Open Access Article
Open Access ArticleDifferentiation properties of 3D scaffolds nanostructured with multi-walled carbon nanotubes on human induced pluripotent stem cells
Federica Cavion†
 a,
Michele Cacioppo†
a,
Michele Cacioppo† bc,
Susanna Bosi
bc,
Susanna Bosi ac,
Michela Carlin
ac,
Michela Carlin a,
Silvio Sosa
a,
Silvio Sosa a,
Maurizio Prato
a,
Maurizio Prato cde and
Marco Pelin
cde and
Marco Pelin *a
*a
aDepartment of Life Sciences, University of Trieste, 34127, Trieste, Italy. E-mail: mpelin@units.it
bDepartment of Biological, Chemical and Pharmaceutical Sciences and Technologies (STEBICEF), University of Palermo, 90128 Palermo, Italy
cDepartment of Chemical and Pharmaceutical Sciences, University of Trieste, 34127, Trieste, Italy
dCenter for Cooperative Research in Biomaterials (CIC biomaGUNE), Basque Research and Technology Alliance (BRTA), 20014, Donostia-San Sebastian, Spain
eBasque Foundation for Science (IKERBASQUE), 48009, Bilbao, Spain
First published on 19th November 2025
Abstract
Implementation of stem cell therapy using novel nanotechnologies is a fruitful approach for regenerative medicine. Induced pluripotent stem cells (iPSC) are considered a gold standard for stem cells research for personalized regenerative medicine, especially for tissues with low regenerative capabilities. To overcome some limitations of existing systems, a novel microporous, self-standing, elastomeric 3D scaffold of polydimethylsiloxane with micrometric cavities of tunable sizes, nanostructured with multi-walled carbon nanotubes (MWCNTs) was developed to investigate its biocompatibility and differentiation potential towards iPSC. Four types of 3D MWCNTs scaffolds were selected to study the role of scaffolds pore size (small: 100–250 µm; large: 250–600 µm) and the level of MWCNTs nanostructuration (3% w/w and 6% w/w) in their effects on iPSC. All scaffolds appeared highly biocompatible with iPSC for up to 7 days, but 3D MWCNTs scaffolds with large pore size (250–600 µm) allowed the most adequate environment for cell growth, increasing cell mass in absence of proliferation stimuli. Only at this porosity, regardless of MWCNTs amount, the iPSC gene expression profile was characterized by a distinct pattern, compatible with a reduced pluripotency and a mesoderm-like differentiation. These results might support possible application of these scaffolds in regenerative medicine, opening new scenarios for stem cell-based approaches.
Introduction
In the last few years, stem cell therapy has become a promising alternative approach for the treatment of various human diseases.1 Due to their innate potential to differentiate into multiple types of cells able to replace damaged parts of human organs and tissues, stem cells can be regarded as the last chance for the therapy of various degenerative diseases and severe tissue damage.2–9 The big limitation in the use of embryonic stem cells (ESC) posed by ethical issues (embryos destruction) has been overcome by induced pluripotent stem cells (iPSC). In general, iPSC have the multiple advantage to: (i) overcome ESC ethical issues; (ii) be pluripotent cells without limited differentiation potential, such as mesenchymal stem cells (MSC); (iii) avoid tissue rejection, observed in the case of heterologous transplants; (iv) bypass problems associated with limited donor sites, being easily expandable in vitro; (v) be characterized by all the donor's genetic inheritance, thus representing a good tool to model any donor- and/or disease-specific different responses, creating novel scenarios for personalized regenerative medicine.10–14In this scenario, the implementation of stem cell therapy by the application of novel nanomaterials and nanotechnologies is a fruitful approach in the field of regenerative medicine. In this view, different nanomaterials (from carbon-based nanomaterials to metal nanoparticles, passing through nanostructured hydrogels and polymers) have been proposed as useful tools in several kinds of therapies.15–18 Among them, carbon nanotubes (CNTs), and in particular multi-walled CNTs (MWCNTs), are certainly some of the most promising nanomaterials. They are constituted by sp2 hybridized elemental carbons forming needle-like tubes, each of them consisting in coaxial cylinders characterized by high length-to-diameter ratio, so that they can be considered as one-dimensional nanostructures.19 CNTs possess unique electrical, optical, thermal and mechanical properties that can be exploited for tissue regeneration purposes.20 Furthermore, their sizes and morphology resemble those of many extracellular matrix (ECM) proteins, such as collagen and laminin, favoring cell adhesion and growth. Intriguingly, the ECM is one of the most important factors affecting stem cell fate, being able to interact with stem cells and providing mechanical strength and biological signals.21,22 Besides their excellent physico-chemical properties, of particular interest for regenerative medicine is the use of MWCNTs as biomimetic substrates to control the differentiation of stem cells. Indeed, several studies have already shown the biocompatibility of MWCNTs with several kinds of stem cells.22,23 However, the majority of these studies were carried out on MSCs. These are multipotent adult stem cells with a limited differentiation capability, mainly towards osteoblasts, chondrocytes and adipocytes.24 In general, the results demonstrated not only a good biocompatibility of MWCNTs with MSC,25–27 but also an increased ability of MSCs differentiation towards neurons28–30 and osteoblasts.31–33 However, unveiling the effects of MWCNTs on pluripotent stem cells, characterized by a complete differentiation ability towards all the three germinative layers (i.e. endoderm, ectoderm and mesoderm), could pave the way to implement their use in regenerative medicine. At best of our knowledge, only few studies on iPSCs of human origin are currently available, demonstrating an acceptable MWCNTs biocompatibility with a tendency of differentiation towards mesoderm cells.23,34
Another important aspect in nanotechnology-based stem cell therapy for tissue regeneration is represented by tridimensional (3D) topography systems, which could mimic an optimal ECM environment for cell adhesion and growth. Topography is a crucial factor driving cellular interaction, biocompatibility and differentiation properties. For instance, micro, nano, and combined micro–nano structures may trigger different stimulatory effects on osteoblasts-like cells.35 In general, embedment of MWCNTs into 3D scaffolds displays a series of fruitful implementations, including: (i) increased scaffold strength and flexibility; (ii) improved biocompatibility; (iii) control of cell division and proliferation; (iv) manipulation of gene expression.36 Several nanostructured scaffolds have been developed to allow cell growth in a 3D fashion. However, some of these systems lack in tissue-like features, such as porosity or elastic properties, preventing their translation into in vivo settings. Hydrogel based materials or 3D electrospun polymers have been also used to obtain more realistic tissue constructs. However, many of these systems are prone to degradation.37 To overcome these limitations, a novel microporous, self-standing, elastomeric 3D scaffold of polydimethylsiloxane (PDMS) with micrometric cavities of tunable sizes nanostructured with MWCNTs has been developed. The use of PDMS as inert polymer for generating implantable scaffolds is traditionally accepted.38 In addition, this novel nanostructured scaffold already showed high in vitro and in vivo biocompatibility with neurons,39 boosting their synaptic activity.37 Intriguingly, the same 3D construct showed an excellent biocompatibility also with neonatal rat ventricular myocytes, promoting their viability, growth, proliferation and functional maturation.40
Hence, on the basis of these observations, this study was carried out to investigate the biocompatibility of this novel 3D MWCNTs-nanostructured PDMS scaffold (3D MWCNTs scaffolds) with iPSC. A particular focus was posed on their differentiation potential, exploring a possible application in regenerative medicine. In particular, four types of 3D MWCNTs scaffolds were tested with the aim to study the role of scaffolds pore size and the degree of nano-structuration with MWCNTs in the effects on iPSC.
Results
3D MWCNT-nanostructured PDMS scaffolds synthesis and characterization
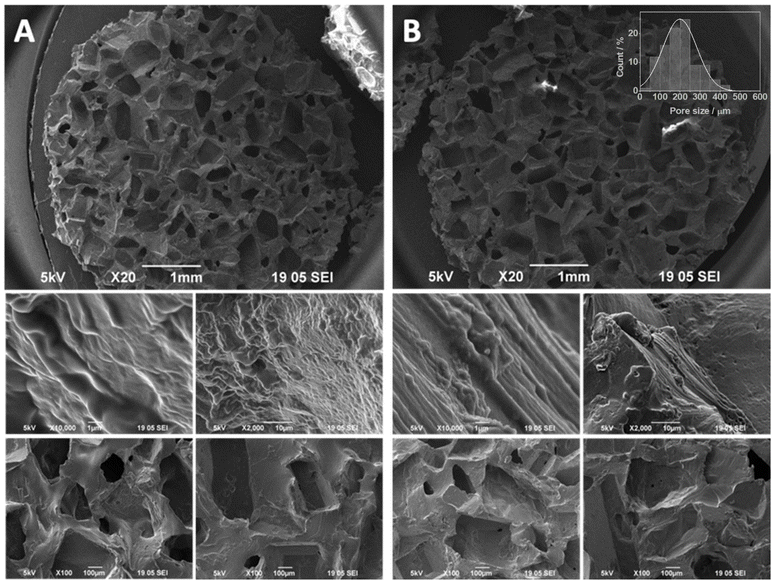
The 3D MWCNTs scaffolds were produced by exploiting the curing of an organic polymer (PDMS), in presence of MWCNTs and made porous through the use of sugar with known grain sizes (100–250 µm and 250–600 µm). These materials were previously synthesized by our group to investigate the possible applications of CNT-based scaffolds as a support in neuronal regeneration.41 The synthesis of these materials was performed starting from the covalent functionalization of MWCNTs by the Tour reaction.41 The mechanism of this reaction involves the formation of diazonium salts through the Sandmayers reaction starting from aromatic amines, in this case aniline, with consequent formation of the C–C bond between the CNT and the aromatic ring (Fig. S1). This particular functionalization has been reported to be very advantageous for an optimal interaction with PDMS and for a favorable cell growth environment formation.42 Indeed, the functionalization of MWCNTs was carried out by introducing a benzylamino group on pristine CNTs surface through a two-step synthesis (Methods; Fig. S1). The first step consisted of reacting the primary amine of 4-[(N-Boc)aminomethyl]aniline by a Tour reaction, followed by the removal of the Boc group by an acid treatment. Preliminary Kaiser Test, with a positive result, confirmed the successful introduction of the amino group in product 3 resulting in good agreement with previously obtained results (Fig. S2).37,43 Subsequently, primary amines on product 3 were quantified by calculating the functional group weight loss trough thermogravimetric analysis (TGA).44 In good agreement with the Kaiser Test and previously published data, the TGA of product 3 lead to determine the degree of the material functionalization (Fig. S3), quantified at ca. 331 µmol g−1, corresponding to 3.5% weight percentage of the functional group. MWCNTs employed in this work have been previously characterized by Raman and XPS spectroscopy by our group, confirming the reproducibility of the Tour-based covalent functionalization.45,46 In those studies, pristine MWCNTs showed the typical Raman D/G ratio of graphitic materials, while functionalized MWCNTs exhibited an increased D/G ratio and the appearance of an N 1s XPS signal, consistent with the presence of surface amine groups. The XPS elemental composition measured for the functionalized MWCNTs used in the present study (C = 95.44%, O = 3.44%, N = 1.12%) further supports the effective and reproducible surface functionalization of the nanotubes.Amino functionalized MWCNTs (3) were then used for scaffolds production through a solidification reaction of the PDMS polymer using granular sugar of specific size to generate pores in the final product (size ranges: 100–250 µm and 250–600 µm). After a thermal treatment, followed by repeated washing in water to completely remove the sugar, porous solids with a white (Fig. S4, in the case of PDMS only) or black (Fig. S4, in the case of PDMS in the presence of product 3) color were obtained. Scanning electron microscopy (SEM) images of the scaffolds showed the typical porosity after the sugar grains removal both for control 3D PDMS scaffolds not nanostructured with MWCNTs (Fig. S5 and S6) and 3D PDMS scaffolds nanostructured with 3% or 6% MWCNTs (Fig. 1 and S7). The slightly irregular and polygonal shape of the pores is attributed to the faceted morphology of the sugar crystals used as porogens. During the curing step, the sugar grains maintain their crystalline geometry, which is subsequently replicated in the PDMS matrix after dissolution. This feature is intrinsic to the templating process and is not influenced by the presence of MWCNTs. On the whole, four types of 3D MWCNT scaffolds, characterized by two porosities (100–250 µm or 250–600 µm) and two quantities of MWCNTs (3% and 6% w/w MWCNTs) were prepared. To quantitatively validate the scaffold porosity, the pore size distribution was analyzed from SEM images using ImageJ (n = 100 pores per scaffold). The resulting data confirmed the two expected porosity ranges (100–250 µm and 250–600 µm), with mean pore diameters of 87 ± 32 µm and 199 ± 85 µm, respectively (see Fig. 1 and S7). These values are in excellent agreement with the designed porosity ranges determined by the sugar sieving procedure. The mechanical response of the 3D PDMS/Ba-MWCNT scaffolds was assessed by uniaxial compression tests performed at room temperature. The stress–strain exhibited the characteristic nonlinear elastic profile of PDMS-based materials, with progressively higher slopes upon increasing MWCNTs content. The Young's modulus, evaluated within the 7–12% strain interval, increased from 230 ± 120 kPa (pure PDMS, 100–250 µm pores) to 896 ± 440 kPa (6% w/w MWCNTs, 100–250 µm pores), confirming that the incorporation of nanotubes effectively reinforces the polymeric matrix without compromising its elasticity (Fig. S8).
3D MWCNT-nanostructured PDMS scaffolds biocompatibility with iPSC
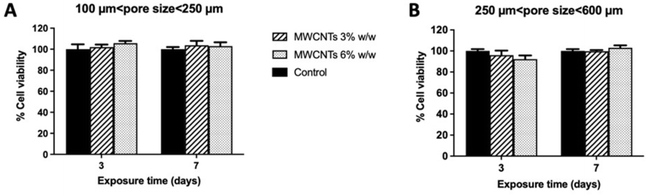
Biocompatibility of the four types of 3D MWCNT scaffolds was evaluated in terms of iPSC viability (CCK-8 assay, Fig. 2) and cell mass (SRB assay, Fig. 3) after 3 and 7 days exposure from cell seeding. As compared to control scaffolds not nanostructured with MWCNTs, none of the scaffolds nanostructured with MWCNTs significantly influenced cell viability, that was maintained at about 100% both after 3 and 7 days exposure. Thus, pore size and percentage of MWCNTs nanostructuration of the 3D MWCNT scaffolds did not influence cell viability.Biocompatibility of iPSC was also evaluated in terms of cell mass by the SRB assay, able to quantify proteins of adhered cells, after 3 and 7 days exposure to each 3D nanostructured scaffold (Fig. 3). Results shown in Fig. 3A demonstrate that the 3D MWCNTs scaffolds characterized by the smaller pore size (100–250 µm) did not significantly influence cell mass, regardless the amount of MWCNTs nano-structuration.
Only a modest and not significant reduction of cell mass was recorded after 7 days exposure. On the other hand, both 3D MWCNTs scaffolds with the larger pore size (250–600 µm) increased cell mass with an effect dependent on the amount of MWCNTs (Fig. 3B). In particular, after 3 days exposure, a non-significant trend to increase cell mass was observed, in particular for the 3D scaffold nanostructured with 6% MWCNTs. More interestingly, after 7 days exposure, both 3D MWCNTs scaffolds significantly increased cell mass with respect to control scaffolds not nanostructured with MWCNTs, the scaffold nanostructured with 6% MWCNT exerting the highest effect (182% cell mass increase; p < 0.01) as compared to that nanostructured with 3% MWCNT (142% cell mass increase; p < 0.01).
Effects of the 3D MWCNT-nanostructured PDMS scaffolds on iPSC proliferation
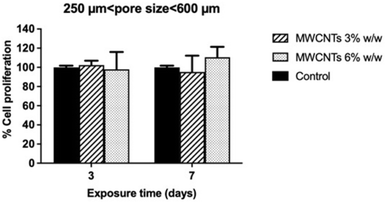
To verify whether the increase of cell mass after 7 days iPSC culture on 3D MWCNTs scaffolds characterized by the larger pore size range (250–600 µm) could be ascribed to an increased cell number consequent to a proliferative stimulus, cell proliferation was evaluated by BrdU incorporation (Fig. 4). No significant alterations in cell proliferation were observed for iPSC exposed to 3D scaffolds nanostructured with 3% or 6% MWCNTs for 3 or 7 days, as compared to those cultured on control scaffolds not nanostructured with MWCNTs. Furthermore, to confirm this result, the number of cells present in each scaffold was evaluated after the longest exposure time (7 days; Fig. S9). After 7 days culture, the number of cells adhered to each nanostructured scaffold (3% or 6% MWCNTs) was not significantly increased, being respectively 83.8 ± 8.8% and 88.0 ± 9.5% as compared to that of cells cultured on control scaffolds not nanostructured with MWCNTs. This result confirms the absence of a proliferative stimulus at the basis of cell mass increase.Effects of 3D MWCNT-nanostructured PDMS scaffolds on iPSC differentiation
On the basis of the biocompatibility results, 7 days culture of iPSC on 3D MWCNTs scaffolds with the larger pore size range (250–600 µm) was selected as the best exposure condition to assess whether these scaffolds could influence pluripotency and/or induce a selective iPSC differentiation. By real time qPCR, expression analysis on a panel of 13 genes was carried out: 5 genes markers of pluripotency (C-MYC, OCT4, NANOG, KLF4 and SOX2), 3 genes markers of endoderm embryonic layer (FOXA2, SOX17, AFP), 3 genes related to mesoderm (ACTA, BRACHYURY, CXCR4) and 2 genes related to ectoderm (PAX6, SOX1). Gene expression results were expressed as 2−ΔΔCt, with reference to iPSC maintained under standard culture conditions, not exposed to the scaffolds. Results reported in Fig. 5 (panel A) shows that control scaffolds not nanostructured with MWCNTs (control) did not significantly influence any gene expression, while the 3D MWCNTs scaffolds led to significant variations of gene expression.In particular, scaffolds nanostructured with 3% w/w MWCNTs induced a significant change in all 5 stemness marker genes, with a down-regulation of OCT4 gene expression (fold change = 0.66; p < 0.01), NANOG (fold change = 0.35; p < 0.001), KLF4 (fold change = 0.65; p < 0.01) and SOX2 (fold change = 0.60; p < 0.01), and a slight up-regulation of the C-MYC gene (fold change = 1.6; p < 0.01). An increased amount of MWCNTs (6% w/w) in the scaffolds induced only a variation in the expression of two genes markers of stemness (OCT4 and NANOG, down-regulated by 0.80-fold (p < 0.05) and 0.40-fold (p < 0.001), respectively). Overall, these results suggest a loss of stemness and/or pluripotency following 7 days contact with both the 3D MWCNTs scaffolds.
The ability of the 3D MWCNTs scaffolds to trigger differentiation of the stem cells was assessed analysing the expression of gene markers of the three embryonic layers, in comparison to iPSCs cultured in standard conditions not exposed to the scaffolds (Fig. 5, panel B). As positive controls, iPSC were differentiated towards endoderm, mesoderm or ectoderm using a specific kit (Fig. S10). Scaffolds not nanostructured with MWCNTs (control) induced a significant up-regulation of only three genes, including SOX1 (fold change = 3.7; p < 0.05), AFP (fold change = 5.6; p < 0.05), and BRACHYURY (fold change = 13.1; p < 0.05). On the contrary, 3D MWCNTs scaffolds increased also the expression of other genes. Indeed, the scaffolds nanostructured with 3% MWCNTs significantly increased the expression of the majority of analysed genes as compared to iPSCs cultured under standard conditions and not exposed to the scaffolds. In particular, a mild but significant increase of the ectoderm gene marker SOX1 (fold change = 10.6; p < 0.05) was recorded, at a level comparable to the ectoderm positive control (Fig. S10), while PAX6 expression, that was highly increased in the positive control, was unaffected by the presence of scaffolds nanostructured with 3% MWCNTs. A higher expression of endoderm gene markers was observed in iPSC cultured with scaffolds nanostructured with 3% MWCNTs, with significantly increased expression of SOX17 (fold change = 46.7; p < 0.01) and AFP (fold change = 23.1; p < 0.01), respectively at levels far lower or comparable to those induced by the endoderm positive control (Fig. S10). The major effect was observed for mesoderm gene markers, in particular for BRACHYURY that was increased even by 756.7 folds (p < 0.001), beside a mild up-regulation of CXCR4 (fold change = 3.8; p < 0.01), both levels being comparable to those induced by the mesoderm positive control (Fig. S10). As regards the scaffolds nanostructured with 6% MWCNTs, a slight weaker, albeit comparable, alteration of gene expression was recorded. In particular, a slight up-regulation of SOX1 ectoderm gene marker was found (fold change = 4.6; p < 0.05), at a level comparable to the ectoderm positive control (Fig. S10). For endoderm gene markers, a significant expression increase of SOX17 (fold change = 40.6; p < 0.01) and AFP (fold change = 5.2; p < 0.05), at levels far lower or comparable to those induced by the endoderm positive control, was observed (Fig. S10). Also for the scaffolds nanostructured with 6% MWCNTs, the major effect was observed for the expression of mesoderm gene markers, in particular for BRACHYURY, which expression was increased by 493.0 folds (p < 0.001), besides a slight up-regulation of CXCR4 (fold change = 3.6; p < 0.05), at levels comparable to those induced by the mesoderm positive control (Fig. S10).
To better understand whether the pattern of gene expression induced by the 3D MWCNTs scaffolds could be ascribed to a selective differentiation of iPSCs into one of the three germ layers, data on gene expression (expressed as 2−ΔΔCt) in iPSCs exposed for 7 days to control scaffolds not nanostructured with MWCNTs (control) or to each of the 3D MWCNTs scaffolds were represented in a heatmap together with the gene expression data recorded after selective differentiation of iPSC into endoderm, mesoderm or ectoderm as positive differentiation controls (Fig. 5, panel C).
The clustering analysis on the heatmap highlighted a primary cluster at high affinity grouping the 3D scaffolds nanostructured with both 3% and 6% MWCNTs, suggesting that the amount of MWCNTs does not impact iPSC differentiation profile. The clustering analysis also showed a secondary cluster at high similarity between both 3D MWCNTs scaffolds and the mesoderm positive control. This result suggests that the 3D MWCNTs scaffolds, regardless of the amount of MWCNTs, seem to primarily stimulate iPSC differentiation towards mesoderm, mainly triggered by BRACHYURY gene.
Discussion and conclusions
In last years, stem cell therapy has become a promising alternative approach for the treatment of various human diseases. In this scenario, the implementation of stem cell therapy through the application of novel nanomaterials and nanotechnologies is a fruitful approach in the field of regenerative medicine.1 Among the different nanomaterials, carbon nanotubes (CNTs), especially multi-walled CNTs (MWCNTs), possess unique electrical, optical, thermal and mechanical properties that can be exploited for their ability to interact with biological systems and promote tissue regeneration.20The materials used in this study are solid elastomeric porous 3D scaffolds made of an organic polymer (PDMS), containing different quantities of MWCNTs, with tunable pore size using sugar with known grain sizes. The use of PDMS as a polymeric material at the base of the 3D matrix is supported by literature data demonstrating its efficacy in promoting growth and proliferation of stem cells, also thanking to its microporous and elastomeric properties resembling those of ECM.47 These 3D MWCNT-nanostructured scaffolds were previously prepared to investigate the possible applications as supports in neuronal regeneration,41 resulting in good biocompatibility with the central nervous system39 as well as with cardiomyocytes derived from rat ventricles.40 This high biocompatibility was given also by the use of a particular class of short MWCNTs (Nanoamor Short MWCNT) that, due to their short dimensions (outside diameter: 20–30 nm), are characterized by a relatively high water dispersibility and biodegradation and, therefore, by a lower cytotoxicity.48 These observations support the potential application of these scaffolds in the field of regenerative medicine and tissue engineering. In this field, iPSC are considered a gold standard for stem cells research in the frame of personalized regenerative medicine, especially for tissues with a low regenerative capability, due to their ability to differentiate into any adult cell type while avoiding ethical issues related to the use of ESCs.
Herein, we evaluated the biocompatibility and differentiation potential on iPSCs of four 3D MWCNT-nanostructured PDMS scaffolds characterized by small (100–250 µm sugar grain size) and large (250–600 µm sugar grain size) pore sizes, and by two amounts of MWCNTs (3% and 6% w/w MWCNTs), with the aim to study the role of scaffolds pore size and the degree of nano-structuration with MWCNTs in these biological properties.
To evaluate the biocompatibility of the 3D MWCNTs scaffolds with iPSC, cell viability (WST-8 assay) and cell mass (SRB assay) were initially evaluated after 3 and 7 days of contact. Both assays showed no significant alterations of these cellular parameters after 3 days exposure. No significant influence on cell viability by the four scaffolds was observed even up to 7 days of contact. This result is in line with literature data demonstrating good biocompatibility with stem cells of different matrices structured with CNTs. For instance, Chao and colleagues reported that viability of human ESC was not affected by 5 days exposure to a thin film of polyacrylic acid coated with CNTs.49 Intriguingly, SRB assay showed an increased cell mass only after cells exposure to 3D MWCNTs scaffolds with the large pore size (250–600 µm) for 7 days, significant with respect to control scaffolds. Interestingly, this effect appears dependent on the amount of MWCNTs used to nanostructure the scaffold, being higher for 6% MWCNTs (182% as compared to control) with respect to 3% MWCNTs (142% as compared to control). These results suggest that the 3D MWCNTs scaffolds may increase cell mass, and that this effect seems to be dependent on the pore size and MWCNTs amount. This phenomenon might be related to a more comfortable environment for iPSC growth due to the larger pore size and the ECM-like properties given by the higher MWCNTs content. In addition, it has to be underlined that these data suggest a good (and increased) adhesion of iPSC to the nanostructured scaffolds, confirming their biocompatibility. Similar observations have been previously recorded for P19 cells (mouse embryonal carcinoma stem cells) cultured for 24 h on glass supports coated with MWCNTs: an increased stem cells adhesion (measured by the SRB assay) was observed as compared to the control cells, in absence of cell viability variations.50
It should be noted that increased cell mass could be compatible either with an increased number of cells and/or an increased protein content of the cell sample. Thus, to determine whether the increased cell mass was dependent on a proliferative stimulus (and therefore on a greater number of cells per sample), proliferation of iPSC was evaluated by BrdU incorporation assay. Our data demonstrated that 3D MWCNTs scaffolds did not stimulate cell proliferation up to 7 days of contact, in line with results obtained by counting the cells in the same experimental conditions, which number was not significantly affected with respect to control scaffolds not nanostructured with MWCNTs. These observations lead to conclude that the increased cell mass was not related to an increased cells number as consequence of a proliferative stimulus, but it seems to be related to an increased protein content, as indirectly suggested by SRB assay.
Since this phenomenon could be compatible with a differentiation process,26 we assessed whether the scaffolds could influence pluripotency properties of iPSC while inducing a selective differentiation. To verify this hypothesis, a qPCR gene expression analysis was performed considering a panel of 13 genes, including 5 pluripotency marker genes (C-MYC, OCT4, NANOG, KLF4 and SOX2), 3 marker genes of endoderm (FOXA2, SOX17, AFP), 3 marker genes of mesoderm (ACTA, BRACHYURY, CXCR4) and 2 marker genes of ectoderm (PAX6, SOX1). For this purpose, iPSC were exposed to the scaffolds for 7 days, considering the above-reported biocompatibility data. Our results show that control scaffolds not nanostructured with MWCNTs did not significantly influence the expression of the 5 pluripotency gene markers, as compared to iPSC maintained in standard culture conditions. This result suggests that control scaffolds did not affect the pluripotency of iPSC after 7 days contact. However, the 3D MWCNTs scaffolds led to significant variations in the expression of these genes. The highest effect was observed for the 3D scaffolds nanostructured with 3% MWCNTs that significantly influenced the expression of all the pluripotency genes, up-regulating C-MYC and down-regulating OCT4, NANOG, KLF4 and SOX2. On the other hand, 3D scaffolds nanostructured with 6% MWCNTs induced a significant down-regulation of OCT4 and NANOG expression. The observed increase in C-MYC expression alongside the reduction of OCT4, SOX2, NANOG, and KLF4 suggests a metabolic and transcriptional transition associated with early stages of differentiation, in which C-MYC expression often persists or even rises during early differentiation when pluripotency genes decline.51 While C-MYC plays a central role in promoting biosynthetic and energetic programs supporting increased mitochondrial activity and cell growth,52–54 the downregulation of OCT4, SOX2, and NANOG reflects the dismantling of the core pluripotency network.55 Overall, these results suggest a loss of iPSC pluripotency after 7 days contact with large-pore 3D MWCNTs scaffolds, with a slightly higher effect for 3% MWCNTs.
These results are in line with those related to 3D MWCNTs scaffolds ability to increase the expression of the majority of differentiation genes considered. The highest effect was recorded for the 3D scaffolds nanostructured with 3% MWCNTs, increasing the expression of the marker genes of ectoderm (1 out of 2), endoderm (3 out of 3) and mesoderm (2 out of 3), with different potencies. The highest up-regulation was measured for the BRACHYURY mesoderm gene marker. In the case of the 3D scaffolds nanostructured with 6% MWCNTs, a significant expression increase was recorded for genes markers of endoderm (2 out of 3) and mesoderm (2 out of 3), but not for ectoderm gene markers. Also in this case, the highest up-regulation was observed for the BRACHYURY mesoderm gene marker. In general, the down-regulation of genes markers of pluripotency and the up-regulation of some genes markers of differentiation, especially those related to endoderm and mesoderm, suggest an induction of iPSC differentiation by 3D MWCNTs scaffolds. However, if taken alone, these results cannot identify a selective differentiation towards a single embryonic layer by the 3D MWCNTs scaffolds. This problem is probably due to the genetic interconnection between the three layers, where the expression of a germ layer gene marker can influence the expression of another gene specific of a different layer. Indeed, these markers are representative but not exclusive of the relevant germ layer.56 Furthermore, many of the genes controlling pluripotency in the early stages of embryonic development can also regulate various phases of differentiation within a very complex network of gene expression that, to date, is not fully clarified.57 Hence, to better understand the biological meaning of the gene expression data, results were visualized through a heatmap on which a clustering analysis was carried out to group gene expression patterns by similarity. The length of the arms of the dendrograms connecting the analyzed samples within each cluster determines the degree of similarity, short branches representing high similarities.58 The clustering analysis showed a first cluster grouping both the scaffolds nanostructured with 3% and 6% MWCNTs, suggesting that cells differentiation may not be influenced by the amount of MWCNTs. The clustering analysis also showed a second grouping with high similarity between both the 3D MWCNTs scaffolds and the positive control for mesoderm differentiation. This result suggests that the 3D MWCNTs scaffolds, regardless of the amount of MWCNTs, seem to stimulate iPSC to a direct differentiation towards mesoderm cells. This conclusion is in agreement with previous findings showing the ability of MWCNTs arrays deposited on silicone substrates to differentiate both human ESC and iPSC towards mesoderm cells.23
In conclusion, the results of this study demonstrate a high biocompatibility of the 3D MWCNT-nanostructured PDMS scaffolds with iPSC up to 7 days contact. In addition, these materials seem to induce a iPSC differentiation characterized by a distinct gene expression profile, mainly compatible with a mesoderm-like pattern. These properties seem to be tunable by modulating porosity size of the scaffolds, large pores (250–600 µm) allowing a more adequate environment for cell growth, while the nanostructuration with MWCNTs, regardless of MWCNTs amount (3% or 6% w/w), triggers this selective differentiation. Altogether, these results would encourage novel applications of these 3D nanostructured scaffolds in regenerative medicine, opening innovative scenarios for stem cell-based approaches. The main text of the article should appear here with headings as appropriate.
Methods
3D MWCNTs scaffolds synthesis
As previously reported by our group, MWCNTs were surface covalently functionalized with aniline pendants by means of Tour reaction, that consists in the reaction between nanotubes and diazo compounds leading to amino functionalities on the surface (Fig. S1).45,46,59 Specifically, 250 mg of MWCNTs (1, Nanostructured & Amorphous Materials Inc.; USA; diameter 20–30 nm, length 0.5–2 µm, purity > 95%) were dispersed in 250 mL of dimethylformamide (DMF; Sigma-Aldrich; Milan, Italy) by ultrasonic treatment (ultrasonic bath; Branson 2510) for 10–15 min in a 500 mL round bottom flask. Subsequently, 2 g of 4-[(N-Boc)aminomethyl]aniline (8.9 mmol; Sigma-Aldrich; Milan, Italy) and 2 mL of isoamyl nitrite (14.8 mmol, Sigma-Aldrich; Milan, Italy) were added to the resulting suspension and the reaction mixture was stirred at 80 °C for 12 h. The suspension was filtered and washed with 250 mL of DMF, 250 mL of methanol, and diethyl ether. Then, the sample was dried under vacuum for 12 h to yield 248 mg of black solid (2). The Boc group was then removed by treating the product with 200 mL of 4 M HCl (Sigma-Aldrich; Milan, Italy) in 1,4-dioxane (Sigma-Aldrich; Milan, Italy), and stirring the reaction mixture overnight at room temperature. Finally, the material was filtered, washed with distilled water until neutral pH was reached (checked with litmus paper), and dried under vacuum overnight to yield 245 mg of black solid (3).3D MWCNTs scaffolds were prepared by exploiting our previously reported protocol (Fig. 6).37 The synthesis was performed using the polymer PDMS (polydimethylsiloxane; Sigma-Aldrich; Milan, Italy) in the presence of sucrose (Eridania Table Sugar) in micrograins mixed with product 3 in different weight ratios. Specifically, sucrose grinded by a ceramic mortar and a pestle was passed through mesh sieves (Fisher Scientific; Milan, Italy) to obtain the proper sizes ranges (small size range: 100–250 µm; large size range: 250–600 µm): 600 µm sieve, 250 µm sieve and 100 µm sieve. Subsequently, each sugar powder was used as it was obtained (500 mg) or premixed overnight with MWCNTs (MWCNTs concentrations: 3% w/w or 6% w/w, Fig. S4), for the scaffold production. Such MWCNTs concentrations were selected to ensure optimal biocompatibility and scaffolds stability as observed in literature.37 In a glass vial, sugar was mixed with MWCNTs by gently stirring overnight through a magnetic stirrer, supported by a magnetic stir bar directly connected to the container (Fig. S4). The powder (500 mg) was then treated with PDMS (2 mL), in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with a curing agent (Commercial Kit Sylgard®184; Dow Corning Co.; Midland, USA), by percolating the liquid into a syringe. Percolation was performed by applying vacuum to the end of the syringe containing the powder, until the solid was completely covered. The syringe containing the mixture was then placed in an oven at 85 °C for 2 h. Once hardened, the product was removed from the syringe by cutting the plastic wrap with a surgical blade. The resulting cylinder of crude scaffold was cut, with the same blade, to obtain 3 mm thick discs. The slices produced were placed in MilliQ water (50 mL, 18.2 mΩ, Milli-Q Plus Purifier 185) for 7 days, replacing the washing solution daily, to completely remove the sugar. Once cleaned, the slices were dried in a plastic Petri placed in a desiccator. In this way, four 3D MWCNTs scaffolds differing for pore size range (small size range: 100–250 µm; large size range: 250–600 µm) and the level of MWCNTs nanostructuration (3% w/w and 6% w/w) were obtained.
1 ratio with a curing agent (Commercial Kit Sylgard®184; Dow Corning Co.; Midland, USA), by percolating the liquid into a syringe. Percolation was performed by applying vacuum to the end of the syringe containing the powder, until the solid was completely covered. The syringe containing the mixture was then placed in an oven at 85 °C for 2 h. Once hardened, the product was removed from the syringe by cutting the plastic wrap with a surgical blade. The resulting cylinder of crude scaffold was cut, with the same blade, to obtain 3 mm thick discs. The slices produced were placed in MilliQ water (50 mL, 18.2 mΩ, Milli-Q Plus Purifier 185) for 7 days, replacing the washing solution daily, to completely remove the sugar. Once cleaned, the slices were dried in a plastic Petri placed in a desiccator. In this way, four 3D MWCNTs scaffolds differing for pore size range (small size range: 100–250 µm; large size range: 250–600 µm) and the level of MWCNTs nanostructuration (3% w/w and 6% w/w) were obtained.
 | ||
| Fig. 6 Schematic representation of the 3D MWCNTs scaffolds production. For schematic purposes, SWCNT surfaces are depicted in CNTs instead of MWCNTs. | ||
Scaffolds characterization
Thermogravimetric analysis (TGA) was used to analyze the degree of MWCNTs surface functionalization. TGA was performed using a TGA Q500 (TA Instruments; New Castle, USA) under inert atmosphere (N2). Samples were subjected to a heating ramp that included a pre-heating up to 100 °C followed by a temperature increase of 10 °C per min up to 800 °C. In line with the previously published protocol, the percentage of weight loss of the functional group was recorded at 500 °C.37 This value was used to calculate the degree of functionalization (FD) of each MWCNT using the following equation:
 | (1) |
The Kaiser Test (KT) was used as secondary analysis to assess the degree of MWCNTs functionalization, and was performed using a commercial kit (Sigma-Aldrich; Milan, Italy).
Scanning electron microscopy (SEM) analysis was employed to study the scaffolds morphology. SEM samples were prepared by fixing scaffold sections (3 mm × 3 mm) on a stub with copper tape. Scaffolds composed of PDMS only were treated by gold sputter-coating using a sputtering machine (ALT1000). The materials were then analyzed using a SEM JSM-6490LV microscope in secondary electron imaging mode. SEM images analysis and pore size assessment were performed using “ImageJ” software. Quantitative pore size distribution analysis was performed on representative SEM micrographs of 3D PDMS/MWCNT (6% w/w) scaffolds. Pore diameters were manually measured using ImageJ software (n = 100 pores per sample) after image thresholding. The resulting histograms were fitted with a distribution curve using OriginPro 9 software to obtain the average pore size and standard deviation. Uniaxial compression tests were conducted at room temperature using a Galdabini Sun 500 universal testing machine equipped with a 100 N load cell. Cylindrical scaffolds (≈5 mm height) were tested at a constant compression rate of 1 mm min−1. Prior to testing, the height and diameter of each specimen were measured with a 0.02 mm-resolution calliper, and the maximum displacement was limited to 20% of the initial height. The Young's modulus (E) was calculated from the linear region between 7% and 12% strain of the stress–strain curve. Six scaffold types were analysed: PDMS controls and PDMS-MWCNTs composites (3% and 6% w/w of MWCNTs) with pore size ranges of 100–250 µm and 250–600 µm. The corresponding modulus values ranged from 230 ± 120 kPa for pure PDMS to 896 ± 440 kPa for 6% w/w MWCNTs scaffolds, confirming the reinforcement effect of the nanotubes. XPS analyses were performed using a SPECS Sage HR 100 spectrometer equipped with a non-monochromatic Mg Kα X-ray source (photon energy = 1261.2 eV, power = 253 W). An electron flood gun was used to compensate for sample charging during data acquisition. The pass energy was set to 30 eV for survey spectra and 15 eV for high-resolution spectra, with energy steps of 0.5 eV and 0.15 eV, respectively. All measurements were carried out under ultra-high vacuum (ca. 8 × 10−8 mbar). Data was processed using CasaXPS 2.3.16 PR 1.6 software.
Scaffolds sanification for the in vitro assay
Before exposure to cell cultures, the 3D PDMS scaffolds (with or without MWCNTs) were sanitized by treatment with ultraviolet rays and ozone using a UV/ozone cleaner (BioForce Nanosciences) for 45 min per side. Then, the scaffolds were fixed to the bottom of the wells of 96- or 12-well plates using a microvolume of PDMS mixed with a curing agent (Sylgard™ 184 Silicone Elastomer Curing Agent; Dow Corning Co.; Midland, USA) in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. After drying for 24 h at room temperature, before cell seeding, the scaffolds were further sterilized by a UV/ozone cleaner cycle (2 h) and a UV irradiation cycle (overnight). The scaffolds were then treated with 1% penicillin–streptomycin (Sigma-Aldrich; Milan, Italy) for 24 h. Subsequently, the scaffolds were again subjected to an UV cycle (overnight) and washed twice with PBS before cell seeding.
1 ratio. After drying for 24 h at room temperature, before cell seeding, the scaffolds were further sterilized by a UV/ozone cleaner cycle (2 h) and a UV irradiation cycle (overnight). The scaffolds were then treated with 1% penicillin–streptomycin (Sigma-Aldrich; Milan, Italy) for 24 h. Subsequently, the scaffolds were again subjected to an UV cycle (overnight) and washed twice with PBS before cell seeding.
Induced-pluripotent stem cells (iPSC) culture
The induced pluripotent stem cell line 253G1 was kindly provided by Prof. Katsunori Sasaki and Fengming Yue (Shinshu University; Matsumoto, Japan). The cell line was maintained at 37 °C and 5% CO2 in basal medium Stem Macs iPS Brew XF added with 2% supplements Stem Macs iPS Brew XF 50 X (Miltenyi Biotec; Bergisch Gladbach, Germany) under feeder-free conditions using cell culture plate coated with Geltrex® solution (LDEV-Free Reduced Growth Factor Basement Membrane Matrix; ThermoFisher Scientific; Milan, Italy) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 1X (DMEM/F12; Sigma-Aldrich; Milan, Italy). Culture was passed twice a week, at a 70–80% of cell confluence, avoiding the complete disintegration of formed clusters. Briefly, after incubation at 37 °C and 5% CO2 for 3 min with Versene solution (ThermoFisher Scientific; Milan, Italy), cells were detached by gentle circular motions using culture medium. The required amount of cells for subculture was transferred to a new plate and 10 µM ROCK inhibitor (Y-27632; Miltenyi Biotec; Bergisch Gladbach, Germany) was added for 24 h to facilitate cell adhesion. In the case of cell seeding for in vitro assays, iPSC were passed into single cells to allow cell counting in a Bürker chamber. The number of cells required for each assay was transferred on the 3D nanostructured scaffolds attached to the wells of 96- or 12-well plates in absence of feeder-free coating (Geltrex®) and in presence of 10 µM ROCK inhibitor for 24 h.
100 in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 1X (DMEM/F12; Sigma-Aldrich; Milan, Italy). Culture was passed twice a week, at a 70–80% of cell confluence, avoiding the complete disintegration of formed clusters. Briefly, after incubation at 37 °C and 5% CO2 for 3 min with Versene solution (ThermoFisher Scientific; Milan, Italy), cells were detached by gentle circular motions using culture medium. The required amount of cells for subculture was transferred to a new plate and 10 µM ROCK inhibitor (Y-27632; Miltenyi Biotec; Bergisch Gladbach, Germany) was added for 24 h to facilitate cell adhesion. In the case of cell seeding for in vitro assays, iPSC were passed into single cells to allow cell counting in a Bürker chamber. The number of cells required for each assay was transferred on the 3D nanostructured scaffolds attached to the wells of 96- or 12-well plates in absence of feeder-free coating (Geltrex®) and in presence of 10 µM ROCK inhibitor for 24 h.
Cell viability
The main text of the article should appear here with headings as appropriate. Biocompatibility of the 3D MWCNTs scaffolds was evaluated in terms of iPSC viability by the WST-8 assay using the Cell Counting Kit-8 (CCK-8) and cell mass using the sulforhodamine B (SRB) assay. These assays were carried out in a 96-well plate considering two time of cell exposure: 3 days (5000 cells per well) and 7 days (500 cells per well) after cells seeding.The CCK-8 assays (Sigma-Aldrich; Milan, Italy) was carried out as previously described.60,61 Briefly, at the end of cells exposure, the CCK-8 reagent was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in each well of a 96-well plate and incubated at 37 °C and 5% CO2 for 4 h. Then, absorbance was measured at 450 nm using a microplate reader (FLUOstar Omega Automated Microplate Reader; Bio-Tek Instruments; Ortenberg, Germany). Results were reported as percentage of cell viability with respect to control scaffolds not nanostructured with MWCNTs (3D PDMS scaffolds).
10 in each well of a 96-well plate and incubated at 37 °C and 5% CO2 for 4 h. Then, absorbance was measured at 450 nm using a microplate reader (FLUOstar Omega Automated Microplate Reader; Bio-Tek Instruments; Ortenberg, Germany). Results were reported as percentage of cell viability with respect to control scaffolds not nanostructured with MWCNTs (3D PDMS scaffolds).
Mass of iPSC cultured for 3 and 7 days on the 3D MWCNTs scaffolds was evaluated by the SRB assay, based on a reagent able to bind the basic residues of proteins of adhered cells.62 Briefly, cells were washed twice in PBS and fixed in 50% trichloroacetic acid (Sigma-Aldrich; Milan, Italy) for 1 h at 4 °C. After two washings with MilliQ water, a 0.4% solution of SRB (Sigma-Aldrich; Milan, Italy) in 1% acetic acid (Sigma-Aldrich; Milan, Italy) was added and the plate was incubated for 30 min at room temperature. Cells were washed three times in 1% acetic acid, and incorporated SRB was solubilized using 200 µL of 10 mM TRIS (Sigma-Aldrich; Milan, Italy). Then, absorbance was measured at 570 nm using a microplate reader (FLUOstar Omega Automated Microplate Reader; Bio-Tek Instruments; Ortenberg, Germany). Results were reported as percentage of cell adhesion with respect to control scaffolds not nanostructured with MWCNTs (3D PDMS scaffolds).
Cell proliferation
Proliferation of iPSC was evaluated after 3 and 7 days exposure to the 3D MWCNTs scaffolds in a 96-well plate using the Cell Proliferation ELISA (Sigma-Aldrich; Milan, Italy), based on incorporation of non-radioactive 5-bromo-2′-deoxyuridine (BrdU) in proliferating cells. According to manufacturer's instructions, after treatment, 100 µM of BrdU solution was added to each well and the plate was incubated for 2 h at 37 °C and 5% CO2. Cells were fixed for 30 min at room temperature, and incubated with the Anti-BrdU-POD (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) for 90 min at room temperature, protected from light. After three washes with a washing solution provided by the kit, the substrate was added (100 µL per well) and the reaction was stopped after 5 min with 25 µL per well of 1 M sulfuric acid. The absorbance was measured at 450 nm using a microplate reader (FLUOstar Omega Automated Microplate Reader; Bio-Tek Instruments; Ortenberg, Germany). Results were reported as percentage of cell proliferation with respect to control scaffolds not nanostructured with MWCNTs (3D PDMS scaffolds).
100) for 90 min at room temperature, protected from light. After three washes with a washing solution provided by the kit, the substrate was added (100 µL per well) and the reaction was stopped after 5 min with 25 µL per well of 1 M sulfuric acid. The absorbance was measured at 450 nm using a microplate reader (FLUOstar Omega Automated Microplate Reader; Bio-Tek Instruments; Ortenberg, Germany). Results were reported as percentage of cell proliferation with respect to control scaffolds not nanostructured with MWCNTs (3D PDMS scaffolds).
Gene expression analysis
The effects of the 3D MWCNTs scaffolds on iPSC stemness and pluripotency were evaluated by means of gene expression analysis by real time qPCR. A panel of 13 genes was considered: five genes markers of stemness (OCT4, C-MYC, KLF4, SOX2 and NANOG)63 and eight genes as differentiation markers for the three germ layers (SOX17, AFP and FOXA2 for endoderm; ACTA2, BRACHYURY and CXCR4 for mesoderm; PAX6 and SOX1 for ectoderm).64,65 As positive differentiation controls, iPSC were differentiated to endoderm, mesoderm or ectoderm using a commercial kit (StemMACSTM Trilineage Differentiation Kit; Miltenyi Biotec; Bergisch Gladbach, Germany), following the provided instructions.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C, the upper aqueous phase containing RNA was transferred to an extraction column with an equal volume of absolute ethanol (Sigma-Aldrich; Milan, Italy). The column was firstly centrifuged at 12
000g for 15 min at 4 °C, the upper aqueous phase containing RNA was transferred to an extraction column with an equal volume of absolute ethanol (Sigma-Aldrich; Milan, Italy). The column was firstly centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 1 min at room temperature and then washed twice with 500 µL of the kit washing solution, centrifuging at 12
000g for 1 min at room temperature and then washed twice with 500 µL of the kit washing solution, centrifuging at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 s at room temperature. The column was allowed to dry for 1 min at room temperature before adding 30 µL of DNAse/RNAse-free water, followed by centrifugation at 12
000g for 30 s at room temperature. The column was allowed to dry for 1 min at room temperature before adding 30 µL of DNAse/RNAse-free water, followed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 2 min at room temperature. For each sample, 100 ng of RNA (quantified using a NanoDrop UV spectrophotometer; NanoDrop 2000 spectrophotometer; Thermo Fisher Scientific; Milan, Italy) was reverse transcripted to cDNA using the High-Capacity RNA-to-cDNA™ Kit (ThermoFisher Scientific; Milan, Italy). To start the reaction, 10 µL buffer and 1 µL of enzyme solution were added to the extracted RNA in a final volume of 20 µL with DNAse/RNAse-free water. Samples were loaded into the thermal cycler (One-Personal; EuroClone; Milan, Italy) by setting the following program: 37 °C for 60 min; 95 °C for 5 min; 4 °C ∞.
000g for 2 min at room temperature. For each sample, 100 ng of RNA (quantified using a NanoDrop UV spectrophotometer; NanoDrop 2000 spectrophotometer; Thermo Fisher Scientific; Milan, Italy) was reverse transcripted to cDNA using the High-Capacity RNA-to-cDNA™ Kit (ThermoFisher Scientific; Milan, Italy). To start the reaction, 10 µL buffer and 1 µL of enzyme solution were added to the extracted RNA in a final volume of 20 µL with DNAse/RNAse-free water. Samples were loaded into the thermal cycler (One-Personal; EuroClone; Milan, Italy) by setting the following program: 37 °C for 60 min; 95 °C for 5 min; 4 °C ∞.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in DNAse/RNAse-free water), 5 µL SYBR Green (SYBR® Green Quantitative RT-qPCR Kit; Sigma-Aldrich; Milan, Italy), 2.1 µL DNAse/RNAse-free water, 0.45 µL forward primer and 0.45 µL reverse primer. All primers were purchased precast from Sigma-Aldrich (Milan, Italy; Table 1). The plate was loaded into the thermal cycler (RealTime PCR Detection System; BIO-RAD; Milan, Italy) and the following program was set for the analysis of stemness and differentiation genes: 95 °C for 3 min; 95 °C for 10 s; 60 °C for 30 s, the second and third step were repeated for 45 cycles. Data were analysed with the 2−ΔΔCt method,66 where the ΔCt represents the difference between the Ct of the gene of interest and the Ct of the housekeeping gene (ACT), while the ΔΔCt indicates the difference between the ΔCt of a treated sample (3D MWCNTs-nanostructured scaffold or 3D PDMS scaffold) and that of the reference sample (iPSC cultured in standard condition, in a feeder-free coating). A heatmap was created on the obtained 2−ΔΔCt values on which a clustering analysis was carried out, which made it possible to cluster the gene expression patterns with high similarity.
2 in DNAse/RNAse-free water), 5 µL SYBR Green (SYBR® Green Quantitative RT-qPCR Kit; Sigma-Aldrich; Milan, Italy), 2.1 µL DNAse/RNAse-free water, 0.45 µL forward primer and 0.45 µL reverse primer. All primers were purchased precast from Sigma-Aldrich (Milan, Italy; Table 1). The plate was loaded into the thermal cycler (RealTime PCR Detection System; BIO-RAD; Milan, Italy) and the following program was set for the analysis of stemness and differentiation genes: 95 °C for 3 min; 95 °C for 10 s; 60 °C for 30 s, the second and third step were repeated for 45 cycles. Data were analysed with the 2−ΔΔCt method,66 where the ΔCt represents the difference between the Ct of the gene of interest and the Ct of the housekeeping gene (ACT), while the ΔΔCt indicates the difference between the ΔCt of a treated sample (3D MWCNTs-nanostructured scaffold or 3D PDMS scaffold) and that of the reference sample (iPSC cultured in standard condition, in a feeder-free coating). A heatmap was created on the obtained 2−ΔΔCt values on which a clustering analysis was carried out, which made it possible to cluster the gene expression patterns with high similarity.
| Gene | Primer | 5′ → 3′ sequence | |
|---|---|---|---|
| Housekeeping | ACT | Forward | GACGACATGGAGAAAATCTG |
| Reverse | ATGATCTGGGTCATCTTCTC | ||
| Marker of stemness | SOX2 | Forward | ATAATAACAATCATCGGCGG |
| Reverse | AAAAAGAGAGAGGCAAACTG | ||
| OCT4 | Forward | GATCACCCTGGGATATACAC | |
| Reverse | GCTTTGCATATCTCCTGAAG | ||
| KLF4 | Forward | TCTTGAGGAAGTGCTGAG | |
| Reverse | ATGAGCTCTTGGTAATGGAG | ||
| NANOG | Forward | CCAGAACCAGAGAATGAAATC | |
| Reverse | TGGTGGTAGGAAGAGTAAAG | ||
| c-MYC | Forward | TGAGGAGGAACAAGAAGATG | |
| Reverse | ATCCAGACTCTGACCTTTTG | ||
| Marker of endoderm | FOXA2 | Forward | CGAGTTAAAGTATGCTGGG |
| Reverse | CATGTACGTGTTCATGCC | ||
| SOX17 | Forward | ATCTCTTTACACTCCTCGAC | |
| Reverse | CCTTTATCTTAAACCCAGCG | ||
| AFP | Forward | GATCCCACTTTTCCAAGTTC | |
| Reverse | TTTGTTCATGAATGTCTCCC | ||
| Marker of mesoderm | ACTA2 | Forward | AGATCAAGATCATTGCCCC |
| Reverse | TTCATCGTATTCCTGTTTGC | ||
| BRACHYURY | Forward | GAATCCACATAGTGAGAGTTG | |
| Reverse | TCACTTCTTTCCTTTGCATC | ||
| CXCR4 | Forward | AACTTCAGTTTGTTGGCTG | |
| Reverse | GTGTATATACTGATCCCCTCC | ||
| Marker of ectoderm | PAX6 | Forward | AGAGAATACCAACTCCATCAG |
| Reverse | GATAATGGGTTCTCTCAAACTC | ||
| SOX1 | Forward | TGCTTGTTCTGTTAACTCAC | |
| Reverse | AAAGAACCTCAGAGAGAGTC |
Author contributions
F. Cavion and M. Cacioppo conducted experiments, analyzed data, and contributed to manuscript drafting. S. Bosi, M. Carlin, and S. Sosa participated in data analysis and performed validation. M. Pelin acquired funding. M. Pelin and M. Prato conceptualized ideas, provided supervision, oversaw the research, and finalized the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra06688c.Acknowledgements
This study was supported by a grant from the University of Trieste (Microgrants 2018).References
- S. Suman, A. Domingues, J. Ratajczak and M. Z. Ratajczak, in Stem Cells: Therapeutic Applications, ed. M. Z. Ratajczak, Springer International Publishing, Cham, 2019, pp. 1–22 Search PubMed.
- S. Ye and Z. Lixian, Curr. Ther. Res., 2022, 96, 100665 CrossRef PubMed.
- T. Maeda, M. Mandai, S. Sugita, C. Kime and M. Takahashi, Trends Mol. Med., 2022, 28, 388–404 CrossRef CAS PubMed.
- X.-Y. Liu, L.-P. Yang and L. Zhao, World J. Stem Cell., 2020, 12, 787–802 CrossRef PubMed.
- S. Chen, K. Du and C. Zou, Stem Cell Res. Ther., 2020, 11, 275 CrossRef PubMed.
- N. H. Goradel, F. G. Hour, B. Negahdari, Z. V. Malekshahi, M. Hashemzehi, A. Masoudifar and H. Mirzaei, J. Cell. Biochem., 2018, 119, 95–104 CrossRef CAS PubMed.
- B. Genc, H. R. Bozan, S. Genc and K. Genc, in Tissue Engineering and Regenerative Medicine, ed. P. V. Pham, Springer International Publishing, Cham, 2019, pp. 145–174 Search PubMed.
- P. Müller, H. Lemcke and R. David, Cell. Physiol. Biochem., 2018, 48, 2607–2655 CrossRef PubMed.
- M. Gazdic, V. Volarevic, C. R. Harrell, C. Fellabaum, N. Jovicic, N. Arsenijevic and M. Stojkovic, Int. J. Mol. Sci., 2018, 19, 1039 CrossRef PubMed.
- M. A. M. Aboul-Soud, A. J. Alzahrani and A. Mahmoud, Cells, 2021, 10, 2319 CrossRef CAS PubMed.
- J. Hu and J. Wang, Clin. Transplant., 2019, 33, e13573 CrossRef PubMed.
- E. Genova, M. Pelin, K. Sasaki, F. Yue, G. Lanzi, S. Masneri, A. Ventura, G. Stocco and G. Decorti, Curr. Med. Chem., 2018, 25, 2826–2839 CrossRef CAS PubMed.
- E. Genova, F. Cavion, M. Lucafò, L. D. Leo, M. Pelin, G. Stocco and G. Decorti, World J. Stem Cell., 2019, 11, 1020–1044 CrossRef PubMed.
- D. A. Robinton and G. Q. Daley, Nature, 2012, 481, 295–305 CrossRef CAS PubMed.
- Y. Dong, X. Wu, X. Chen, P. Zhou, F. Xu and W. Liang, Biomed. Pharmacother., 2021, 137, 111236 CrossRef CAS PubMed.
- F. He, J. Cao, J. Qi, Z. Liu, G. Liu and W. Deng, Front. Bioeng. Biotechnol., 2021, 9, 721581 CrossRef.
- P. Kerativitayanan, J. K. Carrow and A. K. Gaharwar, Adv. Healthcare Mater., 2015, 4, 1600–1627 CrossRef CAS PubMed.
- H. Chen, Y. Zeng, W. Liu, S. Zhao, J. Wu and Y. Du, Biotechnol. Adv., 2013, 31, 638–653 CrossRef CAS PubMed.
- S. Iijima, Nature, 1991, 354, 56–58 CrossRef CAS.
- P. A. Tran, L. Zhang and T. J. Webster, Adv. Drug Delivery Rev., 2009, 61, 1097–1114 CrossRef CAS PubMed.
- J. Ramón-Azcón, S. Ahadian, R. Obregón, H. Shiku, M. Ramalingam and T. Matsue, J. Biomed. Nanotechnol., 2014, 10, 2539–2561 CrossRef PubMed.
- L. Krishna, K. Dhamodaran, C. Jayadev, K. Chatterjee, R. Shetty, S. S. Khora and D. Das, Stem Cell Res. Ther., 2016, 7, 188 CrossRef PubMed.
- M. V. Pryzhkova, Stem Cells Transl. Med., 2013, 2, 376–383 CrossRef CAS.
- I. Ullah, R. B. Subbarao and G. J. Rho, Biosci. Rep., 2015, 35, e00191 CrossRef PubMed.
- V. C. Bitirim, G. Kucukayan-Dogu, E. Bengu and K. C. Akcali, Mater. Sci. Eng. C, 2013, 33, 3054–3060 CrossRef CAS PubMed.
- D. D. Deligianni, Cell Adhes. Migr., 2014, 8, 558–562 CrossRef PubMed.
- G. Lalwani, M. D'agati, A. Gopalan, S. C. Patel, Y. Talukdar and B. Sitharaman, J. Biomed. Mater. Res., Part A, 2017, 105, 1927–1939 CrossRef CAS PubMed.
- J. A. Kim, E. Y. Jang, T. J. Kang, S. Yoon, R. Ovalle-Robles, W. J. Rhee, T. Kim, R. H. Baughman, Y. H. Kim and T. H. Park, Integr. Biol., 2012, 4, 587–594 CrossRef CAS PubMed.
- Y.-S. Chen and G.-H. Hsiue, Biomaterials, 2013, 34, 4936–4944 CrossRef CAS PubMed.
- S. Y. Park, B.-S. Kang and S. Hong, Nanomedicine, 2013, 8, 715–723 CrossRef CAS PubMed.
- A. A. Kroustalli, S. N. Kourkouli and D. D. Deligianni, Ann. Biomed. Eng., 2013, 41, 2655–2665 CrossRef PubMed.
- B. Xu, Y. Ju, Y. Cui and G. Song, Mater. Sci. Eng. C, 2015, 51, 182–188 CrossRef CAS PubMed.
- E.-S. Kang, D.-S. Kim, I. R. Suhito, S.-S. Choo, S.-J. Kim, I. Song and T.-H. Kim, Nano Convergence, 2017, 4, 2 CrossRef PubMed.
- R. Arambula-Maldonado and K. Mequanint, Biomimetics, 2024, 9, 338 CrossRef CAS PubMed.
- S. Jafarkhani, E. Amiri, S. Moazzeni, T. Zohoorian-Abootorabi, M. Eftekhary, S. Aminnezhad and M. Khakbiz, Colloids Surf., A, 2023, 675, 131872 CrossRef CAS.
- E. L. Hopley, S. Salmasi, D. M. Kalaskar and A. M. Seifalian, Biotechnol. Adv., 2014, 32, 1000–1014 CrossRef CAS PubMed.
- S. Bosi, R. Rauti, J. Laishram, A. Turco, D. Lonardoni, T. Nieus, M. Prato, D. Scaini and L. Ballerini, Sci. Rep., 2015, 5, 9562 CrossRef CAS PubMed.
- J.-W. Han, B. Kim, J. Li and M. Meyyappan, Appl. Phys. Lett., 2013, 102, 051903 CrossRef.
- E. R. Aurand, S. Usmani, M. Medelin, D. Scaini, S. Bosi, F. B. Rosselli, S. Donato, G. Tromba, M. Prato and L. Ballerini, Adv. Funct. Mater., 2018, 28, 1700550 CrossRef.
- V. Martinelli, S. Bosi, B. Peña, G. Baj, C. S. Long, O. Sbaizero, M. Giacca, M. Prato and L. Mestroni, ACS Appl. Bio Mater., 2018, 1, 1530–1537 CrossRef CAS PubMed.
- R. Rauti, N. Secomandi, C. Martín, S. Bosi, F. P. U. Severino, D. Scaini, M. Prato, E. Vázquez and L. Ballerini, Adv. Biosyst., 2020, 4, 1900233 CrossRef CAS PubMed.
- K. Das, A. P. Madhusoodan, B. Mili, A. Kumar, A. C. Saxena, K. Kumar, M. Sarkar, P. Singh, S. Srivastava and S. Bag, Int. J. Nanomed., 2017, 12, 3235–3252 CrossRef CAS PubMed.
- M. Garrido, L. Gualandi, S. D. Noja, G. Filippini, S. Bosi and M. Prato, Chem. Commun., 2020, 56, 12698–12716 RSC.
- E. Mansfield, A. Kar and S. A. Hooker, Anal. Bioanal. Chem., 2010, 396, 1071–1077 CrossRef CAS.
- M. Quintana and M. Prato, Chem. Commun., 2009, 6005–6007 RSC.
- L. Maggini, F. M. Toma, L. Feruglio, J. M. Malicka, T. Da Ros, N. Armaroli, M. Prato and D. Bonifazi, Chem.–Eur J., 2012, 18, 5889–5897 CrossRef CAS PubMed.
- S. Usmani, E. R. Aurand, M. Medelin, A. Fabbro, D. Scaini, J. Laishram, F. B. Rosselli, A. Ansuini, D. Zoccolan, M. Scarselli, M. De Crescenzi, S. Bosi, M. Prato and L. Ballerini, Sci. Adv., 2016, 2, e1600087 CrossRef PubMed.
- J. Russier, C. Ménard-Moyon, E. Venturelli, E. Gravel, G. Marcolongo, M. Meneghetti, E. Doris and A. Bianco, Nanoscale, 2011, 3, 893–896 RSC.
- T.-I. Chao, S. Xiang, C.-S. Chen, W.-C. Chin, A. J. Nelson, C. Wang and J. Lu, Biochem. Biophys. Res. Commun., 2009, 384, 426–430 CrossRef CAS PubMed.
- J. Holy, E. Perkins and X. Yu, Annu. Int. Conf. IEEE Eng. Med. Biol. Soc., 2009, 2009, 6022–6025 Search PubMed.
- N. V. Varlakhanova, R. F. Cotterman, W. N. deVries, J. Morgan, L. R. Donahue, S. Murray, B. B. Knowles and P. S. Knoepfler, Differentiation, 2010, 80, 9–19 CrossRef CAS PubMed.
- C. V. Dang, Cell, 2012, 149, 22–35 CrossRef CAS PubMed.
- F. Li, Y. Wang, K. I. Zeller, J. J. Potter, D. R. Wonsey, K. A. O'Donnell, J. Kim, J. T. Yustein, L. A. Lee and C. V. Dang, Mol. Cell. Biol., 2005, 25, 6225–6234 CrossRef CAS PubMed.
- F. Morrish and D. Hockenbery, Cold Spring Harbor Perspect. Med., 2014, 4, a014225 CrossRef PubMed.
- J. Kim, J. Chu, X. Shen, J. Wang and S. H. Orkin, Cell, 2008, 132, 1049–1061 CrossRef CAS PubMed.
- I. S. Schroeder, S. Sulzbacher, T. Nolden, J. Fuchs, J. Czarnota, R. Meisterfeld, H. Himmelbauer and A. M. Wobus, Cells Tissues Organs, 2011, 195, 507–523 CrossRef PubMed.
- D.-E. Parfitt and M. M. Shen, Philos. Trans. R. Soc. B, Biol. Sci., 2014, 369, 20130542 CrossRef PubMed.
- B. Gorain, H. Choudhury, M. Pandey, P. Kesharwani, M. M. Abeer, R. K. Tekade and Z. Hussain, Biomed. Pharmacother., 2018, 104, 496–508 CrossRef CAS PubMed.
- B. K. Price and J. M. Tour, J. Am. Chem. Soc., 2006, 128, 12899–12904 CrossRef CAS PubMed.
- M. Pelin, L. Fusco, V. León, C. Martín, A. Criado, S. Sosa, E. Vázquez, A. Tubaro and M. Prato, Sci. Rep., 2017, 7, 40572 CrossRef CAS PubMed.
- M. Pelin, H. Lin, A. Gazzi, S. Sosa, C. Ponti, A. Ortega, A. Zurutuza, E. Vázquez, M. Prato, A. Tubaro and A. Bianco, Nanomaterials, 2020, 10, 1602 CrossRef CAS PubMed.
- V. Vichai and K. Kirtikara, Nat. Protoc., 2006, 1, 1112–1116 CrossRef CAS PubMed.
- K. Takahashi and S. Yamanaka, Cell, 2006, 126, 663–676 CrossRef CAS PubMed.
- P. Wang, K. D. McKnight, D. J. Wong, R. T. Rodriguez, T. Sugiyama, X. Gu, A. Ghodasara, K. Qu, H. Y. Chang and S. K. Kim, Stem Cells Dev., 2012, 21, 2273–2287 CrossRef CAS PubMed.
- C. Bock, E. Kiskinis, G. Verstappen, H. Gu, G. Boulting, Z. D. Smith, M. Ziller, G. F. Croft, M. W. Amoroso, D. H. Oakley, A. Gnirke, K. Eggan and A. Meissner, Cell, 2011, 144, 439–452 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
Footnote |
| † These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |