DOI:
10.1039/D5RA03831F
(Paper)
RSC Adv., 2025,
15, 22285-22310
From silico to benchtop: cosmosiin as a PD-1/PDL-1 immune checkpoint inhibitor revealed through DFT, network pharmacology analysis, and molecular docking integrated experimental verification
Received
30th May 2025
, Accepted 25th June 2025
First published on 2nd July 2025
Abstract
This study investigated the anti-PD-1/PD-L1 inhibition potential of the flavonoid cosmosiin against breast cancer (BC) using computational chemistry, network pharmacology, bioinformatics, and validated by experimental assays. Execution of Density Functional Theory (DFT) calculations was achieved by GAUSSIAN 09 with the 6-311G/B3LYP formalism to assess cosmosiin's physicochemical properties. Potential targets of cosmosiin were identified through SuperPred, Stitch, and SwissTargetPrediction databases, while BC-allied targets were sourced by accessing GeneCards. Overlapping analysis using Venny 2.0 identified 25 common targets between 38 targets of cosmosiin and 1314 targets of breast cancer. CytoHubba analysis highlighted 10 hub genes, with PTGS2, NFKB1, and CDK5 being the most significant. Molecular docking revealed stable binding of cosmosiin to CDK5 (−8.5 kcal mol−1), NFKB1 (−7.6 kcal mol−1), and PTGS2 (−9.6 kcal mol−1), confirmed by molecular dynamics simulations. GEPIA analysis linked the expression of these genes to survival outcomes and disease stage in breast cancer. Experimentally, cosmosiin was tested on MCF-7 (breast cancer) and MCF-10 (non-tumorigenic) cells. Cytotoxicity was evaluated using the MTT assay, showing dose-dependent viability reduction in MCF-7 cells with minimal impact on MCF-10 cells, thus exhibiting selective cytotoxicity. Phase-contrast microscopy revealed altered morphology in treated MCF-7 cells. Annexin V/PI flow cytometry confirmed increased early and late apoptosis, while EdU incorporation assays indicated decreased DNA synthesis and reduced proliferation. Transwell assays demonstrated up to 81% inhibition of cell migration at higher concentrations. Western blotting validated downregulation of CDK5, NFKB1, and PTGS2, aligning with computational predictions. These findings highlight selective, multi-targeted anticancer activity of cosmosiin, particularly through PTGS2, NFKB1, and CDK5, supporting its therapeutic potency for breast cancer with favorable effects on apoptosis, proliferation, and cell migration, while correlating with survival and immune infiltration outcomes.
1. Introduction
Breast cancer is a complex, multifaceted disease originating in breast tissue cells. It is a kind of metastatic cancer that may disseminate to several organs, including the lungs, bones, liver, and brain, rendering it incurable. However, early detection can lead to a good prognosis and high survival rates. BC is frequently diagnosed among US women. Additionally, it leads in UK, with 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases annually (15%), a figure expected to increase by 2% by 2035.1,2 Breast Cancer predominantly affects women, with incidence rates increasing significantly with age including the diagnosis in about >80% of instances for those over 50 years. According to cancer statistics for 2023, the number of new BC cases has increased to 297
000 new cases annually (15%), a figure expected to increase by 2% by 2035.1,2 Breast Cancer predominantly affects women, with incidence rates increasing significantly with age including the diagnosis in about >80% of instances for those over 50 years. According to cancer statistics for 2023, the number of new BC cases has increased to 297![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780 among females and 2800 among males.3 Worldwide fatalities have increased to 685
780 among females and 2800 among males.3 Worldwide fatalities have increased to 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in 2020 and thus, it is among the top causes of cancer mortality. It is the leading cause of cancer deaths, resulting in fatalities worldwide in 2020.4 Fat, lack of exercise, carcinogen exposure, smoking and alcohol use are modifiable factors that are associated with about 30% of BC incidences, making these cases potentially preventable and non-modifiable factors such as female sex, density of breast tissues, previous radiation therapy, and genetic mutations.5,6 With 15% developing in the lobular epithelium and 85% in the breast ducts, adenocarcinomas make up the bulk of breast cancers. Cancers of the ducts may range from those that remain in situ inside the ducts to those that have invaded the breast parenchyma and spread beyond the foundation membrane.7 The treatments of BC with surgery, chemotherapy, radiotherapy (RT), endocrine therapy, targeted therapy, modify based on specific cancer traits and patient profiles; yet, these cause adverse events, invasiveness, slow wound healing, body shape changes, liver and kidney damage, metabolic changes, severe hair loss, which impact women both physically and mentally and limited efficacy for certain cancer types. Furthermore, drug resistance continues to be a major challenge, with up to 50% of patients developing tamoxifen resistance and 70% to trastuzumab within a year.8 These limitations highlight the need for newer treatments such as immunotherapy, which offers more targeted and potentially less harmful options.9 The immunomodulation represents a promising strategy to unleash the power of the immune system against cancer. The PD-1 (Programmed Death-1) and PD-L1 (Programmed Death-Ligand 1) pathway plays a crucial role in putting the brakes on the immune system, enabling cancer cells to evade immune surveillance. By targeting and inhibiting this pathway, the immune response can be reactivated, allowing it to spot and destroy more malignant cells effectively. Thus, this approach has the potential to revolutionize cancer treatment, turning the body's own defense mechanisms into powerful allies in the fight against malignancy.
000 in 2020 and thus, it is among the top causes of cancer mortality. It is the leading cause of cancer deaths, resulting in fatalities worldwide in 2020.4 Fat, lack of exercise, carcinogen exposure, smoking and alcohol use are modifiable factors that are associated with about 30% of BC incidences, making these cases potentially preventable and non-modifiable factors such as female sex, density of breast tissues, previous radiation therapy, and genetic mutations.5,6 With 15% developing in the lobular epithelium and 85% in the breast ducts, adenocarcinomas make up the bulk of breast cancers. Cancers of the ducts may range from those that remain in situ inside the ducts to those that have invaded the breast parenchyma and spread beyond the foundation membrane.7 The treatments of BC with surgery, chemotherapy, radiotherapy (RT), endocrine therapy, targeted therapy, modify based on specific cancer traits and patient profiles; yet, these cause adverse events, invasiveness, slow wound healing, body shape changes, liver and kidney damage, metabolic changes, severe hair loss, which impact women both physically and mentally and limited efficacy for certain cancer types. Furthermore, drug resistance continues to be a major challenge, with up to 50% of patients developing tamoxifen resistance and 70% to trastuzumab within a year.8 These limitations highlight the need for newer treatments such as immunotherapy, which offers more targeted and potentially less harmful options.9 The immunomodulation represents a promising strategy to unleash the power of the immune system against cancer. The PD-1 (Programmed Death-1) and PD-L1 (Programmed Death-Ligand 1) pathway plays a crucial role in putting the brakes on the immune system, enabling cancer cells to evade immune surveillance. By targeting and inhibiting this pathway, the immune response can be reactivated, allowing it to spot and destroy more malignant cells effectively. Thus, this approach has the potential to revolutionize cancer treatment, turning the body's own defense mechanisms into powerful allies in the fight against malignancy.
Density Functional Theory (DFT) represents an advanced tool for evaluating theoretical aspects in material and medicinal sciences. The application of DFT in predicting and establishing conformational analysis in association with several biophysical parameters is conspicuous from the scientific reports made from the last two decades.10 To correlate DFT studies with biological interaction frontier orbital analysis, electron density mapping, charge distribution exploration, and hyperpolarizability play an important role. These aspects, in turn, help in understanding the non-covalent interaction (NCI) meant for explaining various mechanistic approaches encompassing the strategies to propose treatment for dreadful diseases like cancer, diabetes, AD, etc. Meanwhile, tremendous interest has been developed towards DFT-based investigations on some proposed PD-1-PD-L1 checkpoint inhibitors, viz, natural products, to find the suitability of such compounds for the treatment of breast cancer by invoking their global reactive descriptors and NCI characterization.11–13
Network pharmacology, an emerging field that leverages systems biology, integrates polypharmacology, molecular network data, bioinformatics, and computer simulations. This approach has garnered increasing interest due to the limitations of traditional drug development methods, which focus on a single drug targeting one disease through one pathway and often encounter difficulties with safety, effectiveness, and long-term viability. These methods involve using comprehensive biological data to create drugs that can target multiple pathways simultaneously. Recognizing that complex diseases like cancer involve intricate networks of genes, proteins, and biochemical pathways, network pharmacology systematically maps these interactions to identify key nodes for therapeutic targeting. By utilizing molecular targets and mapping cancer-related pathways, it elucidates the mechanisms by which multi-target phytochemicals like cosmosiin exert their effects, providing deeper insights into their therapeutic potential.14
Cosmosiin (“5-hydroxy-2-(4-hydroxyphenyl)-7-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxychromen-4-one”) also known as apigenin-7-glucoside and apigetrin, is a naturally occurring flavanone glycoside compound with a chemical formula of C21H20O10 and molecular weight 432.4g mol−1 is commonly present in various plants, including yarrow (Achillea millefolium) and Salvia plebeia.15,16 Cosmosiin has been utilized in conventional treatment owing to its health benefits. Initial research indicates that cosmosiin demonstrates a broad spectrum of bioactivities, encompassing antioxidant and immunomodulatory properties. Research on natural substances, such as cosmosiin, has received increased interest because to the growing need for safe and effective medicinal candidates.17,18 Its chemical structure has demonstrated pharmacological properties with significant therapeutic potential.19 Utilizing the therapeutic potential of cosmosiin in diverse disorders requires a thorough understanding of its action mechanisms and an assessment of its safety profile. Cosmosiin is an interesting natural chemical with several potential medicinal uses; it has just come to be known in this regard. Its attractive properties make it a promising contender for regulating cancer cell PD-L1 expression. Since PD-L1 overexpression in cancer cells is associated with immune evasion and tumor growth, it presents an excellent possibility for therapeutic intervention.20 As it pertains to cancer immunotherapy, there is a growing fascination with investigating natural candidates as potent modulators of PD-L1 expression.21
2. Material and methods
2.1. DFT based optimization, FMO analysis, charge distribution, and hyperpolarizability
Cosmosiin, the target molecule, was optimized at the DFT level to get its optimal structural shape or energy minimum state using a B3LYP as functional and 6-311G as basis set. This was followed by a frequency calculation to notice the absence of negative frequency. For the computations, the software package utilized was GAUSSIAN 09 (Revision C.01) with the graphical user interface GaussView 5.0.22,23
Using a combination of non-planar group symmetry and theoretical force constants, a number of systematic algorithms are able to derive practical results from the computation. Molecular orbital analysis and the inclusion of Koopman's theorem-based results were further analyzed using various derivative formulae.24 Electronegativity in terms of FMO energies, ionization energy (IE), electron affinity (EA), and the related parameters, absolute electronegativity (χabs), absolute hardness (η), electrophilicity index (ω), global softness (S)were computed:
| | |
χabs = (IE + IA)/2 = (EHOMO + ELUMO)/2
| (iii) |
| | |
η = (IE − IA)/2 = (EHOMO − ELUMO)/2
| (iv) |
The dipole moment (μ), mean polarizability (α) and the total first static Hyperpolarizability (β0) [69, 70] for the compound in terms of x, y, z components and is given by following equations:
| | |
μ = (μx2 + μy2 + μz2)1/2
| (vii) |
| | |
α = 1/3(αxx + αyy + αzz)
| (viii) |
| |
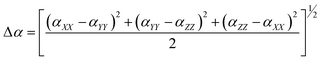 | (ix) |
| | |
β0 = (βx2 + βy2 + βz2)1/2
| (x) |
| and βx = βxxx + βxyy + βxzz |
(or)
| | |
β0 = [(βxxx + βxyy + βxzz)2 + (βyyy + βyzz + βyxx)2 + (βzzz + βzxx + βzyy)2]1/2
| (xi) |
2.2. Compound screening, cosmosiin target prediction, and breast carcinoma target genes
In network pharmacological analysis for compounds screening, crucial parameters include the Blood–Brain Barriers (BBB), drug-likeness, gastrointestinal absorption, and bioavailability. Drug likeness evaluates a compound's potential based on structural, molecular, and physicochemical properties. These metrics were obtained by inputting cosmosiin's SMILES formula into a server. Cosmosiin targets were identified using Super-PRED (“https://prediction.charite.de/index.php”) and the SwissTargetPrediction database. Uniprot provided official names and IDs for the predicted targets. Breast cancer-associated target genes were sourced from the GeneCards (https://www.genecards.org/) database using “breast cancer” as the keyword, and gene names were standardized with UniProt data. Venn diagrams created with venny 2.1 tool intersected cosmosiin targets with breast cancer targets. Overlapping targets were identified as potential anti-breast carcinoma targets for further analysis.
