Open Access Article
Open Access ArticleRational design and synthesis of potent active antimicrobial peptides based on American oyster defensin analogue A3†
Jiawei Zhaoa,
Xin Zhanga,
Yu Liua,
Zhixing Genga,
Jili Daib and
Ye Guo *a
*a
aSchool of Pharmacy, Baotou Medical College, Baotou 014060, China. E-mail: 102015128@btmc.edu.cn
bSchool of Basic Medicine and Forensic Medicine, Baotou Medical College, Baotou 014060, China
First published on 12th June 2025
Abstract
The rise of drug-resistant microbes is increasingly recognized as a significant global health challenge. Microbial drug resistance diminishes the efficacy of existing treatment options, underscoring the urgent necessity for the development of novel antibiotic candidates to effectively address infections. Antimicrobial peptides, owing to their distinctive antibacterial mechanisms, are considered a promising alternative to conventional antibiotics. In this work, we modified the structure of the American oyster defensin (AOD) analogue A3 to obtain four novel antimicrobial peptides (D-A3, A3-C4, A3-C5, A3-C6). These synthesized peptides exhibited broad-spectrum antibacterial activity. Notably, it was demonstrated that A3-C4, A3-C5, and A3-C6 showed enhanced glutathione (GSH) stability compared to A3, while D-A3 exhibited superior protease stability. Importantly, none of the peptides displayed hemolytic toxicity. Mechanistic investigations suggested that the synthesized peptides exert their anti-bacterial effects primarily through membrane disruption. D-A3 and A3-C6 were peptides with the best antibacterial activity and enzymatic stability among the synthesized derived peptides. These studies of D-A3 and A3-C6 will contribute to the development of new candidate drugs for the treatment of microbial infections.
1. Introduction
Severe infections caused by microorganisms have posed significant challenges to humanity for centuries. Prior to the advent of antibiotics, infections such as pneumonia, plague, and syphilis were associated with high mortality rates, leading to substantial human casualties and economic losses.1–3 The discovery and subsequent utilization of antibiotics, exemplified by penicillin, have been instrumental in saving innumerable lives over the past century.4,5 The rapid advancement of technology has facilitated the successive development of numerous antibiotics, empowering healthcare professionals to effectively combat infections. Today, antibiotics are employed across various sectors, including healthcare, food production, and aquaculture.6–8The extensive and pervasive utilization of antibiotics has resulted in a multitude of consequences. Primarily, prolonged antibiotic use contributes to the escalation of antimicrobial resistance (AMR), which progressively diminishes the efficacy of these agents.9 Additionally, antibiotic resistance can render certain antibiotics entirely ineffective against specific pathogens, necessitating a return to previously discontinued medications that were abandoned due to toxicity or adverse side effects.10 The presence of severe infections caused by resistant bacteria not only extends treatment durations and complicates patient treatment but also exacerbates both the economic and physical burdens on affected individuals.11 Currently, the swift emergence of highly resistant bacterial strains has attracted considerable attention.12 Predictions indicates that by the mid-21st century, antibiotic resistance could result in over 10 million fatalities annually. Should this trend persist without intervention, humans may revert to a period where microbial infections are largely unmanageable.13 Consequently, microbial resistance has emerged as a significant global health threat, thereby incentivizing researchers to develop new anti-infective therapeutics.14
In recent years, antimicrobial peptides (AMPs) have been recognized as significant therapeutic agents in combating infections. Several antimicrobial peptides, including Polymyxin, Teicoplanin, Bacitracin, and Vancomycin (often regarded as the “last line of defense” against infections), have been approved for commercial use.15,16 AMPs are ubiquitously found across various organisms and serve as the primary defense mechanism in the innate immune system, with a broad spectrum of antibacterial activity and low toxicity.17 Because they have a positively charged surface, most AMPs exert their bactericidal effects by disrupting bacterial cell membranes. This mode of action is distinct from that of conventional antibiotics, rendering AMPs less susceptible to the development of drug resistance.18–20 Due to their superior antibacterial efficacy and minimal toxic side effects, AMPs hold promises as potent agents for the clinical management of drug-resistant bacterial infections.21,22
American oyster defensin (AOD) is an AMP consisting of 38 amino acids and three disulfide bonds, showing significant antibacterial efficacy against both Gram-positive (G+) and Gram-negative (G−) bacteria. Nonetheless, its intricate structure results in elevated synthesis costs, thereby restricting its practical application.23 To develop AOD as a potential antibiotic, Seo et al. engineered and synthesized five arginine-rich cyclic 9-peptide according to AOD. Among these analogues, the A3 exhibited superior activity with minimum inhibitory concentration (MIC) values ranging from 0.4 to 6.5 μg mL−1 against B. subtilis, S. epidermidis, S. mutans, A.hydrophila, E. coli, P. aeruginosa, and S. enterica. Subsequent validation indicated that the A3 peptide had low hemolytic toxicity towards human red blood cells and minimal cytotoxicity against human dermal fibroblast (HDF) cells, rendering it a promising candidate for novel antibiotic development.24 Despite the considerable potential of the A3 as an AMP, several challenges impede the application of this candidate molecule. Primarily, as a cyclic peptide, its disulfide bond is prone to degradation in reducing environments within the body, which may result in diminished or lost activity. Potential strategy to address this issue involves substituting disulfide bond with triazole ring, synthesized through click chemistry reactions, thereby mitigating the negative effects associated with disulfide bond. Additionally, the rapid degradation of peptides by proteases in the body can lead to their inactivation. To prevent such enzymatic degradation, peptides can be synthesized using D-amino acids. The specificity of proteases prevents them from degrading peptides composed of D-amino acids.25–28
In this work, we improved the stability of the A3 through structural modifications, including substitution of all L-amino acids with D-amino acids or the replacement of the disulfide bond with a triazole ring. Furthermore, we synthesized the D-A3 (consisting of D-amino acids) and three triazole-containing peptides (A3-C4, A3-C5, A3-C6). We further examined the antibacterial activity, mechanism of action, stability, and toxicity of these peptides to acquire lead peptides with best antibacterial activity, enzymatic stability and serum stability for the development of novel antibiotic candidates.
