Open Access Article
Open Access ArticleIdentification of prochlorperazine dimaleate as a Sortase A inhibitor from FDA libraries for MRSA infection treatment†
Abhinit Kumarab,
Sonali Chhabraab and
Raman Parkesh *ab
*ab
aGNRPC, CSIR – Institute of Microbial Technology, Chandigarh, 160036, India. E-mail: raman.parkesh@csir.res.in
bAcademy of Scientific and Innovation Research (AcSIR), Ghaziabad, 201002, India
First published on 25th June 2025
Abstract
Staphylococcus aureus is acknowledged as an essential contributor to global disease burden, particularly with the emergence of methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA) strains. This study employs a systematic computational and experimental strategy to screen and validate FDA-approved drugs to target Sortase A, an essential enzyme involved in MRSA virulence. Herein, we have identified six molecules exhibiting antimicrobial potency against MRSA and reduced biofilm formation. Among the hits obtained, prochlorperazine dimaleate showed potent activity against MRSA while proving non-cytotoxic to hepatocellular Carcinoma (HepG2) cells at inhibitory concentration. Further, it also disrupts the membrane potential and creates pores inside the membrane of MRSA. In vivo thigh infection studies in mice showed that prochlorperazine dimaleate successfully reduced the MRSA infection load. Taken together, we herein report that prochlorperazine dimaleate attenuated the pathogenicity of S. aureus, thus reducing MRSA infection by directly targeting Sortase A. Therefore, prochlorperazine dimaleate can be utilized as an adjuvant therapy for treating MRSA infection.
1. Introduction
Staphylococcus aureus (S. aureus) is a highly concerning and rapidly spreading pathogen that causes soft tissue infection, bacteremia, pneumonia, endocarditis, osteomyelitis, toxic shock syndrome, and skin infection.1,2 Methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA) have already posed a hazard to human health. This is further exacerbated by the biofilm-forming nature of S. aureus, which exhibits a high propensity for resistance to conventional antibiotics, posing a significant challenge in treating wound infections. MRSA now accounts for more than 60% of all isolated S. aureus.3 Hence, there is a critical need for novel treatment approaches that address drug resistance while simultaneously decreasing bacterial infections.4,5 The pathogenicity of Staphylococcus aureus is closely linked to the virulence factors it secretes, encompassing surface proteins, toxins, and enzymes.6,7 These factors aid in bacterial adherence, tissue invasion and degradation, and evasion of host defense mechanisms. Surface-associated adhesins such as clumping factor A/clumping factor B (ClfA/ClfB), fibronectin-binding proteins (FnBPs), and collagen adhesin (CNA) are involved in the pathogenesis of S. aureusinfection.8–10 Sortase A (SrtA), a cysteine transpeptidase, covalently attaches crucial virulence factors of S. aureus to the bacterial surface, promotes bacterial adherence, facilitates biofilm formation, mediates host cell penetration, acquires vital nutrients from the host, and contributes to immune evasion and suppression.11–13 Further, SrtA stands out as a promising drug target due to its location on the extracellular side of the cell membrane, and an additional advantage is the absence of human homologs,14 suggesting that selective inhibitors of this enzyme should exhibit lower toxicity.15 Consequently, inhibiting this enzyme is anticipated to exert reduced selection pressure on developing resistance.16,17 Over the last decade, several natural products such as flavonoids,18 salvinolic acid,19 chalcone,20 peptide analogs, and synthetic small molecules have been reported as Sortase A inhibitors (Table S1†). However, potential toxicity and antimicrobial resistance are persistent concerns.To address this, we have employed computational and experimental strategies to target Sortase A in MRSA. We have screened Food and Drug Administration (FDA)-approved drug libraries against Sortase A and identified 60 hit molecules. Phenotypic screening of the top virtual screening hits by antimicrobial assay identified six promising molecules against clinically isolated MRSA. Among these, prochlorperazine dimaleate exhibited potent antibacterial activity, biofilm eradication, membrane potential disruption, and was non-cytotoxic in the HepG2 cell line. Further, it also reduced the MRSA infection load in the thigh infection murine model. In summary, we have identified novel chemical scaffolds promising novel therapeutic agents for combating MRSA infections.
2. Experimental
2.1. Materials and methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) with the 0.1% glucose supplement. This diluted culture was added to a flat-bottom 96-well microtiter plate, followed by compounds at MIC and 2 × MIC, and further incubation at 37 °C for 24 hours.32 Following the incubation, the planktonic cells were removed from the wells, and the wells were washed with 1 × PBS (pH 7.4).35 The microtiter plate was dried for 10 minutes, and biofilm was stained with 0.1% crystal violet for 10 minutes at 25 °C. The wells were washed with distilled water to remove any excess stain. To solubilize the stain, glacial acetic acid (33%) was added and incubated at room temperature for 30 minutes on an orbital shaker.36 The absorbance of the stained cells was measured at 600 nm with a plate reader, and the percentage of biofilm growth inhibition relative to the untreated control was calculated. The experiment was conducted in triplicate with three biological replicates, and the data were analyzed utilizing GraphPad Prism 8.
