalyses the formation or depletion of 1 µmol of product or substrate per minute. To determine enzymatic activity, 20 mg of immobilized enzyme was added to 10 mL of a substrate mixture (containing substrate and cofactor concentrations identical to the free enzyme assays). The progress of the reaction was monitored by sampling the mixture at 2-minute intervals and measuring the absorbance of NAD(P)H at 340 nm.
Enzyme immobilisation
Enzymes were immobilised using two primary strategies: directed immobilisation, through heterogenous supports modified with metal-chelates and epoxy functionalities and covalent attachment, using either epoxy or aldehyde modified resins. The cobalt-chelated and aldehyde-functionalised supports were prepared as previously described.21For immobilisation via metal affinity and epoxy chemistry, the respective metal-chelated resin was suspended in an enzyme solution prepared in potassium phosphate buffer (50 mM, pH 8.0) to reach the desired loading of enzyme to support in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 resin-to-solution ratio (w/v). All suspensions were incubated for 16 hours at RT with orbital shaking. Following incubation, the immobilised preparations were washed with 3 volumes of desorption buffer (EDTA (50 mM), NaCl (500 mM) in phosphate buffer (50 mM, pH 7.2)) to remove the metal and incubated overnight with a 3 M glycine solution (3 M) to block any unreacted epoxy groups. For the aldehyde immobilisation, the solution of protein was added to the support and incubated at room temperature for at least 4 h. After the incubation, 5 volumes of NaBH4 (1 mg mL−1) were used to reduce the formed imines, and the resin subsequently washed with abundant water.
10 resin-to-solution ratio (w/v). All suspensions were incubated for 16 hours at RT with orbital shaking. Following incubation, the immobilised preparations were washed with 3 volumes of desorption buffer (EDTA (50 mM), NaCl (500 mM) in phosphate buffer (50 mM, pH 7.2)) to remove the metal and incubated overnight with a 3 M glycine solution (3 M) to block any unreacted epoxy groups. For the aldehyde immobilisation, the solution of protein was added to the support and incubated at room temperature for at least 4 h. After the incubation, 5 volumes of NaBH4 (1 mg mL−1) were used to reduce the formed imines, and the resin subsequently washed with abundant water.
BmGDH was immobilised following a previously reported protocol.19
All immobilised biocatalysts were washed thoroughly with their respective buffers after immobilisation and stored at 4 °C until used.
The immobilisation yield (IY) was determined by measuring the protein concentration via the Bradford assay before and after the immobilisation procedure. The initial protein concentration (Ei) was measured from the enzyme solution prior to adding the support. After immobilisation, the supernatant containing the unbound enzyme was collected, and its protein concentration was measured (Ef). The immobilisation yield was expressed as a percentage using the following formula:
| IY (%) = 100 × (Ei − Ef)/Ei |
Recovered activity (RA) measures the percentage of the enzyme's original catalytic activity that is retained after it has been immobilised onto the support.