2.3. Protein-interaction network construction
The STRING database (“https://string-db.org/) was used to generate a protein–protein interaction (PPI) network for Homo sapiens by importing the overlapping genes. We applied a composite score in Cytos threshold to ensure the quality of interactions included in the network. After then, Cytoscape 3.10.2 software was used to analyze and visualize this PPI network in order to find important genes. Using the CytoHubba plugin in Cytoscape, we constructed and analysed a diagram highlighting the hub genes.
2.4. Enrichment analysis
GO and KEGG pathways analysis were performed to pinpoint potential targets of cosmosiin against breast cancer. GO functional enrichment analysis elucidated the toles of target proteins affected by cosmosiin gene function through differential gene enrichment. KEGG pathway enrichment provided insights into the involved genes and their corresponding pathways. The common targets shared between cosmosiin and breast carcinoma were uploaded and analysed through the ShinyGO 0.80 online tool (“https://bioinformatics.sdstate.edu/go/”). The pathways were prioritized based on their significance.
2.5. Differential expression
The differential-expression of the top three hub genes—NFKB1, PTGS2, and CDK5—was evaluated using an online tool called GEPIA2 (http://gepia2.cancer-pku.cn/), for exploring cancer transcriptomics. This gave insights into the expression of these genes across different stages of breast cancer; it is possible to find out the stage-specific nature of expression for these genes with GEPIA2, and is aware of how the gene is regulated during disease progression. In addition, survival analysis was used to evaluate their predictive significance. The DFS and OS metrics were used to measure the association amid the expressions of these hub genes and the clinical outcomes of BC patients. This revealed the potential role of the biomarkers NFKB1, PTGS2, and CDK5, whose expression could be related to the progression of BC and prognosis in BC patients. The comprehensive evaluation through GEPIA2 underscores the translational relevance of these hub genes in therapeutic and prognostic applications of BC.
2.6. Immune infiltration
The immune infiltration analysis of the hub genes NFKB1, PTGS2, and CDK5, we utilized the TIMER2.0 (“http://timer.cistrome.org/”). In a number of cancer types, the TIMER 2.0 methodically investigates the correlation between gene-expressions and the invasion of innate and adaptive immune cells. It utilizes several algorithms to infer the relative abundance of six subsets of immune cells: B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells in the tumor microenvironment. This research looked at the levels of immune infiltration into BC tissues and how their expression correlated with the hub genes. It implies how these genes may regulate the tumor immune microenvironment and which way the immune cell recruitment and function will be affected. TIMER2.0 also provides facilities for statistical analysis and visualization of the relationship, including the estimation of tumor purity and immune interactions, thereby highlighting the importance of these genes in breast cancer immunology.
2.7. In Silico molecular docking
The optimized structure of cosmosiin generated through DFT calculation was used for the docking study. The crystal structures of its three primary protein targets, NFKB1, CDK5, and PTGS2 and one CTLA4 protein as interference with other immune checkpoint-related pathways, were retrieved from the Protein Data Bank (PDB) with IDs 1NFK, 7VDR, 1CX2, and 3OSK, respectively.25–27 These targets, along with cosmosin, underwent molecular docking to evaluate the interactions between the ligand and the proteins' active sites. Before docking, the protein structures were optimized using BIOVIA Discovery Studio 2022 software to remove water molecules, heteroatoms, and ligands. AutoDock Vina version 1.2 was used for the docking process, and BIOVIA Discovery Studio 2022 was utilized for visualization and analysis. Additionally, the CB-DOCK2 online server was employed to create cartoon surface representations of the target proteins and identify their active cavities.
2.8. Molecular dynamics simulations
CABS-flex (“https://biocomp.chem.uw.edu.pl/CABSflex2/about”) is a web server for modeling protein flexibility, which is very useful in generating near-native protein dynamics. It uses coarse-graining to reduce the computational cost of predicting structural fluctuations, compared to classical molecular dynamics simulations. Protein structures in the PDB format are used as input, and models and fluctuation profiles are generated as output to analyze protein dynamics. The new CABS-flex 2.0 handles larger and multimeric proteins, with additional functionality for contact maps and customized restraints. This study uses CABS-flex to model interactions between the hub genes CDK5, NFKB1, PTGS2, and the complex formed with cosmosiin, which revealed insights into their dynamics.
2.9. Chemicals and reagents
All reagents and chemicals were of analytical grade and consisted of Cosmosiin (Sigma-Aldrich Co. LLC, USA), dimethyl sulfoxide (DMSO; Sigma-Aldrich Co. LLC, USA), RPMI-1640 medium (Gibco, Thermo Fisher Scientific, USA), fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, USA), penicillin–streptomycin solution (Gibco, Thermo Fisher Scientific, USA), MTT (thiazolyl blue tetrazolium bromide; Sigma-Aldrich Co. LLC, USA), Click-iT™ EdU Imaging Kit (Thermo Fisher Scientific, USA), Annexin V-FITC/PI Apoptosis Detection Kit (BD Biosciences, USA), Transwell® inserts (8.0 μm PET; Corning Life Sciences, USA), RIPA buffer (Thermo Fisher Scientific, USA), BCA protein assay reagents (Thermo Fisher Scientific, USA), PVDF membranes (Immobilon-P; Merck KGaA, Germany), and ECL substrate (Bio-Rad Laboratories, USA); primary antibodies were anti-CDK5 (rabbit monoclonal; Cell Signaling Technology, Inc., USA), anti-NFKB1 (rabbit polyclonal; Abcam Ltd, UK), anti-PTGS2 (rabbit polyclonal; Santa Cruz Biotechnology, Inc., USA), and anti-GAPDH (mouse monoclonal; Sigma-Aldrich Co. LLC, USA), with HRP-conjugated goat anti-rabbit IgG secondary antibody (Jackson ImmunoResearch Laboratories, Inc., USA).
2.10. Cell culture and conditions
MCF-7 (breast adenocarcinoma) and MCF-10 (non-tumorigenic mammary epithelial) cell lines were obtained from ATCC (10801 University Boulevard, Manassas, VA 20110, USA). Upon receipt, mycoplasma-free cell authentication was achieved by the use of short tandem repeat profiling. Cultures were maintained in RPMI-1640 supplemented with 10% FBS and 100 U per mL penicillin–100 μg mL−1 streptomycin, at 37 °C in a humidified incubator with 5% CO2.
2.11. MTT assay against MCF-10 and MCF-7 cells
Cells were seeded at 1 × 104 cells per well in 96-well plates and allowed to adhere for 24 h. Cosmosiin (0, 10, 50, 100, 200 μM) was prepared in DMSO (final DMSO ≤ 0.1%) and added to each well in triplicate. After 48 h of treatment, 20 μL of 5 mg mL−1 MTT solution was added and plates were incubated for an additional 4 h at 37 °C. The medium was carefully removed and formazan crystals were solubilized in 150 μL DMSO. Absorbance at 570 nm was measured with a BioTek ELx800 microplate reader, and cell viability percentages were calculated relative to vehicle controls.
2.12. Phase-contrast microscopy
MCF-7 cells were plated in 6-well plates at 2 × 105 cells per well and treated with cosmosiin concentrations for 24 h. Morphological alterations—such as changes in cell size, shape, and adherence—were documented using an Olympus IX73 inverted phase-contrast microscope (Olympus Corporation, Tokyo, Japan) fitted with a 10× objective. Representative fields were imaged and analyzed for evidence of cytotoxicity and cell death.
2.13. EdU (5-ethynyl-2′-deoxyuridine) assay
Proliferative capacity was assessed by 5-ethynyl-2′-deoxyuridine (EdU) incorporation. After 24 h of cosmosiin treatment, cells were incubated with 10 μM EdU for 2 h, then fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Click-iT™ reaction cocktail was applied according to the manufacturer's protocol (Thermo Fisher Scientific) to fluorescently label incorporated EdU. Nuclei were counterstained with DAPI, and images were captured on the Olympus IX73. The percentage of EdU-positive cells was quantified from five random fields per sample.
2.14. Annexin V/PI assay
Apoptosis was assessed using dual staining with Annexin V-FITC and propidium iodide. Following 24 h of treatment, cells were harvested, washed twice in cold PBS, and resuspended in binding buffer. Annexin V-FITC (5 μL) and PI (5 μL) were added to 1 × 105 cells and incubated in the dark for 15 min at room temperature. Samples were analyzed on a BD FACSCanto™ II flow cytometer (BD Biosciences) within 1 h, and data were processed using FlowJo software to distinguish viable, early and late apoptotic, and necrotic populations.
2.15. Transwell migration assay
Cell migratory potential was evaluated using Corning Transwell inserts (8 μm pore size). Serum-starved MCF-7 cells (5 × 104) in 200 μL serum-free medium containing cosmosiin were seeded into the upper chambers, while lower chambers contained 600 μL medium supplemented with 10% FBS as chemoattractant. After 24 h at 37 °C, non-migrated cells were excised from the top surface using a cotton swab. Cells that migrated to the bottom membrane were fixed with methanol, stained with 0.1% crystal violet for 15 min, washed thoroughly, and counted under a Nikon Eclipse TS2-IN inverted microscope in five random fields per insert.