2. Materials and methods
2.1. Materials
2.2. Synthesis of A3 and its derived peptides
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HCTU
HCTU![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DIEA molar ratio for coupling was 4 eq.
DIEA molar ratio for coupling was 4 eq.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.8 eq.
3.8 eq.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 eq. After swelling the resin and removing the protecting group, amino acids were coupled, with each amino acid requiring two coupling steps (45 min each) to ensure complete reaction. The coupling and deprotection steps were repeated, connecting all amino acids in sequence. The Kaiser test was used for deprotection and coupling verification after each step. After all amino acids were coupled, the protecting group of the last amino acid was removed. The resin was washed, and then 10 mL of cleavage reagent (TFA
8 eq. After swelling the resin and removing the protecting group, amino acids were coupled, with each amino acid requiring two coupling steps (45 min each) to ensure complete reaction. The coupling and deprotection steps were repeated, connecting all amino acids in sequence. The Kaiser test was used for deprotection and coupling verification after each step. After all amino acids were coupled, the protecting group of the last amino acid was removed. The resin was washed, and then 10 mL of cleavage reagent (TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIPS = 95%
TIPS = 95%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5%
2.5%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5%) was added for cleavage, with a reaction time of 3 hours. After concentrating by nitrogen bubbling and precipitating via centrifugation with ice-cold methyl tert-butyl ether, the yellow crude peptide was obtained.
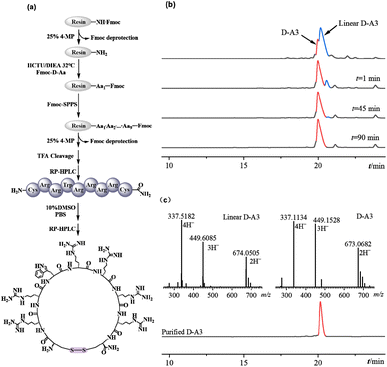
2.5%) was added for cleavage, with a reaction time of 3 hours. After concentrating by nitrogen bubbling and precipitating via centrifugation with ice-cold methyl tert-butyl ether, the yellow crude peptide was obtained.Oxidation of peptides was carried out using dimethyl sulfoxide (DMSO) as the oxidant. Specifically, 3 mg of crude peptide was dissolved in 0.9 mL of phosphate buffer solution (PBS, pH 7.4), and then mixed with 100 μL of DMSO. The mixture was placed on a magnetic stirrer and allowed to react at room temperature. The progress of the reaction was monitored using HPLC. After 90 minutes of reaction, the cysteine residues on both sides of the A3 peptide were oxidized to form a cyclic peptide with a disulfide bond. Subsequently, the linear peptides and cyclic peptides were separated and purified using semi-preparative RP-HPLC. The peptides were then analyzed using ESI-MS.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HATU
HATU![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DIEPA used was 2 eq.
DIEPA used was 2 eq.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.9 eq.
1.9 eq.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 eq. After completing all amino acid couplings, the protecting groups were removed, and the resin was washed and cleaved. Nitrogen was then bubbled through to concentrate and remove excess solvent, followed by precipitation with ice-cold methyl tert-butyl ether and centrifugation to obtain the yellow crude peptide.