The recovered activity is then calculated as a percentage using the following formula:
| RA (%) = [Aimmo/(Asol × loading × IY)] × 100 |
Batch biotransformations
Initial screenings of ER-mediated reduction of cinnamaldehyde in batch were performed for 24 hours under shaking at 25 °C in phosphate buffer (100 mM, pH 7.4) containing 20% of DMSO in a total reaction volume of 1 mL, in the presence of cinnamaldehyde (1 mM), NAD(P)H (2 mM) without cofactor recycling system. 0.1 mg mL−1 of the free enzymes was used, and equivalent amounts of enzyme units was used for both soluble and heterogenous biotransformations. At the 10 mM scale, NAD(P)H (1 mM) was used with BmGDH as recycling enzyme partner and glucose (40 mM) as sacrificial substrate using the same conditions.Biotransformations aiming to convert indole into indigo were performed for 2–24 hours under shaking at 25 °C in citrate buffer (100 mM, pH 4.4) or phosphate buffer (100 mM, pH 7.4) containing 10% of acetonitrile in a total reaction volume of 1 mL, in the presence of indole (1–50 mM), 1–5 equivalents of H2O2 or cumene hydroperoxide, and 0.05 mg mL−1 of rAaeUPO (3 U). Controls were run in the presence of all reaction components, except the enzyme. Indigo dyeing was performed by prewetting the tissue with water, soaking it into the reducing bath for 2–24 hours at 25 °C under shaking, removing it and letting it air oxidise, washing it and finally neutralising by soaking it with acetic acid. For chemical reductions, the oxidating bath was composed of sodium dithionite (Na2S2O4, 40 mM) or glucose as reducing agents at pH 12 (NaOH, 200 mM), whereas enzymatic reductions were performed at pH 7.4 using 1 mg mL−1 of each ER, 0.5 mg mL−1 of glucose dehydrogenase from Bacillus megaterium (BmGDH) and glucose (40 mM) to recycle 1 mM of NAD(P)H, keep the system under reduced conditions and potentially outcompete air oxidation. A layer of mineral oil was also added on the top of each reaction before each solution was bubbled with nitrogen to help in this purpose. Both chemical and enzymatic reductions were performed using 0.5 or 1 mM of indigo or indigo carmine.
Flow biotransformations
For flow biotransformations, 1.5 g of immobilised HeOYE was mixed with 1.4 g of BmGDH (activity ratio 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in a packed-bed reactor, and flow reactions were performed at 30 °C in phosphate buffer (100 mM, pH 7.4) containing 20% of DMSO with 30 minutes residence time (0.1 mg mL−1 flow rate in a 3 mL mixed bed reactor), in the presence of cinnamaldehyde (10 mM), NAD(P)H (1 mM) and glucose (40 mM). Space-time yield (STY, [g L−1 h−1]) was calculated by dividing the amounts of grams produced in batch or in flow by the time required to reach steady-state conversion. Reactions were performed at each enzyme's optimal working temperature: 30 °C for McOYE and HeOYE, and 75 °C for TtENR.
1) in a packed-bed reactor, and flow reactions were performed at 30 °C in phosphate buffer (100 mM, pH 7.4) containing 20% of DMSO with 30 minutes residence time (0.1 mg mL−1 flow rate in a 3 mL mixed bed reactor), in the presence of cinnamaldehyde (10 mM), NAD(P)H (1 mM) and glucose (40 mM). Space-time yield (STY, [g L−1 h−1]) was calculated by dividing the amounts of grams produced in batch or in flow by the time required to reach steady-state conversion. Reactions were performed at each enzyme's optimal working temperature: 30 °C for McOYE and HeOYE, and 75 °C for TtENR.
Analytics
HPLC samples were prepared by adding 50 µL of the reaction mixture to 225 µL of HCl (0.1%) and 225 µL of acetronitrile. The samples were filtered (0.45 µm) and analysed by HPLC (Dionex UltiMate 3000, Waters X-Bridge C18 (3.5 µm, 2.1 Å ∼100 mm)), eluent: A (water + 0.1% trifluoroacetic acid) and B (MeCN + 0.1% trifluoroacetic acid). The sample was run at 0.8 mL min−1 at 45 °C with a gradient from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 A
95 A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B to 95
B to 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 B
5 B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A over 4 minutes. While indole and 2-oxindole could be easily analysed by HPLC, indigo proved to be a very challenging compound to analyse because of its insolubility in almost all organic solvents. Its conversion was therefore determined by simply weighting the precipitate (10–50 mM scale) or dissolving it in DMSO followed by absorbance measurements at 620 nm (0.1–5 mM scales).
A over 4 minutes. While indole and 2-oxindole could be easily analysed by HPLC, indigo proved to be a very challenging compound to analyse because of its insolubility in almost all organic solvents. Its conversion was therefore determined by simply weighting the precipitate (10–50 mM scale) or dissolving it in DMSO followed by absorbance measurements at 620 nm (0.1–5 mM scales).