2.16. Western blotting
Total protein extracts were obtained by lysing cells in ice-cold RIPA buffer containing protease and phosphatase inhibitors. Lysates were clarified by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 15 min at 4 °C, and protein concentration was determined by BCA assay. Equal amounts (30 μg) of protein were separated on 10% SDS–PAGE gels using the Bio-Rad Mini-PROTEAN® Tetra System and transferred onto PVDF membranes (MilliporeSigma, Merck KGaA) with the Trans-Blot® Turbo Transfer System (Bio-Rad). Membranes were blocked in 5% BSA for 1 h, then incubated overnight at 4 °C with primary antibodies: anti-CDK5 (1
000×g for 15 min at 4 °C, and protein concentration was determined by BCA assay. Equal amounts (30 μg) of protein were separated on 10% SDS–PAGE gels using the Bio-Rad Mini-PROTEAN® Tetra System and transferred onto PVDF membranes (MilliporeSigma, Merck KGaA) with the Trans-Blot® Turbo Transfer System (Bio-Rad). Membranes were blocked in 5% BSA for 1 h, then incubated overnight at 4 °C with primary antibodies: anti-CDK5 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-NFKB1 (1
1000), anti-NFKB1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), anti-PTGS2 (1
500), anti-PTGS2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), and anti-GAPDH (1
500), and anti-GAPDH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000). We probed the membranes for 1 hour at room temperature with HRP-conjugated goat anti-rabbit or anti-mouse secondary antibodies (1
2000). We probed the membranes for 1 hour at room temperature with HRP-conjugated goat anti-rabbit or anti-mouse secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) after washings in TBS-T. A combination of increased chemiluminescence and densitometry in ImageJ allowed for the visualization of protein bands and their subsequent quantification.
5000) after washings in TBS-T. A combination of increased chemiluminescence and densitometry in ImageJ allowed for the visualization of protein bands and their subsequent quantification.
2.17. Statistical analysis
All experiments were performed in triplicate, and data are expressed as mean ± standard deviation (SD). Statistical analysis was done using GraphPad Prism version 10.4.2 (GraphPad Software, San Diego, CA). One-way analysis of variance (ANOVA) was used to compare groups, followed by Tukey's multiple comparisons test to determine specific group differences. A p-value below 0.05 was regarded as statistically significant, and p-values below 0.01 were regarded as highly significant. Significance levels are noted in the figures and their respective legends.
3. Results and discussion
3.1. DFT (Density Functional Theory) aspects
The optimized cosmosiin structure generated through density functional level of theory under the application of 6311 G/B3LYP formalism is given in Fig. 1. The respective labels and symbols are highlighted on the model with the respective color codes of different atoms present in the target complex. The results indicate that the given structure is energy minimal with the respective nuclear–nuclear, electron-nuclear and kinetic energy (au) respectively as 3.023, −9.706 and 1.56. The absence of imaginary frequency in the respective IR-spectrum (Fig. 2) computed through the same level of theory supports that the suggested structure bears the lowest energy among several possible conformations.
 |
| | Fig. 1 Optimized structure of cosmosiin. | |
 |
| | Fig. 2 Theoretical IR spectrum to show absence of imaginary frequency. | |
Similarly, the molecular orbital analysis of the compound has been given in Fig. 3. With the respective global reactive descriptors calculated through the application of Koopman's theorem. The results indicate 4.08 eV HOM-LUMO gap, absolute electronegativity (χabs) 4.337, absolute hardness (η) 2.047, electrophilicity index (ω) 13.00 and global softness (S) 0.488.
 |
| | Fig. 3 HOMO–LUMO representation of cosmosiin. | |
Likewise, electrostatic potential surface represented by Fig. 4, displays electronegative, electropositive and neutral zones present on the computed topography of the complex under +7e and −7e extreme fields. The resulting mesh shows both positive and negative potentials with the red and blue coloration, respectively, but the major portion is green suggesting neutral or halfway potential of the compound. These surface potentials are important for discussing intermolecular interactions. This is further explained by the Mulliken charge analysis displayed in Table 1 and graphically represented in Fig. 5. The hyperpolarizability tensors given in Table 2 are first, second and third order dipole tensors showing most of the values to be negative.
 |
| | Fig. 4 MESP diagram of cosmosiin. | |
Table 1 Charge distribution analysis of the compound
| Atom assignment |
Charge |
Atom assignment |
Charge |
| 1-O |
−0.512 |
26-C |
−0.160 |
| 2-O |
−0.510 |
27-C |
−0.038 |
| 3-O |
−0.611 |
28-C |
−0.038 |
| 4-O |
−0.578 |
29-C |
−0.208 |
| 5-O |
−0.596 |
30-C |
−0.188 |
| 6-O |
−0.582 |
31-C |
0.288 |
| 7-O |
−0.566 |
32-H |
0.157 |
| 8-O |
−0.608 |
33-H |
0.205 |
| 9-O |
−0.472 |
34-H |
0.200 |
| 10-O |
−0.609 |
35-H |
0.160 |
| 11-C |
0.044 |
36-H |
0.186 |
| 12-C |
0.014 |
37-H |
0.196 |
| 13-C |
0.065 |
38-H |
0.178 |
| 14-C |
−0.010 |
39-H |
0.383 |
| 15-C |
0.279 |
40-H |
0.358 |
| 16-C |
−0.065 |
41-H |
0.385 |
| 17-C |
0.262 |
42-H |
0.377 |
| 18-C |
−0.171 |
43-H |
0.216 |
| 19-C |
−0.161 |
44-H |
0.198 |
| 20-C |
0.296 |
45-H |
0.189 |
| 21-C |
−0.208 |
46-H |
0.395 |
| 22-C |
0.342 |
47-H |
0.178 |
| 23-C |
0.329 |
48-H |
0.196 |
| 24-C |
0.253 |
49-H |
0.161 |
| 25-C |
−0.160 |
50-H |
0.182 |
| 51-H |
0.377 |
 |
| | Fig. 5 Mulliken charge analytical graph of cosmosiin. | |
Table 2 Hyperpolarizability tensors of the compound
| Dipole moment (μ) |
Hyperpolarizability (β) |
| μx |
−3.293 |
βxxx |
−463.3123 |
| μy |
−6.371 |
βyyy |
−146.3697 |
| μz |
−1.348 |
βzzz |
−2.7883 |
| μ total |
7.297 |
βxyy |
126.3531 |
| Polarizability(α) |
βxxy |
−113.0148 |
| αxx |
−173.048 |
βxxz |
66.4833 |
| αyy |
−203.683 |
βxzz |
31.0647 |
| αzz |
184.462 |
βyzz |
1.6420 |
| αxy |
−5.004 |
βyyz |
−35.5793 |
| αxz |
3.018 |
βxyz |
32.8099 |
| αyz |
14.391 |
|
|
| α total |
282.494 |
|
|
DFT based studies in the geometry optimization indicate that the compound bears more positive charge bearing capacity or hydrogen accepting property which is a very important character in QSAR studies. This is also supported by the FMO analysis associated with the global reactive descriptors. One of the most significant result derived from DFT based characterization is the intermolecular interaction supported MESP and Mulliken analysis. Such properties represent drug likeness of the compound stemmed by the analysis derived from ADME and NP discussed later. To prove or infer whether a compound is able to interact non-covalently DFT proves significant. In the present case electrostatic mapping and partial charge results also suggest that the cosmosiin bears potent drug properties.28,29
3.2. Drug likeness characteristics of cosmosiin
Cosmosiin has desirable drug-like properties for breast cancer immunotherapy (Table 3). The Consensus Log Po/w is 0.52, showing moderate lipophilicity. It has four rotatable bonds, ensuring structural flexibility. It possesses 10 hydrogen bond acceptors and 6 hydrogen bond donors, allowing for strong interactions with biological target molecules. The topological polar surface area measures 170.05 Å2, which indicates a target-sensitive compound. Cosmosiin is soluble in water; its gastrointestinal absorption is however very low, and it is not capable of penetrating the blood–brain barrier. Cosmosiin has a bioavailability score of 0.55, thus being suggesting druglikeness characteristics. Furthermore, no toxicities were noted to be significant with cosmosiin, such as hepatotoxicity, neurotoxicity, cytotoxicity, cardiotoxicity, or nephrotoxicity, which makes this compound an interesting candidate for further study in breast cancer immunotherapy.
Table 3 The physicochemical properties essential for a potential drug candidate were predicted using SwissADME, while toxicity was assessed with Pro-Tox 3.0. Cosmosiin was found to have moderate water solubility, good bioavailability, low gastrointestinal absorption, and no BBB permeability
| S. no. |
Descriptor |
Value |
| 1 |
Formula |
C21H20O10 |
| 2 |
Molecular weight |
432.38 g mol−1 |
| 3 |
Consensus log Po/w |
0.52 |
| 4 |
Rotatable bonds |
4 |
| 5 |
Hydrogen bond acceptors |
10 |
| 6 |
Hydrogen bond donors |
6 |
| 7 |
TPSA |
170.05 Å2 |
| 8 |
Water solubility |
Soluble |
| 9 |
GI absorption |
Low |
| 10 |
Blood brain barrier (BBB) permeability |
No |
| 11 |
Bioavailability score |
0.55 |
| 12 |
Drug likeness |
Yes |
| 14 |
Hepatotoxicity |
No |
| 15 |
Neurotoxicity |
No |
| 16 |
Cytotoxic |
No |
| 17 |
Cardiotoxicity |
No |
| 18 |
Nephrotoxicity |
No |
The physicochemical profile and safety of cosmosiin place it as a promising small-molecule modulator of the PD-1/PD-L1 axis with advantages over current checkpoint inhibitors. This stands in stark contrast to many approved PD-1/PD-L1 monoclonal antibodies and emerging small-molecule inhibitors, which frequently trigger immune-related adverse events such as colitis, hepatitis or endocrine dysfunction, or exhibit off-target liabilities that manifest as cardiac or renal injury. By combining targeted checkpoint modulation with an exceptionally clean safety profile, cosmosiin holds the potential to deliver anti-tumor immune activation with substantially reduced risk of the severe toxicities that limit dosing and patient quality of life with existing therapies.
3.3. Predicted biological targets for cosmosiin and intersection with breast cancer targets
The search for biological targets of cosmosiin was performed using the databases including SwissTargetPrediction and SuperPred. After removing the duplicates and applying screening criteria, a total of 38 targets remained. The breast cancer-related targets were sourced from the GeneCards database by searching the term “Breast cancer” leading to the identification of 1314 target genes. The 38 cosmosiin targets and 1314 disease targets were searched for intersection using Venny 2.1, leading to the recognition of 25 overlapping targets for cosmosiin with breast cancer (Fig. 6).
 |
| | Fig. 6 Venn diagram depicting the intersection of predicted biological targets of cosmosiin (38) with breast cancer-related targets (1314), identifying 25 overlapping targets. | |
3.4. PPI (protein–protein interaction) network and hub genes
A PPI network for 25 identified intersecting genes was created using the STRING database, which relies on experimental evidence and probabilistic prediction of PPIs (Fig. 7A). The PPI network was exported in a .tsv format with a confidence score set at 0.4, securing that only reliable interactions were used; unconnected nodes were removed to increase cohesion, and the final PPI was visualized using Cytoscape. The network showed 25 and 135, nodes and edges respectively, with an average number of neighbours per node of 10.8 and a network density of 0.225, indicating moderate connectivity (Fig. 7B). Using the CytoHubba plugin, the top 10 hub genes-key interaction targets were identified: PTGS2, NFKB1, PTPN1, NTRK3, CYP3A4, CDK4, CDK5, CXCR4, NTRK1, and HIF1A (Fig. 7C), highlighting their central roles in the regulatory architecture of the network.