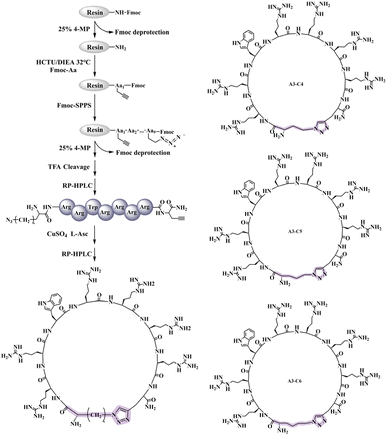
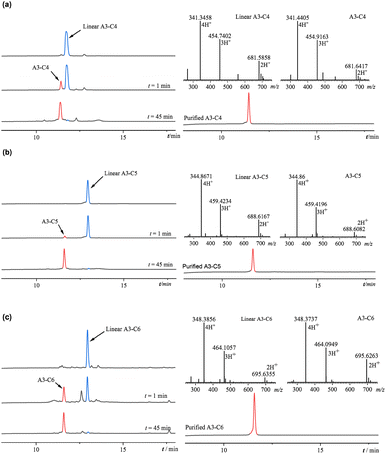
4 eq. After completing all amino acid couplings, the protecting groups were removed, and the resin was washed and cleaved. Nitrogen was then bubbled through to concentrate and remove excess solvent, followed by precipitation with ice-cold methyl tert-butyl ether and centrifugation to obtain the yellow crude peptide.The click chemistry reaction was catalyzed using copper sulfate. Specifically, 0.5 mM (approximately 7 mg) of linear A3-C4 crude peptide was dissolved in 0.9 mL of pure water. In a 2 mL centrifuge tube, 4 mg of anhydrous copper sulfate (CuSO4) and 11.8 mg of sodium ascorbate were weighed and dissolved in 2 mL of pure water. Initially, the solution appeared dark brown, but upon vigorous shaking, it rapidly turned orange-yellow. At this point, 100 μL of the mixed solution was promptly added to the peptide solution using a pipette, and the mixture was stirred and allowed to react at room temperature. The reaction progress was monitored using HPLC. After reacting for 90 minutes, the linear and cyclic peptides were separated and purified using semi-preparative RP-HPLC. The purified peptides were then obtained through freeze-drying. Both the purified linear A3-C4, linear A3-C5, linear A3-C6 and A3-C4, A3-C5, A3-C6 were analyzed using ESI-MS.
2.3. Circular dichroism
The secondary structural characteristics of A3 and A3-derived peptides (D-A3, A3-C4, A3-C5, A3-C6) in aqueous solution (pH 5.60) were tested using a circular dichroism spectrometer. The testing method involved preparing 1 mg mL−1 peptide solutions of the five peptides using pure water. A quartz cuvette with a 1 mm path length was used, with a scanning bandwidth of 1 nm, a scanning wavelength ranging from 300 nm to 170 nm, and a scanning speed of 200 nm min−1. Each sample's CD spectrum was measured three times, and the average value was taken.2.4. Antibacterial activity of A3 and derived peptides
The minimum inhibitory concentrations (MIC) of A3 and its four derivative peptides against S. aureus, S. aureus ATCC-25923, B. subtilis CMCC-63501, B.cereus, E. coli ATCC-25922, P. aeruginosa ATCC-27853, S. enterica, and S. para-typhi B were determined using the agar well diffusion method.29 Bacterial suspensions at a concentration of 1 × 108 CFU mL−1 were prepared from logarithmic phase bacteria and used to inoculate nutrient agar plates containing 5% bacteria. Sterile 3 mm punchers were used to create uniform wells in the agar. The peptide solution was prepared at a maximum concentration of 1 mg mL−1 in 0.01% acetic acid aqueous solution, followed by two-fold serial dilutions. Subsequently, 10 μL of each peptide solution at different concentrations was carefully added to the wells, with 0.01% acetic acid aqueous solution serving as the negative control. Ten microliters of each peptide solution at different concentrations were carefully added to the wells, and 0.01% acetic acid water was used as a negative control. Three replicates were performed for each bacterial strain. The plates were incubated at 37 °C for 24 hours. After incubation, the plates were examined for the presence of zones of inhibition, and the corresponding peptide concentrations were recorded as the MIC. After the incubation period, check whether there are inhibition zones on the plates, and take the concentration corresponding to the appearance of the smallest inhibition zone as the MIC.2.5. Antibacterial mechanism of A3 and its derived peptides
2.6. Stability of A3 and its derived peptides
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was carried out at room temperature. The reaction was monitored using HPLC at 0 min, 1 min, 5 min, 10 min, and 20 min. The reaction was then quenched by adding 50 μL of 2 M HCl to 10 μL reaction mixture. The residual peptide at each time point was monitored by HPLC. The degradation curve was plotted based on the residual amount.
1) was carried out at room temperature. The reaction was monitored using HPLC at 0 min, 1 min, 5 min, 10 min, and 20 min. The reaction was then quenched by adding 50 μL of 2 M HCl to 10 μL reaction mixture. The residual peptide at each time point was monitored by HPLC. The degradation curve was plotted based on the residual amount.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. In a 96-well plate, 180 μL of the bacterial culture medium was added to the second well, and 100 μL was added to wells 3 through 11. A 20 μL aliquot of the drug solution, pre-incubated with fetal bovine serum, was introduced into the first well, followed by successive two-fold gradient dilutions up to the 10th well. The 11th well served as the negative control. The 96-well plate was incubated on a shaker for 18–24 hours. Finally, the optical density at 600 nm (OD600) was measured using an enzyme-linked immunosorbent assay reader.
