Open Access Article
Open Access ArticleDesign, synthesis and antifungal activity of novel matrine-hydroxamic acid derivatives containing benzene sulfonamide†
Suzhen Yana,
Jamal A. H. Kowahb,
Qingfeng Longa,
Qian Liua,
Hanqing Zhanga,
Siying Lua,
Lisheng Wang *a and
Haixia Yu*c
*a and
Haixia Yu*c
aSchool of Medicine, Guangxi University, Nanning, 530004, China. E-mail: lswang@gxu.edu.cn
bSchool of Chemistry and Chemical Engineering, Guangxi University, Nanning, 530004, China
cSchool of Chemical Engineering, Jilin Vocational College of Industry and Technology, Jilin, 132013, China. E-mail: yhx2006304022@126.com
First published on 19th May 2025
Abstract
To address the urgent need for novel antibacterial drugs, herein, a series of 27 novel matrine derivatives incorporating hydroxamic acid and benzene sulfonamide moieties were designed and synthesized. Antimicrobial testing demonstrated exceptional inhibitory activity against Candida albicans, with the most potent compound (10g) showing a MIC value of 0.062 mg mL−1, which was significantly lower than that of the clinical antibiotic fluconazole (8.590 mg mL−1). 3D-QSAR analysis identified the phenylsulfonyl group as crucial for activity, particularly when substituted with a 4-(CH3)3 group. The hydroxamic acid moiety was also found to contribute positively to the antifungal effects. Mechanistic studies indicated that these compounds act by both preventing biofilm formation and disrupting established biofilms. Furthermore, molecular docking studies of compounds 9j and 10g with fungal proteins (PDB: 2QZX) revealed that their antifungal activity involves multiple interactions, including hydrogen bonding, hydrophobic interactions, and van der Waals forces. These findings position compound 10g as a particularly promising lead candidate for the development of new antifungal agents.
Introduction
Fungal infections represent a significant cause of morbidity, mortality, and healthcare burden in critically ill patients.1 Candida albicans is the predominant causative agent of candidiasis in clinical settings and represents the third most commonly isolated pathogen in hospital-acquired bloodstream infections.2 This organism typically colonizes the human oral cavity, gastrointestinal tract, and vaginal mucosa, but in immunocompromised individuals, it can disseminate hematogenously to invade multiple organs, leading to life-threatening systemic infections.3 While polyenes, azoles, and echinocandins remain the primary therapeutic options,4 the emergence of drug resistance in C. albicans due to prolonged antimicrobial exposure has created substantial challenges in clinical management.5 These pressing concerns underscore the urgent need for novel antifungal agents with improved efficacy.Nitrogen-containing heterocyclic alkaloids have attracted much attention for their superior biological activities, rich structural diversity, and significant chemical effects. Matrine, a natural tetracyclic quinolizidine alkaloid derived from ginseng, possesses diverse pharmacological activities including antitumor,6–8 antifibrotic,9,10 anti-inflammatory,11,12 antibacterial,13,14 and antiviral15,16 properties. Current literature demonstrates that matrine can significantly enhance antimicrobial efficacy and reverse bacterial resistance to drugs when combined with antibiotics or other therapeutic agents.17,18 Moreover, matrine derivatives exhibit broad-spectrum antibacterial activity through multi-target mechanisms.13,14 The natural abundance and accessibility of matrine have also enhanced its potential as a lead compound.
In recent years, compounds containing hydroxamic acid groups have emerged as a promising focus in antimicrobial drug development due to their unique mechanisms of action. These compounds can form stable chelates with metal ions (e.g., Zn2+, Fe3+), selectively inhibiting bacterial metalloenzymes,19 an approach that offers high specificity and low toxicity, given the structural differences between bacterial and human metalloenzymes.20 Additionally, the hydroxamic acid moiety competitively disrupts bacterial siderophore-mediated iron uptake, inducing iron starvation.21 Since iron metabolism is essential for bacterial growth and difficult to bypass, this mechanism effectively suppresses bacterial proliferation while minimizing the risk of resistance development.22 The benzene sulfonamide moiety represents a privileged pharmacophore in medicinal chemistry, extensively incorporated in various classes of antimicrobial agents. These compounds exert antibacterial effects through multiple mechanisms, including competitive inhibition of bacterial dihydropteroate synthase23 and suppression of carbonic anhydrase catalytic activity.24
In this work, we implemented a rational pharmacophore fusion approach for antimicrobial discovery, utilizing matrine, a naturally occurring alkaloid, as the central scaffold functionalized with two mechanistically distinct antibacterial motifs: a hydroxamic acid group and a benzene sulfonamide unit (Fig. 1). The rationally designed dual-pharmacophore strategy is expected to substantially enhance the antimicrobial potency of these compounds through synergistic multi-target mechanisms of action. To optimize the drug design, we adopted a structure–activity relationship (SAR)-driven strategy involving: (i) systematic variation of linker chain length and composition, and (ii) strategic modification of substituents to probe electronic and steric effects, thereby generating a comprehensive library of structural analogs.
Results and discussion
Chemistry
As shown in Fig. 2, commercially available matrine (1) was used as the starting material. Heating and refluxing in an aqueous solution of potassium hydroxide afforded intermediate 2. Treatment with SOCl2 yielded the ester derivative 3,13 followed by N-phenylsulfonylation to obtain phenylsulfonyl-containing ester 4. This key intermediate was then divergently transformed: (i) reaction with alkalinized hydroxylamine solution produced target compounds 9a–9q,25 while (ii) hydrolysis of 4 followed by EDCI/HOBT-mediated condensation with methyl 4-(aminomethyl) benzoate generated the extended-chain methyl benzoate intermediate,26 which was similarly converted to target compounds 10a–10j. | ||
| Fig. 2 Synthetic routes and the substituent for the derivatives. (a) KOH, H2O; (b) SOCl2, MeOH; (c) K2CO3, CH3CN; (d) 3 N HCl; (e) NH2OH·HCl, CH3ONa; (f) EDCI, HOBT, TEA. | ||
Stepwise yield evaluation revealed that the terminal hydroxamic acid formation constituted the principal bottleneck in the synthesis, exhibiting both incomplete reactant consumption and undesirable side-product generation. To optimize the reaction efficiency, we systematically investigated several critical factors using compound 10g as a model substrate, including: pH, temperature, reaction time, and feed ratio (ester![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hydroxylamine hydrochloride). The detailed optimization results are presented in the Table 1.
hydroxylamine hydrochloride). The detailed optimization results are presented in the Table 1.
| Number | Factors | Efficiency (%) | |||
|---|---|---|---|---|---|
| pH | Temperature (°C) | Reaction time (h) | Feed ratio | ||
| 1 | 5 | 25 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0 |
| 2 | 7 | 25 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6% |
| 3 | 10 | 25 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
40% |
| 4 | 10 | 0 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
19% |
| 5 | 10 | 0 | 6 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
35% |
| 6 | 10 | 70 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
14% |
| 7 | 10 | 25 | 1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
37% |
| 8 | 10 | 25 | 4 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
41% |
| 9 | 10 | 25 | 6 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
38% |
| 10 | 10 | 25 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
50% |
| 11 | 10 | 25 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 1.5 |
64% |
| 12 | 10 | 25 | 2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2 1.2 |
63% |
The table data demonstrate that proper pH control is essential for successful reaction progression. Under acidic conditions, the protonation of hydroxylamine diminishes its nucleophilicity while simultaneously promoting ester hydrolysis, ultimately leading to reaction failure. Lower temperatures significantly reduce the reaction rate, whereas excessive temperatures facilitate over-attack by hydroxylamine, generating undesired byproducts. Prolonged reaction times increase the risk of either over-substitution or decomposition of the hydroxamic acid product. Furthermore, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 feed ratio (ester
2 feed ratio (ester![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hydroxylamine hydrochloride) leads to disubstituted byproducts and complicates purification. Based on comprehensive consideration of cost efficiency and workup feasibility, an optimal 1
hydroxylamine hydrochloride) leads to disubstituted byproducts and complicates purification. Based on comprehensive consideration of cost efficiency and workup feasibility, an optimal 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2 molar ratio was selected for the process.
1.2 molar ratio was selected for the process.
A total of 27 novel matrine derivatives were successfully synthesized and structurally characterized by comprehensive spectroscopic analyses, including 1H NMR, 13C NMR, and HR-MS. The purity of all compounds was verified by HPLC to be ≥97%, confirming the high quality of the synthesized derivatives for subsequent biological evaluation (ESI†).
Antimicrobial activity
The minimum inhibitory concentrations (MIC, mg mL−1) of the synthesized matrine derivatives, negative control, and positive control drugs against each bacterial strain are shown in Table 2. Analysis of the tabulated data reveals that all derivatives exhibited superior inhibitory activity compared to matrine against all five bacterial species. Notably, as anticipated, every derivative demonstrated lower MIC values against Candida albicans than the positive control drug fluconazole, with compound 10g showing the most potent activity (MIC = 0.0621 mg mL−1).| Compounds | MIC(mg mL−1) | ||||
|---|---|---|---|---|---|
| Gram-negative bacterium | Gram-positive bacterium | Fungus | |||
| MICE.co | MICP.ae | MICP.ac | MICS.au | MICC.al | |
| a Kanamycin.b Ceftazidime.c Doxycycline hydrochloride.d Penicillin G sodium.e Fluconazole.f SD = Standard deviation. Std = Standard. S. au: Staphylococcus aureus; P. ac: Propionibacterium acnes; P. ae: Pseudomonas aeruginosa; E. co: Escherichia coli and C. al: Candida albicans. | |||||
| 9a | 2.162 | 1.674 | 1.385 | 1.625 | 1.149 |
| 9b | 2.910 | 1.989 | 1.158 | 2.778 | 0.945 |
| 9c | 0.667 | 1.338 | 1.568 | 0.800 | 1.022 |
| 9d | 1.258 | 2.157 | 1.274 | 1.853 | 0.891 |
| 9e | 2.349 | 2.805 | 1.016 | 2.486 | 0.944 |
| 9f | 0.766 | 2.482 | 1.599 | 2.319 | 1.313 |
| 9g | 3.138 | 3.104 | 1.941 | 3.201 | 1.203 |
| 9h | 1.432 | 1.373 | 1.823 | 1.418 | 3.438 |
| 9i | 1.433 | 0.980 | 1.017 | 0.759 | 0.977 |
| 9j | 1.458 | 1.362 | 0.241 | 0.780 | 0.234 |
| 9k | 1.205 | 1.431 | 5.858 | 0.712 | 0.988 |
| 9l | 0.778 | 1.381 | 2.916 | 1.601 | 1.808 |
| 9m | 1.468 | 1.545 | 10.674 | 1.439 | 1.968 |
| 9n | 1.404 | 2.263 | 1.347 | 1.420 | 1.366 |
| 9o | 0.696 | 3.236 | 1.826 | 1.836 | 0.678 |
| 9p | 1.747 | 1.843 | 1.532 | 1.787 | 1.371 |
| 9q | 1.076 | 1.253 | 2.853 | 1.941 | 2.414 |
| 10a | 2.858 | 0.580 | 1.841 | 2.365 | 0.484 |
| 10b | 2.713 | 1.098 | 1.640 | 4.858 | 0.822 |
| 10c | 1.629 | 1.981 | 1.577 | 5.683 | 0.788 |
| 10d | 2.078 | 2.906 | 2.485 | 5.146 | 0.356 |
| 10e | 1.362 | 1.436 | 2.784 | 0.770 | 1.194 |
| 10f | 1.347 | 1.382 | 0.327 | 0.781 | 0.249 |
| 10g | 4.798 | 3.936 | 0.249 | 1.288 | 0.062 |
| 10h | 2.573 | 1.374 | 6.646 | 1.274 | 1.720 |
| 10i | 0.715 | 1.399 | 4.201 | 0.664 | 1.254 |
| 10j | 1.318 | 1.675 | 3.016 | 0.745 | 0.419 |
| SD | 0.108 | 0.121 | 0.092 | 0.110 | 0.062 |
| Matrine | 5.459 | 6.731 | 15.962 | 7.432 | 14.849 |
| Std | 0.145a | 0.238b | 0.031c | 0.508d | 8.590e |
Notably, between the two series of compounds sharing identical R1 groups (series 9 and 10), those from series 10 demonstrated superior inhibitory potency. While compounds bearing electron-withdrawing substituents (–CF3, –X, –NO2) on the benzene sulfonamide moiety generally showed lower MIC values compared to their electron-donating counterparts (–CH3, –OCH3), the most potent analog 10g (MIC = 0.0621 mg mL−1) paradoxically contained a 4-(CH3)3 group. This unexpected observation prompted further consideration of steric effects imposed by bulky substituents on the aromatic ring, suggesting that spatial constraints may play a more significant role in antimicrobial activity than electronic properties alone.