Results and discussion
Ene-reductases selection, expression and purification
Among the significant diversity of available ERs, we focused on those stemming from extremophilic organisms, or showing homology with thermophilic ERs identified based on three-dimensional constellations of functional groups in active sites by Steinkellner et al.,11 aptly termed “thermophilic like” OYEs or ERs.2 First, the isoform 1 of a thermophilic-like OYE from Mucor circinelloides (McOYE) was selected because of its ability to efficiently reduce classical OYE substrates in whole-cell biotransformations (Table S1, entry 1).22 Then, as previous examples of enzymes extracted from the halophile Halomonas elongata showed excellent expression profiles and good performance in our group,23 we identified an ER gene found in the genome of H. elongata that we named HeOYE (Table S1, entry 2).24 Finally, we selected the two thermophilic ERs from Pyrococcus horikoshii (PhENR) and Thermus thermophilus (TtENR) with completely different fold but similar active sites as OYE because of their extremophilic character and inverted enantiopreference as compared to classical OYEs (Table S1, entries 3 and 4).11 While PhENR and TtENR were previously characterised, McOYE was never purified and HeOYE never expressed nor purified. The four ERs were successfully expressed and purified (Fig. S1A and B). While McOYE, HeOYE and PhENR could be expressed using E. coli BL21, TtENR had to be expressed in E. coli BL21 Lemo21 (Fig. S3). Because it was found in the insoluble fraction when E. coli BL21 was used as expression strain. While good to excellent expression yields were obtained for McOYE and HeOYE, only moderate expression yields were obtained for PhENR and TtENR (Fig. S2C).Substrate scope
Optimal reactions conditions to establish standard activity assays were identified (Fig. S4). While McOYE and TtENR were found to be NADPH-dependent as in previous studies,11,22 with only traces of activity detected with NADH (<10%), HeOYE strongly preferred NADH with no activity detected with NADPH. Surprisingly, even though a small activity was detected in the crude extract of PhENR, no activity was detected using the purified PhENR with either cofactors, nor with any of the tested substrates (Table 1). Activity of TtENR and PhENR was previously reported at pH of 7.11 Our investigations revealed maximum activity at pH 7.4 for both McOYE and HeOYE, in agreement with the growth pHs of their respective organisms (Fig. S4).23,24 As expected, the optimal temperature for McOYE and HeOYE stemming from mesophilic/halophilic organisms was found to be 30 °C, while the thermophilic TtENR exhibited maximum activity at 75 °C. With the purified ERs in our hands, we investigated their respective substrate scope using optimised conditions. While several substrate concentrations were tested, only the ones resulting in the best activity are highlighted in Table 1. Not surprisingly, substrates activated with the most EWGs such as trans-β-nitrotyrosol (Table 1, entry 10) and maleimide (Table 1 entry 7) resulted in higher activity, and HeOYE tends to have higher activity than McOYE, followed by TtENR which is generally less active. Surprisingly, HeOYE showed no activity with trans-β-nitrotyrosol, while only minimal activities were detected with 4-phenylbut-3-yn-2-one.Heterogenous biocatalyst preparation
To design an efficient immobilized system, we analysed the surface properties of McOYE, TtENR, and HeOYE using CapiPy25 to guide the choice of immobilisation chemistry. We focused on the lysine coverage (percentage of the surface occupied by lysine residues) as well as the position of the His-tag, which is in all cases located at the N-terminal (Fig. S5). McOYE (9% lysine coverage, Fig. S5A) contains two peripheral lysine clusters, as well as three negative clusters (Fig. S5). TtENR (7% lysine coverage) has lysines distributed across the surface, with the His-tag positioned far from both active sites in this dimeric enzyme (Fig. S5B). HeOYE has only four surface lysines (1%), one near the active site, limiting covalent attachment options and risking catalytic site obstruction and the His-tag is located at the N-terminus, and is positioned close the active site (Fig. S5B).Given the presence of the His-tag and favourable positioning of it, especially in McOYE and TtENR, all enzymes were immobilised on epoxy-metal support. This enables a first rapid His-tag coordination to cobalt followed by a slower covalent bond of the surface lysines in the proximity to the epoxy moiety of the support. Table 2 presents the results of the selected optimal immobilisation for each enzyme. TtENR and McOYE achieved >99% immobilization yield at 1 mg g−1 loading. TtENR showed full activity retention with EP400/SS, likely due to pre-orientation that left both active sites accessible (immobilisation screening in Table S5). McOYE showed 75% retained activity with HFA403/S, a long arm amino-epoxy methacrylate resin, which will position the protein guided by the His-tag as well as the previously mentioned negative clusters specially those located down from the active site and in close proximity to the N-terminal (Fig. S6b, immobilisation screening Table S4).