100. In a 96-well plate, 180 μL of the bacterial culture medium was added to the second well, and 100 μL was added to wells 3 through 11. A 20 μL aliquot of the drug solution, pre-incubated with fetal bovine serum, was introduced into the first well, followed by successive two-fold gradient dilutions up to the 10th well. The 11th well served as the negative control. The 96-well plate was incubated on a shaker for 18–24 hours. Finally, the optical density at 600 nm (OD600) was measured using an enzyme-linked immunosorbent assay reader.2.7. Toxicity of A3 and its derived peptides
| Hemolysis rate (%) = (Aexperimental group − Ablank group)/(Apositive group − Ablank group) × 100% |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) were placed in a mating tank and separated by a divider. On the second day, after the appearance of light, the divider was removed to allow zebrafish mating. Following spawning, dead embryos were discarded, and healthy zebrafish embryos were collected and placed in a shaking incubator at 28 °C overnight. Well-developed embryos were then selected and placed into a 6-well plate, with 20 embryos per well. Peptides were added to the wells at concentrations of 0 μg mL−1, 1.56 μg mL−1, 3.12 μg mL−1, 6.25 μg mL−1, 12.5 μg mL−1, 25.0 μg mL−1, and 50.0 μg mL−1. The number of hatched zebrafish was recorded using a stereo microscope every 24 hours. The body length of zebrafish was measured using a stereo microscope every 24 hours. This process was repeated for 5 days.
1 ratio) were placed in a mating tank and separated by a divider. On the second day, after the appearance of light, the divider was removed to allow zebrafish mating. Following spawning, dead embryos were discarded, and healthy zebrafish embryos were collected and placed in a shaking incubator at 28 °C overnight. Well-developed embryos were then selected and placed into a 6-well plate, with 20 embryos per well. Peptides were added to the wells at concentrations of 0 μg mL−1, 1.56 μg mL−1, 3.12 μg mL−1, 6.25 μg mL−1, 12.5 μg mL−1, 25.0 μg mL−1, and 50.0 μg mL−1. The number of hatched zebrafish was recorded using a stereo microscope every 24 hours. The body length of zebrafish was measured using a stereo microscope every 24 hours. This process was repeated for 5 days.3. Results
3.1. Synthesis of peptides
3.2. Secondary structure of A3 and its derived peptides
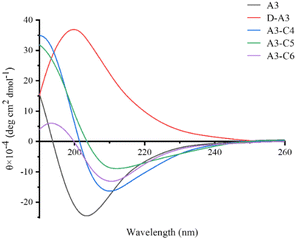
Circular dichroism (CD) spectrum serves as a robust technique for assessing the secondary structure of peptides. In this study, we analyzed the secondary structures of A3 and its derivative peptides (D-A3, A3-C4, A3-C5, A3-C6) in an aqueous environment at pH 5.6. As depicted in Fig. 4, the CD spectrum of A3 in water displayed a negative absorption peak near 205 nm, indicative of a disordered structure. Conversely, D-A3 exhibited a positive absorption peak at 205 nm, which is symmetrical to the absorption peak of the A3, suggesting a mirror-image relationship. This observation aligned with the CD characteristics typical of D-peptide. The symmetrical nature of the peaks for both peptides implied structural similarity, thereby confirming the successful synthesis of the D-peptide. Furthermore, we explored the impact of introducing a triazole ring into the A3 structure using CD spectrum. The CD spectra of the triazole-containing peptides showed a positive absorption peak around 190 nm and a negative absorption peak around 210 nm. The positions of absorption peaks were consistent with the characteristic absorption peak of β-sheet structure documented by Greenfield,29 suggesting that the three triazole-containing peptides had β-sheet conformation. CD spectrum revealed that the A3 underwent structural alterations after disulfide bond substitution, shifting from a disordered conformation to a β-sheet structure.3.3. Antibacterial activity of A3 and its derived peptides
The antimicrobial efficacy of A3 and its derivative peptides was assessed utilizing the radial diffusion method at 1 mg mL−1. In the qualitative antibacterial assay (Fig. 5), we evaluated the antibacterial efficacy of A3 peptide and its derivatives against S. aureus, B. subtilis, B.cereus, E. coli, P. aeruginosa, S. enterica and S. paratyphi B, respectively. All five peptides exhibited antibacterial activity against bacteria, as evidenced by the formation of clear inhibition zones. Notably, D-A3 exhibited the highest activity, characterized by the largest inhibition zone, whereas the inhibition zones of the three triazole-containing peptides were marginally smaller than that of A3, among the 7 types of bacteria mentioned above. | ||
| Fig. 5 Qualitative antibacterial test of A3 and its derived peptides against Gram-negative bacteria and Gram-positive bacteria. | ||
In addition, we assessed the minimum inhibitory concentration (MIC) of the A3 and its derivative peptides utilizing the agar well diffusion method. The MIC was determined by evaluating five peptides against four Gram-positive bacteria (B. subtilis,B. cereus, and two strains of S. aureus) and four Gram-negative bacteria (E. coli, P. aeruginosa, and two strains of S. enterica) at various concentrations, using a twofold dilution method (Table 1). Our findings indicated that A3 had significant bactericidal activity against both Gram-positive and Gram-negative bacteria, corroborating previous research by Seo. All derivative peptides exhibited broad-spectrum antibacterial activity, with MIC values ranging from 0.9 to 250 μg mL−1 for the strains tested. A3 and D-A3 showed the same antibacterial activity against S. aureus ATCC-25923 and B. subtilis CMCC-63501. For S. aureus, B. cereus, S. enterica, and S. para-typhi B, the antibacterial efficacy of D-A3 was twice higher than that of A3. Notably, the MIC value of D-A3 was 3.9 μg mL−1 for E. coli; this value for A3 was only 15.62 μg mL−1. The antibacterial efficacy of D-A3 against P. aeruginosa (15.6 μg mL−1) was markedly superior to that of A3 (125 μg mL−1), demonstrating an eight-fold enhancement in activity. The triazole-containing peptides preserved their broad-spectrum antibacterial properties, exhibiting MIC values ranging from 7.8 to 250 μg mL−1. Nevertheless, their antibacterial potency against certain bacterial strains was reduced compared to A3, with decreases ranging from 2 to 8 folds. For P. aeruginosa ATCC-27853 and S. enterica, A3-C6 exhibited equivalent antibacterial activity to A3 (125 μg mL−1). Among the three triazole-containing compounds, similar antibacterial activity was observed for S. aureus ATCC-25923 and S. paratyphi B, with MIC values of 125 μg mL−1 and 15.6 μg mL−1, respectively. In the case of S. aureus and E. coli ATCC-25922, the antibacterial efficacy of A3-C4 and A3-C6 was equivalent and surpassed that of A3-C5. A3-C6 was demonstrated to have the highest antimicrobial activity among the triazole-containing peptides. A3-C4, A3-C5 and A3-C6 prepared from different azido amino acid precursors, showed different antibacterial activities. The activity of A3-C6 prepared from azido amino acid precursors with longer linker was stronger than that of peptides prepared from azido amino acid precursors with shorter linker. The antibacterial activity of triazole-containing peptides showed that the linker length of azido amino acids precursors would affect the antibacterial activity by changing the size of cyclic peptide ring. These results suggest that the activity of antimicrobial peptides can be regulated by the length of linker of azido amino acid precursors. Overall, the antibacterial activity of D-A3 was comparable to or exceeded that of A3.
| Microbe | Gram stain | MIC (μg ml−1) | ||||
|---|---|---|---|---|---|---|
| A3 | D-A3 | A3-C4 | A3-C5 | A3-C6 | ||
| S. aureus | + | 31.2 | 15.6 | 62.5 | 125.0 | 62.5 |
| S. aureus ATCC-25923 | + | 1.9 | 1.9 | 7.8 | 7.8 | 7.8 |
| B. subtilis CMCC-63501 | + | 1.9 | 1.9 | 31.2 | 31.2 | 15.6 |
| B. cereus | + | 31.2 | 15.6 | 125.0 | 125.0 | 62.5 |
| E. coli ATCC-25922 | − | 15.62 | 3.9 | 125 | 250 | 125 |
| P. aeruginosa ATCC-27853 | − | 125.0 | 15.6 | 250.0 | 250.0 | 125.0 |
| S. enterica | − | 125.0 | 62.5 | 250.0 | 250.0 | 125.0 |
| S. para-typhi B | − | 1.9 | 0.9 | 15.6 | 15.6 | 15.6 |
CD data suggested that the triazole-containing peptides adopted a β-sheet conformation in aqueous solution, whereas the A3 remained in a disordered structure. The D-A3 maintained excellent antibacterial activity against certain bacteria due to its structural similarity. The enhanced antibacterial efficacy of D-A3 against specific bacteria, such as E. coli ATCC-25922 and P. aeruginosa ATCC-27853, compared to A3, may result from the improved compatibility of the D-peptide with specific bacterial targets, thereby augmenting antibacterial effectiveness. Modifying A3 by replacing disulfide bonds with a triazole ring could alter its structure, leading to diminished antibacterial activity for the three triazole-containing peptides. In comparing the antibacterial activities of the three triazole-containing peptides, it was observed that A3-C6 demonstrated slightly superior antibacterial activity compared to A3-C4 and A3-C5. It probably is that the extended linker of A3-C6 enabled its structure to more closely mimic the structure of the A3, thereby enhancing its antibacterial activity.
3.4. Antibacterial mechanism of A3 and its derived peptides
To elucidate the mode of action of A3 and its derivative peptides on bacteria, crystal violet staining was performed. Crystal violet is an alkaline dye that can bind to bacterial intracellular DNA or extracellular polysaccharides to form dark blue crystals. Crystal violet is used for bacterial DNA or biofilm staining. In order to understand the effect of A3 and its derived peptides on bacterial biofilm, the absorption of crystal violet by S. aureus ATCC-25923 (treated with different concentrations of peptides) was measured. According to the absorption of crystal violet in different samples (Fig. S13†), the inhibition rate of different concentrations of A3 and its derived peptides on biofilm was calculated by using the inhibition rate calculation.32 As illustrated in Fig. 6, the inhibitory effect of peptides on biofilm formation is concentration dependent. At a concentration of 1/2 MIC, A3, D-A3 and A3-C6 inhibited biofilm formation of S. aureus ATCC-25923 by about 50% or more, comparing to the untreated group. At a concentration of 2 MIC, A3, D-A3 and A3-C6 inhibited bio-film formation of S. aureus ATCC-25923 by about 80%. Seo et al. reported the action mode of A3 was that it interacted with intracellular components such as DNA or DNA amplification reactions24. These results suggest that an additional action mode of A3 and its derivative peptides is that they can inhibit the biofilm formation of bacteria. | ||
| Fig. 6 Biofilm inhibition rate of S. aureus ATCC-25923 in the presence of different concentrations of A3, D-A3 and A3-C6. | ||
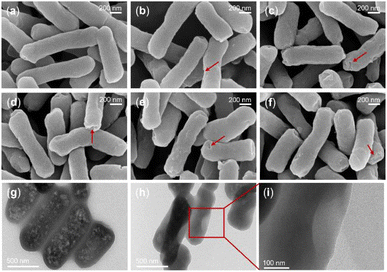
To confirm the effect of A3 and its derivative peptides on bacteria, scanning electron microscopy (SEM) was used. The study focused on three bacterial strains: S. enterica, S. aureus ATCC-25923, and P. aeruginosa ATCC-27853. Initially, SEM was utilized to observe morphological alterations in S. enterica following peptide treatment, with phosphate-buffered saline (PBS)-treated bacteria serving as a negative control. As illustrated in Fig. 7a–f, the results demonstrated that PBS-treated S. enterica maintained a rod-shaped appearance with uniform size, intact morphology, and a smooth surface, exhibiting no signs of depression, shrinkage, or damage. Conversely, treatment with A3, D-A3, A3-C4, A3-C5, and A3-C6 resulted in S. enterica exhibiting irregular morphologies and varying degrees of surface depression compared to the PBS group. Notably, the D-A3 treatment group displayed the most pronounced shrinkage, while S. enterica treated with A3-C4, A3-C5, and A3-C6 exhibited similar degrees of morphological shrinkage. The D-A3 and A3-C6 have already been demonstrated to have significant antibacterial activity against P. aeruginosa ATCC-27853. SEM was employed to examine the effects of these peptides on P. aeruginosa ATCC-27853. As depicted in Fig. S13,† the PBS-treated P. aeruginosa ATCC-27853 exhibited a relatively smooth surface with no significant damage. In contrast, peptide-treated bacteria showed degrees of membrane damage, including shrinkage and, in some cases, rupture. Subsequently, another potential antimicrobial peptide A3-C6 was utilized to treat S. aureus ATCC-25923. The morphological alterations of S. aureus ATCC-25923 were assessed using SEM. As illustrated in Fig. S14,† the S. aureus ATCC-25923 in the control group maintained a spherical shape with an intact external morphology and a smooth surface.
We utilized transmission electron microscopy (TEM) to investigate the impact of the D-A3, which had exceptional antibacterial activity, on P. aeruginosa ATCC-27853. As illustrated in Fig. 7g, in the absence of the peptide, P. aeruginosa ATCC-27853 retained an intact morphological structure with a consistent distribution of intracellular materials. Conversely, following peptide treatment, the bacteria exhibited pronounced alterations, including ruptured or collapsed surfaces, uneven intracellular material distribution, potential vacuolation, and compromised cell membrane integrity.
3.5. Stability of A3 and its derived peptides
| MIC (μg mL−1) | |||
|---|---|---|---|
| Control | 12.5% serum | 50% serum | |
| A3 | 1.9 | 3.9 | 7.8 |
| D-A3 | 1.9 | 1.9 | 1.9 |
| A3-C6 | 7.8 | 15.6 | 31.2 |
3.6. Toxicity of A3 and its derived peptides
4. Conclusions
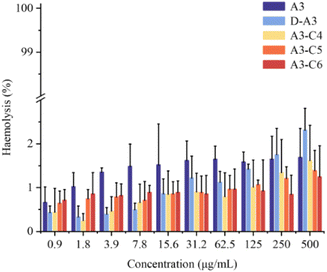
The rapid emergence of drug-resistant bacteria has heightened due to the misuse of traditional antibiotics. In response, researchers are actively seeking novel antibiotics to address these infections. AMPs have emerged as a promising alternative due to their distinctive antibacterial mechanisms. The structures of antimicrobial peptides, constrained by disulfide bonds, endow them with diverse spatial conformations, enabling them to perform a wide range of functions. Consequently, AMPs represent a potentially valuable class of therapeutic agents. However, despite the fact that many AMPs have been discovered, they are vulnerable to rapid degradation by reducing agents and various proteases. This instability has resulted in a limited number of peptide drugs successfully advancing from laboratory research to clinical application, thereby posing a significant challenge to the development of peptide-based antibiotics.In this study, we engineered and synthesized four novel derivative peptides based on the A3 by incorporating D-amino acids or replacing the disulfide bond with a triazole ring, with the objective of enhancing their stability and biological activity under reducing conditions. Initially, we synthesized D-A3, A3-C4, A3-C5, and A3-C6 utilizing Fmoc SPPS. The successful synthesis of the target peptides was verified via liquid-phase reaction monitoring and mass spectrometry. Subsequently, we assessed the antibacterial activity of the synthesized peptides using the pore diffusion method. The antibacterial experimental data revealed that the derivative peptides exhibited broad-spectrum antibacterial activity. Notably, D-A3 displayed superior antibacterial efficacy compared to A3, particularly against specific bacteria such as E. coli ATCC-25922 and P. aeruginosa ATCC-27853. However, the antibacterial activity of the triazole-containing peptides showed a slight decrease compared to A3. Through investigation of the antibacterial mechanism of the peptides, we found an additional mechanism that they inhibit biofilm formation. Stability assays demonstrated that the triazole-containing peptides exhibited substantial stability under reducing conditions, whereas D-A3 showed significant resistance to trypsin or serum degradation. Toxicity assessments revealed that D-A3, A3-C4, A3-C5, and A3-C6 exhibited minimal hemolytic toxicity (<5%). Furthermore, zebrafish toxicity assays indicated that D-A3 and A3 had comparable toxicity levels, while A3-C6 had the least impact on zebrafish embryo hatching. In summary, our results suggest that the A3 derivative peptides exert antibacterial effects through membrane disruption. Notably, the D-A3 and A3-C6 demonstrate promising antibacterial activity, enhanced enzymatic stability, and low toxicity, thereby presenting themselves as potential candidates for the development of efficient AMPs for bacterial infection treatment.
Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Baotou Medical College and approved by the Animal Ethics Committee of Baotou Medical College.Data availability
All supporting data can be founding the ESI file.†Author contributions
Methodology, conceptualization, data curation, writing original draft, Jiawei Zhao. Methodology, data curation, Xin Zhang. Methodology, data curation, Yu Liu and Zhixing Geng. Methodology, Jili Dai. Supervision and review, Ye Guo. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge Meiying Zhang for her help in providing various bacteria required for the experiment. This research was funded by the National Natural Science Foundation of China, grant numbers 21967017, and the Natural Science Foundation of Inner Mongolia, grant number 2023MS02001, and 2022 Inner Mongolia Autonomous Region “Grassland Talents” Project Youth Innovation and Entrepreneurship Talent Project, grant numbers 2022CYYC1C88.References
- N. Yadav and V. S. Chauhan, Adv. Colloid Interface Sci., 2024, 333, 103282 CrossRef CAS PubMed.
- A. Mercer, Pathog. Global Health, 2021, 115(3), 151–167 CrossRef CAS PubMed.
- R. M. Olson and D. M. Anderson, PLoS One, 2019, 14(5), e0217440 CrossRef CAS PubMed.
- S. Paul, S. Verma and Y.-C. Chen, ACS Infect. Dis., 2024, 10(4), 1034–1055 CrossRef CAS PubMed.
- X. Gong, Y. Han, T. Wang, G. Song, H. Chen, H. Tang, X. Huang, K. Deng, S. Wang and Y. Wang, Adv. Mater., 2025, 37(4), e2414357 Search PubMed.
- T. Pulingam, T. Parumasivam, A. M. Gazzali, A. M. Sulaiman, J. Y. Chee, M. Lakshmanan, C. F. Chin and K. Sudesh, Eur. J. Pharm. Sci., 2022, 170, 106103 CrossRef CAS PubMed.
- C. Xu, L. Kong, H. Gao, X. Cheng and X. Wang, Front. Microbiol., 2022, 13, 822689 CrossRef PubMed.
- K. Xu, X. Zhao, Y. Tan, J. Wu, Y. Cai, J. Zhou and X. Wang, Biomater. Adv., 2023, 155, 213684 CrossRef CAS PubMed.
- Y. Hou, T. Tan, Z. Guo, Y. Ji, J. Hu and Y. Zhang, Biomater. Sci., 2022, 10, 3831–3844 Search PubMed.
- Y. Zhu, W. Xu, W. Chen, B. Li, G. Li, H. Deng, L. Zhang, C. Shao and A. Shan, Sci. Adv., 2025, 11(6), eads3844 CrossRef CAS PubMed.
- X. Y. Luo, C. M. Hu, Q. Yin, X. M. Zhang, Z. Z. Liu, C. K. Zhou, J. G. Zhang, W. Chen and Y. J. Yang, Adv. Sci., 2024, 11(30), e2401793 CrossRef PubMed.
- B. Li, Y. Liu, P. Yan, X. Ouyang, Z. Ba, Y. Wang, T. Yang, Z. Yu, B. Ren, C. Zhong, H. Liu, Y. Zhang, S. Gou and J. Ni, Eur. J. Med. Chem., 2025, 283, 117149 CrossRef CAS PubMed.
- G. Subramaniam and M. Girish, Indian J. Pediatr., 2020, 87(11), 937–944 CrossRef PubMed.
- K. Botelho Sampaio de Oliveira, M. Lopes Leite, V. Albuquerque Cunha, N. Brito da Cunha and O. Luiz Franco, Drug Discovery Today, 2023, 28(8), 103629 CrossRef CAS PubMed.
- C. H. Chen and T. K. Lu, Antibiotics, 2020, 9(1), 24 CrossRef CAS PubMed.
- A. M. Turner, J. Y. H. Lee, C. L. Gorrie, B. P. Howden and G. P. Carter, Front. Microbiol., 2021, 12, 637656 CrossRef PubMed.
- N. Zhang, X. Gu, D. Song, P. Zhang, N. Zhang, W. Chen, S. Ji, Y. Qi and S. Ma, Bioorg. Chem., 2022, 119, 105583 CrossRef CAS PubMed.
- Y. Tian, Y. Hou, J. Tian, J. Zheng, Z. Xiao, J. Hu and Y. Zhang, J. Mater. Chem. B, 2024, 12(33), 8122–8132 RSC.
- M. Chen, N. Lin, X. Liu, X. Tang, Z. Wang and D. Zhang, Front. Immunol., 2023, 14, 1168517 CrossRef CAS PubMed.
- C. Bucataru and C. Ciobanasu, Microbiol. Res., 2024, 286, 127822 CrossRef CAS PubMed.
- C. Tian, N. Zhao, L. Yang, F. Lin, R. Cai, Y. Zhang, J. Peng and G. Guo, Front. Cell. Infect. Microbiol., 2024, 14, 1334378 CrossRef CAS PubMed.
- M. V. Arasu and N. A. Al-Dhabi, J. Infect. Public Health, 2023, 16(12), 2031–2037 CrossRef PubMed.
- J.-K. Seo, J. M. Crawford, K. L. Stone and E. J. Noga, Biochem. Biophys. Res. Commun., 2005, 338(4), 1998–2004 CrossRef CAS PubMed.
- J. K. Seo, D. G. Kim, J. E. Lee, K. S. Park, I. A. Lee, K. Y. Lee, Y. O. Kim and B. H. Nam, Mar. Drugs, 2021, 19(8), 451 CrossRef CAS PubMed.
- R. Mourtada, H. D. Herce, D. J. Yin, J. A. Moroco, T. E. Wales, J. R. Engen and L. D. Walensky, Nat. Biotechnol., 2019, 37(10), 1186–1197 CrossRef CAS PubMed.
- Z. Lai, X. Yuan, H. Chen, Y. Zhu, N. Dong and A. Shan, Biotechnol. Adv., 2022, 59, 107962 CrossRef CAS PubMed.
- C. Bechtler and C. Lamers, RSC Med. Chem., 2021, 12(8), 1325–1351 RSC.
- A. M. White, S. J. de Veer, G. Wu, P. J. Harvey, K. Yap, G. J. King, J. E. Swedberg, C. K. Wang, R. H. P. Law, T. Durek and D. J. Craik, Angew Chem. Int. Ed. Engl., 2020, 59(28), 11273–11277 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1(6), 2876–2890 CrossRef CAS PubMed.
- N. Akram, M. Usman, S. Haider, M. S. Akhtar and K. Gul, Polymers, 2022, 14(17), 3701 CrossRef CAS PubMed.
- A. Aspatwar, M. M. Hammaren, M. Parikka and S. Parkkila, J. Visualized Exp., 2019, 25, 150 Search PubMed.
- Y. Liu, Y. Jiang, J. Zhu, J. Huang and H. Zhang, Carbohydr. Polym., 2019, 206, 412–419 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra02745d |
| This journal is © The Royal Society of Chemistry 2025 |