Toxicity predictions
To evaluate potential toxicity risks, compounds 9j and 10g were subjected to toxicity prediction using the pkCSM platform. Fluconazole as a control. Their respective SMILES strings were input to assess mutagenicity and biological toxicity parameters. The computational toxicity profiles of compounds are comprehensively summarized in Table 3.| Property | Compound 9j | Compound 10g | Fluconazole |
|---|---|---|---|
| AMES toxicity | No | No | No |
Max. tolerated dose (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg per kg per day) mg per kg per day) |
−0.95 | −0.594 | 0.627 |
| Oral rat acute toxicity (mol kg−1) | 2.763 | 2.782 | 2.272 |
Oral rat chronic toxicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg per kg per day) mg per kg per day) |
1.051 | 1.24 | 1.081 |
| Skin sensitisation | No | No | No |
T.Pyriformis toxicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μg L−1) μg L−1) |
0.419 | 0.294 | 0.288 |
Minnow toxicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM) mM) |
4.331 | 2.939 | 2.989 |
The comparative toxicity assessment reveals that compound 10g (MIC = 0.0621 mg mL−1) exhibits superior antifungal potency compared to fluconazole (MIC = 8.590 mg mL−1), while maintaining a favorable safety profile. Although 10g's maximum tolerated dose in humans (−0.594 log mg per kg per day) is lower than fluconazole (0.627 log mg per kg per day), its 100-fold greater efficacy may allow for lower therapeutic doses, potentially mitigating this difference. Both compounds show no mutagenicity (AMES-negative) or skin sensitization risk, supporting their clinical potential. Notably, 10g demonstrates comparable acute toxicity, similar chronic toxicity and lower ecological impact. Further in vivo pharmacodynamic/toxicity validation is required to confirm its clinical translational potential.
3D-QSAR
To elucidate the structure–activity relationship, we performed 3D-QSAR studies using the MIC data of derivatives against Candida albicans. For analytical convenience, MIC values were converted to pMIC13 (pMIC = −log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MIC), where pMIC > 0 indicates MIC < 1 mg mL−1, and pMIC < 0 corresponds to MIC > 1 mg mL−1. Three-dimensional conformations of all derivatives were generated, and after selecting the common backbone (Fig. 3A), manual molecular alignment was performed using 10g as the template molecule, yielding the superimposed structure (Fig. 3B).27
MIC), where pMIC > 0 indicates MIC < 1 mg mL−1, and pMIC < 0 corresponds to MIC > 1 mg mL−1. Three-dimensional conformations of all derivatives were generated, and after selecting the common backbone (Fig. 3A), manual molecular alignment was performed using 10g as the template molecule, yielding the superimposed structure (Fig. 3B).27
The partial least squares (PLS) statistics of the CoMFA and CoMSIA models for Candida albicans are shown in Table 4. The q2 values for both models were higher than 0.5, indicating satisfactory prediction accuracy. High N values of 20 and 15 further support the reliability of the prediction models. The correlation coefficients (r2) for the CoMFA and CoMSIA models are 0.999 and 0.983, respectively, which are very close to 1, indicating a robust constitutive relationship. The standard errors (SE) of 0.020 (CoMFA) and 0.073 (CoMSIA) suggest that the models exhibit stable predictions. Notably, the test value of the CoMFA model was as high as 417.012, reflecting an excellent model fit.
| Fraction (field contribution) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| q2 | N | r2 | SE | F | Steric | Electrostatic | Hydrophobic | Donor | Acceptor | |
| COMFA | 0.56 | 20 | 0.999 | 0.020 | 417.012 | 0.506 | 0.494 | — | — | — |
| COMSIA | 0.555 | 15 | 0.983 | 0.073 | 41.254 | 0.089 | 0.314 | 0.366 | 0.087 | 0.143 |
In the CoMFA model, the spatial and electrostatic field contributions were 50.6% and 49.4%, respectively, highlighting the nearly equal importance of steric hindrance and charge distribution on activity. For the CoMSIA model, the contributions were 8.9% (steric), 31.4% (electrostatic), 36.6% (hydrophobic), 8.7% (hydrogen bond donor), and 14.3% (hydrogen bond acceptor), indicating that charge distribution and hydrophobic interactions dominate, followed by hydrogen bonding, with steric effects playing a minor role. Both models consistently emphasize the significant influence of charge distribution on activity.
To further validate the models' predictive ability, Table 5 lists the derivative pMIC values alongside their predicted values. Fig. 4 shows the linear correlation between experimental and predicted activities for both models, with minimal deviations confirming their high reliability and accuracy. Notably, the CoMFA model exhibited superior predictive performance. Fig. 5 visualizes the CoMFA (A: steric field; B: electrostatic field) and CoMSIA (C: steric; D: electrostatic; E: hydrophobic; F: hydrogen-bond donor/acceptor) contour maps, using 10g as the reference molecule. These maps intuitively link structural features to activity. While both models highlight electrostatic and steric effects, CoMSIA provides a more comprehensive analysis by incorporating hydrophobic and hydrogen-bonding interactions. 3D-QSAR visualization analysis reveals key structure–activity relationships. (1) Steric effects: both models display prominent green contour maps around the R1 substituent region, with the CoMFA model additionally showing extensive yellow contours. These features indicate that introducing substituents with moderate bulk (e.g., halogens, methyl groups) on the benzene ring enhances activity, but the steric tolerance has an optimal range. (2) Electrostatic properties: the coexistence of red (negative charge-favorable) and blue (positive charge-favorable) electrostatic fields near the R1 position suggests a bidirectional electronic effect. This implies that either small electron-withdrawing groups (e.g., –F, –Cl) or bulky electron-donating groups (e.g., –C(CH3)3, –NMe2) can be beneficial at this site. (3) Hydrophobic and hydrogen-bonding features: yellow hydrophobic contours localize around both the R1 group and hydroxamic acid moiety, while hydrogen-bonding fields (blue donor/magenta acceptor) precisely map onto the hydroxamic acid group. This confirms that (i) hydrophobic interactions significantly contribute to activity, (ii) the hydroxamic acid acts as a critical pharmacophore via hydrogen-bond networks, and (iii) the linker region must balance hydrophobicity with hydrogen-bonding capacity.
| Compound | pMICc.al | Residues | pMICc.al | Residues | |
|---|---|---|---|---|---|
| (Experimental) | COMFA (predicted) | COMSIA (predicted) | |||
| 9a | −0.0603 | −0.06 | −0.0005 | −0.050 | −0.0104 |
| 9b | 0.0246 | 0.024 | 0.0004 | 0.045 | −0.0201 |
| 9c | −0.0095 | −0.011 | 0.0016 | −0.021 | 0.0117 |
| 9d | 0.0501 | 0.051 | −0.0005 | 0.052 | −0.0018 |
| 9e | 0.0250 | 0.027 | −0.0015 | 0.029 | −0.0039 |
| 9f | −0.1183 | −0.116 | −0.0018 | −0.153 | 0.0349 |
| 9g | −0.0803 | −0.082 | 0.0012 | −0.086 | 0.0053 |
| 9h | −0.5363 | −0.538 | 0.0012 | −0.546 | 0.0101 |
| 9i | 0.0101 | 0.012 | −0.0017 | 0.009 | 0.0014 |
| 9j | 0.6308 | 0.631 | −0.0002 | 0.635 | −0.0045 |
| 9k | 0.0052 | 0.004 | 0.0014 | 0.002 | 0.0032 |
| 9l | −0.2572 | −0.257 | −0.0004 | −0.242 | −0.0150 |
| 9m | −0.294 | −0.287 | −0.0070 | −0.361 | 0.0674 |
| 9n | −0.1355 | −0.1650 | 0.0291 | −0.041 | −0.0948 |
| 9o | 0.1688 | 0.191 | −0.0219 | 0.157 | 0.0118 |
| 9p | −0.1370 | −0.137 | −0.0005 | −0.144 | 0.0071 |
| 9q | −0.3827 | −0.383 | 0.0004 | −0.380 | −0.0027 |
| 10a | 0.3152 | 0.316 | −0.0010 | 0.299 | 0.0164 |
| 10b | 0.0851 | 0.086 | −0.0004 | 0.097 | −0.0121 |
| 10c | 0.1035 | 0.103 | 0.0009 | 0.112 | −0.0080 |
| 10d | 0.4486 | 0.447 | 0.0015 | 0.460 | −0.0112 |
| 10e | −0.077 | −0.076 | −0.0011 | −0.072 | −0.0052 |
| 10f | 0.6038 | 0.604 | −0.0002 | 0.605 | −0.0008 |
| 10g | 1.2069 | 1.207 | 0.0003 | 1.199 | 0.0083 |
| 10h | −0.2355 | −0.241 | 0.0055 | −0.252 | 0.0165 |
| 10i | −0.0983 | −0.074 | −0.0245 | 0.047 | −0.1454 |
| 10j | 0.3778 | 0.359 | 0.0019 | 0.236 | 0.1418 |
 | ||
| Fig. 4 Calculated pMICC.al and experimental pMICC.al values for target compounds obtained by PLS analysis using CoMFA (A) and CoMSIA (B) models. | ||
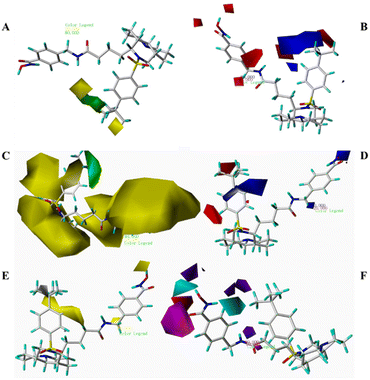 | ||
| Fig. 5 CoMFA steric field (A) and electrostatic field (B); CoMSIA steric field (C), electrostatic field (D), hydrophobic field (E), hydrogen bond acceptor and donor field (F). | ||
The analysis provides clear optimization strategies. (1) The R1 position should incorporate substituents with balanced steric bulk and electronic properties (electron-withdrawing/donating). (2) The hydroxamic acid moiety's structural integrity and spatial orientation must be preserved. (3) The linker region requires tailored hydrophobicity to maintain optimal activity.
Biofilm experiments
The high MIC value of fluconazole (positive control) is likely due to drug resistance,28 which is closely associated with biofilm formation. When bacteria adhere to surfaces and develop biofilms, they become more resistant to antimicrobials as they are encased in a protective extracellular polymeric matrix.29 To explore the derivatives' antimicrobial mechanism, we selected compounds 9j and 10g (the lowest-MIC derivatives from series 9 and 10, respectively) for biofilm assays, evaluating both biofilm prevention and mature biofilm disruption.30As shown in Fig. 6A and C, biofilm formation increased as the concentration of compounds decreased. Similarly, Fig. 6B and D demonstrate reduced biofilm disruption at lower compound concentrations. This confirms that compounds 9j and 10g exhibit dose-dependent inhibition of biofilm formation and disruption of mature biofilms. Notably, both compounds showed significant anti-biofilm effects at MIC concentrations, with 10g demonstrating particularly strong activity.
To visualize trends, Fig. 7 plots the biofilm formation rates calculated from OD values using the formula
| (A595drug − A595blank)/(A595DMSO − A595blank) × 100%. |
The analysis of biofilm inhibition and disruption activities reveals that compounds 9j and 10g exhibit significant dose-dependent antibiofilm effects. At their respective MICs, 9j and 10g suppressed biofilm formation to 6.43% and 0.01%, with 10g demonstrating near-complete inhibition, while also reducing preformed biofilms to 6.67%and 3.61%, respectively. This dual activity, both preventing biofilm establishment and degrading existing structures, highlights their potential as novel anti-biofilm agents. Notably, 10g showed superior performance in both assays, particularly in biofilm prevention, with its activity strongly correlating with the excellent antifungal MIC values, supporting our hypothesis of biofilm interference as a key mechanism of action.
These findings carry important clinical implications given the critical role of biofilms in Candida albicans drug resistance and persistent infections.31 The exceptional biofilm blockade by 10g at MIC concentrations (Fig. 7) suggests promising applications for preventing device-related infections and treating recalcitrant fungal infections, addressing a major clinical challenge posed by biofilm-associated resistance to conventional antifungals.31 The compounds' ability to simultaneously inhibit biofilm formation and disrupt mature biofilms positions them as valuable leads for developing next-generation anti-biofilm therapeutics.
Building upon the remarkable biofilm inhibition efficacy demonstrated by compound 10g (0.01% formation at MIC), we conducted a systematic comparison with fluconazole to elucidate the structure–activity advantages underlying this performance (Table 6).
Molecular docking studies
We performed molecular docking using Discovery Studio to investigate the binding mechanisms of compounds 9j and 10g with bacterial proteins. The study targeted SAP5 (PDB: 2QZX), a key virulence factor associated with Candida albicans biofilm formation.32SAP (secreted aspartic protease) facilitates fungal pathogenesis by hydrolyzing host cell surface proteins to promote adhesion and tissue invasion, while also disrupting host defense molecules to evade antifungal responses.33 SAP5, predominantly expressed in hyphal cells, plays a crucial role in systemic infections and biofilm formation.34
Fig. 8 reveals that compound 9j interacts with the target protein through hydrogen bonds, van der Waals forces, alkyl interactions, and π-alkyl interactions, enabling stable binding within the protein's cavity. The hydroxamic acid group in the side chain forms hydrogen bonds with residues GLN282 and PHE291, demonstrating its contribution to enhancing matrine's pharmacological activity. The opened-ring structure's phenyl sulfonamide group at the 12-position nitrogen interacts with SER277 via van der Waals forces, while establishing alkyl and π–alkyl interactions with PHE281 and LYS257, further improving antibacterial activity. The 3D structure shows 9j adopting a “sheet-like” conformation at the protein's active center.
 | ||
| Fig. 8 Binding pattern of compound 9j to amino acids in the active site of the 2QZX protein (A), 3D (B), 2D (C) and docking pocket of hydrogen bonding (D), hydrophobic (E), aromatic (F), SAS (G), ionization (H), and interpolated charge (I) interaction diagrams. | ||
Comparative analysis of 10g (Fig. 9) demonstrates similar interaction patterns (hydrogen bonds, π–π stacking, alkyl/π–alkyl interactions), but with key differences. (1) The extended linker positions the hydroxamic acid group deeper into the hydrophobic pocket, forming additional hydrogen bonds with ARG297, ARG312, and ALA11. (2) The inserted benzene ring engages in π–alkyl interactions with LYS257 and ALA162. These features force the side chain into a unique spatial arrangement – initially perpendicular then parallel to the matrine-phenyl sulfonamide core. The 3D visualization clearly shows 10g adopting an “H”-shaped topology that tightly fits the hydrophobic cavity.
 | ||
| Fig. 9 Binding pattern of compound 10g to amino acids in the active site of the 2QZX protein (A), 3D (B), 2D (C) and docking pocket of hydrogen bonding (D), hydrophobic (E), aromatic (F), SAS (G), ionization (H), and interpolated charge (I) interaction diagrams. | ||
This superior binding mode explains 10g's enhanced antibacterial activity over 9j, and consistently accounts for the observed higher potency of series 10 compared to series 9 in antimicrobial assays.
Experimental
Instruments and materials
Matrine 98%, 4-(aminomethyl) benzoic acid 98% and other reagents were analytically pure. Gram-negative bacteria: Pseudomonas aeruginosa #5336, provided by Guangxi Medical University; Escherichia coli, provided by Guangxi University of Traditional Chinese Medicine. Gram-positive bacteria: Staphylococcus aureus, provided by Guangxi University of Traditional Chinese Medicine. Propionibacterium acnes, obtained from Guangdong Microbial Strain Preservation Center. Fungus: Candida albicans, obtained from Guangdong Microbial Species Preservation Center. Melting point meter: XT-4 melting point meter. AVANCE III HD600-type nuclear magnetic resonance instrument. Impact I ultra-high performance liquid chromatography-mass spectrometry (Bruker, Germany). Enzyme labelling instrument: M200PRO, TECAN.Synthesis of target compounds
Compound 9a. 4-(2-((2-Chlorophenyl)sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxybutanamide, C21H30ClN3O4S, white power, yield 78%, mp: 70–72 °C. 1H NMR (600 MHz, DMSO-d6) δ 9.31 (s, 1H), 7.78–7.74 (m, 1H), 7.22 (dd, J = 7.9, 1.6 Hz, 1H), 6.82–6.76 (m, 2H), 6.69 (td, J = 7.5, 1.5 Hz, 1H), 2.88–2.80 (m, 2H), 2.64–2.59 (m, 1H), 1.87 (dd, J = 12.9, 9.3 Hz, 2H), 1.66 (p, J = 1.9 Hz, 2H), 1.29 (t, J = 3.0 Hz, 1H), 1.21–1.17 (m, 1H), 1.07–1.00 (m, 3H), 0.96–0.91 (m, 1H), 0.74 (d, J = 13.6 Hz, 3H), 0.68–0.59 (m, 3H), 0.52 (d, J = 4.2 Hz, 1H), 0.50 (dq, J = 7.0, 3.7, 3.1 Hz, 1H), 0.46 (d, J = 3.8 Hz, 1H), 0.24–0.18 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.39, 139.70, 133.97, 131.90, 131.05, 129.98, 127.71, 63.24, 59.11, 56.49, 56.44, 49.13, 36.30, 31.56, 27.54, 27.41, 27.33, 22.20, 20.51, 20.13. HR-MS (ESI), measured values, m/z: 456.1746 (456.1646) [M + H]+.
Compound 9b. N-Hydroxy-4-(2-((2-(trifluoromethyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C22H30F3N3O4S, white power, yield 72%, mp: 66–67 °C. 1H NMR (600 MHz, dimethyl sulfoxide-d6) δ 10.23 (d, 1H), 8.62 (d, 1H), 8.39 (d, 1H), 7.97 (dd, 1H), 7.87 (td, 1H), 7.82 (t, 1H), 3.71 (q, 1H), 3.45 (dd, 1H), 3.30 (d, 1H), 2.72 (dd, 2H), 2.09 (t, 1H), 1.93 (s, 1H), 1.84 (dd, 3H), 1.69 (m, 3H), 1.58 (q, 2H), 1.51 (d, 2H), 1.44 (td, 2H), 1.33 (m, 3H), 1.28 (m, 1H), 1.17 (m, 1H). 13C NMR (151 MHz, dimethyl sulfoxide-d6) δ 188.54, 140.57, 133.30, 132.93, 130.18, 126.50, 123.78, 121.97, 62.46, 58.24, 56.23, 56.15, 47.49, 40.06, 34.78, 31.84, 30.67, 27.99, 27.38, 22.06, 20.34. HR-MS (ESI), measured values, m/z: 490.1977 (490.1909) [M + H]+.
Compound 9c. N-Hydroxy-4-(2-((2-nitrophenyl)sulfonyl)decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C21H30N4O6S, white power, yield 82%, mp: 60–61 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.28 (s, 1H), 8.64 (s, 1H), 8.11 (dd, J = 7.4, 1.9 Hz, 1H), 7.91 (dd, J = 7.4, 1.8 Hz, 1H), 7.83 (td, J = 7.0, 1.7 Hz, 2H), 3.67 (q, J = 6.9 Hz, 1H), 2.58 (dd, J = 30.1, 10.9 Hz, 2H), 1.99 (s, 1H), 1.87 (d, J = 9.0 Hz, 1H), 1.76 (dt, J = 16.3, 8.1 Hz, 5H), 1.67 (d, J = 11.6 Hz, 1H), 1.57 (ddd, J = 15.3, 11.1, 5.9 Hz, 2H), 1.49 (d, J = 9.7 Hz, 1H), 1.44–1.38 (m, 3H), 1.37–1.28 (m, 6H), 1.23 (s, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.89, 148.01, 134.46, 132.95, 132.21, 130.11, 124.13, 62.23, 57.69, 56.27, 56.16, 46.11, 34.05, 32.22, 31.91, 28.57, 27.82, 22.23, 20.55, 20.33. Measured values, m/z: 467.1948 (467.1886) [M + H]+.
Compound 9d. N-Hydroxy-4-(2-((3-(trifluoromethyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C22H30F3N3O4S, white power, yield 78%, mp: 71–72 °C., 1H NMR (600 MHz, DMSO-d6) δ 10.37 (s, 1H), 8.70 (s, 1H), 8.17 (d, J = 7.9 Hz, 1H), 8.08–8.03 (m, 2H), 7.85 (t, J = 7.8 Hz, 1H), 3.54 (q, J = 6.4 Hz, 1H), 3.10 (dd, J = 12.8, 9.4 Hz, 1H), 2.37–2.29 (m, 1H), 2.21 (d, J = 11.3 Hz, 1H), 1.95 (td, J = 7.1, 1.9 Hz, 2H), 1.89–1.80 (m, 2H), 1.69–1.64 (m, 1H), 1.63–1.58 (m, 4H), 1.56–1.51 (m, 2H), 1.43 (t, J = 7.7 Hz, 1H), 1.36–1.28 (m, 4H), 1.22–1.11 (m, 2H). 13C NMR (151 MHz, DMSO-d6) δ 168.95, 140.94, 131.61, 130.40, 129.19, 124.56, 123.76, 123.73, 122.75, 61.42, 55.79, 55.71, 55.61, 43.66, 38.86, 34.07, 32.36, 32.27, 28.74, 27.56, 21.48, 20.42, 19.80. Measured values, m/z: 490.1969 (490.1909) [M + H]+.
Compound 9e. 4-(2-((3-Bromophenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxybutanamide, C21H30BrN3O4S, white power, yield 80%, mp: 80–83 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.35 (s, 1H), 8.68 (s, 1H), 7.98 (t, J = 1.9 Hz, 1H), 7.84 (ddd, J = 9.1, 7.5, 1.8 Hz, 2H), 7.53 (t, J = 7.9 Hz, 1H), 3.48 (q, J = 6.3 Hz, 1H), 3.34–3.30 (m, 1H), 3.04 (dd, J = 12.6, 9.4 Hz, 1H), 2.48 (d, J = 2.9 Hz, 1H), 2.38 (dt, J = 11.5, 3.5 Hz, 1H), 1.93 (td, J = 7.1, 3.2 Hz, 2H), 1.84 (q, J = 3.7, 3.0 Hz, 2H), 1.67–1.63 (m, 2H), 1.61 (d, J = 3.6 Hz, 1H), 1.59 (tt, J = 8.5, 6.1, 4.8 Hz, 3H), 1.51 (q, J = 7.2 Hz, 2H), 1.44–1.40 (m, 1H), 1.35–1.29 (m, 4H), 1.18 (tdd, J = 12.8, 9.1, 5.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 168.87, 141.45, 135.23, 130.89, 129.81, 126.53, 121.74, 61.37, 55.78, 55.62, 43.87, 38.75, 33.54, 32.46, 32.19, 28.56, 27.50, 21.25, 20.35, 19.80. Measured values, m/z: 500.1205 (500.1140) [M + H]+.
Compound 9f. N-Hydroxy-4-(2-(m-tolylsulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C22H33N3O4S, yellow oily liquid, yield 49%, 1H NMR (600 MHz, DMSO-d6) δ 10.33 (s, 1H), 8.66 (s, 1H), 7.62 (d, J = 2.3 Hz, 1H), 7.60 (dt, J = 6.8, 2.1 Hz, 1H), 7.46 (d, J = 6.7 Hz, 2H), 3.40 (dt, J = 8.0, 5.6 Hz, 1H), 3.10 (dd, J = 12.4, 10.5 Hz, 1H), 2.52 (s, 1H), 2.44 (d, J = 10.9 Hz, 1H), 2.39 (s, 3H), 1.90 (qd, J = 6.2, 4.5, 1.9 Hz, 2H), 1.85 (dd, J = 14.4, 7.1 Hz, 2H), 1.76–1.68 (m, 3H), 1.68–1.65 (m, 3H), 1.61–1.57 (m, 1H), 1.51 (q, J = 6.3 Hz, 1H), 1.48–1.44 (m, 2H), 1.39–1.35 (m, 1H), 1.35–1.33 (m, 1H), 1.28 (dt, J = 12.6, 3.8 Hz, 3H), 1.23 (d, J = 7.3 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.78, 139.30, 138.51, 132.97, 128.62, 127.31, 124.26, 61.91, 56.08, 55.88, 45.78, 38.62, 33.31, 32.04, 31.10, 27.78, 27.40, 20.76, 20.19, 19.99. Measured values, m/z: 436.2265 (436.2192) [M + H]+.
Compound 9g. 4-(2-((4-Chlorophenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxybutanamide, C21H30ClN3O4S, white power, yield 76%, mp: 77–78 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.34 (s, 1H), 8.68 (s, 1H), 7.83–7.81 (m, 2H), 7.67–7.65 (m, 2H), 3.42 (dd, J = 8.0, 5.8 Hz, 1H), 3.36 (d, J = 6.8 Hz, 1H), 3.34 (s, 1H), 3.08 (dd, J = 12.4, 10.2 Hz, 1H), 2.41 (d, J = 11.2 Hz, 1H), 1.96–1.91 (m, 1H), 1.89 (t, J = 5.3 Hz, 2H), 1.86–1.83 (m, 1H), 1.73–1.66 (m, 3H), 1.66–1.61 (m, 3H), 1.61–1.56 (m, 1H), 1.53–1.44 (m, 3H), 1.38–1.34 (m, 1H), 1.30 (d, J = 7.7 Hz, 3H), 1.23 (dd, J = 8.6, 3.9 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.89, 138.29, 137.37, 129.24, 128.98, 61.80, 56.33, 56.14, 55.90, 55.84, 45.45, 38.77, 33.21, 32.16, 31.70, 28.02, 27.52, 20.95, 20.34, 20.05. Measured values, m/z: 456.1708 (456.1646) [M + H]+.
Compound 9h. N-hydroxy-4-(2-tosyldecahydro-1H,4H-pyrido [3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C22H33N3O4S, white power, yield 75%, mp: 67–69 °C. H NMR (600 MHz, DMSO-d6) δ 10.33 (s, 1H), 8.67 (s, 1H), 7.69–7.67 (m, 2H), 7.40 (d, J = 8.1 Hz, 2H), 3.39–3.36 (m, 1H), 3.33–3.31 (m, 1H), 3.07 (t, J = 11.5 Hz, 1H), 2.55 (dd, J = 11.5, 3.2 Hz, 1H), 2.49–2.46 (m, 1H), 2.39 (s, 3H), 1.92 (td, J = 5.8, 3.5 Hz, 2H), 1.88–1.82 (m, 2H), 1.77 (ddd, J = 11.4, 9.2, 4.5 Hz, 1H), 1.69 (td, J = 11.4, 5.5 Hz, 4H), 1.60–1.52 (m, 2H), 1.46 (ddt, J = 12.9, 7.5, 3.1 Hz, 2H), 1.41–1.33 (m, 2H), 1.28 (d, J = 6.8 Hz, 3H), 1.24 (d, J = 11.5 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.96, 142.86, 136.33, 129.47, 127.30, 62.21, 56.33, 56.05, 46.67, 38.65, 33.74, 32.22, 30.35, 27.56, 21.01, 20.55, 20.36, 20.27. Measured values, m/z: 436.2244 (436.2192) [M + H]+.
Compound 9i. N-Hydroxy-4-(2-((4-(trifluoromethyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C22H30F3N3O4S, white power, yield 75%, mp: 71–74 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.34 (s, 1H), 8.68 (s, 1H), 8.03 (d, J = 8.2 Hz, 2H), 7.95 (d, J = 8.2 Hz, 2H), 3.49 (q, J = 6.4 Hz, 1H), 3.39–3.36 (m, 1H), 3.12 (dd, J = 12.7, 9.8 Hz, 1H), 2.43–2.38 (m, 1H), 2.30 (dd, J = 11.2, 3.5 Hz, 1H), 1.91 (dq, J = 17.1, 7.0 Hz, 2H), 1.85 (s, 2H), 1.69–1.63 (m, 2H), 1.61 (q, J = 6.2, 4.5 Hz, 3H), 1.58 (t, J = 5.3 Hz, 1H), 1.50 (qd, J = 11.2, 9.9, 4.4 Hz, 2H), 1.44 (d, J = 13.4 Hz, 1H), 1.34–1.26 (m, 4H), 1.20 (ddt, J = 10.8, 7.7, 3.9 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 168.81, 143.43, 128.21, 125.89, 124.44, 122.63, 61.50, 56.00, 55.74, 55.62, 44.63, 38.82, 32.84, 32.67, 32.09, 28.21, 27.44, 21.18, 20.26, 19.82. Measured values, m/z: 490.1980 (490.1909) [M + H]+.
Compound 9j. 4-(2-((4-(Tert-butyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxy butanamide, C25H39N3O4S, white power, yield 71%, mp: 64–68 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.32 (s, 1H), 8.65 (s, 1H), 7.73–7.71 (m, 2H), 7.59 (d, J = 8.4 Hz, 2H), 3.40 (dt, J = 8.0, 5.5 Hz, 1H), 3.37–3.34 (m, 1H), 3.08 (dd, J = 12.4, 10.4 Hz, 1H), 2.41 (d, J = 11.4 Hz, 1H), 1.91–1.88 (m, 2H), 1.87–1.83 (m, 2H), 1.70 (d, J = 12.0 Hz, 2H), 1.67–1.63 (m, 3H), 1.60–1.56 (m, 1H), 1.47–1.41 (m, 2H), 1.35 (t, J = 6.0 Hz, 1H), 1.30 (s, 11H), 1.28–1.25 (m, 3H), 1.23 (d, J = 5.6 Hz, 1H), 1.22–1.19 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.89, 155.41, 136.57, 127.23, 125.53, 61.87, 56.00, 55.88, 45.49, 38.75, 34.79, 33.30, 32.18, 31.54, 30.78, 27.92, 27.55, 20.89, 20.30, 20.00. Measured values, m/z: 478.2722 (478.2661) [M + H]+.
Compound 9k. N-Hydroxy-4-(2-((4-nitrophenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C21H30N4O6S, white oily liquid, yield 42%, 1H NMR (600 MHz, DMSO-d6) δ 10.32 (d, J = 1.5 Hz, 1H), 8.67 (d, J = 1.6 Hz, 1H), 8.41–8.37 (m, 2H), 8.10–8.06 (m, 2H), 3.52 (q, J = 6.5 Hz, 1H), 3.40 (dd, J = 12.6, 7.0 Hz, 1H), 3.18 (dd, J = 12.7, 9.6 Hz, 1H), 2.45 (d, J = 10.7 Hz, 1H), 2.36 (d, J = 11.3 Hz, 1H), 1.95–1.84 (m, 4H), 1.69–1.61 (m, 5H), 1.60–1.54 (m, 1H), 1.46 (qq, J = 12.8, 7.1, 6.7 Hz, 3H), 1.37–1.27 (m, 5H), 1.23 (d, J = 5.9 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.77, 149.47, 145.30, 128.76, 124.11, 61.64, 56.34, 55.84, 55.73, 45.05, 40.06, 38.90, 33.08, 32.28, 32.09, 28.18, 27.48, 21.30, 20.31, 19.94. Measured values, m/z: 467.1942 (467.1886) [M + H]+.
Compound 9l. N-Hydroxy-4-(2-((4-methoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide, C22H33N3O5S, white oily liquid, yield 45%, 1H NMR (600 MHz, DMSO-d6) δ 10.34 (s, 2H), 7.74–7.72 (m, 2H), 7.12–7.09 (m, 2H), 3.83 (s, 1H), 3.37–3.31 (m, 2H), 3.07–3.02 (m, 1H), 2.56 (dd, J = 11.3, 3.0 Hz, 1H), 1.92 (dt, J = 6.4, 4.3 Hz, 2H), 1.89 (d, J = 5.0 Hz, 2H), 1.87–1.81 (m, 2H), 1.76 (d, J = 9.3 Hz, 1H), 1.69 (s, 6H), 1.56 (q, J = 9.1, 6.2 Hz, 1H), 1.46 (dt, J = 11.5, 3.6 Hz, 2H), 1.38–1.34 (m, 1H), 1.28 (p, J = 3.8, 3.3 Hz, 3H), 1.26–1.21 (m, 2H). 13C NMR (151 MHz, DMSO-d6) δ 166.20, 162.28, 130.82, 129.51, 114.14, 62.20, 56.22, 56.06, 55.69, 46.49, 38.64, 33.66, 32.27, 30.54, 27.67, 27.60, 20.59, 20.40, 20.27.13C NMR (151 MHz, dimethyl sulfoxide-d6). Measured values, m/z: 452.2236 (452.2141) [M + H]+.
Compound 9m. 4-(2-((5-Fluoro-2-methoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxy butanamide, C22H32FN3O5S, white power, yield 85%, mp: 57–59 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.24 (s, 1H), 8.60 (d, J = 1.6 Hz, 1H), 7.62 (dd, J = 8.3, 3.2 Hz, 1H), 7.47–7.43 (m, 1H), 7.24 (dd, J = 9.2, 4.1 Hz, 1H), 3.88 (s, 3H), 3.60–3.55 (m, 1H), 3.53 (dd, J = 13.0, 5.6 Hz, 1H), 3.39 (dd, J = 13.1, 11.1 Hz, 1H), 2.67 (dd, J = 10.6, 5.0 Hz, 2H), 2.05 (t, J = 3.1 Hz, 1H), 1.85–1.80 (m, 3H), 1.74–1.70 (m, 1H), 1.66 (p, J = 7.1 Hz, 2H), 1.60–1.55 (m, 3H), 1.54 (s, 1H), 1.44 (dt, J = 13.4, 4.6 Hz, 1H), 1.39 (dd, J = 14.2, 3.4 Hz, 1H), 1.35 (d, J = 3.9 Hz, 1H), 1.33 (t, J = 4.3 Hz, 1H), 1.29 (dd, J = 13.5, 3.3 Hz, 2H), 1.24–1.20 (m, 1H), 1.13–1.07 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.64, 155.68, 152.95, 130.94, 120.45, 115.67, 115.02, 62.86, 57.85, 56.78, 56.36, 47.90, 35.31, 31.89, 29.02, 27.63, 27.56, 21.61, 20.47, 20.26.13C NMR (151 MHz, dimethyl sulfoxide-d6). Measured values, m/z: 470.2149 (470.2047) [M + H]+.
Compound 9n. 4-(2-((5-Chloro-2-methoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxy butanamide, C22H32ClN3O5S, yield 85%, white power, mp: 75–78 °C. 1H NMR (600 MHz, DMSO-d6) δ 9.42 (s, 1H), 7.78 (s, 1H), 7.00 (d, J = 2.7 Hz, 1H), 6.80 (dd, J = 8.9, 2.7 Hz, 1H), 6.41 (d, J = 8.9 Hz, 1H), 3.07 (s, 3H), 2.74 (dt, J = 8.7, 6.3 Hz, 1H), 2.67 (dd, J = 13.0, 5.8 Hz, 1H), 2.53 (dd, J = 13.0, 10.9 Hz, 1H), 1.84 (dd, J = 10.2, 4.9 Hz, 2H), 1.67 (p, J = 1.8 Hz, 2H), 1.20 (t, J = 3.1 Hz, 1H), 1.03 (d, J = 9.6 Hz, 1H), 0.97 (tt, J = 11.6, 2.8 Hz, 3H), 0.85 (td, J = 7.2, 4.8 Hz, 2H), 0.74 (qd, J = 13.0, 7.1 Hz, 4H), 0.59 (dtd, J = 20.6, 11.5, 10.3, 6.0 Hz, 2H), 0.52–0.44 (m, 3H), 0.39 (dt, J = 14.9, 7.5 Hz, 1H), 0.28 (dp, J = 14.8, 7.5 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.65, 155.38, 133.71, 131.24, 128.21, 123.67, 115.26, 62.72, 57.68, 56.65, 56.33, 56.28, 47.47, 35.05, 31.88, 29.54, 27.81, 27.54, 21.60, 20.41, 20.25. Measured values, m/z: 486.1849 (486.1751) [M + H]+.
Compound 9o. 4-(2-((5-Bromo-2-methoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxy butanamide, C22H32BrN3O5S, white power, yield 87%, mp: 65–68 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.25 (s, 1H), 8.61 (s, 1H), 7.93 (d, J = 2.5 Hz, 1H), 7.75 (dd, J = 8.9, 2.6 Hz, 1H), 7.19 (d, J = 8.9 Hz, 1H), 3.90 (s, 3H), 3.57 (dt, J = 8.8, 6.4 Hz, 1H), 3.50 (dd, J = 13.0, 5.9 Hz, 1H), 3.36 (dd, J = 13.0, 10.9 Hz, 1H), 2.67 (d, J = 11.5 Hz, 2H), 2.03 (t, J = 3.2 Hz, 1H), 1.85 (s, 1H), 1.83–1.77 (m, 3H), 1.69 (qt, J = 7.2, 2.9 Hz, 3H), 1.56 (dt, J = 16.4, 10.5 Hz, 4H), 1.42 (dtd, J = 19.7, 11.1, 9.9, 5.6 Hz, 2H), 1.35–1.28 (m, 3H), 1.24–1.20 (m, 1H), 1.11 (dt, J = 14.1, 7.4 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 168.80, 155.94, 136.76, 131.71, 131.07, 115.84, 111.14, 62.86, 57.79, 56.74, 56.48, 56.42, 47.55, 40.19, 35.15, 32.02, 29.72, 27.96, 27.68, 21.74, 20.54, 20.39. Measured values, m/z: 530.1341 (530.1246) [M + H]+.
Compound 9p. 4-(2-((3-Chloro-2-fluorophenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxy butanamide, C21H29ClFN3O4S, white power, yield 70%, mp: 76–78 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.30 (s, 1H), 8.66 (s, 1H), 7.87 (ddd, J = 8.2, 6.7, 1.6 Hz, 1H), 7.83 (ddd, J = 8.0, 6.2, 1.6 Hz, 1H), 7.38 (t, J = 8.0 Hz, 1H), 3.55 (q, J = 6.7 Hz, 1H), 3.41 (dd, J = 13.1, 7.6 Hz, 1H), 3.33 (s, 1H), 3.29 (dd, J = 13.0, 9.5 Hz, 1H), 2.44–2.38 (m, 2H), 1.85 (q, J = 7.0 Hz, 4H), 1.68–1.55 (m, 6H), 1.45 (d, J = 13.6 Hz, 1H), 1.42–1.38 (m, 1H), 1.35 (dq, J = 13.4, 7.7, 6.8 Hz, 2H), 1.31–1.27 (m, 1H), 1.23 (dq, J = 7.5, 3.4 Hz, 3H). 13C NMR (151 MHz, DMSO-d6) δ 168.82, 154.76, 153.06, 135.10, 130.00, 129.91, 129.21, 125.42, 125.39, 121.61, 121.48, 61.86, 56.37, 56.23, 56.15, 44.60, 39.14, 33.25, 33.20, 32.24, 28.44, 27.66, 21.76, 20.09, 19.84. 13C NMR (151 MHz, dimethyl sulfoxide-d6). Measured values, m/z: 474.1655 (474.1551) [M + H]+.
Compound 9q. 4-(2-((3,4-dimethoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl)-N-hydroxy butanamide, C23H35N3O6S, white power, yield 73%, mp: 68–70 °C. 1H NMR (600 MHz, DMSO-d6) δ 10.36–10.30 (m, 1H), 8.66 (d, J = 1.6 Hz, 1H), 7.39 (dd, J = 8.5, 2.2 Hz, 1H), 7.22 (d, J = 2.2 Hz, 1H), 7.13 (d, J = 8.6 Hz, 1H), 3.83 (d, J = 16.4 Hz, 7H), 3.37 (dd, J = 12.3, 5.8 Hz, 3H), 3.11 (t, J = 11.5 Hz, 1H), 2.58–2.50 (m, 3H), 1.96–1.90 (m, 2H), 1.87 (dq, J = 14.7, 7.4 Hz, 3H), 1.78–1.65 (m, 6H), 1.64–1.53 (m, 2H), 1.48 (dt, J = 11.4, 5.5 Hz, 2H), 1.41–1.35 (m, 2H), 1.28 (dt, J = 16.7, 5.5 Hz, 5H). 13C NMR (151 MHz, DMSO-d6) δ 168.94, 152.05, 148.44, 130.87, 121.05, 111.14, 110.04, 62.29, 56.37, 56.12, 55.84, 46.66, 38.64, 33.62, 32.24, 30.44, 27.66, 27.52, 20.63, 20.37, 20.32. 13C NMR (151 MHz, dimethyl sulfoxide-d6). Measured values, m/z: 482.2349 (482.2247) [M + H]+.
Compound 10a. N-Hydroxy-4-((4-(2-((2-(trifluoromethyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl) benzamide, C30H37F3N4O5S, white power, yield 56%, mp: 77–78 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 9.00 (s, 1H), 8.37 (d, J = 7.9 Hz, 1H), 8.24 (t, J = 6.0 Hz, 1H), 7.97–7.94 (m, 1H), 7.85 (t, J = 7.6 Hz, 1H), 7.80 (t, J = 7.6 Hz, 1H), 7.70–7.68 (m, 2H), 7.25 (d, J = 8.0 Hz, 2H), 4.24–4.21 (m, 2H), 3.73 (q, J = 7.3 Hz, 1H), 3.46 (dd, J = 13.0, 5.9 Hz, 1H), 3.39 (d, J = 11.6 Hz, 1H), 2.72 (t, J = 13.0 Hz, 2H), 2.09 (t, J = 3.2 Hz, 1H), 1.93–1.83 (m, 6H), 1.72 (d, J = 12.5 Hz, 1H), 1.60 (dq, J = 14.7, 5.0, 4.4 Hz, 2H), 1.52 (d, J = 12.6 Hz, 2H), 1.46–1.39 (m, 2H), 1.36–1.29 (m, 4H), 1.18 (dd, J = 13.0, 5.3 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 171.63, 164.06, 143.00, 140.72, 133.27, 132.88, 131.20, 130.17, 128.44, 126.98, 126.85, 123.78, 62.58, 58.44, 56.27, 56.20, 47.79, 41.68, 34.99, 34.83, 30.34, 27.93, 27.38, 22.26, 20.32. HR-MS (ESI), measured values, m/z: 623.2538 (623.2437) [M + H]+.
Compound 10b. N-Hydroxy-4-((4-(2-((2-nitrophenyl)sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl) benzamide, C29H37N5O7S, yellow power, mp: 74–75 °C. yield 52%, 1H NMR (500 MHz, DMSO-d6) δ 11.17 (s, 1H), 8.99 (s, 1H), 8.28 (t, J = 6.0 Hz, 1H), 8.14–8.09 (m, 1H), 7.94–7.88 (m, 1H), 7.81 (tt, J = 7.5, 5.6 Hz, 2H), 7.72–7.67 (m, 2H), 7.26 (d, J = 8.1 Hz, 2H), 4.24 (d, J = 6.1 Hz, 2H), 3.68 (q, J = 7.0 Hz, 1H), 3.36 (d, J = 8.4 Hz, 2H), 2.60 (dd, J = 20.0, 11.2 Hz, 2H), 2.03–1.94 (m, 3H), 1.85 (s, 1H), 1.74 (d, J = 10.2 Hz, 3H), 1.66 (d, J = 11.4 Hz, 1H), 1.61–1.54 (m, 2H), 1.50 (d, J = 11.0 Hz, 1H), 1.44–1.27 (m, 8H). 13C NMR (126 MHz, DMSO-d6) δ 171.85, 164.21, 147.83, 143.18, 134.31, 133.04, 132.12, 131.37, 129.94, 127.15, 127.02, 124.04, 62.23, 57.80, 56.20, 56.10, 46.32, 41.86, 35.09, 34.15, 31.48, 28.42, 27.69, 22.29, 20.41, 20.27. Measured values, m/z: 600.2520 (600.2414) [M + H]+.
Compound 10c. N-hydroxy-4-((4-(2-((3-(trifluoromethyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl) benzamide, C30H37F3N4O5S, white power, yield 55%, mp: 80–83 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 9.00 (s, 1H), 8.37 (t, J = 6.0 Hz, 1H), 8.18–8.11 (m, 1H), 8.07–8.00 (m, 2H), 7.83 (t, J = 7.9 Hz, 1H), 7.73–7.67 (m, 2H), 7.30 (d, J = 8.1 Hz, 2H), 4.28 (d, J = 5.9 Hz, 2H), 3.54 (q, J = 6.4 Hz, 1H), 3.34 (d, J = 7.4 Hz, 1H), 3.11 (dd, J = 12.8, 9.3 Hz, 1H), 2.32 (d, J = 10.8 Hz, 1H), 2.22 (d, J = 10.9 Hz, 1H), 2.13 (t, J = 7.2 Hz, 2H), 1.81 (q, J = 5.6, 3.1 Hz, 2H), 1.69–1.59 (m, 4H), 1.58–1.50 (m, 4H), 1.42 (d, J = 13.2 Hz, 1H), 1.33–1.24 (m, 4H), 1.21–1.12 (m, 2H). 13C NMR (151 MHz, DMSO-d6) δ 171.95, 164.07, 143.06, 140.93, 131.48, 130.33, 129.53, 129.21 (d, J = 32.3 Hz), 127.02, 126.87, 124.46, 123.62. Measured values, m/z: 623.2530 (623.2437) [M + H]+.
Compound 10d. 4-((4-(2-((3-Bromophenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl)-N-hydroxy benzamide, C29H37BrN4O5S, white power, yield 55%, mp: 76–79 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 8.99 (s, 1H), 8.37 (t, J = 6.0 Hz, 1H), 7.99 (s, 1H), 7.84 (t, J = 6.8 Hz, 2H), 7.70 (d, J = 7.8 Hz, 2H), 7.53 (t, J = 7.9 Hz, 1H), 7.30 (d, J = 7.9 Hz, 2H), 4.29 (d, J = 5.7 Hz, 2H), 3.50 (q, J = 6.3 Hz, 1H), 3.34–3.30 (m, 1H), 3.06 (dd, J = 12.6, 9.3 Hz, 1H), 2.41 (d, J = 11.1 Hz, 1H), 2.14 (q, J = 7.4 Hz, 2H), 1.83 (d, J = 14.9 Hz, 2H), 1.75–1.48 (m, 9H), 1.43 (d, J = 13.1 Hz, 1H), 1.37–1.26 (m, 4H), 1.24–1.16 (m, 2H). 13C NMR (151 MHz, DMSO-d6) δ 172.10, 164.19, 143.19, 141.75, 135.34, 131.33, 131.03, 129.89, 127.15, 127.00, 126.59, 121.88, 61.57, 55.94, 55.91, 55.78, 44.20, 41.89, 38.91, 35.31, 33.47, 32.71, 28.68, 27.61, 21.57, 20.45, 19.97. Measured values, m/z: 633.1772 (633.1668) [M + H]+.
Compound 10e. N-Hydroxy-4-((4-(2-tosyldecahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl) benzamide, C30H40N4O5S, white power, brown power, yield 60%, mp: 67–68 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 8.99 (s, 1H), 8.34 (t, J = 6.0 Hz, 1H), 7.70 (d, J = 2.9 Hz, 2H), 7.68 (d, J = 2.9 Hz, 2H), 7.39 (d, J = 8.0 Hz, 2H), 7.29 (d, J = 8.1 Hz, 2H), 4.28 (dd, J = 6.0, 2.0 Hz, 2H), 3.40–3.35 (m, 2H), 3.09 (t, J = 11.6 Hz, 1H), 2.55 (d, J = 11.1 Hz, 1H), 2.38 (s, 3H), 2.12 (dt, J = 13.7, 6.8 Hz, 1H), 2.07 (q, J = 7.3 Hz, 1H), 1.92 (t, J = 3.1 Hz, 1H), 1.83 (d, J = 9.0 Hz, 1H), 1.77 (d, J = 4.3 Hz, 1H), 1.73–1.66 (m, 4H), 1.63–1.55 (m, 2H), 1.51–1.45 (m, 2H), 1.37 (d, J = 10.2 Hz, 2H), 1.31–1.20 (m, 5H). 13C NMR (151 MHz, DMSO-d6) δ 172.04, 164.04, 163.67, 143.08, 142.81, 136.53, 131.18, 129.43, 127.24, 127.00, 126.85, 62.24, 56.47, 56.05, 46.74, 41.73, 38.65, 35.23, 33.76, 30.28, 27.60, 27.52, 20.98, 20.81, 20.30, 20.26. Measured values, m/z: 569.2820 (569.2719) [M + H]+.
Compound 10f. N-Hydroxy-4-((4-(2-((4-(trifluoromethyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl) benzamide. C30H37F3N4O5S, white power, yield 52%, mp: 79–81 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.17 (s, 1H), 8.98 (s, 1H), 8.34 (t, J = 6.0 Hz, 1H), 8.04 (d, J = 8.2 Hz, 2H), 7.96 (d, J = 8.2 Hz, 2H), 7.70–7.67 (m, 2H), 7.31–7.27 (m, 2H), 4.27 (t, J = 5.9 Hz, 2H), 3.50 (t, J = 6.7 Hz, 1H), 3.37 (dd, J = 12.6, 6.9 Hz, 1H), 3.14 (dd, J = 12.7, 9.8 Hz, 1H), 2.42 (d, J = 10.9 Hz, 1H), 2.32 (d, J = 11.2 Hz, 1H), 2.10 (h, J = 7.2 Hz, 2H), 1.86 (t, J = 3.1 Hz, 1H), 1.84 (s, 1H), 1.65 (dt, J = 8.6, 4.5 Hz, 2H), 1.62 (d, J = 4.6 Hz, 2H), 1.60 (d, J = 4.3 Hz, 1H), 1.52 (td, J = 11.8, 6.3 Hz, 2H), 1.46–1.43 (m, 1H), 1.36–1.32 (m, 1H), 1.28 (s, 4H), 1.22 (d, J = 9.2 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 172.05, 143.69, 128.34, 127.13, 126.99, 126.08, 61.72, 56.29, 55.94, 55.83, 44.99, 41.86, 40.20, 38.99, 35.24, 33.10, 32.63, 28.35, 27.59, 21.51, 20.40, 20.02. Measured values, m/z: 623.2539 (623.2437) [M + H]+.
Compound 10g. 4-((4-(2-((4-(Tert-butyl) phenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl)-N-hydroxy benzamide, C33H46N4O5S, white power, yield 63%, mp: 87–88 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.17 (s, 1H), 8.99 (s, 1H), 8.34 (t, J = 6.0 Hz, 1H), 7.72 (d, J = 8.4 Hz, 2H), 7.70–7.68 (m, 2H), 7.59 (d, J = 8.4 Hz, 2H), 7.29 (d, J = 8.0 Hz, 2H), 4.28 (t, J = 6.0 Hz, 2H), 3.42 (q, J = 6.4 Hz, 1H), 3.38–3.34 (m, 1H), 3.11 (t, J = 11.4 Hz, 1H), 2.42 (d, J = 11.1 Hz, 1H), 2.08 (dq, J = 14.4, 7.1 Hz, 2H), 1.90 (q, J = 4.2, 3.2 Hz, 1H), 1.84 (s, 1H), 1.75–1.62 (m, 6H), 1.59 (dd, J = 12.6, 6.8 Hz, 1H), 1.56–1.52 (m, 1H), 1.45 (d, J = 14.4 Hz, 2H), 1.40–1.33 (m, 2H), 1.30 (s, 9H), 1.28–1.24 (m, 3H), 1.24–1.22 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 172.00, 164.03, 155.43, 143.06, 136.70, 131.18, 127.22, 127.00, 126.84, 125.58, 61.97, 56.19, 55.91, 45.76, 41.73, 40.06, 38.76, 35.22, 34.81, 33.45, 31.29, 30.79, 27.89, 27.54, 21.08, 20.28, 20.06. Measured values, m/z: 611.3302 (611.3189) [M + H]+.
Compound 10h. 4-((4-(2-((5-Fluoro-2-methoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl)-N-hydroxy benzamide, C30H39FN4O6S, white power, mp: 66–68 °C. yield 65%, 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 9.00 (s, 1H), 8.25 (t, J = 6.0 Hz, 1H), 7.69 (d, J = 8.1 Hz, 2H), 7.61 (dd, J = 8.3, 3.2 Hz, 1H), 7.43 (td, J = 8.4, 3.2 Hz, 1H), 7.26 (d, J = 8.0 Hz, 2H), 7.22 (dd, J = 9.2, 4.0 Hz, 1H), 4.26–4.20 (m, 2H), 3.87 (s, 3H), 3.60 (td, J = 8.5, 4.7 Hz, 1H), 3.55 (dd, J = 13.1, 5.5 Hz, 1H), 3.40 (dd, J = 13.2, 11.1 Hz, 1H), 2.67 (d, J = 11.1 Hz, 2H), 2.05 (t, J = 3.1 Hz, 1H), 1.91–1.79 (m, 6H), 1.70 (d, J = 12.2 Hz, 1H), 1.58 (dtd, J = 28.0, 9.4, 4.3 Hz, 4H), 1.44 (dt, J = 13.6, 4.5 Hz, 1H), 1.39–1.31 (m, 2H), 1.29–1.21 (m, 3H), 1.13–1.07 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 172.00, 155.95, 154.37, 153.23, 143.34, 131.46, 131.42, 131.38, 127.27, 127.11, 120.83, 120.68, 115.87, 115.70, 115.28, 115.23, 63.24, 58.35, 57.08, 56.67, 48.37, 41.96, 35.78, 35.18, 29.10, 27.90, 27.84, 22.18, 20.78, 20.50. Measured values, m/z: 603.2670 (603.2574) [M + H]+.
Compound 10i. 4-((4-(2-((5-Chloro-2-methoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl)-N-hydroxybenzamide. C30H39ClN4O6S, white power, yield 62%, mp: 77–78 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 9.00 (s, 1H), 8.25 (t, J = 6.0 Hz, 1H), 7.81 (d, J = 2.7 Hz, 1H), 7.71–7.68 (m, 2H), 7.62 (dd, J = 8.9, 2.7 Hz, 1H), 7.26 (d, J = 8.1 Hz, 2H), 7.23 (d, J = 8.9 Hz, 1H), 4.24 (d, J = 6.0 Hz, 2H), 3.89 (s, 3H), 3.60 (td, J = 8.4, 5.0 Hz, 1H), 3.53 (dd, J = 13.2, 5.7 Hz, 1H), 3.38 (dd, J = 13.1, 11.0 Hz, 1H), 2.68 (d, J = 11.0 Hz, 2H), 2.05 (s, 1H), 1.92–1.81 (m, 5H), 1.69 (d, J = 11.9 Hz, 1H), 1.57 (ddt, J = 18.6, 13.5, 4.8 Hz, 4H), 1.47–1.31 (m, 4H), 1.30–1.21 (m, 3H), 1.14–1.08 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 171.54, 163.89, 155.23, 142.92, 133.53, 131.29, 131.04, 127.99, 126.83, 126.69, 123.53, 115.11, 62.70, 57.79, 56.53, 56.20, 47.60, 41.55, 35.13, 34.74, 29.09, 27.63, 27.40, 21.73, 20.30, 20.07. Measured values, m/z: 619.2340 (619.2279) [M + H]+.
Compound 10j. 4-((4-(2-((5-Bromo-2-methoxyphenyl) sulfonyl) decahydro-1H,4H-pyrido[3,2,1-ij] [1,6] naphthyridin-1-yl) butanamide) methyl)-N-hydroxy benzamide, C30H39BrN4O6S, white power, yield 60%, mp: 70–74 °C. 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 8.99 (s, 1H), 8.25 (t, J = 6.0 Hz, 1H), 7.91 (d, J = 2.5 Hz, 1H), 7.74 (dd, J = 8.9, 2.6 Hz, 1H), 7.70–7.68 (m, 2H), 7.26 (d, J = 8.1 Hz, 2H), 7.18 (d, J = 8.9 Hz, 1H), 4.24 (d, J = 6.0 Hz, 2H), 3.89 (s, 3H), 3.61 (d, J = 9.1 Hz, 1H), 3.53 (dd, J = 13.3, 5.7 Hz, 1H), 3.38 (dd, J = 13.1, 11.0 Hz, 1H), 2.68 (d, J = 10.2 Hz, 2H), 2.04 (s, 1H), 1.87 (ddd, J = 26.2, 13.3, 7.7 Hz, 6H), 1.69 (d, J = 12.1 Hz, 1H), 1.57 (ddd, J = 18.9, 12.5, 5.6 Hz, 4H), 1.45–1.34 (m, 3H), 1.30–1.21 (m, 3H), 1.14–1.08 (m, 1H). 13C NMR (151 MHz, DMSO-d6) δ 171.81, 164.16, 155.93, 143.20, 136.73, 131.89, 131.30, 130.98, 127.10, 126.97, 115.83, 111.14, 62.98, 58.04, 56.76, 56.47, 47.83, 41.82, 35.38, 35.02, 27.90, 27.66, 22.00, 20.57, 20.34. Measured values, m/z: 663.1836 (662.1774) [M + H]+.
In vitro antimicrobial activity
The test compound solution was prepared by dissolving 32 mg of compound in 10 μL DMSO followed by dilution with 990 μL of appropriate medium. Antibacterial activity was evaluated through minimum inhibitory concentration (MIC) determination using a two-fold serial dilution method in 96-well microplates. Each well initially received 100 μL medium, followed by sequential transfer of 100 μL drug solution from well 1 through well 8 to create concentration gradients (16–0.125 mg mL−1) in triplicate. After adding 5 μL bacterial culture per well, plates were incubated to logarithmic growth phase, with OD595 measurements determining MIC values. Controls included: (1) culture medium blanks, (2) species-specific positive controls (sodium penicillin G for S. aureus, fluconazole for C. albicans, doxycycline hydrochloride for P. acnes, ceftazidime for P. aeruginosa, and kanamycin sulfate for E. coli), and (3) matrine negative controls. All experiments were performed in triplicate to ensure reliability.
3D-QSAR studies
The compounds for modelling were derived from newly synthesized target compounds (9a–9q, 10a–10j). The CoMFA and CoMSIA models for 3D-QSAR were used to analyze the data based on determining the minimum inhibitory concentration values of the corresponding compounds and calculating the pMIC values as an indicator of the biological activity of the compounds. The formula for converting MIC values to pMIC values was as follows: pMIC = −lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MIC. Molecular datasets including molecular structure and bioactivity data were collected for all target compounds using SYBYL-X 2.0 software; 3D conformations were generated; template molecules and common backbones were selected from the target compounds for manual molecular superposition to obtain the optimal CoMFA and CoMSIA modeling; molecular field calculations were performed for CoMFA and CoMSIA; and PLS (partial least squares) analysis was performed to correlate molecular fields with bioactivity data to generate models. Perform non-cross validation to assess model performance and stability. Visualize analysis results; predict biological activity.
MIC. Molecular datasets including molecular structure and bioactivity data were collected for all target compounds using SYBYL-X 2.0 software; 3D conformations were generated; template molecules and common backbones were selected from the target compounds for manual molecular superposition to obtain the optimal CoMFA and CoMSIA modeling; molecular field calculations were performed for CoMFA and CoMSIA; and PLS (partial least squares) analysis was performed to correlate molecular fields with bioactivity data to generate models. Perform non-cross validation to assess model performance and stability. Visualize analysis results; predict biological activity.
Biofilm experiments
Biofilm inhibition assays were performed by adding 500 μL working suspensions to 24-well plates, followed by treatment with test compounds or 0.1% DMSO (negative control) at 30 °C for 48 h. After incubation, non-adherent cells were removed by gently washing each well twice with PBS. The biofilms were then fixed with 100 μL of 10% methanol for 15 min, air-dried at room temperature, and stained with 500 μL 0.1% crystal violet for 30 min. Excess stain was removed by three PBS washes before solubilizing the biofilm-bound dye with 500 μL absolute ethanol. Absorbance was measured at 595 nm, with culture medium serving as the blank control. Biofilm formation rates were calculated as:| (A595drug − A595blank)/(A595DMSO − A595blank) × 100% |
For mature biofilm disruption assays, 500 μL fungal suspensions were incubated in 24-well plates at 30 °C for 48 h to establish biofilms. After removing planktonic cells with PBS washes, compounds or 0.1% DMSO were added and incubated for an additional 48 h. The subsequent staining and quantification steps mirrored the biofilm inhibition protocol. All experiments were performed in triplicate.
Molecular docking
The molecular structures of target compounds 9j and 10g were prepared for docking studies with SAP5 (PDB: 2QZX), a key virulence factor in Candida albicans biofilm formation. Using Discovery Studio, the protein structure was pre-processed by removing water molecules and ions, repairing structural defects (including hydrogen bonding networks), and adding hydrogen atoms to ensure structural integrity. Small molecule structures were energy-minimized using the CHARMM force field to optimize their geometries.Active site prediction identified potential binding pockets before performing protein-ligand docking with the Dock Ligands algorithm. The resulting complexes were evaluated based on Absolute Energy, Relative Energy, and LibDock Score metrics. The conformation exhibiting the highest docking score was selected for further analysis as it represents the most energetically favorable and biologically relevant binding mode, providing critical insights into the molecular interactions.
Conclusions
In this study, we developed 27 novel matrine-hydroxamic acid derivatives exhibiting significant anti-Candida albicans activity, with lead compound 10g demonstrating remarkable potency (MIC = 0.0621 mg mL−1), approximately 138-fold more effective than fluconazole (MIC = 8.590 mg mL−1). Our 3D-QSAR models identified three critical structural features contributing to this enhanced activity: the electron-donating p-tert-butyl group, the hydroxamic acid pharmacophore, and its conjugated benzene ring system. The derivatives' antifungal mechanism appears multifaceted, as evidenced by their dual capacity to inhibit planktonic growth (MIC assays) and disrupt biofilm formation while degrading mature biofilms at sub-inhibitory concentrations. Molecular docking studies further suggested potential interactions with the key virulence factor SAP5 (PDB: 2ZQX), indicating these compounds may simultaneously target fungal viability and biofilm-mediated pathogenicity.The structural versatility of these matrine derivatives extends their potential applications beyond antifungal therapy. Their broad-spectrum antimicrobial profile against mixed infections, combined with potent biofilm disruption capabilities, positions them as promising candidates for medical device coatings or chronic wound management. Furthermore, the conserved matrine scaffold may retain or enhance known bioactivities against cancer, fibrosis, and inflammation, as reported for the parent compound. Future structure–activity optimization of the R-group substitutions could yield additional therapeutic variants while maintaining the core pharmacophoric elements.
Through integrated experimental and computational approaches, this work establishes a robust foundation for developing matrine-based antimicrobials. The combination of in vitro bioassays, quantitative structure–activity modeling, and molecular docking studies not only validates the observed biological activities but also provides mechanistic insights into the compounds' mode of action. These findings offer valuable design principles for addressing drug-resistant infections while demonstrating a generalizable strategy for modifying natural product scaffolds to combat evolving clinical challenges.
Data availability
All relevant data are included in the manuscript and its additional files. The data are available from the corresponding author on reasonable request.Conflicts of interest
The authors have no conflict of interest regarding manuscript publication.Acknowledgements
This work is supported by Scientific research project of Department of Education of Jilin Province (the project number: JJKH20251817KJ) and the special fund for science and technology development guided by the central government for local areas (Guike ZY21195012).References
- C. Lass-Flörl and S. Steixner, The changing epidemiology of fungal infections, Mol. Aspects Med., 2023, 94, 101215 CrossRef.
- R. Pereira, R. O. Santos Fontenelle and E. H. S. Brito, et al., Biofilm of Candida albicans: formation, regulation and resistance, J. Appl. Microbiol., 2020, 131(1), 11–22 CrossRef.
- M. Swidergall, Candida albicans at Host Barrier Sites: Pattern Recognition Receptors and Beyond, Pathogens, 2019, 8(1), 40 CrossRef CAS PubMed.
- Y. Lee, E. Puumala and N. Robbins, et al., Antifungal Drug Resistance: Molecular Mechanisms in Candida albicans and Beyond, Chem. Rev., 2020, 121(6), 3390–3411 CrossRef PubMed.
- L. Galgóczy and F. Marx, Do Antimicrobial Proteins Contribute to Overcoming the Hidden Antifungal Crisis at the Dawn of a Post-Antibiotic Era?, Microorganisms, 2019, 7(1), 16 CrossRef PubMed.
- L. Wei, H. Wang and X. Ye, et al., Oxymatrine and astragaloside IV co-loaded liposomes: Scale-up purposes and their enhancement of anti-PD-1 efficacy against breast cancer, Mater. Today Bio, 2025, 32, 101634 CrossRef CAS PubMed.
- R.-L. Mo, Z. Li and P. Zhang, et al., Matrine inhibits invasion and migration of gallbladder cancer via regulating the PI3K/AKT signaling pathway, Naunyn-Schmiedeberg's Arch. Pharmacol., 2024, 397(10), 8129–8143 CrossRef CAS PubMed.
- C. Song, H. Li and Z. Huang, et al., Matrine inhibits cell proliferation, invasion, and stem cell formation in hepatocellular carcinoma by regulating the ELAVL1/RBM3-mediated Wnt/β-catenin signaling pathway, Chem. Biol. Drug Des., 2024, 103(2), e14442 CrossRef CAS.
- X. Tan, Y. Hao and N. Ma, et al., M6P-modified solid lipid nanoparticles loaded with matrine for the treatment of fibrotic liver, Drug Delivery, 2023, 30(1), 2219432 CrossRef.
- X. Zhang, C. Hu and N. Zhang, et al., Matrine attenuates pathological cardiac fibrosis via RPS5/p38 in mice, Acta Pharmacol. Sin., 2020, 42(4), 573–584 CrossRef PubMed.
- X. Zhang, J. Huang and J. Zhao, et al., Exosome-mimetic vesicles derived from fibroblasts carrying matrine for wound healing, Burns Trauma, 2024, 12, tkae015 CrossRef.
- D. Luo, N. H. Chen and W. Z. Wang, et al., Structurally Diverse Matrine-Based Alkaloids with Anti-inflammatory Effects from Sophora alopecuroides, Chin. J. Chem., 2021, 39(12), 3339–3346 CrossRef CAS.
- Y. Li, J. A. H. Kowah and M. Jiang, et al., Synthesis, antibacterial activity, and 3D-QASR studies of matrine-indole derivatives as potential antibiotics, Bioorg. Med. Chem. Lett., 2024, 102, 129671 CrossRef CAS PubMed.
- G. Qiu, Y. Long and J. A. H. Kowah, et al., Three-Dimensional Quantitative Structure–Activity Relationship, Antimicrobial Activity, and Molecular Docking Studies of C-14 Chiral Matrine Derivatives as Potential Antibiotics, Chem. Biodiversity, 2025, e20240280 Search PubMed.
- W.-T. Qiao, X. Yao and W.-H. Lu, et al., Matrine exhibits antiviral activities against PEDV by directly targeting Spike protein of the virus and inducing apoptosis via the MAPK signaling pathway, Int. J. Biol. Macromol., 2024, 270, 132408 CrossRef CAS PubMed.
- H. Wang, N. Sun and P. Sun, et al., Matrine regulates autophagy in ileal epithelial cells in a porcine circovirus type 2-infected murine model, Front. Microbiol., 2024, 15, 1455049 CrossRef PubMed.
- L. Hu, X. Zhu and P. Wang, et al., Combining with matrine restores ciprofloxacin efficacy against qnrS producing E. coli in vitro and in vivo, Microb. Pathog., 2025, 1980, 107132 CrossRef.
- J. Meng, J. Ding and W. Wang, et al., Reversal of gentamicin sulfate resistance in avian pathogenic Escherichia coli by matrine combined with berberine hydrochloride, Arch. Microbiol., 2024, 206(7), 292 CrossRef CAS PubMed.
- H. Wu, Y. Ren and J. Zhang, et al., Research progress of LpxC inhibitor on Gram-negative bacteria, Eur. J. Med. Chem., 2025, 289, 117440 CrossRef CAS PubMed.
- S. Mielniczuk, K. Hoff and F. Baselious, et al., Development of Fragment-Based Inhibitors of the Bacterial Deacetylase LpxC with Low Nanomolar Activity, J. Med. Chem., 2024, 67(19), 17363–17391 CrossRef CAS PubMed.
- P. Zhou, Y. Du and K. Ran, et al., Modular Synthesis of Desferrioxamine B and Its Methotrexate Conjugate as a Selective Trojan Horse Antibiotic That Targets Streptococcus pneumoniae, Org. Lett., 2025, 27(7), 1719–1723 CrossRef CAS PubMed.
- D. Al Shaer, O. Al Musaimi and B. G. de la Torre, et al., Hydroxamate siderophores: Natural occurrence, chemical synthesis, iron binding affinity and use as Trojan horses against pathogens, Eur. J. Med. Chem., 2020, 208, 112791 CrossRef CAS.
- M. M. Ghorab, A. M. Soliman and M. S. Alsaid, et al., Synthesis, antimicrobial activity and docking study of some novel 4-(4,4-dimethyl-2,6-dioxocyclohexylidene)methylamino derivatives carrying biologically active sulfonamide moiety, Arabian J. Chem., 2020, 13(1), 545–556 CrossRef CAS.
- M. Fantacuzzi, I. D'Agostino and S. Carradori, et al., Benzenesulfonamide derivatives as Vibrio cholerae carbonic anhydrases inhibitors: a computational-aided insight in the structural rigidity-activity relationships, J. Enzyme Inhib. Med. Chem., 2023, 38(1), 2201402 CrossRef PubMed.
- V. Jakubkiene, G. E. Valiulis and M. Schweipert, et al., Synthesis and HDAC inhibitory activity of pyrimidine-based hydroxamic acids, Beilstein J. Org. Chem., 2022, 18, 837–844 CrossRef CAS.
- X. Liu, B. Fan and S. Huang, et al., Design and synthesis of matrine derivatives for anti myocardial ischemia–reperfusion injury by promoting angiogenesis, Bioorg. Med. Chem., 2024, 108, 117776 CrossRef CAS PubMed.
- Y.-X. Li, J.-Z. Wang and Y. Shimadate, et al., C-6 fluorinated casuarines as highly potent and selective amyloglucosidase inhibitors: Synthesis and structure-activity relationship study, Eur. J. Med. Chem., 2022, 244, 114852 CrossRef CAS.
- C. Xia, R. Liu and S. Zhang, et al., Fluconazole-induced changes in azole resistance and biofilm production in Candida glabrata in vitro, Diagn. Microbiol. Infect. Dis., 2025, 111(3), 116683 CrossRef CAS PubMed.
- M. El-Telbany, G. El-Didamony and A. Askora, et al., Bacteriophages to Control Multi-Drug Resistant Enterococcus faecalis Infection of Dental Root Canals, Microorganisms, 2021, 9(3), 517 CrossRef CAS.
- C. Li, J. Tu and G. Han, et al., Heat shock protein 90 (Hsp90)/Histone deacetylase (HDAC) dual inhibitors for the treatment of azoles-resistant Candida albicans, Eur. J. Med. Chem., 2022, 227, 113961 CrossRef CAS.
- B. P. Singh, S. Ghosh and A. Chauhan, Development, dynamics and control of antimicrobial-resistant bacterial biofilms: a review, Environ. Chem. Lett., 2021, 19(3), 1983–1993 CrossRef CAS.
- M. Alreshidi, R. Badraoui and M. Adnan, et al., Phytochemical profiling, antibacterial, and antibiofilm activities of Sargassum sp. (brown algae) from the Red Sea: ADMET prediction and molecular docking analysis, Algal Res., 2023, 69, 102912 CrossRef.
- A. Hartanto, F. G. Naibaho, D. Panjaitan, A. Lutfia and E. Munir, Molecular docking analysis of Allium chinense compounds as Secreted Aspartyl Proteinase-5 (SAP5) inhibitor [Z], IOP Conf. Ser.: Earth Environ. Sci., 2022, 012017 CrossRef.
- A. Priya and S. K. Pandian, Piperine Impedes Biofilm Formation and Hyphal Morphogenesis of Candida albicans, Front. Microbiol., 2020, 11, 756 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra01689d |
| This journal is © The Royal Society of Chemistry 2025 |