| Enzyme | Support | Loading (mg g−1) | IY | RA | U per g |
|---|---|---|---|---|---|
| TtENR | EP400/SS | 1 | >99% | 100% | 2.6 ± 0.1 |
| McOYE | HFA403/S | 1 | >99% | 75% | 6.6 ± 0.2 |
| HeOYE | 6BCL | 1 | 60% | 23% | 5.5 ± 0.3 |
For the immobilisation of HeOYE, on the other hand, both aldehyde and epoxy-metal supports were tested (Table S6). Low immobilisation yields were obtained with high loading (5 mg g−1), possibly due to the presence of only 3 exposed lysines. Lowering the loading to 1 mg g−1, allowed for yields over 60% with all different resins, but the immobilised enzyme was less active, achieving our maximum with agarose support and a 23% recovered activity. Noteworthy, silica materials showed a very good immobilisation yield and recovered activity, but the reusability of the catalyst was compromised because of abrasion of the material (Table S6, entries 8 and 17).
These results confirm that dual-mode affinity-covalent immobilisation efficiency depends strongly on surface architecture, with optimal His-tag placement and distal lysines being key to high yield and activity preservation.
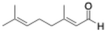
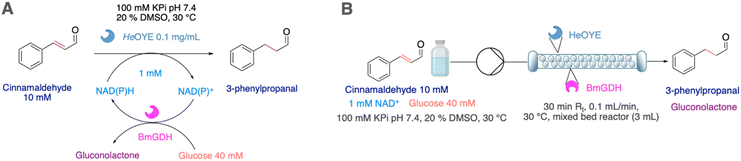
Flow biocatalytic synthesis of 3-propionaldehyde
To test the efficiency of ER-mediated biocatalysis in flow, we chose the readily available and easy to monitor cinnamaldehyde, whose reduced product 3-phenylpropionaldehyde (or hydrocinnamal-dehyde) is used in perfumes, as flavouring agent, as building block in organic synthesis, in agrochemicals or for pharmaceuticals,26 and has even shown interesting antibacterial properties.27 Initial screenings of batch reactions using the free and immobilised enzymes at a 1 mM scale, using NAD(P)H (2 mM) without cofactor recycling system revealed that excellent conversions were reached in all cases (89–100%) although with longer time for TtENR and for the immobilised McOYE (Table 3 entries 1–6 and Fig. S7).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 unit of BmGHD ratio and glucose (40 mM) as sacrificial substrate. For flow biotransformations (B), a residence time of 30 minutes was used. Reactions were performed at each enzyme's optimal working temperature: 30 °C for McOYE and HeOYE and 75 °C for TtENR. Conversions were determined by HPLC analysis based on product formation
1 unit of BmGHD ratio and glucose (40 mM) as sacrificial substrate. For flow biotransformations (B), a residence time of 30 minutes was used. Reactions were performed at each enzyme's optimal working temperature: 30 °C for McOYE and HeOYE and 75 °C for TtENR. Conversions were determined by HPLC analysis based on product formation
| Entry | Type | Enzyme | Scale [mM] | Recycling system | Rxn time [h] | Conversion [%] | Space-time-yield [mg L−1 h−1] |
|---|---|---|---|---|---|---|---|
| 1 | Batch | Free TtENR | 1 | — | 18 | 100 | 7.5 |
| 2 | Immobilised TtENR | 24 | 100 | 5.6 | |||
| 3 | Free McOYE | 2 | 89 | 59.7 | |||
| 4 | Immobilised McOYE | 18 | 89 | 6.6 | |||
| 5 | Free HeOYE | 2 | 98 | 65.8 | |||
| 6 | Immobilised HeOYE | 2 | 97 | 65.1 | |||
| 7 | Free HeOYE | 10 | BmGDH | 24 | 62 | 34.7 | |
| 8 | Immobilised HeOYE | 24 | 62 | 34.7 | |||
| 9 | Flow | Immobilised HeOYE | 0.5 | 81 | 2173.9 | ||
HeOYE was further investigated in flow biocatalytic transformations in a mixed-bed reactor co-housing immobilised glucose dehydrogenase from Bacillus megaterium (BmGDH) using a 2 units of HeOYE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 unit of BmGHD ratio, glucose (40 mM) and 0.1 equivalents of NAD(P)H. While batch biotransformations with the free and immobilised enzymes resulted in 60% conversion after 24 hours (Table 3 entries 7 and 8), 80% conversion was reached in flow and the biocatalyst was stable over 6 cycles with residence times of 30 minutes (Table 3 entry 8, Fig. S8). The space time yield (STY) for the production of 3-phenylpropionaldehyde could thus be increased from 34.7 [mg L−1 h−1] in batch to 2173.9 [mg L−1 h−1] in flow, which represents a 62-fold process intensification.