100) with the 0.1% glucose supplement. This diluted culture was added to a flat-bottom 96-well microtiter plate, followed by compounds at MIC and 2 × MIC, and further incubation at 37 °C for 24 hours.32 Following the incubation, the planktonic cells were removed from the wells, and the wells were washed with 1 × PBS (pH 7.4).35 The microtiter plate was dried for 10 minutes, and biofilm was stained with 0.1% crystal violet for 10 minutes at 25 °C. The wells were washed with distilled water to remove any excess stain. To solubilize the stain, glacial acetic acid (33%) was added and incubated at room temperature for 30 minutes on an orbital shaker.36 The absorbance of the stained cells was measured at 600 nm with a plate reader, and the percentage of biofilm growth inhibition relative to the untreated control was calculated. The experiment was conducted in triplicate with three biological replicates, and the data were analyzed utilizing GraphPad Prism 8.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 HepG2 cells seeded in each well. The plate was then incubated at 37 °C with 5% CO2 for 24 hours.49 DMEM medium was removed, and the cells were treated with compounds at MIC and 2 × MIC, further incubated for 24 hours. Culture media was removed, and HepG2 cells were washed with 1 × PBS (pH 7.4) to eliminate residual media components.50 Next, the HepG2 cells were stained with MTT solution (5 mg mL−1) and incubated for 4 hours. Viable cells produced a purple color due to the formation of formazan crystals. After that, 50 μL of DMF (a solubilizing agent) was added to each well, incubating for 6 hours again, and measuring absorbance at 570 nm.51 The percentage of cell viability was calculated in triplicate.
000 HepG2 cells seeded in each well. The plate was then incubated at 37 °C with 5% CO2 for 24 hours.49 DMEM medium was removed, and the cells were treated with compounds at MIC and 2 × MIC, further incubated for 24 hours. Culture media was removed, and HepG2 cells were washed with 1 × PBS (pH 7.4) to eliminate residual media components.50 Next, the HepG2 cells were stained with MTT solution (5 mg mL−1) and incubated for 4 hours. Viable cells produced a purple color due to the formation of formazan crystals. After that, 50 μL of DMF (a solubilizing agent) was added to each well, incubating for 6 hours again, and measuring absorbance at 570 nm.51 The percentage of cell viability was calculated in triplicate.2.1.14.1. Thigh infection in a neutropenic mouse. The effectiveness of prochlorperazine dimaleate was assessed in an in vivo model of thigh infection caused by S. aureus ATCC 43300.52 Female BALB/C mice (n = 4; 6–8 weeks old) were made neutropenic by receiving two doses of cyclophosphamide (Sigma-Aldrich) via intraperitoneal injection, with the first dose given 4 days before infection (150 mg kg−1 of body weight) and the second dose given one day prior (100 mg kg−1 of body weight).53 Following neutropenia induction, the mice were infected with S. aureus ATCC 43300 at a concentration of 5 × 105 CFU per mouse, administered intramuscularly into the right thigh muscles. Four hours after infection, each group of mice (n = 4) received an oral dose of prochlorperazine dimaleate and vancomycin 100 mg kg−1 of body weight, respectively. The remaining group of mice served as the vehicle control. After 24 hours of infection, the mice were sacrificed, and the right thigh muscles were dissected and preserved in 1 × PBS (pH 7.4). Subsequently, the muscles were homogenized, and the number of colony-forming units (CFUs) in the muscles was determined by spreading dilutions on MHA plates.
3. Results
3.1. Identification of novel antimicrobial agents against MRSA
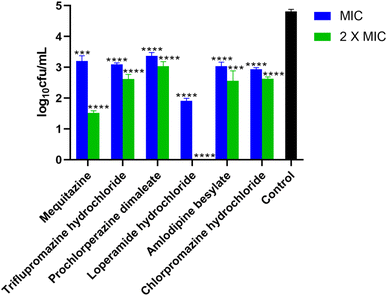
We performed molecular docking of FDA-approved drugs with Sortase A protein to identify novel inhibitors of S. aureus by drug-repurposing. This allows an accelerated drug development pathway for clinical use as the safety and pharmacological profiles of these molecules are already established. Additionally, this further strengthens the existing antimicrobial arsenal. Out of the 3040 molecules screened, 60 compounds were selected (Table S2†), based on binding scores and availability of the chemical molecules for further evaluation by in vitro phenotypic screening against MRSA.We determined the minimum inhibitory concentration and minimum bactericidal concentration (MIC and MBC) of 60 compounds against the S. aureus ATCC 43300 strain. Among them, prochlorperazine dimaleate, loperamide hydrochloride, and chlorpromazine hydrochloride displayed the lowest MIC activity of 32 μg mL−1. Whereas, mequitazine, loperamide hydrochloride, and amlodipine besylate exhibited MICs of 64 μg mL−1. Furthermore, we determined the MBC of these compounds. Triflupromazine hydrochloride, prochlorperazine dimaleate, and chlorpromazine hydrochloride showed the lowest bactericidal activity of 64 μg mL−1. On the other hand, mequitazine, loperamide hydrochloride, and amlodipine besylate demonstrated the lowest bactericidal activity of 128 μg mL−1 (Table 1).
| Compounds | S. aureus ATCC 43300 | |
|---|---|---|
| MIC (μg mL−1) | MBC (μg mL−1) | |
| Mequitazine | 64 | 128 |
| Triflupromazine hydrochloride | 32 | 64 |
| Prochlorperazine dimaleate | 32 | 64 |
| Loperamide hydrochloride | 64 | 128 |
| Amlodipine besylate | 64 | 128 |
| Chlorpromazine hydrochloride | 32 | 64 |
We further explored the binding interaction of these drugs exhibiting potent activity against MRSA with Sortase A enzyme. The binding fingerprints of top most potent molecules are shown in Fig. 1. All the examined drug molecules showed strong binding at their active site, ranging from −5.9 to −7.67 kcal mol−1 (Fig. S1 and Table S3†). The primary amino acids involved in forming hydrogen bonds with the drug molecules include Val108, Asp112, Glu113, Gln114, Trp136, and Arg139.
3.2. Fluorescence quenching assay
A fluorescence quenching assay was used to characterize the binding affinity of Sortase A with the compounds. Sortase A exhibits intrinsic fluorescence due to aromatic amino acids, tryptophan, tyrosine, and phenylalanine. Treatment of Sortase A with these compounds resulted in a dose-dependent quenching of its fluorescence intensity, except loperamide hydrochloride Fig. 2. The binding constants (Ksv) for mequitazine, triflupromazine hydrochloride, prochlorperazine dimaleate, amlodipine besylate, and chlorpromazine hydrochloride with Sortase A were found to be 5.6 × 103 L mol−1, 5.4 × 103 L mol−1, 7.2 × 103 L mol−1, 4.7 × 103 L mol−1, and 6.3 × 103 L mol−1, respectively. Quercetin hydrate has been used as positive control. These results indicate that these compounds significantly interact with Sortase A.3.3. Sortase A activity assay
We determined the percent inhibition of compounds (mequitazine, triflupromazine hydrochloride, prochlorperazine dimaleate, loperamide hydrochloride, amlodipine besylate, chlorpromazine hydrochloride) against Sortase A protein. Sortase A activity was evaluated using the fluorescent substrate peptide Abz-LPATG-Dap(DNP). The enzyme cleaves the peptide specifically between threonine (T) and glycine (G), disrupting fluorescence resonance energy transfer (FRET) and leading to an increase in fluorescence intensity. When incubated with Sortase A, all tested compounds, except loperamide hydrochloride, demonstrated inhibitory activity against the enzyme (Table 2).| Compounds | S. aureus ATCC 43300 Sortase A | |
|---|---|---|
| Concentration (μg mL−1) | % inhibition | |
| Mequitazine | 128 | 64 |
| Triflupromazine hydrochloride | 128 | 48 |
| Prochlorperazine dimaleate | 128 | 47 |
| Amlodipine besylate | 128 | 57 |
| Chlorpromazine hydrochloride | 128 | 49 |
| Quercetin hydrate | 128 | 73 |
3.4. Time-dependent eradication of MRSA by potent compounds
We evaluated these compounds' effectiveness in killing S. aureus ATCC 43300 strains over time. Mequitazine and amlodipine besylate effectively eliminated the bacteria within 12–24 hours and 4–24 hours, respectively. Similarly, loperamide hydrochloride and chlorpromazine hydrochloride eliminated the bacteria within 8–12 hours, and re-growth of the bacteria was observed after 24 hours in the presence of these compounds. All four of these compounds (mequitazine, amlodipine besylate, loperamide hydrochloride, and chlorpromazine hydrochloride) exhibited a ≥3 log reduction in colony-forming units (CFU) from the initial inoculum. These results indicated that the above four drugs exhibit bactericidal characteristics. On the other hand, triflupromazine hydrochloride and prochlorperazine dimaleate achieved a ∼2 log reduction in CFU within 2 to 4 hours. However, re-growth of the bacteria was observed after 24 hours in the presence of these compounds Fig. 3. This suggests that although these two compounds initially reduced the bacterial population, they were ineffective in completely eradicating S. aureus ATCC 43300. In conclusion, the findings highlight the efficient bactericidal activity of mequitazine, amlodipine besylate, loperamide hydrochloride, and chlorpromazine hydrochloride against S. aureus ATCC 43300. These compounds demonstrated rapid and sustained killing of the bacteria, making them promising candidates for further investigation in combating S. aureus infections.3.5. The identified drugs successfully inhibit the MRSA biofilm
The effectiveness of these compounds in inhibiting biofilm formation by the S. aureus ATCC 43300 strain was investigated. When used at MIC and 2 × MIC, the compounds successfully inhibited biofilm formation in S. aureus ATCC 43300 cells. When the compounds mequitazine, triflupromazine hydrochloride, prochlorperazine dimaleate, loperamide hydrochloride, amlodipine besylate, and chlorpromazine hydrochloride were employed at MIC, they significantly inhibited the biofilm growth by 95%, 19%, 67%, 95%, 40%, and 93%, respectively. Whereas, at 2 × MIC, all these compounds inhibit 94% of the biofilm growth Fig. 4. Since biofilms provide a protective shield for pathogenic bacteria from antibiotic treatments and host immune responses, this association often leads to chronic infections. The present study's compounds can eliminate MRSA biofilm formation and are promising candidates for novel drug development.3.6. Membrane potential disruption of MRSA by drugs
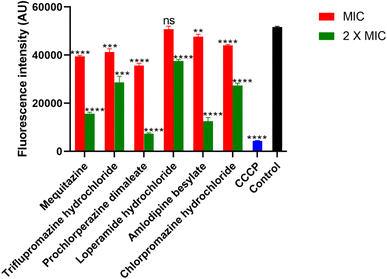
We determined the membrane potential perturbation of the S. aureus ATCC 43300 strain. Gram-positive bacteria have negatively charged structures on their outer surface, such as teichoic acid and polysaccharide components in the cytoplasmic membrane.54 The negative potential is crucial for several membrane complex systems, bacterial cell division and proliferation, signaling, and antibiotic susceptibility.55–59 All these compounds, except loperamide hydrochloride, successfully disrupted the membrane potential at their MIC and 2 × MIC. While loperamide hydrochloride showed significant reduction only at 2 × MIC (Fig. 5), A proton ionophore, Carbonyl cyanide m-chlorophenyl hydrazone (CCCP), 1 mg mL−1, has been used as a positive control. CCCP can shift the membrane potential to ∼ zero mV (proton Nernst potential in the current scenario). These findings indicate that the compounds can interfere with the membrane potential of S. aureus ATCC 43300, disrupting its normal function.3.7. The drugs compromise the MRSA cell membrane integrity
We investigated the membrane disruption capability of compounds by performing a membrane permeability assay. In this scenario, propidium iodide dye (PI) permeates the bacterial cell membrane and binds to double-stranded DNA only when the compounds damage the membrane, which otherwise remains impermeable to the dye.60,61 Nisin was used as a positive control due to its ability to create pores in the cell membrane.62,63 We observed that the fluorescence intensity of PI was increased over time after being treated with these compounds, which can be attributed to membrane permeabilization Fig. 6. Therefore, the results suggest that the compounds have the capability to permeabilize the cell membrane of S. aureus ATCC 43300.3.8. Assessment of macrophage invasion by MRSA
The invasion of S. aureus ATCC 43300 in macrophage (THP1) was determined in the presence of drugs under investigation at MIC and 2 × MIC. The results reveal that the compounds significantly reduce S. aureus ATCC 43300 invasion at MIC and 2 × MIC. Prochlorperazine dimaleate successfully reduced MRSA invasion of macrophages at 2 × MIC Fig. 7.3.9. Toxicity assessment in rabbit blood
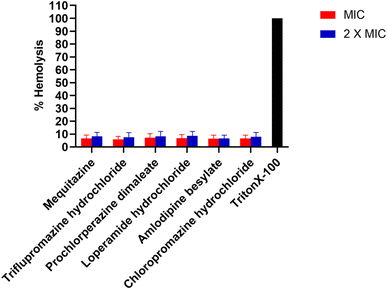
We examined the hemolytic effects of these compounds in rabbit blood. A hemolytic value below 10% indicates non-hemolytic activity, while a value above 25% indicates hemolytic activity.64 We treated the rabbit blood with the compounds at their MIC and 2 × MIC. Remarkably, we observed that all the compounds demonstrated a percentage of hemolysis lower than 10% Fig. 8. Based on our investigation, the compounds mequitazine, triflupromazine hydrochloride, prochlorperazine dimaleate, loperamide hydrochloride, amlodipine besylate, and chlorpromazine hydrochloride have shown acceptable safety profiles in relation to hemolytic activity when tested on rabbit blood. Therefore, these findings suggest that these compounds exhibit a favorable safety profile and can be potentially used for adjuvant therapy.3.10. Cytotoxicity assessment in HepG2 cell line
Cytotoxicity in the HepG2 cell line was assessed utilizing the MTT assay. When tested at their respective minimum inhibitory concentration levels, mequitazine, triflupromazine hydrochloride, loperamide hydrochloride, amlodipine besylate, and chlorpromazine hydrochloride demonstrated a percentage viability of 30%, 63%, 84%, 60%, and 28%. Prochlorperazine dimaleate exhibited a percentage cell viability of 86% at MIC and approximately 50% at 2 × MIC. These results indicate that prochlorperazine dimaleate exhibits low cytotoxicity towards mammalian cells at MIC and 2 × MIC Fig. 9. Hence, these findings suggest that prochlorperazine dimaleate demonstrates a promising safety profile and may have potential applications in biomedicine.3.11. Animal studies
4. Discussion
This study identifies FDA-approved drugs from diverse chemical classes as potent inhibitors of Staphylococcus aureus Sortase A, offering promising candidates against multidrug-resistant MRSA. By integrating computational screening with functional validation, we demonstrate that repurposed neuropharmacologic agents-including phenothiazines (triflupromazine, prochlorperazine, chlorpromazine), mequitazine, amlodipine, and loperamide-target SrtA's catalytic domain with submicromolar affinity. Their established clinical usage underscores the feasibility of drug repurposing for infectious diseases and raises the prospect of rapid clinical translation. These compounds expand the chemotherapeutic arsenal against multidrug-resistant MRSA by exploiting non-growth-dependent antivirulence mechanisms.Sortase A anchors virulence-associated surface proteins to the bacterial cell wall, thereby enhancing MRSA's capacity to colonize and persist in host tissues. The identified compounds demonstrated high-affinity binding to the Sortase A active site, as confirmed by molecular docking, fluorescence quenching, and enzyme inhibition assays. All molecules, except loperamide hydrochloride, inhibited Sortase A activity in vitro, supporting their mechanism-based antimicrobial potential. Among the six candidates, mequitazine and amlodipine besylate exhibited bactericidal activity, achieving ≥3 − log reduction in MRSA colony-forming units, while the remaining compounds displayed bacteriostatic effects. This differentiation is critical for tailoring therapeutic strategies, especially in life-threatening infections requiring rapid bacterial clearance.
In addition to antimicrobial activity, these compounds also inhibited MRSA biofilm formation, a major contributor to chronic and device-associated infections. Several hits notably compromised the extracellular matrix, a hallmark of biofilm resilience, suggesting their potential in addressing recalcitrant infections that evade standard antibiotic therapy. Mechanistically, five of the six compounds induced membrane depolarization in MRSA, collapsing the proton gradient critical for ATP generation and nutrient uptake. This disruption likely underpins their antibacterial efficacy. Furthermore, increased membrane permeability was observed, consistent with irreversible damage and bacteriolysis.
The ability of these compounds to reduce intracellular MRSA burden in macrophage (THP-1) models suggests additional utility in eradicating bacterial reservoirs that contribute to persistence and relapse. This is particularly relevant given the emerging recognition of S. aureus's capacity for transient intracellular survival.
Importantly, the compounds exhibited minimal hemolytic activity (<10%), indicating low risk of erythrocyte toxicity. Cytotoxicity profiling in HepG2 cells confirmed favorable safety margins, with prochlorperazine dimaleate demonstrating high cell viability even at 2 × MIC concentrations. In vivo validation in a neutropenic thigh infection model confirmed that prochlorperazine dimaleate significantly reduced MRSA burden in murine muscle tissue, showing efficacy comparable to vancomycin. These findings highlight its translational potential as a repurposed anti-MRSA agent with a favorable safety and pharmacological profile.
Among the phenothiazine scaffold, mequitazine exhibits weaker antimicrobial potency. This decreased activity may be attributed to the presence of bridgehead ring system, which decreases the molecule lipophilicity, and thus may hinder its penetration into the peptidoglycan layer of MRSA (Fig. S2†). Advancing our understanding of the structure-activity relationship (SAR) of SrtA inhibitors and the enzyme structural features will enable the development of potent antibiotic agents against MRSA.
5. Conclusion
In conclusion, docking studies together with experimental evaluation suggest that mequitazine, triflupromazine hydrochloride, prochlorperazine dimaleate, loperamide hydrochloride, amlodipine besylate, and chlorpromazine hydrochloride are potential novel lead molecules that bind with Sortase A and exhibit antibacterial activity against the S. aureus ATCC 43300 strain. The prochloroperazine showed comparative in vivo potency against S. aureus. However, it is essential to acknowledge the constraints in the present study. The antimicrobial compounds evaluated in the present study, apart from prochloperazine dimaleate, displayed toxic potential in mammalian cells. Hence, the identification of toxicophores and rational scaffold optimization may be carried out to enrich the antimicrobial arsenal against MRSA. These molecules have remarkable potential for future medicinal chemistry and represent the lead candidates in adjuvant therapy for fatal MRSA infection.Data availability
The data supporting this article have been included in the ESI.†Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors express their gratitude to the CSIR-Institute of Microbial Technology for the financial support (projects: OLP0136 and OLP0186). A. K. is grateful to the University Grants Commission (UGC) for awarding the Senior Research Fellowship (SRF). S. C. is thankful to the Council of Scientific and Industrial Research (CSIR) for awarding the Senior Research Fellowship (SRF).References
- F. D. Lowy, N. Engl. J. Med., 1998, 339, 520–532 CrossRef CAS PubMed.
- F. D. Lowy, N. Engl. J. Med., 2011, 364, 1987–1990 CrossRef CAS PubMed.
- Z. Bai, M. Chen, Q. Lin, Y. Ye, H. Fan, K. Wen, J. Zeng, D. Huang, W. Mo, Y. Lei and Z. Liao, Front. Cell Dev. Biol., 2021, 9, 629681 CrossRef PubMed.
- A. Kali, Pharmacogn. Rev., 2015, 9, 29–34 CrossRef CAS.
- S. S. Boswihi and E. E. Udo, Current Medicine Research and Practice, 2018, 8, 18–24 CrossRef.
- M. S. Smeltzer, Trends Microbiol., 2016, 24, 681–682 CrossRef CAS.
- D. Oliveira, A. Borges and M. Simões, Toxins, 2018, 10, 252 CrossRef PubMed.
- J. M. Patti and M. Höök, Curr. Opin. Cell Biol., 1994, 6, 752–758 CrossRef CAS PubMed.
- Y.-A. Que, P. François, J.-A. Haefliger, J.-M. Entenza, P. Vaudaux and P. Moreillon, Infect. Immun., 2001, 69, 6296–6302 CrossRef CAS PubMed.
- R. Heying, J. van de Gevel, Y.-A. Que, P. Moreillon and H. Beekhuizen, Thromb. Haemostasis, 2007, 97, 617–626 CrossRef CAS.
- T. Spirig, E. M. Weiner and R. T. Clubb, Mol. Microbiol., 2011, 82, 1044–1059 CrossRef CAS PubMed.
- L. A. Marraffini, A. C. DeDent and O. Schneewind, Microbiol. Mol. Biol. Rev., 2006, 70, 192–221 CrossRef CAS PubMed.
- S. Cascioferro, M. Totsika and D. Schillaci, Microb. Pathog., 2014, 77, 105–112 CrossRef CAS.
- S. K. Mazmanian, G. Liu, H. Ton-That and O. Schneewind, Science (New York, N.Y.), 1999, 285, 760–763 CrossRef CAS PubMed.
- S. Cascioferro, D. Raffa, B. Maggio, M. V. Raimondi, D. Schillaci and G. Daidone, J. Med. Chem., 2015, 58, 9108–9123 CrossRef CAS PubMed.
- K. R. V. Thappeta, L. N. Zhao, C. E. Nge, S. Crasta, C. Y. Leong, V. Ng, Y. Kanagasundaram, H. Fan and S. B. Ng, Int. J. Mol. Sci., 2020, 21, 8601 CrossRef CAS.
- S. Alharthi, S. E. Alavi, P. M. Moyle and Z. M. Ziora, Drug discovery today, 2021, 26, 2164–2172 CrossRef CAS.
- M. Chamlagain, J. Hu, R. V. Sionov and D. Steinberg, Front. Microbiol., 2024, 15, 1333274 CrossRef PubMed.
- R. Boudjemaa, C. Cabriel, F. Dubois-Brissonnet, N. Bourg, G. Dupuis, A. Gruss, S. Lévêque-Fort, R. Briandet, M. P. Fontaine-Aupart and K. Steenkeste, Antimicrob. Agents Chemother., 2018, 62, e00023 CrossRef CAS PubMed.
- G. Wang, Y. Gao, H. Wang, X. Niu and J. Wang, Front. Cell. Infect. Microbiol., 2018, 8, 418 CrossRef CAS PubMed.
- Y. Zong, T. W. Bice, H. Ton-That, O. Schneewind and S. V. Narayana, J. Biol. Chem., 2004, 279, 31383–31389 CrossRef CAS.
- C. P. Guimaraes, M. D. Witte, C. S. Theile, G. Bozkurt, L. Kundrat, A. E. Blom and H. L. Ploegh, Nat. Protoc., 2013, 8, 1787–1799 CrossRef PubMed.
- T. Cheeseright, M. Mackey, S. Rose and A. Vinter, J. Chem. Inf. Model., 2006, 46, 665–676 CrossRef CAS PubMed.
- M. R. Bauer and M. D. Mackey, J. Med. Chem., 2019, 62, 3036–3050 CrossRef CAS.
- M. Kuhn, S. Firth-Clark, P. Tosco, A. Mey, M. Mackey and J. Michel, J. Chem. Inf. Model., 2020, 60, 3120–3130 CrossRef CAS PubMed.
- T. Ginex, E. Madruga, A. Martinez and C. Gil, Front. Pharmacol., 2023, 14, 1244317 CrossRef CAS PubMed.
- D. Mu, Y. Luan, L. Wang, Z. Gao, P. Yang, S. Jing, Y. Wang, H. Xiang, T. Wang and D. Wang, Emerging Microbes Infect., 2020, 9, 169–179 CrossRef CAS.
- X. Wang, J. L. Chen, G. Otting and X. C. Su, Sci. Rep., 2018, 8, 16371 CrossRef PubMed.
- J. S. Lewis Ii, Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing, Clinical and Laboratory Standards Institute, 33rd edn, 2023 Search PubMed.
- M. A. T. Blaskovich, K. A. Hansford, Y. Gong, M. S. Butler, C. Muldoon, J. X. Huang, S. Ramu, A. B. Silva, M. Cheng, A. M. Kavanagh, Z. Ziora, R. Premraj, F. Lindahl, T. A. Bradford, J. C. Lee, T. Karoli, R. Pelingon, D. J. Edwards, M. Amado, A. G. Elliott, W. Phetsang, N. H. Daud, J. E. Deecke, H. E. Sidjabat, S. Ramaologa, J. Zuegg, J. R. Betley, A. P. G. Beevers, R. A. G. Smith, J. A. Roberts, D. L. Paterson and M. A. Cooper, Nat. Commun., 2018, 9, 22 CrossRef PubMed.
- E. V. K. Ledger and A. M. Edwards, mBio, 2023, 14, e0355822 Search PubMed.
- W. Liang, X. F. Liu, J. Huang, D. M. Zhu, J. Li and J. Zhang, BMC Infect. Dis., 2011, 11, 109 CrossRef CAS.
- G. A. McKay, S. Beaulieu, F. F. Arhin, A. Belley, I. Sarmiento, T. Parr, Jr. and G. Moeck, J. Antimicrob. Chemother., 2009, 63, 1191–1199 CrossRef CAS PubMed.
- A. Ogunsile, N. Songnaka, S. Sawatdee, M. Lertcanawanichakul, S. Krobthong, Y. Yingchutrakul, J. Uchiyama and A. Atipairin, PeerJ, 2023, 11, e16143 CrossRef PubMed.
- G. A. McKay, S. Beaulieu, F. F. Arhin, A. Belley, I. Sarmiento, T. Parr, Jr. and G. Moeck, J. Antimicrob. Chemother., 2009, 63, 1191–1199 CrossRef CAS PubMed.
- A. Ogunsile, N. Songnaka, S. Sawatdee, M. Lertcanawanichakul, S. Krobthong, Y. Yingchutrakul, J. Uchiyama and A. Atipairin, PeerJ, 2023, 11, e16143 CrossRef PubMed.
- M. A. Hudson, D. A. Siegele and S. W. Lockless, Antimicrob. Agents Chemother., 2020, 64, e00910 CrossRef CAS PubMed.
- H. Cao, Y. Qiao, X. Liu, T. Lu, T. Cui, F. Meng and P. K. Chu, Acta Biomater., 2013, 9, 5100–5110 CrossRef CAS PubMed.
- A. Masood, N. Ahmed, M. F. M. Razip Wee, A. Patra, E. Mahmoudi and K. S. Siow, Polymers, 2023, 15, 307 CrossRef CAS PubMed.
- N. P. Kalia, P. Mahajan, R. Mehra, A. Nargotra, J. P. Sharma, S. Koul and I. A. Khan, J. Antimicrob. Chemother., 2012, 67, 2401–2408 CrossRef CAS PubMed.
- L. Kovacs, R. A. Davis, T. Ganguly, R. Chammas and J. L. Sutcliffe, Biomed. Pharmacother., 2022, 145, 112469 CrossRef CAS PubMed.
- F. Richter, L. Martin, K. Leer, E. Moek, F. Hausig, J. C. Brendel and A. Traeger, J. Mater. Chem. B, 2020, 8, 5026–5041 RSC.
- F. Tramer, T. Da Ros and S. Passamonti, Methods Mol. Biol., 2012, 926, 203–217 CrossRef CAS.
- C. Xu, R. Song, P. Lu, J. Chen, Y. Zhou, G. Shen, M. Jiang and W. Zhang, Int. J. Nanomed., 2020, 15, 65–80 CrossRef CAS PubMed.
- M. Jangra, M. Kaur, R. Tambat, R. Rana, S. K. Maurya, N. Khatri, A. Ghafur and H. Nandanwar, Antimicrob. Agents Chemother., 2019, 63, 00338 CrossRef PubMed.
- S. Sreelatha, N. Kumar and S. Rajani, Front. Microbiol., 2022, 13, 1085113 CrossRef PubMed.
- S. Afrasiabi, A. Partoazar and N. Chiniforush, Sci. Rep., 2023, 13, 11552 CrossRef CAS PubMed.
- J. O'Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 267, 5421–5426 CrossRef PubMed.
- D. Seleem, B. Benso, J. Noguti, V. Pardi and R. M. Murata, PloS one, 2016, 11, e0157188 CrossRef PubMed.
- A. A. Javia, J. Maharaja Sayajirao Univ. Baroda, 2022 Search PubMed.
- M. V. Abdul Jaleel, Petroleum ether extract of Ophiorrhiza eriantha Wight induces apoptosis in human breast cancer MCF-7 cell line, J. Appl. Pharm. Sci., 2022, 12, 134–142 Search PubMed.
- P. A. Smith, M. F. T. Koehler, H. S. Girgis, D. Yan, Y. Chen, Y. Chen, J. J. Crawford, M. R. Durk, R. I. Higuchi, J. Kang, J. Murray, P. Paraselli, S. Park, W. Phung, J. G. Quinn, T. C. Roberts, L. Rougé, J. B. Schwarz, E. Skippington, J. Wai, M. Xu, Z. Yu, H. Zhang, M. W. Tan and C. E. Heise, Nature, 2018, 561, 189–194 CrossRef CAS PubMed.
- X. Zhang, J. Cao, Y. Pei, J. Zhang and Q. Wang, Oncol. Lett., 2016, 11, 3465–3470 CrossRef CAS PubMed.
- J. G. Hurdle, A. J. O'Neill, I. Chopra and R. E. Lee, Nat. Rev. Microbiol., 2011, 9, 62–75 CrossRef CAS PubMed.
- H. Strahl and L. W. Hamoen, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12281–12286 CrossRef CAS PubMed.
- J. P. Stratford, C. L. A. Edwards, M. J. Ghanshyam, D. Malyshev, M. A. Delise, Y. Hayashi and M. Asally, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 9552–9557 CrossRef CAS PubMed.
- A. Prindle, J. Liu, M. Asally, S. Ly, J. Garcia-Ojalvo and G. M. Süel, Nature, 2015, 527, 59–63 CrossRef CAS PubMed.
- H. Zhang, Y. Pan, L. Hu, M. A. Hudson, K. S. Hofstetter, Z. Xu, M. Rong, Z. Wang, B. V. V. Prasad, S. W. Lockless, W. Chiu and M. Zhou, Nat. Commun., 2020, 11, 547 CrossRef CAS PubMed.
- C. Y. Yang, M. Bialecka-Fornal, C. Weatherwax, J. W. Larkin, A. Prindle, J. Liu, J. Garcia-Ojalvo and G. M. Süel, Cell Syst., 2020, 10, 417–423.e413 CrossRef CAS PubMed.
- A. Müller, M. Wenzel, H. Strahl, F. Grein, T. N. V. Saaki, B. Kohl, T. Siersma, J. E. Bandow, H. G. Sahl, T. Schneider and L. W. Hamoen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E7077–e7086 CrossRef PubMed.
- S. M. Stocks, Cytom. J. Int. Soc. Anal. Cytol., 2004, 61, 189–195 CrossRef CAS PubMed.
- T. Schneider and H. G. Sahl, Curr. Opin. Invest. Drugs, 2010, 11, 157–164 CAS.
- E. Breukink and B. de Kruijff, Nat. Rev. Drug Discov., 2006, 5, 321–332 CrossRef CAS PubMed.
- I. Greco, N. Molchanova, E. Holmedal, H. Jenssen, B. D. Hummel, J. L. Watts, J. Håkansson, P. R. Hansen and J. Svenson, Sci. Rep., 2020, 10, 13206 CrossRef CAS PubMed.
- W. Wirth, R. Goesswald and W. Vater, Arch. Int. Pharmacodyn. Ther., 1959, 123, 78–114 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra01781e |
| This journal is © The Royal Society of Chemistry 2025 |