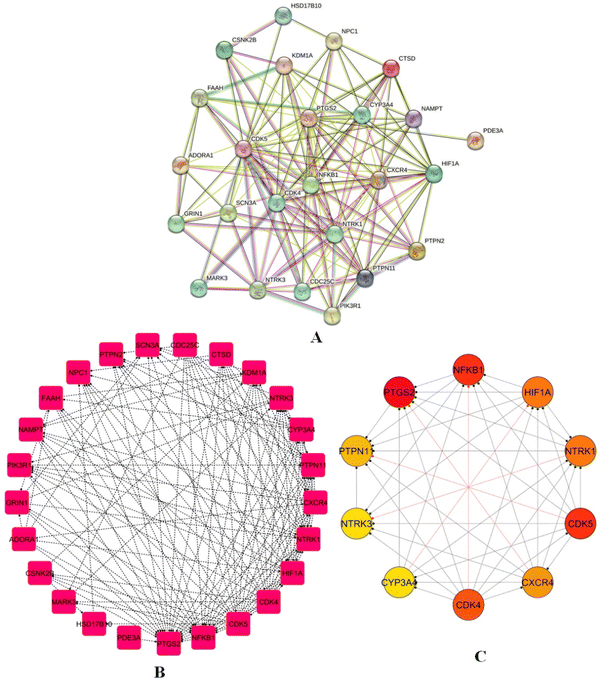 |
| | Fig. 7 PPI network (A) PPI network for 25 intersecting genes using the STRING database, illustrating interactions with a confidence score ≥ 0.4. (B) Refined PPI network visualized in cytoscape, comprising 25 nodes and 135 edges, demonstrating connectivity metrics such as average neighbours (10.8) and network density (0.225). (C) Top 10 hub genes: PTGS2, NFKB1, PTPN1, NTRK3, CYP3A4, CDK4, CDK5, CXCR4, NTRK1, and HIF1A, highlighting central regulatory roles. | |
3.5. Gene ontology and pathway enrichment
GO and KEGG enrichment of the intersecting targets of cosmosiin with breast cancer targets using ShinyGo 0.80 highlighted their involvement in critical BP, CC and MF. Key BPs of intersecting targets of cosmosiin include regulation of protein serine/threonine phosphatase activity, positive regulation of activin receptor signalling, adiponectin-activated pathways, endothelial morphogenesis, and blood vessel endothelial cell migration, pointing to roles in angiogenesis and cellular signalling (Fig. 8A). The cellular components which include the protein kinase CK2 complex, PcG protein complex, secretory granules, and vesicle lumens were implicated in intracellular signalling and vesicle-mediated transport (Fig. 8B). The enriched MFs included kinase regulation, DNA-binding transcription-factor binding, and transferase activity highlighting the role of the targets in phosphorylation and also in transcriptional regulation (Fig. 8C). KEGG pathway analysis found significant enrichment in cancer-related pathways, including PD-L1 expression and regulation of the PD-1 checkpoint, central carbon metabolism in cancer, neurotrophin signalling, and cAMP signalling, along with chronic myeloid leukemia, small cell lung cancer, and Kaposi sarcoma-associated herpesvirus infection, as well as Alzheimer's (Fig. 9A). These findings relate potential of cosmosiin to modulate key oncogenic and immune-related mechanisms in the therapeutic role of breast cancer. Additionally, a gene map was constructed for the biological targets of cosmosiin in the enriched pathway PD-L1 and regulation of the PD-1 checkpoint showing their active involvement in its modulation (Fig. 9B).
 |
| | Fig. 8 Gene ontology (A) enrichment analysis of biological processes for intersecting targets, emphasizing roles in angiogenesis and cellular signalling. (B) GO cellular components analysis of intersecting targets, showing involvement in intracellular signalling and vesicle-mediated transport. (C) GO molecular functions analysis, identifying enrichment in kinase regulation, phosphorylation, and transcriptional regulation. | |
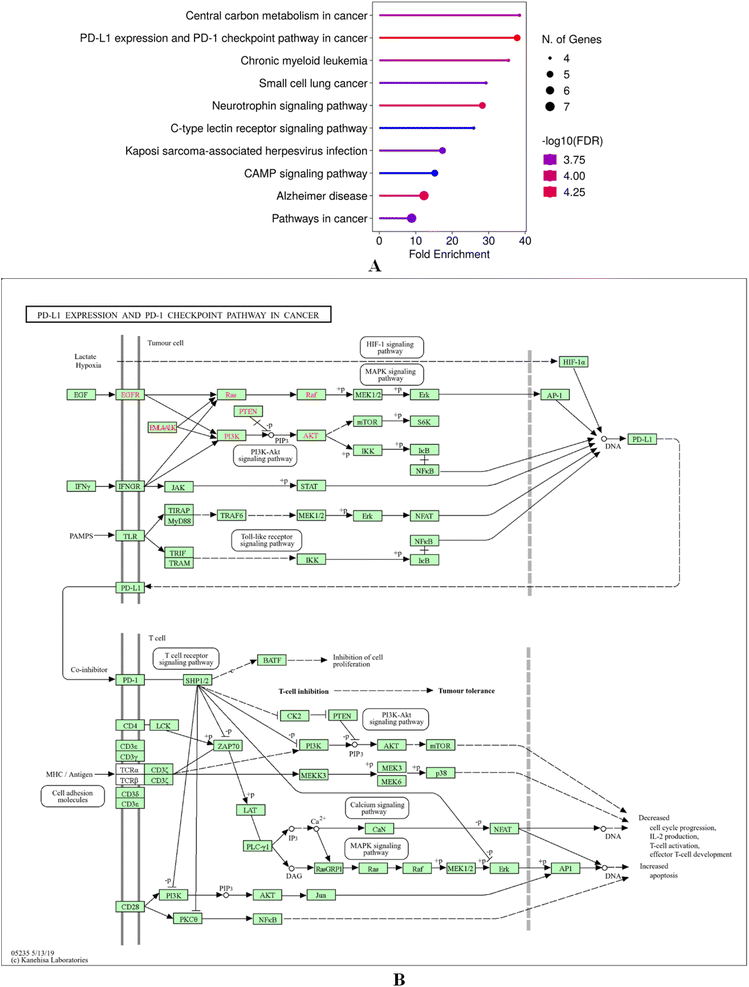 |
| | Fig. 9 (A) KEGG pathway enrichment analysis highlighting cancer-related pathways, including PD-L1 regulation, neurotrophin signalling, and central carbon metabolism in cancer. (B) Gene map of cosmosiin targets enriched in the PD-L1/PD-1 pathway, showcasing their regulatory roles. | |
3.6. Gene targets and enriched pathway interactions through sankey plot
A Sankey plot was created to visually represent the interactions of intersecting targets between cosmosiin and breast cancer, mapped into the involvement of these targets with enriched pathways (Fig. 10). The diagram well represents the flow of targets associated with different pathways, where PD-L1/PD-1 pathway shown to be the most significant. Visually, this is highlighted using the node and flow properties of the Sankey plot, which depicts larger and more vibrant nodes to represent higher counts and more significant p-values. These visualizations take attention to the critical pathways supporting the biological relevance of cosmosiin in the way it modulates immune checkpoints in regulation of breast cancer. The plot integrates these data with pathway analysis and provides an intuitive understanding of the target distribution across pathways, allowing insight into therapeutic potential and pathway prioritization.
 |
| | Fig. 10 Sankey plot depicting interactions between intersecting targets of cosmosiin and enriched pathways, with emphasis on PD-L1/PD-1 checkpoint involvement. | |
3.7. Differential gene expression, survival and disease stage analysis
The analysis of the hub genes CDK5, NFKB1, and PTGS2 in the case of breast cancer revealed distinct expression patterns and prognostic implications. Box plots in CDK5 displayed a strongly upregulated expression in tumor vs. normal tissues (Fig. 11A). Such upregulation was observed across all stages of breast cancer, as shown by stage plots (Fig. 11B). In addition, survival studies indicated higher CDK5 expression was correlated with low OS and DFS, which makes it a good prognostic biomarker with poor outcome (Fig. 11C and 6D). NFKB1 also showed upregulation in cancer tissues compared with normal tissues (Fig. 12A). Stage-wise analysis showed steady upregulation across the progression stages of cancer, and its expression profile was much like that of CDK5 (Fig. 12B). Survival analyses indicated its participation in disease progression and relevance in the prognosis of breast cancer (Fig. 12C and D). PTGS2 showed completely opposite patterns with considerable lower mRNA expression in tumor tissues compared to normal tissues (Fig. 13A–D). Such behavior suggests a potentially unique role in tumor biology, possibly as a tumor suppressor or a factor that becomes aberrant following the progression of cancer.
 |
| | Fig. 11 Expression and survival analysis of (A) hub gene CDK5 in tumor vs. normal tissues, (B) upgradation in stage plots, (C) and (D) low OS and DFS, respectively at higher CDK5 expression. | |
 |
| | Fig. 12 (A) NFKB1 upregulation in cancer tissues versus normal tissues, (B) stage-wise analysis in the expression profile, (C and D) survival analyses and disease progression in the prognosis of breast cancer. | |
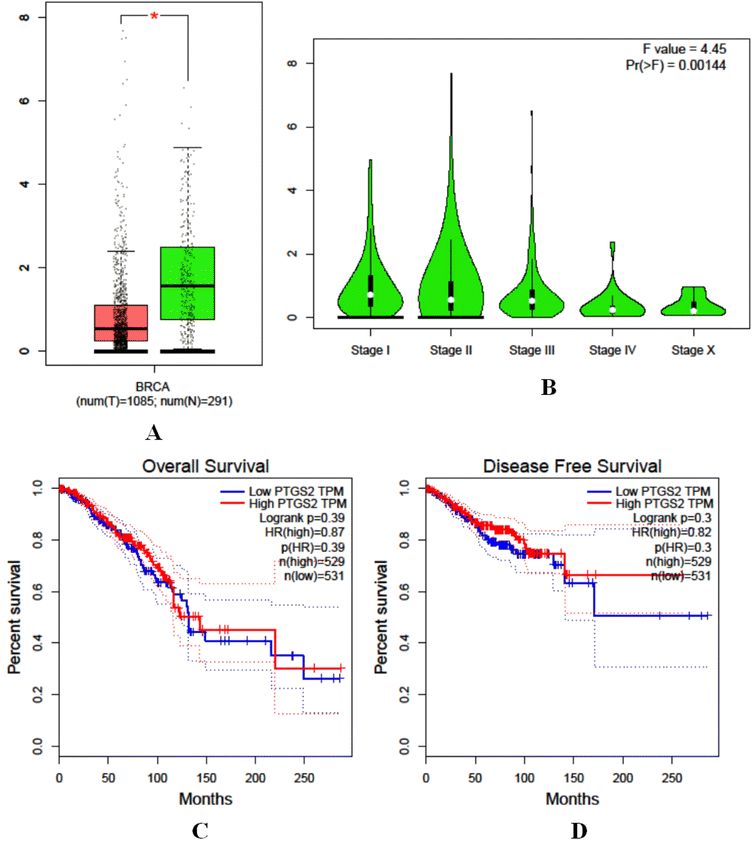 |
| | Fig. 13 Expression and survival analysis of hub gene PTGS2, (A) lower mRNA expression in tumour tissues than normal tissue, (B) stage analysis, (C) survival analysis and (D) progression/relevance insights. | |
3.8. Correlation between hub gene expression and immune infiltration
The TIMER analysis indicated the relationship between gene expression and the immune system penetration by different types of immune cells, and the findings demonstrated that the production of NFKB1 and PTGS2 showed a strong correlation with the immune infiltration by various immune cells especially CD8+ cells, CD4+ cells, macrophages, neutrophil and dendritic cells. For CDK5 hub gene, no such correlation was observed for any of the immune cells (Fig. 14).
 |
| | Fig. 14 Correlation between hub genes (NFKB1, PTGS2, CDK5) and immune infiltration in BC, evaluated using TIMER2.0. | |
3.9. Molecular docking and dynamic insights into top hub genes and cosmosiin interaction
Molecular docking studies elucidated binding affinities and interaction profiles of cosmosiin with the CDK5, NFKB1, and PTGS2 hub proteins and CTLA4. For the cosmosiin-CDK5 complex, Vina docking score is as high as −8.5 kcal mol−1, indicating strong binding affinity (Table 4). The interaction occurs within a pocket of 3358 Å3 volume, and the coordinates are situated at (53, −45, 65) with docking size as (23, 35, 23). Although the donor–donor interactions appearing at the terminal end are unfavorable, van der Waals forces, conventional hydrogen-bonds, and Pi-based interactions like Pi-Sigma and Pi-Alkyl contribute to the overall stability of the complex (Fig. 15A).
Table 4 Molecular docking outcomes obtained from CB-Dock2 server by performing cavity based blind docking
| Complex |
CurPocket ID |
Vina score |
Cavity volume (Å3) |
Center (x, y, z) |
Docking size (x, y, z) |
| Cosmosiin-CDK5 |
C1 |
−8.5 |
3358 |
53, −45, 65 |
23, 35, 23 |
| Cosmosiin-NFKB1 |
C1 |
−7.6 |
1118 |
−19, −24, 30 |
23, 23, 23 |
| Cosmosiin-PTGS2 |
C1 |
−9.8 |
20723 |
36, 34, 58 |
35, 32, 32 |
| Cosmosiin-CTLA4 |
C2 |
−6.6 |
110 |
−5, −8, 31 |
23, 23, 23 |
| Curcumin-CDK5 |
C1 |
−7.7 |
3358 |
53, −45, 65 |
26, 35, 26 |
| Curcumin-NFKB1 |
C1 |
−7.5 |
1118 |
−19, −24, 30 |
26, 26, 26 |
| Curcumin-PTGS2 |
C1 |
−8.6 |
20723 |
36, 34, 58 |
35, 32, 32 |
| Curcumin-CTLA4 |
C1 |
−5.8 |
141 |
−9, 1, 34 |
26, 26, 26 |
 |
| | Fig. 15 Molecular docking of cosmosiin with hub genes represented by ribbon models and 2D interaction models (A) CDK5 (−8.5 kcal mol−1) interactions: van der Waals, hydrogen bonds, Pi-Sigma, Pi-Alkyl, and unfavorable donor–donor. (B) NFKB1 (−7.6 kcal mol−1) interactions: van der Waals, Pi-Cation, and Pi-Alkyl. (C) PTGS2 (−9.8 kcal mol−1) interactions: van der Waals, hydrogen bonds, and Pi-Alkyl. (D) Ribbon structure of the docked complex with the possible interactions. | |
In the cosmosiin-NFKB1 complex, the binding score was −7.6 kcal mol−1, with the interaction localized in a smaller cavity of 1118 Å3, centered at (−19, −24, 30), and dimensions of (23, 23, 23) (Table 4). The binding is stabilized by van der Waals forces, Pi-Cation, and Pi-Alkyl interactions, showcasing the compound's ability to engage hydrophobic and charged regions within the binding pocket (Fig. 15B).
The cosmosiin-PTGS2 complex showed highest docking score corresponding to the binding affinity of −9.8 kcal mol−1 indicating good binding in a large cavity of 20723 Å3 in size (Table 4). It falls centered at (36, 34, 58) with dimensions of (35, 32, 32). van der Waals interactions along with conventional hydrogen bonds and Pi-Alkyl play a role here, indicating a very well-optimized fit within the active site of this protein (Fig. 15C). This leads to a meaningful indication that cosmosiin has major binding potential to these target proteins and it is therapeutically important in breast cancer.
The cosmosiin-CTLA4 complex exhibited a moderate binding affinity with a docking score of −6.6 kcal mol−1, indicating stable engagement within a compact C2 pocket of 110 Å3 (Table 4). This cavity is centered at (−5, −8, 31) and spans dimensions of 23 × 23 × 23 Å, reflecting a snug fit. Key van der Waals contacts anchor the flavonoid core to the pocket walls, while conventional hydrogen bonds, particularly between hydroxyl groups of cosmosiin and backbone carbonyls, and Pi-Alkyl interactions with surrounding hydrophobic residues further stabilize the complex (Fig. 15D). Together, these interactions suggest cosmosiin is well-optimized to occupy this confined region of CTLA-4, potentially obstructing its ligand-binding interface.
The structural dynamics of the CDK5, NFKB1, and PTGS2 hub proteins and CTLA-4 in complex with cosmosiin are promising according to the CABS-flex analysis (Fig. 16–19). Models showed stability in their conformational states with cosmosiin binding consistently at regions critical for protein activity. A contact map detailed the persistent interactions found, especially in the active or regulatory domains, suggesting that cosmosiin could modulate the functional attributes of these proteins. Furthermore, RMSF analysis proved the reduced flexibility in particular functional domains on cosmosiin binding, showing enhanced structural rigidity and possible stabilization effects. These results highlight the promise of the use of cosmosiin in the targeting of such hub genes as therapeutic drugs.
 |
| | Fig. 16 CABS-flex 2.0 MD simulations of the CDK5-cosmosiin complex display structural dynamics. MD Model Created by CABS-flex: According to the CABS coarse-grained method, the CDK5-cosmosiin complex's main conformational states show persistent binding of cosmosiin to crucial interaction sites. Contact map: this displays residue–residue interactions in CDK5-cosmosiin. The stabilize cosmosiin-induced contacts within the CDK5 protein; thus, the darker patches denote strong and persistent contacts, particularly within the binding interface. RMSF Graph: This measures residue flexibility during simulation. Lower RMSF findings in crucial CDK5 areas imply decreased mobility following binding with cosmosiin, suggesting the chemical might stabilize functionally relevant domains. | |
 |
| | Fig. 17 CABS-flex 2.0 MD simulations of the NFKB1-cosmosiin complex display structural dynamics. MD Model Created by CABS-flex: according to the CABS coarse-grained method, the NFKB1-cosmosiin complex's main conformational states show persistent binding of cosmosiin to crucial interaction sites. Contact map: this displays residue–residue interactions in NFKB1-cosmosiin. The stabilize cosmosiin-induced contacts within the NFKB1 protein; thus, the darker patches denote strong and persistent contacts, particularly within the binding interface. RMSF graph: this measures residue flexibility during simulation. Lower RMSF findings in crucial NFKB1 areas imply decreased mobility following binding with cosmosiin, suggesting the chemical might stabilize functionally relevant domains. | |
 |
| | Fig. 18 CABS-flex 2.0 MD simulations of the PTGS2-cosmosiin complex display structural dynamics. MD Model Created by CABS-flex: according to the CABS coarse-grained method, the PTGS2-cosmosiin complex's main conformational states show persistent binding of cosmosiin to crucial interaction sites. Contact map: this displays residue–residue interactions in PTGS2-cosmosiin. The stabilize cosmosiin-induced contacts within the PTGS2 protein; thus, the darker patches denote strong and persistent contacts, particularly within the binding interface. RMSF graph: this measures residue flexibility during simulation. Lower RMSF findings in crucial PTGS2 areas imply decreased mobility following binding with cosmosiin, suggesting the chemical might stabilize functionally relevant domains. | |
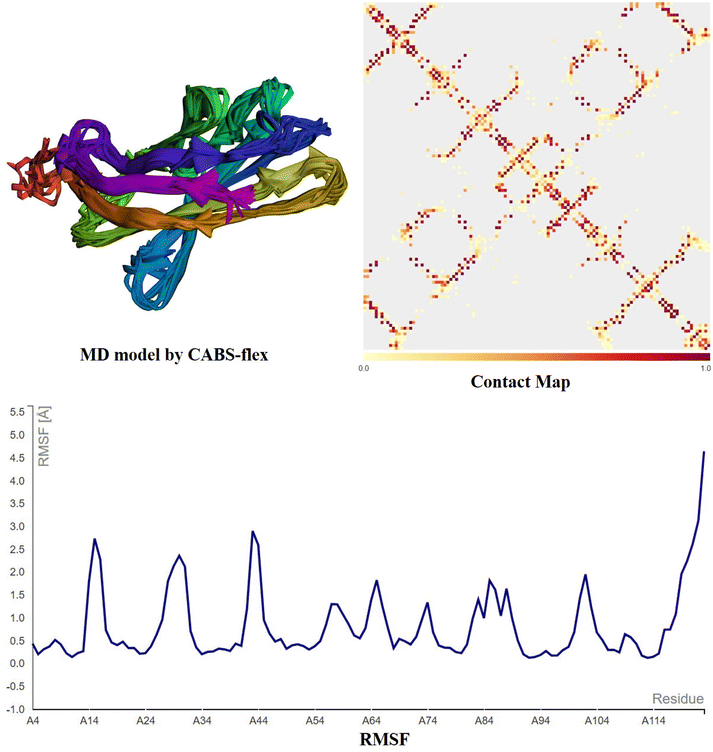 |
| | Fig. 19 CABS-flex 2.0 MD simulations of the CTLA4-cosmosiin complex display structural dynamics. MD Model Created by CABS-flex: according to the CABS coarse-grained method, the CTLA4-cosmosiin complex's main conformational states show persistent binding of cosmosiin to crucial interaction sites. Contact map: this displays residue–residue interactions in CTLA4-cosmosiin. The stabilize cosmosiin-induced contacts within the CTLA4 protein; thus, the darker patches denote strong and persistent contacts, particularly within the binding interface. RMSF graph: this measures residue flexibility during simulation. Lower RMSF findings in crucial CTLA4 areas imply decreased mobility following binding with cosmosiin, suggesting the chemical might stabilize functionally relevant domains. | |
The strong binding affinity of cosmosiin to CDK5, NFKB1, and PTGS2, demonstrated in the docking studies, implies its ability to modulate their activity significantly. The complex cosmosiin-CDK5 has a good binding score of −8.5 kcal mol−1 with a variety of stabilizing interactions, including van der Waals forces and Pi-based interactions. A binding score of −7.6 kcal mol−1 was similarly observed for cosmosiin with NFKB1, which indicates moderate but important interaction potential. The strongest binding is with PTGS2 (−9.8 kcal mol−1), which very well optimizes the fit in its active site. The cosmosiin-CTLA4 complex exhibited a moderate binding affinity with a docking score of −6.6 kcal mol−1. Cosmosiin binds to the binding sites of CDK5, NFKB1 and PTGS2 with very favorable complementarity, occupying pockets with combination of different interactions. Since these three targets directly feed into the PD-1/PD-L1 checkpoint signalling pathway, such strong docking indicates cosmosiin may significantly interfere with or modulate the pathway. By comparison, its binding to the far smaller CTLA-4 pocket is less apparent, stable nonetheless, but with fewer, more limited interactions, indicating only a modest effect on CTLA-4-mediated immune control. Collectively, these results suggest a strong likelihood of cosmosiin on PD-1/PD-L1-driven immunity, with relatively less severe impact on the CTLA-4 pathway. Finally, their expression profile in normal vs. tumor tissues is essential for stratification of patients and designs for tailored therapies.
3.10. Cosmosiin induces selective cell proliferation inhibition
A substantial decrease in MCF-7 cell viability was detected by the MTT assay following 24-hour treatment with cosmosiin (Fig. 20A). At concentrations of 10, 50, 100, and 200 μM, cell viability decreased to 76%, 31.5%, 15.2%, and 4.8%, respectively (p < 0.05 and 0.01 vs. vehicle control). In contrast, non-tumorigenic MCF-10 cells exhibited no significant cytotoxicity, indicating selective activity against cancer cells. Vehicle controls (0.1% DMSO) showed no adverse effects on viability in either cell line.
 |
| | Fig. 20 Cosmosiin reduces viability and alters morphology of MCF-7 cells (A) MTT assay quantification of MCF-7 and non-tumorigenic MCF-10 cell viability after 24 h treatment with 0 (vehicle), 10, 50, 100, or 200 μM cosmosiin. Data are mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus vehicle control. (B) Representative phase-contrast micrographs of MCF-7 cells following 24 h cosmosiin exposure. At 50 μM, cells show cytoplasmic shrinkage and membrane blebbing; at 200 μM, extensive rounding, detachment, and debris indicate late apoptosis/necrosis. | |
3.11. Cosmosiin induced morphological alterations reminiscent of cell cytotoxicity
Phase-contrast microscopy of MCF-7 cells treated with cosmosiin for 24 h demonstrated dose-dependent morphological changes characteristic of apoptosis and cell stress. At 50 μM, cells exhibited reduced adherence, cytoplasmic shrinkage, and membrane blebbing (Fig. 20B). At 200 μM, extensive cell rounding, detachment, and fragmented cellular debris were observed, consistent with late-stage apoptosis or necrosis. Untreated cells retained typical epithelial morphology with intact cell–cell contacts.
3.12. Cosmosiin suppresses DNA synthesis in MCF-7 cells
EdU incorporation assay revealed a marked inhibition of proliferation in cosmosiin-treated MCF-7 cells (Fig. 21A). The percentage of EdU-positive cells decreased from 85.5% in controls to 57.3%, 40.4%, and 17.2% at 50, 100, and 200 μM, respectively (p < 0.05 vs. control) (Fig. 21B). Fluorescent imaging confirmed diminished nuclear EdU staining, correlating with the observed anti-proliferative effects.
 |
| | Fig. 21 EDU fluorescence intensity in MCF-7 cancer cells relative to the untreated cells. This corresponds to inhibition of DNA synthesis and hence decreased cell proliferation. (A) Representative fluorescent images of EdU incorporation (red) and DAPI nuclear counterstain (blue) in MCF-7 cells treated for 24 h with control, 50, 100, and 200 μM cosmosiin. (B) Quantification of EdU-positive cells (% of total nuclei) which decreases with increased cosmosiin concentration. (*p < 0.05 versus control). | |
3.13. Early and late apoptotic cell death is triggered by cosmosiin
Flow cytometry demonstrated a dose-dependent increase in apoptosis (Fig. 22A). At 200 μM cosmosiin, 8.16% of cells showed Annexin V+/PI− and 34.17% showed Annexin V+/PI+, compared to 2.22% and 4.21% in controls (p < 0.05 and 0.01) revealing high early and late apoptotic cell populations (Fig. 22B). Necrotic populations (Annexin V−/PI+) also showed gradual increase across increasing concentrations of cosmosiin.
 |
| | Fig. 22 Cosmosiin induces apoptosis and inhibits cell migration in MCF-7 Cells (A) representative Annexin V–FITC/PI dot plots of MCF-7 cells after 24 h treatment with control or 50, 100, 200 μM cosmosiin. Quadrants indicate viable (Q3) (Annexin V−/PI−), early apoptotic (Annexin V+/PI−) (Q4), late apoptotic (Annexin V+/PI+) (Q2), and necrotic (Annexin V−/PI+) populations (Q1). (B) Quantification of early and late apoptotic populations. (*p < 0.05, **p < 0.01 versus vehicle) (C) representative crystal-violet-stained Transwell membranes showing migrated MCF-7 cells after treatment. (D) Quantification of migrated cells per field. Data are mean ± SD (n = 3). (*p < 0.05 versus vehicle). | |
3.14. Cosmosiin inhibits MCF-7 cell migration
Transwell migration assays showed a 81% reduction in migrated cells at 200 μM cosmosiin (65 cells per field) versus control (345 cells per field; p < 0.05) (Fig. 22C and D). Intermediate concentrations (50 and 100 μM) reduced migration by 22% and 55%, respectively (p < 0.05; Fig. 22C and D). Crystal violet staining revealed sparse, irregularly shaped cells in treated groups, contrasting with the dense, organized clusters in untreated wells.
3.15. Cosmosiin targets key hub targets as revealed by western blot
Western blot analysis demonstrated significant downregulation of CDK5 (43% of control), NFKB1 (60%), and PTGS2 (70%) protein expression in MCF-7 cells (Fig. 23A and B) treated with 200 μM cosmosiin (p < 0.05 and 0.01 vs. control). GAPDH levels remained consistent across groups. These findings suggest cosmosiin suppresses pro-survival signaling via NF-κB and COX-2 pathways while inhibiting cell cycle progression through CDK5 modulation.
 |
| | Fig. 23 Cosmosiin modulates expression of cell cycle and inflammatory markers (A) western blots for CDK5, NFKB1, PTGS2, and loading control GAPDH in MCF-7 treated and control cells. (B) Densitometric analysis normalized to GAPDH. (*p < 0.05, **p < 0.01 versus vehicle control). | |
Immunotherapy in oncology has been a ground-breaking approach to tackling cancerous cells by targeting the immune system to identify and clear the cells.30 Such a therapy is specific as it targets the cells through immune checkpoints, modulating T-cell responses, or monoclonal antibodies, unlike chemotherapy and radiation that target cancerous and healthy tissues.31 This is particularly the case in BC, where specified subtypes, like triple-negative BC (TNBC), lack hormone or HER2 targets, and thus there are fewer options for therapeutic intervention.32 The CTLA-4 and PD-1/PD-L1 immune checkpoint inhibitor targets have been promising, improving patients' survival status for those cases of advanced or metastatic breast cancer. Adoptive-T-cell and vaccines therapies are additional under study for eliciting long-term immunity and reducing recurrence.33 The complexity of the TME in BC is one reason it suppresses immune responses as a part of promoting resistance to standard treatments. Clinical trials have shown that adding immunotherapy to the standard treatments leads to improved efficacy, especially in immune-responsive cancers.34 Outcomes are still very dynamic and hold promise for more individualized, long-lasting, and effective treatments as breast cancer treatment continues to evolve and patients continue to better improve in quality of life. The three hub genes, NFKB1, PTGS2, and CDK5, are prominent in cancer biology, especially in breast cancer through the intersection of cosmosiin targets and breast cancer-related genes. Each modulates the cellular pathways and the tumor microenvironment, and for this reason, they are significant therapeutic targets.
NFKB1 is a pivotal transcription factor of inflammation and immunity.35,36 Dysregulation of NFKB1 has been seen in a number of cancers, among which is breast cancer.37 It can enhance tumor progression through upregulating angiogenic, proliferating, and metastatic activities.35 In the case of breast cancer, elevated NFKB1 levels, as shown in this study, are associated with disease progression and poor prognosis. This function also extends into the modulation of immune checkpoint pathways, which include PD-L1 expression-an extremely important pathway for immune evasion mechanisms. Additionally, the gene encodes NFKB1, directly correlated with resistance against chemotherapy and radiotherapy therapy: hence, it has become a critical biomarker for the plans of personalized therapies.38
PTGS2, also known as COX-2, is a key enzyme in the synthesis of prostaglandins and is often upregulated in inflammation-associated cancers.39 However, in this study, PTGS2 displayed a significantly lower expression in BC vs. normal tissues, manifesting unique tumor-suppressive functions under certain conditions. PTGS2 inhibition represents one of the most common therapeutic approaches in cancer, given that its overexpression is usually associated with angiogenesis, invasion, and metastasis.40,41 However, differential expression of PTGS2 in breast cancer suggests a complex interplay between oncogenic and tumor-suppressive functions that demands further exploration to make its role clear in a specific context.
CDK5, known mainly for functions in the development of neurons, has emerged as a key member that acts in cancer.42 The pathways involve cell cycle regulation and apoptosis, which drive tumorigenesis of CDK5 in breast cancer.43 The increased expression seen in tumor tissues corresponds with low survival rates, since patients belonging to high expression groups showed decreased OS and DFS. CDK5 also has been implicated in modulating the tumor microenvironment with immune evasion and enhanced metastatic potential.44 Cosmosiin, which acts by interacting with CDK5, is likely to emerge as a treatment candidate that inhibits its activity to prevent further progression of the tumor. Their significance goes beyond breast cancer. NFKB1 has been linked to the pathogenesis of inflammatory cancers in the form of colorectal and pancreatic cancer, whereas PTGS2 overexpression is a well-established feature of gastric and lung cancers.45–48 CDK5 is established as a protein that promotes aggressive phenotypes in glioblastoma and prostate cancers.49,50 Such universal roles for these genes in tumor progression underscore broader applicability of these targets for therapy.
CDK5, NFKB1, and PTGS2 each feed into PD-L1 regulation at distinct but complementary levels: CDK5 is required for IFN-γ-mediated PD-L1 up-regulation by phosphorylating PD-L1 repressors (IRF2/IRF2BP2) to permit IRF1-driven transcription and by modulating PD-L1 turnover via chaperone-mediated autophagy.51,52 The NFKB1 encodes the p50 subunit of NF-κB, which upon activation by inflammatory signals (e.g., TNF-α, IFN-γ) forms p50:p65 heterodimers that bind κB sites in the CD274 promoter to directly enhance PD-L1 transcription and sustain an immunosuppressive microenvironment.53 The PTGS2 (COX-2) produces prostaglandin E2, which through EP-receptor signaling activates STAT3 and NF-κB pathways converging on the PD-L1 promoter to increase its expression in both tumor and myeloid cells.54 Targeting these nodes (e.g. CDK5 inhibitors, NF-κB blockers, COX-2 inhibitors) can therefore synergize with PD-1/PD-L1 blockade to reawaken anti-tumor immunity.55–58
4. Conclusion
This study explored the anti-PD-1/PD-L1 inhibition potential of cosmosiin, a naturally occurring flavonoid, using computational chemistry, network pharmacology, and bioinformatics approaches. Drug-likeness and physicochemical properties were assessed through DFT calculations and SwissADME tools, revealing favorable properties for cosmosiin. Among 25 overlapping targets between cosmosiin and breast cancer, 10 key hub genes (e.g., PTGS2, NFKB1, CDK5) were identified as central players in cancer-related pathways. KEGG pathway enrichment highlighted role of cosmosiin in critical pathways, including the PD-L1 expression and PD-1 checkpoint pathway. Molecular docking and dynamics confirmed stable interactions between cosmosiin and hub genes, especially CDK5, NFKB1, and PTGS2, with strong docking scores. These collective computational predictions were validated by experimental outcomes, reinforcing role of Cosmosiin as a multi-target agent with anticancer potential through apoptosis induction, proliferation inhibition, migration suppression, and modulation of pro-survival signalling pathways in MCF-7 breast cancer cells. The findings suggest cosmosiin's potential as a promising therapeutic agent, supported by its correlations with survival outcomes and immune infiltration in breast cancer.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
Conflicts of interest
The authors declare that there is no conflict of interest to indicate.
Acknowledgements
The authors declare that financial support was received for this research by Taif University, Saudi Arabia, project No. (TU-DSPP-2024-86). The authors extend their appreciation to Taif university, Saudi Arabia, for supporting this work.
References
- A. N. Giaquinto, K. D. Miller, K. Y. Tossas, R. A. Winn, A. Jemal and R. L. Siegel, Cancer statistics for African American/black people 2022, Ca-Cancer J. Clin., 2022, 72(3), 202–229 CrossRef PubMed.
- A. N. Giaquinto, H. Sung and K. D. Miller, et al., Breast cancer statistics, 2022, Ca-Cancer J. Clin., 2022, 72(6), 524–541 CrossRef PubMed.
- E. Clancy, ACS Report Shows Prostate Cancer on the Rise, Cervical Cancer on the Decline, Ren Urol News, 2023 Search PubMed.
- Organization WH, WHO Consultation to Adapt Influenza Sentinel Surveillance Systems to Include COVID-19 Virological Surveillance: Virtual Meeting, 6–8 October 2020, World Health Organization, 2022 Search PubMed.
- F. Islami, A. Goding Sauer and K. D. Miller, et al., Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States, Ca-Cancer J. Clin., 2018, 68(1), 31–54 CrossRef PubMed.
- S. Łukasiewicz, M. Czeczelewski, A. Forma, J. Baj, R. Sitarz and A. Stanisławek, Breast cancer—epidemiology, risk factors, classification, prognostic markers, and current treatment strategies—an updated review, Cancers, 2021, 13(17), 4287 CrossRef PubMed.
- Y. Feng, M. Spezia and S. Huang, et al., Breast cancer development and progression: risk factors, cancer stem cells, signalling pathways, genomics, and molecular pathogenesis, Genes Dis., 2018, 5(2), 77–106 CrossRef CAS PubMed.
- N. J. Niklaus, I. Tokarchuk, M. Zbinden, A. M. Schläfli, P. Maycotte and M. P. Tschan, The multifaceted functions of autophagy in breast cancer development and treatment, Cells, 2021, 10(6), 1447 CrossRef CAS PubMed.
- F. Cardoso, S. Kyriakides and S. Ohno, et al., Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up, Ann. Oncol., 2019, 30(8), 1194–1220 CrossRef CAS PubMed.
- Quantum Medicinal Chemistry, ed. Carloni, P., F. U. Alber and F. Alber, John Wiley & Sons, 2003, vol. 17 Search PubMed.
- N. Ye, Z. Yang and Y. Liu, Applications of density functional theory in COVID-19 drug modeling, Drug discovery today, 2022, 27(5), 1411–1419 CrossRef CAS PubMed.
- P. S. Sobral, V. C. Luz, J. M. Almeida, P. A. Videira and F. Pereira, Computational approaches drive developments in immune-oncology therapies for PD-1/PD-L1 immune checkpoint inhibitors, Int. J. Mol. Sci., 2023, 24(6), 5908 CrossRef CAS PubMed.
- N. Ye, Z. Yang and Y. Liu, Applications of density functional theory in COVID-19 drug modeling, Drug discovery today, 2022, 27(5), 1411–1419 CrossRef CAS PubMed.
- J. W. S. Ho, Naturally Occurring Small Molecules for Disease and Cancer Treatment: Therapeutic Benefits in Combination Therapy, World Scientific, 2019.
- S. M. Kim, P. Vetrivel, S. E. Ha, H. H. Kim, J.-A. Kim and G. S. Kim, Apigetrin induces extrinsic apoptosis, autophagy and G2/M phase cell cycle arrest through PI3K/AKT/mTOR pathway in AGS human gastric cancer cell, J. Nutr. Biochem., 2020, 83, 108427 CrossRef CAS PubMed.
- V. A. Kurkin, A. I. Vaskova and I. V. Sokolova, Determination of the Cosmosiin Content in Yarrow (Achillea millefolium L.) Herb by HPLC, Pharm. Chem. J., 2023, 57(9), 1454–1459 CrossRef CAS.
- J.-G. Jeon, P. L. Rosalen, M. L. Falsetta and H. Koo, Natural products in caries research: current (limited) knowledge, challenges and future perspective, Caries Res., 2011, 45(3), 243–263 CrossRef PubMed.
- H. Yuan, Q. Ma, L. Ye and G. Piao, The traditional medicine and modern medicine from natural products, Molecules, 2016, 21(5), 559 CrossRef PubMed.
- M. V. Lakshmi and T. S. Swapna, A computational study on Cosmosiin, an antiviral compound from Memecylon randerianum SM Almeida & MR Almeida, Med. Plants - Int. J. Phytomed. Relat. Ind., 2021, 13(3), 515–523 CrossRef.
- S. P. Patel and R. Kurzrock, PD-L1 expression as a predictive biomarker in cancer immunotherapy, Mol. Cancer Ther., 2015, 14(4), 847–856 CrossRef CAS PubMed.
- L.-J. Deng, M. Qi, N. Li, Y.-H. Lei, D.-M. Zhang and J.-X. Chen, Natural products and their derivatives: Promising modulators of tumor immunotherapy, J. Leukoc. Biol., 2020, 108(2), 493–508 CrossRef CAS PubMed.
- Garnegieoffice. Park, GaussView 5.0, Gaussian Inc., Pittsburgh. PA, USA Search PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski, and D. J. Fox, GAUSSIAN 09 (Revision C.01), Gaussian, Inc., Wallingford CT, 2010 Search PubMed.
- C. J. Brabec and N. S. Sarici, Ci and JC Hummelen, Adv. Funct. Mater., 2001, 11, 15–26 CrossRef CAS.
- G. Ghosh, G. V. Duyne, S. Ghosh and P. B. Sigler, Structure of NF-κB p50 homodimer bound to a κB site, Nature, 1995, 373(6512), 303–310 CrossRef CAS PubMed.
- M. H. Daniels, G. Malojcic, S. L. Clugston, B. Williams, M. Coeffet-Le Gal, X. R. Pan-Zhou, S. Venkatachalan, J. C. Harmange and M. Ledeboer, Discovery and optimization of highly selective inhibitors of CDK5, J. Med. Chem., 2022, 65(4), 3575–3596 CrossRef CAS PubMed.
- R. G. Kurumbail, A. M. Stevens, J. K. Gierse, J. J. McDonald, R. A. Stegeman, J. Y. Pak, D. Gildehaus, J. M. Iyashiro, T. D. Penning, K. Seibert and P. C. Isakson, Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents, Nature, 1996, 384(6610), 644–648 CrossRef CAS PubMed.
- T. C. Ngo, D. Q. Dao, M. T. Nguyen and P. C. Nam, A DFT analysis on the radical scavenging activity of oxygenated terpenoids present in the extract of the buds of Cleistocalyx operculatus, RSC Adv., 2017, 7(63), 39686–39698 RSC.
- S. A. Majid, J. M. Mir, S. Paul, M. Akhter, H. Parray, R. Ayoub and A. H. Shalla, Experimental and molecular topology-based biological implications of Schiff base complexes: a concise review, Rev. Inorg. Chem., 2019, 39(2), 113–128 CAS.
- C. Barbari, T. Fontaine, P. Parajuli, N. Lamichhane, S. Jakubski, P. Lamichhane and R. R. Deshmukh, Immunotherapies and combination strategies for immuno-oncology, Int. J. Mol. Sci., 2020, 21(14), 5009 CrossRef CAS PubMed.
- D. E. Magee, A. E. Hird, Z. Klaassen, S. S. Sridhar, R. K. Nam, C. J. Wallis and G. S. Kulkarni, Adverse event profile for immunotherapy agents compared with chemotherapy in solid organ tumors: a systematic review and meta-analysis of randomized clinical trials, Ann. Oncol., 2020, 31(1), 50–60 CrossRef CAS PubMed.
- Y. Zhu, X. Zhu, C. Tang, X. Guan and W. Zhang, Progress and challenges of immunotherapy in triple-negative breast cancer, Biochim. Biophys. Acta, Rev. Cancer, 2021, 1876(2), 188593 CrossRef CAS PubMed.
- M. Nagasaka, B. Potugari, A. Nguyen, A. Sukari, A. S. Azmi and S. H. Ou, KRAS inhibitors–yes but what next? Direct targeting of KRAS–vaccines, adoptive T cell therapy and beyond, Cancer Treat. Rev., 2021, 101, 102309 CrossRef CAS PubMed.
- D. R. Wang, X. L. Wu and Y. L. Sun, Therapeutic targets and biomarkers of tumor immunotherapy: response versus non-response, Signal Transduction Targeted Ther., 2022, 7(1), 331 CrossRef CAS PubMed.
- N. Fathi, M. Nirouei, Z. Salimian Rizi, S. Fekrvand, H. Abolhassani, F. Salami, A. H. Ketabforoush, G. Azizi, A. Saghazadeh, M. Esmaeili and A. Almasi-Hashiani, Clinical, Immunological, and Genetic Features in Patients with NFKB1 and NFKB2 Mutations: a Systematic Review, J. Clin. Immunol., 2024, 44(7), 160 CrossRef CAS PubMed.
- J. Chen, R. S. Liang and B. B. Zhuang, et al., Cordycepin inhibits glioma growth by downregulating PD-L1 expression via the NOD-like receptor/NFKB1/STAT1 axis, Chem.-Biol. Interact., 2024, 400, 111178 CrossRef CAS PubMed.
- O. M. Bilyy, N. A. Mitriaieva, M. V. Krasnoselskyi and L. V. Grebinyk, Features of the expression of the transcription factor NF-kB in patients with secondary edematous breast cancer with various pro-inflammatory cytokines, Ukr. J. Radiol. Oncol., 2024, 32(2), 134–142 Search PubMed.
- K. A. Gaptulbarova, M. M. Tsyganov, A. M. Pevzner, M. K. Ibragimova and N. V. Litviakov, NF-kB as a potential prognostic marker and a candidate for targeted therapy of cancer, Exp. Oncol., 2020, 42(4), 263–269 CrossRef CAS PubMed.
- K. S. Chun, E. H. Kim, D. H. Kim, N. Y. Song, W. Kim, H. K. Na and Y. J. Surh, Targeting cyclooxygenase-2 for chemoprevention of inflammation-associated intestinal carcinogenesis: an update, Biochem. Pharmacol., 2024, 116259 CrossRef CAS PubMed.
- Y. Zhang, S. Tighe and Y. T. Zhu, COX-2 signalling in the tumor microenvironment, Tumor Microenvironment: Molecular Players–Part B, 2020, pp. 87–104 Search PubMed.
- P. Rodrigues, H. Bangali, A. Hammoud, Y. F. Mustafa, H. R. Al-Hetty, A. T. Alkhafaji, M. M. Deorari, M. M. Al-Taee, R. S. Zabibah and A. Alsalamy, COX 2-inhibitors; a thorough and updated survey into combinational therapies in cancers, Med. Oncol., 2024, 41(1), 41 CrossRef CAS PubMed.
- P. A. Do and C. H. Lee, The role of CDK5 in tumours and tumour microenvironments, Cancers, 2020, 13(1), 101 CrossRef PubMed.
- N. F. Lisany, M. A. Jamal, H. J. Chung, S. T. Hong and M. S. Rahman, Prognostic significance of the Cdk5 gene in breast cancer: an in silico study, Netw. Model. Anal. Health Inform. Bioinform., 2020, 9, 1 CrossRef.
- H. F. Chen, C. W. Xu, W. X. Wang, P. Chen, X. F. Li, G. Lan, Z. Q. Zhai, Y. C. Zhu, K. Q. Du and L. C. Huang, CDK5 affects the resistance of
lung cancer to radiotherapy by altering the tumor microenvironment, J. Radiat. Res. Appl. Sci., 2024, 17(4), 101104 CAS.
- J. Silke and L. A. O'Reilly, NF-κB and pancreatic cancer; chapter and verse, Cancers, 2021, 13(18), 4510 CrossRef CAS PubMed.
- M. Dobre, B. Trandafir, E. Milanesi, A. Salvi, I. A. Bucuroiu, C. Vasilescu, A. M. Niculae, V. Herlea, M. E. Hinescu and G. Constantinescu, Molecular profile of the NF-κB signalling pathway in human colorectal cancer, J. Cell. Mol. Med., 2022, 26(24), 5966–5975 CrossRef CAS PubMed.
- W. Jiang, Q. Wei, H. Xie, D. Wu, H. He and X. Lv, Effect of PTGES3 on the Prognosis and Immune Regulation in Lung Adenocarcinoma, Anal. Cell. Pathol., 2023, 2023(1), 4522045 CrossRef PubMed.
- I. Bednarz-Misa, P. Fortuna, D. Diakowska, N. Jamrozik and M. Krzystek-Korpacka, Distinct local and systemic molecular signatures in the esophageal and gastric cancers: possible therapy targets and biomarkers for gastric cancer, Int. J. Mol. Sci., 2020, 21(12), 4509 CrossRef CAS PubMed.
- E. Tabouret, H. Wang, N. Amin, J. Jung, R. Appay, J. Cui, Q. Song, A. Cardone, D. M. Park, M. R. Gilbert and H. Pant, TP5, a peptide inhibitor of aberrant and hyperactive CDK5/p25: a novel therapeutic approach against glioblastoma, Cancers, 2020, 12(7), 1935 CrossRef CAS PubMed.
- S. Sharma and P. Sicinski, A kinase of many talents: non-neuronal functions of CDK5 in development and disease, Open Biol., 2020, 10(1), 190287 CrossRef CAS PubMed.
- X. Lin, K. Kang, P. Chen, Z. Zeng, G. Li, W. Xiong, M. Yi and B. Xiang, Regulatory mechanisms of PD-1/PD-L1 in cancers, Mol. Cancer, 2024, 23(1), 108 CrossRef PubMed.
- R. Zhang, J. Wang, Y. Du, Z. Yu, Y. Wang, Y. Jiang, Y. Wu, T. Le, Z. Li, G. Zhang and L. Lv, CDK5 destabilizes PD-L1 via chaperon-mediated autophagy to control cancer immune surveillance in hepatocellular carcinoma, J. immunotherap. cancer, 2023, 11(11), e007529 CrossRef PubMed.
- F. Antonangeli, A. Natalini, M. C. Garassino, A. Sica, A. Santoni and F. Di Rosa, Regulation of PD-L1 Expression by NF-κB in Cancer, Front. Immunol., 2020, 11, 584626 CrossRef CAS PubMed.
- V. Prima, L. N. Kaliberova, S. Kaliberov, D. T. Curiel and S. Kusmartsev, COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(5), 1117–1122 CrossRef CAS PubMed.
- H. Liu, R. Deng and C. W. Zhu, et al., Rosmarinic acid in combination with ginsenoside Rg1 suppresses colon cancer metastasis via co-inhibition of COX-2 and PD1/PD-L1 signaling axis, Acta Pharmacol. Sin., 2024, 45(1), 193–208 CrossRef CAS PubMed.
- D. L. Cecil, E. A. Gad, L. R. Corulli, N. Drovetto, R. A. Lubet and M. L. Disis, COX-2 Inhibitors Decrease Expression of PD-L1 in Colon Tumors and Increase the Influx of Type I Tumor-infiltrating Lymphocytes, Cancer Prev. Res., 2022, 15(4), 225–231 CrossRef CAS PubMed.
- H. Hu, L. Kang and J. Zhang, et al., Neoadjuvant PD-1 blockade with toripalimab, with or without celecoxib, in mismatch repair-deficient or microsatellite instability-high, locally advanced, colorectal cancer (PICC): a single-centre, parallel-group, non-comparative, randomised, phase 2 trial, Lancet Gastroenterol. Hepatol., 2022, 7(1), 38–48 CrossRef CAS PubMed.
- T. Suriyo, M. Fuangthong and C. Artpradit, et al., Inhibition of T-cell-mediated immune response via the PD-1/PD-L1 axis in cholangiocarcinoma cells, Eur. J. Pharmacol., 2021, 897, 173960 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2025 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article b,
Manzoor A. Ratherc,
Jan M. Mir
b,
Manzoor A. Ratherc,
Jan M. Mir *c,
N. A. Dangroo
*c,
N. A. Dangroo c,
Faisal A. Almalki
c,
Faisal A. Almalki d,
Taibi Ben Haddade,
Rabab S. Jassasf,
Sultan I. Alkubaysi
d,
Taibi Ben Haddade,
Rabab S. Jassasf,
Sultan I. Alkubaysi g and
Saleh A. Ahmed
g and
Saleh A. Ahmed *bh
*bh
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases annually (15%), a figure expected to increase by 2% by 2035.1,2 Breast Cancer predominantly affects women, with incidence rates increasing significantly with age including the diagnosis in about >80% of instances for those over 50 years. According to cancer statistics for 2023, the number of new BC cases has increased to 297
000 new cases annually (15%), a figure expected to increase by 2% by 2035.1,2 Breast Cancer predominantly affects women, with incidence rates increasing significantly with age including the diagnosis in about >80% of instances for those over 50 years. According to cancer statistics for 2023, the number of new BC cases has increased to 297![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780 among females and 2800 among males.3 Worldwide fatalities have increased to 685
780 among females and 2800 among males.3 Worldwide fatalities have increased to 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in 2020 and thus, it is among the top causes of cancer mortality. It is the leading cause of cancer deaths, resulting in fatalities worldwide in 2020.4 Fat, lack of exercise, carcinogen exposure, smoking and alcohol use are modifiable factors that are associated with about 30% of BC incidences, making these cases potentially preventable and non-modifiable factors such as female sex, density of breast tissues, previous radiation therapy, and genetic mutations.5,6 With 15% developing in the lobular epithelium and 85% in the breast ducts, adenocarcinomas make up the bulk of breast cancers. Cancers of the ducts may range from those that remain in situ inside the ducts to those that have invaded the breast parenchyma and spread beyond the foundation membrane.7 The treatments of BC with surgery, chemotherapy, radiotherapy (RT), endocrine therapy, targeted therapy, modify based on specific cancer traits and patient profiles; yet, these cause adverse events, invasiveness, slow wound healing, body shape changes, liver and kidney damage, metabolic changes, severe hair loss, which impact women both physically and mentally and limited efficacy for certain cancer types. Furthermore, drug resistance continues to be a major challenge, with up to 50% of patients developing tamoxifen resistance and 70% to trastuzumab within a year.8 These limitations highlight the need for newer treatments such as immunotherapy, which offers more targeted and potentially less harmful options.9 The immunomodulation represents a promising strategy to unleash the power of the immune system against cancer. The PD-1 (Programmed Death-1) and PD-L1 (Programmed Death-Ligand 1) pathway plays a crucial role in putting the brakes on the immune system, enabling cancer cells to evade immune surveillance. By targeting and inhibiting this pathway, the immune response can be reactivated, allowing it to spot and destroy more malignant cells effectively. Thus, this approach has the potential to revolutionize cancer treatment, turning the body's own defense mechanisms into powerful allies in the fight against malignancy.
000 in 2020 and thus, it is among the top causes of cancer mortality. It is the leading cause of cancer deaths, resulting in fatalities worldwide in 2020.4 Fat, lack of exercise, carcinogen exposure, smoking and alcohol use are modifiable factors that are associated with about 30% of BC incidences, making these cases potentially preventable and non-modifiable factors such as female sex, density of breast tissues, previous radiation therapy, and genetic mutations.5,6 With 15% developing in the lobular epithelium and 85% in the breast ducts, adenocarcinomas make up the bulk of breast cancers. Cancers of the ducts may range from those that remain in situ inside the ducts to those that have invaded the breast parenchyma and spread beyond the foundation membrane.7 The treatments of BC with surgery, chemotherapy, radiotherapy (RT), endocrine therapy, targeted therapy, modify based on specific cancer traits and patient profiles; yet, these cause adverse events, invasiveness, slow wound healing, body shape changes, liver and kidney damage, metabolic changes, severe hair loss, which impact women both physically and mentally and limited efficacy for certain cancer types. Furthermore, drug resistance continues to be a major challenge, with up to 50% of patients developing tamoxifen resistance and 70% to trastuzumab within a year.8 These limitations highlight the need for newer treatments such as immunotherapy, which offers more targeted and potentially less harmful options.9 The immunomodulation represents a promising strategy to unleash the power of the immune system against cancer. The PD-1 (Programmed Death-1) and PD-L1 (Programmed Death-Ligand 1) pathway plays a crucial role in putting the brakes on the immune system, enabling cancer cells to evade immune surveillance. By targeting and inhibiting this pathway, the immune response can be reactivated, allowing it to spot and destroy more malignant cells effectively. Thus, this approach has the potential to revolutionize cancer treatment, turning the body's own defense mechanisms into powerful allies in the fight against malignancy.
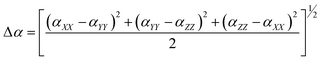
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 15 min at 4 °C, and protein concentration was determined by BCA assay. Equal amounts (30 μg) of protein were separated on 10% SDS–PAGE gels using the Bio-Rad Mini-PROTEAN® Tetra System and transferred onto PVDF membranes (MilliporeSigma, Merck KGaA) with the Trans-Blot® Turbo Transfer System (Bio-Rad). Membranes were blocked in 5% BSA for 1 h, then incubated overnight at 4 °C with primary antibodies: anti-CDK5 (1
000×g for 15 min at 4 °C, and protein concentration was determined by BCA assay. Equal amounts (30 μg) of protein were separated on 10% SDS–PAGE gels using the Bio-Rad Mini-PROTEAN® Tetra System and transferred onto PVDF membranes (MilliporeSigma, Merck KGaA) with the Trans-Blot® Turbo Transfer System (Bio-Rad). Membranes were blocked in 5% BSA for 1 h, then incubated overnight at 4 °C with primary antibodies: anti-CDK5 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), anti-NFKB1 (1
1000), anti-NFKB1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), anti-PTGS2 (1
500), anti-PTGS2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), and anti-GAPDH (1
500), and anti-GAPDH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000). We probed the membranes for 1 hour at room temperature with HRP-conjugated goat anti-rabbit or anti-mouse secondary antibodies (1
2000). We probed the membranes for 1 hour at room temperature with HRP-conjugated goat anti-rabbit or anti-mouse secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) after washings in TBS-T. A combination of increased chemiluminescence and densitometry in ImageJ allowed for the visualization of protein bands and their subsequent quantification.
5000) after washings in TBS-T. A combination of increased chemiluminescence and densitometry in ImageJ allowed for the visualization of protein bands and their subsequent quantification.