1 unit of BmGHD ratio, glucose (40 mM) and 0.1 equivalents of NAD(P)H. While batch biotransformations with the free and immobilised enzymes resulted in 60% conversion after 24 hours (Table 3 entries 7 and 8), 80% conversion was reached in flow and the biocatalyst was stable over 6 cycles with residence times of 30 minutes (Table 3 entry 8, Fig. S8). The space time yield (STY) for the production of 3-phenylpropionaldehyde could thus be increased from 34.7 [mg L−1 h−1] in batch to 2173.9 [mg L−1 h−1] in flow, which represents a 62-fold process intensification.
Sustainable indigo dyeing
Among substrates containing an α,β-unsaturated motif activated in Cα by EWGs, indigo is certainly one of the most widely used worldwide for textile dyeing. To intercalate into the fibres, this insoluble pigment needs to be reduced to the water soluble alkaline leuco-indigo that will subsequently be air oxidised back to the everlasting indigo.17,18 Typical chemical reducing agents used in this process such as sodium dithionite result in environmentally hazardous sulfate by-products that can also corrode pipes. Promising microbial route using recombinant whole-cell biocatalysts28,29 or electrochemical methods reduction processes30 have emerged more recently, and biocatalysis in particular could offer a greener and less energy-intensive alternative.31,32 The combination of an unspecific peroxygenase with reductases is a promising route. In fact, compared to other enzymes such as P450 monooxygenases that can perform similar functions, UPOs do not require expensive electron donors and use H2O2 as co-substrate and oxidant agent, which make them particularly appealing for the synthesis of valuable industrial products (Fig. 2).33UPO-mediated synthesis of indigo and 2-oxindole from indole
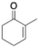
Following the method of Ullrich et al.33 with used the unspecific fungal oxidase from Agrocybe aegerita (rAeUPO, PaDa-I variant) available in house. The reaction could be followed by TLC as well as visually, with the solution rapidly turning blue to amber-greenish with a blue precipitate appearing after 30 minutes (Fig. S9A). We performed the reaction at different scales, however, indigo conversions were low (<5%) (Table S7). Almost complete indole depletion was observed in all cases (Fig. S9C and D), and while the presence of indigo was confirmed with standard samples (Fig. S9B), the main product was identified to be 2-oxindole (64–85% conversion, Table S7). In an attempt to increase indigo's yield, we screened different H2O2 equivalents, pHs (4.4 and 7.4) as well as an alternative oxidant (Table S8), but none of the tested conditions resulted in any improvement of indigo conversion. Ullrich et al.33 reported that different UPOs showed quite different preference regarding the formation of 2-oxindole or indoxyl (3-oxindole), suggesting a direct influence of the UPOs active site on the 2-3-epoxide opening. However, 2-oxindole itself has been reported to be a valuable precursor for many pharmaceutically-relevant compounds such as nintedanib, ziprasidone or ropinirole.34ER-mediated reduction of indigo into leuco-indigo
Reduction of indigo into leuco-indigo was previously reported with azo-reductases (AZs)31,35,36 but never using ERs. ERs sequence alignments with these enzymes resulted in low dope scores (0.006–0.03), <15% identity, and different residues composing their respective active sites, thereby indicating that AZRs and ERs are fundamentally different enzymes. While no activity was detected in the control in the absence of indigo and indigo carmine, low (0.1–0.3 U per mg) and moderate (0.7–2.5 U per mg) activities were detected using McOYE, HeOYE and TtENR with indigo and indigo-carmine, respectively (Table S9). Even though these activities are lower than the ones reported using AZRs (30–66 U per mg), the latter mainly work at basic pH whereas the reduction with ERs is performed at neutral pH.To compare the efficacy of ERs with conventional indigo dyeing processes, in the absence of better analytical methods, we dyed cotton cloth chemically at pH 12 using sodium dithionite (Na2S2O4, 40 mM) or glucose (40 mM) as reducing agents, since both were previously reported to reduce indigo.37 Using indigo, only the chemical reduction at basic pH resulted in dyeing (Fig. S10A and Table S10 entries 1–6), most probably because it could not be solubilised enough to reach the active site of ERs. When the more soluble indigo carmine was used instead, reduction into the soluble yellow form was observed in all cases after 2 hours (Fig. S10B), but in the case of chemical dyeing, this colouration was show to be due to dye degradation (Table S10 entries 7–9), whereas the system stayed yellow with the ER-mediated reactions, even after 1 week (Fig. S10C and Table S10 entries 10–12). The three enzymes (TtENR, McOYE and HeOYE) were indeed able to efficiently dye the cotton pieces at neutral pH, while chemical methods failed (Fig. 3). While indigo dyeing itself would require optimisation of the solubility conditions using for instance a biphasic system, this process could be very promising for textile dyeing using indigo derivatives with many colour shades that could be synthetised using indole derivatives (Fig. S11).33
Conclusions
In conclusion, in this work, we expanded the repertoire of enzymes available for the synthesis of chiral molecules by successfully expressing and purifying a novel ene-reductase from the halophilic organism Halomonas elonga. Besides being expressed with excellent yields and exhibiting moderate activity towards almost all of the substrates, HeOYE was shown to perform well in flow, with a 62-fold process intensification compared to similar batch synthesis of 2-phenylpropionaldehyde from cinnamaldehyde and an increase of conversion from 62 to 81%, with the biocatalyst being stable over 3 hours. These first step into ER-mediated flow biocatalysis could pave the way to further studies at higher scale or to their integration in flow biocatalytic cascades. Moreover, while attempts of reducing indigo-carmine into leuco-indigo carmine using chemical methods resulted in dye degradation, three of the four extremophilic/extremophilic-like ERs proved to efficiently perform that reaction at neutral pH. This is the first example of the ER-mediated dyeing. By being able to produce small quantities of indigo from indole with rAaeUPO and co-producing the valuable 2-oxindole precursor of many pharmaceutically relevant molecules, we provide a proof-of-concept of a sustainable enzymatic cascade's feasibility for indigo derivates dyeing at neutral pH. Many possibilities remain open for further enzyme engineering, optimisation/scale-up of the process or ways to assemble the cascade, such as the co-immobilisation of the three enzymes (UPOs, ERs and GDH) for an in situ one pot dyeing process in batch, or even in flow. Our strategy could even be applied to other dyes with different colors starting from functionalised indole. Overall, both flow biocatalytic and dyeing approaches using HeOYE are innovative, safe, green and versatile strategies for the reduction of carbon–carbon double bonds, which could pave the way to the effective implementation of biocatalysis at the industrial scale, while alleviating the ecological imbalance of conventional (electro)chemical reductions.Author contributions
F. P. supervised and guided the project, M. B. performed initial selection of ERs, InSEIT rationally immobilised all enzymes, L. P. conceptualised the idea, performed most of the experimental work and wrote the initial draft. All authors discussed and agreed to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra06869j.Acknowledgements
This project was supported by the SNSF (200021_192274, as well as 200021_232199 F.P.)Notes and references
- G. Brown, T. S. Moody, M. Smyth, S. J. C. Taylor, in Biocatalysis: an Industrial Perspective, The Royal Society Of Chemistry, 2018, 29, pp. 229–256 Search PubMed.
- H. S. Toogood, J. M. Gardiner and N. S. Scrutton, ChemCatChem, 2010, 2, 892–914 CrossRef CAS.
- T. Kumar Roy, R. Sreedharan, P. Ghosh, T. Gandhi and D. Maiti, Chem. Eur. J., 2022, 28(21), e202103949 CrossRef CAS PubMed.
- F. Hollmann, D. J. Opperman and C. E. Paul, Angew. Chem., Int. Ed., 2021, 60(11), 5644–5665 CrossRef CAS PubMed.
- M. Hall, RSC. Chem. Biol., 2021, 2(4), 958–989 RSC.
- D. Karrer, M. Gand and M. Rühl, ChemCatChem, 2021, 13(7), 2191–2199 CrossRef CAS.
- X. Huang, B. Wang, Y. Wang, G. Jiang, J. Feng and H. Zhao, Nature, 2020, 584, 69–74 CrossRef CAS PubMed.
- C. Peters and R. Buller, Z. Naturforsch., C:J. Biosci., 2019, 74(3–4), 63–70 CrossRef CAS PubMed.
- S. Debarge, P. McDaid, P. O'Neill, J. Frahill, J. W. Wong, D. Carr, A. Burrell, S. Davies, M. Karmilowicz and J. Steflik, Org. Process. Res. Dev., 2014, 18(1), 109–121 CrossRef CAS.
- E. Eichhorn, C. Baumgartner and M. Biermann, Chimia, 2023, 77, 384–389 CrossRef CAS PubMed.
- G. Steinkellner, C. C. Gruber, T. Pavkov-Keller, A. Binter, K. Steiner, C. Winkler, A. Łyskowski, O. Schwamberger, M. Oberer, H. Schwab, K. Faber, P. MacHeroux and K. Gruber, Nat. Commun., 2014, 5, 4150 CrossRef CAS PubMed.
- J. Feng, Y. Xue, J. Wang, X. Xie, C. Lu, H. Chen, Y. Lu, L. Zhu, D. Chu and X. Chen, Process Biochem., 2023, 126, 108–116 CrossRef CAS.
- N. Nett, S. Duewel, L. Schmermund, G. E. Benary, K. Ranaghan, A. Mulholland, D. J. Opperman and S. Hoebenreich, Mol. Catal., 2021, 502, 111404 CAS.
- H. Li, W. Xiao, P. Xie and L. Zheng, Enzyme Microb. Technol., 2018, 109, 66–73 CrossRef CAS PubMed.
- R. Villa, C. Ferrer-Carbonell and C. E. Paul, Catal. Sci. Technol., 2023, 13, 5530–5535 RSC.
- F. Nagy, I. Gyujto, G. Tasnádi, B. Barna, D. Balogh-Weiser, K. Faber, L. Poppe and M. Hall, J. Biotechnol., 2020, 323, 246–253 CrossRef CAS PubMed.
- K. Y. Choi, Dyes and Pigm., 2020, 181, 108570 CrossRef CAS.
- C. Y. Huang and S. Hecht, Chem. Eur. J., 2023, 29(43), e202300981 CrossRef CAS PubMed.
- A. Benítez-Mateos, D. Lim, D. Roura Padrosa, V. Marchini, H. Wu, F. Buono and F. Paradisi, ACS Sustainable Chem. Eng., 2025, 13(13), 5009–5018 CrossRef.
- Expasy, “ProtParam”, can be found under https://web.expasy.org/protparam/, accessed 8 August 2025.
- C. Mateo, J. M. Palomo, M. Fuentes, L. Betancor, V. Grazu, F. López-Gallego, B. Pessela, A. Hidalgo, G. Fernández-Lorente, R. Fernández-Lafuente and J. Guisan, Enzyme Microb. Technol., 2006, 39(2), 274–280 CrossRef CAS.
- A. Romagnolo, F. Spina, A. Poli, S. Risso, B. Serito, M. Crotti, D. Monti, E. Brenna, L. Lanfranco and G. C. Varese, Sci. Rep., 2017, 7, 12093 CrossRef PubMed.
- A. I. Benítez-Mateos and F. Paradisi, Appl. Microbiol. Biotechnol., 2023, 107, 3183–3190 CrossRef PubMed.
- J. I. Mamani, K. B. Pacheco, P. Elorrieta, P. Romoacca, H. Castelan, S. Davila, J. L. Sierra and M. A. Quispe-Ricalde, Microbiol. Resour. Announc., 2019, 8(1), e00934–18 Search PubMed.
- D. Roura Padrosa, V. Marchini and F. Paradisi, Bioinformatics, 2021, 37(17), 2761–2762 CrossRef PubMed.
- K. N. Patil, P. Manikanta, P. M. Srinivasappa, A. H. Jadhav and B. M. Nagaraja, J. Environ. Chem. Eng., 2023, 11(1), 109168 CrossRef CAS.
- A. A. Doyle and J. C. Stephens, Fitoterapia, 2019, 139, 104405 CrossRef CAS PubMed.
- H. Yin, H. Chen, M. Yan, Z. Li, R. Yang, Y. Li, Y. Wang, J. Guan, H. Mao, Y. Wang and Y. Zhang, ACS Omega, 2021, 6(31), 20569–20576 CrossRef CAS PubMed.
- L. Du, J. Yue, Y. Zhu and S. Yin, Foods, 2022, 11(14), 2117 CrossRef CAS PubMed.
- C. Yi, X. Tan, B. Bie, H. Ma and H. Yi, Sci. Rep., 2020, 10, 4927 CrossRef CAS PubMed.
- H. Suzuki, T. Abe, K. Doi and T. Ohshima, Appl. Microbiol. Biotechnol., 2018, 102(21), 9171–9181 CrossRef CAS PubMed.
- M. P. Haaf, K. M. Piemonte, D. T. McQuade, L. Cotton and R. S. Blackburn, Color. Technol., 2019, 135(2), 127–132 CAS.
- R. Ullrich, M. Poraj-Kobielska, O. M. Herold-Majumdar, J. Vind and M. Hofrichter, Catalysts, 2021, 11(12), 1495 CrossRef CAS.
- S. Sharma, Y. Monga, A. Gupta and S. Singh, RSC Adv., 2023, 13(21), 14249–14267 RSC.
- K. Yoneda, H. Sakuraba, T. Araki and T. Ohshima, FEBS. Open Bio., 2021, 11(7), 1981–1986 CrossRef CAS PubMed.
- K. Yoneda, M. Yoshioka, H. Sakuraba, T. Araki and T. Ohshima, Int. J. Biol. Macromol., 2020, 164, 3259–3267 CrossRef CAS PubMed.
- L. Saikhao, J. Setthayanond, T. Karpkird, P. Suwanruji, MATEC Web Conf., 2017, vol. 108, p. 03001 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |