Open Access Article
Open Access ArticleTripeptides inhibit dual targets AChE and BACE-1: a computational study†
Anh Tuan Do ab,
Trung Hai Nguyenab,
Minh Quan Pham
ab,
Trung Hai Nguyenab,
Minh Quan Pham cd,
Huy Truong Nguyenb,
Nguyen Phuoc Longe,
Van Van Vuf,
Huong Thi Thu Phung
cd,
Huy Truong Nguyenb,
Nguyen Phuoc Longe,
Van Van Vuf,
Huong Thi Thu Phung *f and
Son Tung Ngo
*f and
Son Tung Ngo *ab
*ab
aLaboratory of Biophysics, Institute for Advanced Study in Technology, Ton Duc Thang University, Ho Chi Minh City, Vietnam. E-mail: ngosontung@tdtu.edu.vn
bFaculty of Pharmacy, Ton Duc Thang University, Ho Chi Minh City, Vietnam
cInstitute of Chemistry, Vietnam Academy of Science and Technology, Hanoi, Vietnam
dGraduate University of Science and Technology, Vietnam Academy of Science and Technology, Hanoi, Vietnam
eDepartment of Pharmacology and PharmacoGenomics Research Center, Inje University College of Medicine, Busan, Republic of Korea
fNTT Hi-Tech Institute, Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam. E-mail: ptthuong@ntt.edu.vn
First published on 22nd April 2025
Abstract
Alzheimer's disease (AD) is a progressive neurodegenerative disorder characterized by cognitive decline and memory loss, with amyloid-beta (Aβ) plaques and acetylcholine deficits being central pathological features. Inhibition of dual targets including acetylcholinesterase (AChE) and beta-site amyloid precursor protein cleaving enzyme 1 (BACE-1) represents a promising strategy to address cholinergic deficits and amyloid pathology. In this study, we used computational approaches to evaluate 8000 tripeptides as potential dual inhibitors of AChE and BACE-1. Machine learning models revealed the four top-lead tripeptides including WHM, HMW, WMH, and HWM. Molecular docking simulations indicated that WHM possessed the most favorable interactions through hydrogen bonds, π–π stacking, and salt bridges with key catalytic residues in both enzymes. Molecular dynamics simulations confirmed the stability of the protein–ligand complexes, with WHM exhibiting the most consistent conformations and significant disruption of catalytic residue geometries. Free energy perturbation analysis further supported WHM's superior stability across both targets. ADMET predictions suggested moderate oral absorption and limited brain penetration, consistent with the typical behavior of peptide-based compounds. Overall, WHM demonstrated the strongest potential as a dual inhibitor of AChE and BACE-1, offering a promising lead for future therapeutic development in AD.
Introduction
Alzheimer's disease (AD) is a debilitating and progressive neurodegenerative disorder characterized by the deterioration of cognitive functions, memory impairment, and significant behavioral alterations.1–3 The impact of AD extends beyond individual patients, placing substantial emotional, social, and economic burdens on families, caregivers, and healthcare systems internationally. The pathogenesis of AD is multifaceted and not yet fully elucidated, involving a complex interplay of genetic, environmental, and lifestyle factors.4 Central to its pathology is the accumulation of extracellular amyloid-beta (Aβ) plaques and intracellular neurofibrillary tangles composed of hyperphosphorylated tau protein.5–7 These aggregates disrupt neuronal communication and synaptic function, leading to neuronal death and brain atrophy.4 Additionally, processes such as oxidative stress, mitochondrial dysfunction, metal ion dysregulation, and chronic neuroinflammation contribute to the progression of neurodegeneration.8 Despite significant research efforts, the intricate mechanisms underlying AD remain a subject of intense investigation, and effective disease-modifying therapies are still lacking.9–13 Current therapeutic options for AD are limited and primarily offer symptomatic relief without altering the disease's progression.13Recent advancements in immunotherapy targeting Aβ peptides and tau proteins have shown promise but have faced challenges related to efficacy and safety in clinical trials.14–17 A critical therapeutic strategy in AD management focuses on inhibiting key enzymes involved in the disease's pathology, such as acetylcholinesterase (AChE) and beta-site amyloid precursor protein cleaving enzyme 1 (BACE-1).18 AChE is responsible for the hydrolysis of acetylcholine, a neurotransmitter essential for learning and memory processes.19 Inhibition of AChE leads to increased acetylcholine levels in the synaptic cleft, enhancing cholinergic transmission and potentially improving cognitive function.20–22 BACE-1 is the rate-limiting enzyme in the amyloidogenic pathway of amyloid precursor protein (APP) processing, initiating the production of neurotoxic Aβ peptides by cleaving APP at the β-site.23–25 Inhibiting BACE-1 can reduce Aβ production, addressing one of the central pathological features of AD.24,26,27
Peptides, particularly short-chain peptides like tripeptides, have attracted attention for their potential as enzyme inhibitors.28–32 Tripeptides, composed of three amino acid residues, possess favorable pharmacokinetic properties, including low molecular weight and the ability to penetrate biological membranes.33–35 Their structural simplicity allows for ease of synthesis and modification, enabling the exploration of structure–activity relationships. Recent studies have highlighted the potential of certain tripeptides to inhibit AChE and BACE-1 activities effectively31,36 For example, tripeptides derived from natural sources or designed based on active site interactions have demonstrated the ability to bind to these enzymes, blocking substrate access and catalytic activity.37–39 The mechanism of inhibition often involves interactions with key amino acid residues within the active sites, such as forming hydrogen bonds, hydrophobic interactions, and coordination with catalytic residues. These findings suggest that tripeptides could serve as dual inhibitors, targeting both cholinergic deficits and amyloid pathology in AD.40,41
Advancements in computational techniques have revolutionized the drug discovery process, particularly in the early stages of lead identification and optimization.42–44 Molecular docking is a computational method that predicts the preferred orientation of a ligand when bound to a target protein, allowing for the estimation of binding affinities and the identification of key interacting residues.45,46 This technique helps prioritize compounds based on their potential efficacy and guides modifications to improve binding.47,48 Molecular dynamics (MD) simulations extend these insights by modeling the physical movements of atoms and molecules over time, providing a dynamic view of protein–ligand interactions under simulated physiological conditions.49–52 MD simulations can reveal the stability of the complex, conformational changes, and the persistence of key interactions, which are critical for inhibitory activity.53–55
In this study, we used a comprehensive computational approach to evaluate how 8000 tripeptides interact with the active sites of AChE and BACE-1. Initially, pre-trained machine learning (ML) models were applied to narrow down the candidates, resulting in the selection of four top-leading tripeptides. Molecular docking analyses then provided predictions of binding modes, affinities, and key interacting residues for these tripeptides in each enzyme. The results of these docking studies guided the choice of complexes for subsequent MD simulations, which offered detailed insights into the dynamic behavior and stability of each complex under physiologically relevant conditions. Free energy perturbation (FEP) calculations further refined these findings, ultimately identifying promising tripeptide candidates for future development. By simultaneously targeting both cholinergic deficits and amyloid pathology, these tripeptides have the potential to modify disease progression rather than merely alleviating symptoms. Furthermore, the insights gained from this work may inform the broader field of neurodegenerative disease therapeutics, where multi-targeted strategies are increasingly crucial given the complex, multifactorial nature of such disorders.
Materials and methods
Target proteins
The three-dimensional structures of AChE and BACE-1 were retrieved from the Protein Data Bank (PDB) (http://www.rcsb.org/pdb) with their respective PDB IDs of 4M0E56 for AChE and 6EQM57 for BACE-1, respectively. These structures, determined by X-ray diffraction, were selected based on their resolution and relevance for docking studies. The structures were preprocessed by removing water molecules and non-essential ligands, followed by the addition of hydrogens, merging of all non-polar hydrogens, and the application of Kollman charges to prepare them for molecular docking.Ligand generation
Each tripeptide was generated and refined via a multi-step procedure to ensure chemically complete and stable three-dimensional models. Initially, the amino acid sequence (e.g., “WHM” for tryptophan–histidine–methionine) was defined in a simple text format. The PeptideBuilder library (https://github.com/clauswilke/PeptideBuilder) was then used to construct the tripeptide's backbone and side-chain coordinates, noting that PeptideBuilder does not include hydrogen atoms by default. To address this omission, the pdbfixer tool (https://github.com/openmm/pdbfixer) was employed to add all missing hydrogens, resulting in a chemically complete structure. Next, each hydrogen-complete tripeptide was parameterized using the Amber14 force field, which assigned atom types, bonded parameters, and partial charges. Finally, to validate stability and correctness, each peptide underwent a brief (1000-step) MD simulation in OpenMM (http://openmm.org/), where we monitored energy terms and root-mean-square deviation (RMSD). These simulations were performed in implicit solvent to approximate aqueous conditions while maintaining computational efficiency. This step confirmed that all tripeptides converged to stable conformations without significant distortions.Machine learning model
We employed two ML models to predict ligand binding free energy to AChE and to BACE1, respectively. The first version of ML model for AChE was previously trained on 600 compounds using XGBoost method and tested on 162 compounds.58 The latest version of the AChE model, which was used for this work, was trained on 1046 compounds and tested on 362 compounds.59 Here we briefly describe the training and testing procedure of the AChE model. The SMILES and experimental binding constant of the training and test compounds were collected from binding database.60,61 Four different kinds of regression models, namely linear regression (LR), random forest (RF), XGBoost62 and graph-convolutional networks (Graph-Conv)63 were trained. Features input into LR, RF and XGBoost were calculated by using RDKitDescriptors toolkit, implemented in DeepChem,64 which generates 200 physicochemical descriptors, including molecular weight, number of valence electrons, numbers of HB donors and acceptors, and maximum and minimum partial charge. The set of features was reduced to 108 by excluding those that were highly correlated or had predominantly zero value. Features for GraphConv did not need to be manually computed as they were learned on the fly. Instead, the input to this model is a graph of the ligands in which nodes represent atoms with node features such as atom type and charge, and edges represent chemical bonds. It should be noted that our ML models were based only on features extracted from ligands. Therefore they did not need inputs from the receptor, neither the receptor's structure nor its sequence. Among the four models, XGBoost showed the best performance on the randomly selected test set of 362 compounds with the lowest RMSE = 1.357 ± 0.096 kcal mol−1 and the highest correlation coefficients (Pearson's R = 0.813 ± 0.027, Spearman's ρ = 0.808 ± 0.026).59 Therefore XGBoost was selected to predict AChE binding free energy for 8000 tripeptides.The ML models for BACE1 were trained using similar approaches,65 and we briefly mention the most essential information here. The BACE1 models were trained on 2137 compounds and tested on 400 compounds. Both training and testing compounds were collected from binding database60,61 with their SMILES and experimental binding constants. We similarly trained four models, LR, RF, XGBoost and GraphConv using features extracted from ligands. XGBoost again showed the best performance on the random test set of 400 compounds (RMSE = 1.01 ± 0.05 kcal mol−1, Pearson's R = 0.77 ± 0.02, Spearman's ρ = 0.78 ± 0.02)65 and was selected to predict binding free energies for the same library of 8000 tripeptides.
Molecular docking simulations
AutoDockTools66 was utilized to prepare receptor and ligand parameter files for molecular docking studies. Molecular docking simulations were performed using a modified version of AutoDock Vina (mVina)67,68 to assess the binding interactions between the target proteins and four tripeptide inhibitors. The binding sites of AChE (PDB ID: 4M0E)69 and BACE-1 (PDB ID: 6EQM)57 were selected as the enzymic residues contacting with the native inhibitors (Fig. S1†). The docking process involved optimizing the spatial orientation and conformational arrangement of the tripeptides to predict their binding affinities and key molecular interactions within the active sites of the target proteins The docking grid dimensions were set to 22.5 × 22.5 × 22.5 Å, ensuring complete coverage of the active site. The Vina “exhaustiveness” parameter was retained at its default value to balance accuracy and computational efficiency. The docking pose corresponding to the lowest binding energy was selected for further MD simulations.MD simulations
MD simulations were performed to validate the results obtained from molecular docking outcomes. All simulations were conducted using GROMACS 2019.6.70 The protein–ligand complexes were described using the Amber99SB-iLDN force field71 for the protein, peptide and ions, and the TIP3P model72 for water molecules. Each AChE/BACE-1-ligand complex was placed in a dodecahedral simulation box, ensuring a minimum distance of 16.0 Å between the complex and the box boundaries. Under these conditions, the AChE simulation box had a volume of approximately 937.53 nm3 and contained around 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms (the protein complex, a single ligand, water molecules, and neutralizing Na+ ions). The corresponding BACE-1 system occupied 769.62 nm3 and included about 76
000 atoms (the protein complex, a single ligand, water molecules, and neutralizing Na+ ions). The corresponding BACE-1 system occupied 769.62 nm3 and included about 76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms. Nonbonded interactions were treated with a 1.0 nm cutoff. Electrostatic interactions were evaluated using the particle mesh Ewald (PME) method, while van der Waals interactions were computed via a cutoff approach. The LINCS algorithm, order 4,73 was applied to constrain all bonds throughout the simulation.
000 atoms. Nonbonded interactions were treated with a 1.0 nm cutoff. Electrostatic interactions were evaluated using the particle mesh Ewald (PME) method, while van der Waals interactions were computed via a cutoff approach. The LINCS algorithm, order 4,73 was applied to constrain all bonds throughout the simulation.
Following parameterization, the system was energy-minimized using the steepest descent method. The minimized structure then underwent a two-step equilibrations involving an NVT ensemble simulation for 100 ps, and then followed by an NPT ensemble simulation for 100 ps, with positional restraints, spring constant 1000 kJ mol−1 nm−2, applied to all heavy atoms of the protein and ligand. After equilibration, the restraints were removed, and the production MD run was carried out for 200 ns. To ensure adequate sampling and reproducibility, the entire simulation protocol was repeated three times.
FEP simulations
The ligand binding free energy to AChE or BACE-1 was calculated using the double-annihilation binding free energy method.74 The equilibrium conformation derived from MD simulations served as the initial structure for FEP calculations,75 consistent with previously established approach.76 FEP simulations used a coupling parameter (λ) varying from 0 (fully interactive) to 1 (fully decoupled), enabling the calculation of the free energy change (ΔG) as a ligand transition from complete interaction with its surroundings to none—a process known as ligand annihilation. These transitions were driven by systematic modifications in the system's Hamiltonian. To isolate the contributions of Coulomb (Cou) and vdW forces, distinct λ values were strategically selected as 0.00, 0.10, 0.20, 0.35, 0.50, 0.65, 0.80, and 1.00 for Cou; and 0.00, 0.10, 0.25, 0.35, 0.50, 0.65, 0.75, 0.90, and 1.00 for vdW—yielding sixteen simulations in total. Each λ-alteration simulation lasted 3.0 ns. The Bennett's Acceptance Ratio (BAR) method then integrated these data,77 providing a comprehensive measure of the free energy associated with ligand annihilation.To determine a ligand's absolute binding free energy, two annihilation processes were evaluated: one where the ligand is removed from its solvated state, and another where it is removed from the solvated protein–ligand complex. The difference between these two processes provides the absolute binding free energy, as described by the equation:
 | (1) |
Analysis tools
Protonation states of the ligands during the MD simulations were determined using the ChemAxon webserver (https://www.chemicalize.com). The RMSD of atomic positions throughout the MD simulation was calculated with the “gmx rms” tool in GROMACS.70 The free energy landscape (FEL) of non-hydrogen atoms of AChE/BACE-1 active site and ligands was generated using the principal component analysis (PCA).78 The first and second principal components were utilized as reaction coordinates to define the system's conformational space. Representative structures of the AChE/BACE-1-tripeptide complexes were then identified through clustering analysis.79,80 Intermolecular vdW and Coulomb interaction energies between the tripeptide and its surroundings were computed using the “gmx energy” function in GROMACS.70 Additionally, the PreADMET web server81 was used to evaluate key metrics related to the compounds' ability. All molecular graphics and protein–ligand interaction diagrams were prepared using the Maestro free version.82Results and discussion
Machine learning model calculation
The use of ML models,54,59,65 particularly those based on the XGBoost algorithm, to predict ligand-binding free energy for AChE and BACE-1 provides a highly effective starting point for tripeptide screening. The high accuracy of these models—demonstrated by low RMSE values and strong correlation coefficients in previous studies,54,59,65 – establishes their reliability in identifying promising candidates from the large dataset of 8000 tripeptides used in this study. The obtained value ΔGML ranges from −5.09 to −9.13 kcal mol−1, with an average binding free energy of −6.80 ± 0.65 kcal mol−1 for AChE target (see ESI file 1†). For BACE-1 target, the average binding free energy is of −7.43 ± 0.53 kcal mol−1, which ranges from −6.45 to −9.27 kcal mol−1. Drawing from our analysis, 4 top-lead tripeptides including WHM, HMW, WMH, and HWM (Table 1) were selected for further examination. Notably, all candidates are composed of the same three amino acid residues—tryptophan (W), histidine (H), and methionine (M). This suggests that these residues may play a crucial role in driving strong interactions with both AChE and BACE-1, likely due to their unique physicochemical properties, such as hydrophobicity (W and M), aromatic stacking (W and H), and potential for hydrogen bonding (H). The consistency of this pattern underscores the importance of these specific amino acids in enhancing binding affinity and specificity across both targets.| No. | Ligand | AChE | BACE-1 | ||
|---|---|---|---|---|---|
| ΔGML | ΔGmVina | ΔGML | ΔGmVina | ||
| a The unit is of kcal mol−1. | |||||
| 1 | WHM | −9.13 | −16.6 | −9.03 | −14.4 |
| 2 | HMW | −9.10 | −16.3 | −9.03 | −15.1 |
| 3 | WMH | −9.03 | −16.9 | −9.07 | −14.5 |
| 4 | HWM | −8.94 | −16.9 | −9.11 | −15.2 |
Molecular docking simulations
The speed and computational efficiency of AutoDock Vina (Vina)83 in predicting ligand binding have led to its widespread application. Its uses range from modeling binding poses of large substrates against protein targets84 to evaluating the binding affinities of smaller molecules, including peptides, proteins, and genes.85,86 More recently, mVina, an altered version of Vina that incorporates experimentally optimized parameters,68 has demonstrated improved correlation coefficients and higher docking success rates.68 Consequently, we utilized mVina to predict the binding affinities of the selected tripeptides against AChE and BACE-1.The molecular docking simulations provided detailed insights into the binding affinities and interaction patterns of the top-lead tripeptides including WHM, HMW, WMH, and HWM with the active sites of AChE and BACE-1 (Table S1 and Fig. S2†). The binding free energies, calculated using mVina, revealed strong interactions between the tripeptides and the enzymes, with ΔGmVina values ranging from −16.9 to −16.3 kcal mol−1 for AChE and −15.2 to −14.4 kcal mol−1 for BACE-1 (Table 1). Meanwhile, ΔGML values ranging from −9.13 to −8.94 kcal mol−1 for AChE and −9.11 to −9.03 kcal mol−1 for BACE-1 (Table 1). These results indicate that mVina-based docking tends to overestimate ligand-binding affinities compared to ML predictions. This finding is consistent with prior studies, which have observed that mVina often yields overestimated binding free energies relative to experimental and ML results.59,68
The docking binding pose analysis highlights the potential of the four as dual inhibitors targeting AChE and BACE-1 (Fig. 1). These tripeptides demonstrated stable binding interactions with the active sites of both enzymes, forming critical hydrogen bonds, π–π stacking, π–cation interactions, and salt bridges. For AChE (Fig. 1A), WHM and HWM showed strong binding affinities, with WHM forming multiple π–π stacking interactions with Trp286, Tyr124, and Tyr341, and a π–cation interaction with Phe295, enhancing its stability within the active site. HWM exhibited notable π–π stacking with His447, a key catalytic residue, potentially disrupting AChE's enzymatic function. In comparison, WMH and HMW relied more on hydrogen bonding with residues like Tyr337 and Ser293, providing stable yet slightly weaker interactions.
 | ||
| Fig. 1 2D interaction of four tripeptides with AChE/BACE-1 obtained via molecular docking simulations. (A) Interaction of AChE with ligands. (B) Interaction of BACE-1 with ligands. | ||
Regarding BACE-1 (Fig. 1B), WHM and WMH emerged as the most promising inhibitors, forming extensive hydrogen bonds with catalytic residues Asp228 (ref. 87) and Gly230 and π–π stacking with Tyr71. These interactions directly involve residues essential for BACE-1's enzymatic activity, suggesting effective inhibition of amyloidogenic processing. HWM also displayed a strong profile by forming a salt bridge with key residue Asp32,87 along with hydrogen bonding and π–π interactions, indicating its potential to stabilize within the binding pocket and inhibit BACE-1 function. Across both targets, WHM exhibited the strongest and most balanced interaction profile, making it a standout candidate for dual inhibition.
MD simulations
Because molecular docking relies on approximations to streamline computational requirements, additional verification using more precise methods is essential.66,83,88 To this end, MD simulations were performed to confirm the docking results and assess the dynamic stability of the protein–ligand complexes under physiological conditions.59,89 Each system achieved an equilibrium state within approximately 25–50 ns, with RMSD values stabilizing around 0.2 nm (Fig. S3†). This degree of stability indicates that the presence of the tripeptide induced only minimal structural perturbations, thereby maintaining the overall integrity of AChE/BACE-1. Furthermore, the observed binding event appeared to hinder the access of other potential ligands to the enzyme's active site, implying that the peptides likely function as inhibitors rather than promoters. Conducting three independent MD simulations for each system provided additional confidence in these findings by ensuring that the conformational space was adequately sampled and that the results were not unduly influenced by specific initial conditions.The structural impact of tripeptide binding on the catalytic triad in AChE and the catalytic dyad in BACE-1 were evaluated by combining quantitative distance measurements (Table 2) with the corresponding distribution profiles (Fig. 2), providing a detailed view of how each tripeptide influences key structural elements essential for enzymatic activity. For AChE, the ligand-free state of the catalytic triad, represented by distances between Ser203–His447 (0.42 ± 0.07 nm) and His447–Glu334 (0.29 ± 0.06 nm), served as a reference point. Among the tested tripeptides, WHM elicited the most pronounced and stable changes. The His447–Glu334 interaction increased to 0.45 ± 0.08 nm (Table 2), and the corresponding distribution (red line in Fig. 2) shifted distinctly to a new peak at approximately 0.45 nm. Similarly, the Ser203–His447 distance rose to about 0.45 ± 0.07 nm, producing a narrow, well-defined peak (black line in Fig. 2). This combination of larger distance values and sharper peaks indicates that WHM effectively disrupts the catalytic triad and stabilizes a new, altered conformation.
| Average distance (nm) | ||||||
|---|---|---|---|---|---|---|
| No ligand | WHM | HMW | WMH | HWM | ||
| a Error represents the standard deviation. | ||||||
| AChE catalytic triad atoms | Ser203 OG – His447 NE2 | 0.42 ± 0.07 | 0.45 ± 0.07 | 0.52 ± 0.17 | 0.43 ± 0.09 | 0.44 ± 0.05 |
| His447 ND1 – Glu334 OE1/OE2 | 0.29 ± 0.06 | 0.45 ± 0.08 | 0.35 ± 0.09 | 0.37 ± 0.11 | 0.38 ± 0.09 | |
| BACE-1 catalytic dyad atoms | Asp32 CG – Asp228 CG | 0.59 ± 0.05 | 0.51 ± 0.04 | 0.60 ± 0.05 | 0.52 ± 0.04 | 0.62 ± 0.04 |
 | ||
| Fig. 2 Distribution of catalytic triad distances in AChE (A) and catalytic dyad distances in BACE-1 (B). | ||
HMW, by contrast, induced the largest absolute change in the Ser203–His447 distance (0.52 ± 0.17 nm) but failed to achieve a stable rearrangement. The corresponding histogram displays a broad secondary peak at 0.52 nm (Fig. 2), coexisting with remnants of the original state and suggesting that the enzyme samples multiple conformations. This indicates that while HMW can disrupt the triad, it does not stabilize a single, well-defined structural arrangement. WMH and HWM exerted comparatively weaker effects. WMH shifted the Ser203–His447 distance slightly to 0.43 ± 0.09 nm and the His447–Glu334 distance to 0.37 ± 0.11 nm, while HWM produced distances of 0.44 ± 0.05 nm and 0.38 ± 0.09 nm, respectively (Table 2). Both led to mild changes without forming new, distinct peaks, reflecting minimal restructuring of the catalytic triad (Fig. 2).
A parallel assessment of the BACE-1 catalytic dyad (Asp32–Asp228) further highlights the superior performance of WHM. Starting from the unbound state (0.59 ± 0.05 nm), WHM reduced this distance to 0.51 ± 0.04 nm (Table 2). Fig. 2B shows a new peak at 0.51 nm, replacing the original distribution, thereby indicating a stable reconfiguration of the dyad. WMH also induced a shift, albeit to a lesser degree (0.52 ± 0.04 nm), with a new peak emerging near 0.52 nm. In contrast, HWM and HMW failed to disrupt the dyad meaningfully. HWM slightly increased the distance to 0.62 ± 0.04 nm without forming a new peak, suggesting mere expansion rather than rearrangement, while HMW remained essentially near the original state at 0.60 ± 0.05 nm (Table 2 and Fig. 2).
Taken together, these results position WHM as the most effective tripeptide in reshaping the structural landscape of both AChE and BACE-1 active sites. The emergence of distinct new peaks and stable conformations under WHM treatment supports its potential as a promising inhibitor. Although HMW demonstrates the capacity to create large conformational shifts, its inability to stabilize these changes limits its overall effectiveness. WMH and HWM elicit only minor modifications and thus appear less suitable for achieving meaningful catalytic site inhibition.
Finding popular conformation using PCA/FEL
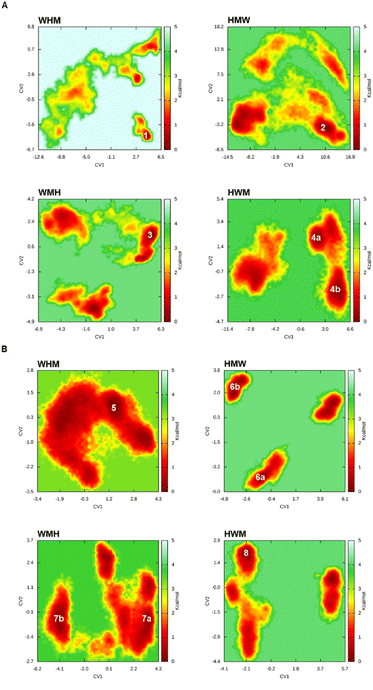
FEL analyses provided insights into the free-energy landscapes of AChE and BACE-1 complexes with each of the four tripeptides by mapping two key collective variables (CV1 and CV2) that capture essential motions of the protein–ligand system (Fig. 3 and Table S2†). In the AChE system, WHM occupied a single dominant energy basin at (CV1, CV2) coordinates of (4.18, −4.87), indicating a tightly bound conformation that is both energetically and conformationally stable. In BACE-1, WHM similarly formed a deep low-energy basin at (1.50, 0.06), underscoring its potential as a robust inhibitor for both enzymes. Biologically, a single low-energy basin can signify a strong affinity for the active site and reduced likelihood of destabilizing conformational shifts, which together enhance inhibitory efficacy.By contrast, the other tripeptides exhibited more dispersed energy basins, reflecting varying degrees of flexibility and potentially lower specificity. For example, HWM balanced stability with adaptability, featuring two notable basins in AChE at (1.62, 2.22) (4a) and (4.59, −1.94) (4b) (Fig. 3A). Although these basins may allow HWM to accommodate enzyme conformational changes, its overall stability was weaker than that of WHM, which may affect its inhibitory consistency. Likewise, WMH and HMW demonstrated higher conformational freedom but lacked the deep, singular basin seen in WHM. Notably, WMH in BACE-1 displayed two basins (7a and 7b), suggesting significant flexibility but lower specificity (Fig. 3B). Meanwhile, BACE-1-HMW had two distinct minima (6a and 6b) separated by energy barriers that limited interconversion, thus conferring only moderate adaptability. Taken together, these observations link the conformational stability captured by FEL to potential inhibitory strength. WHM's consistently stable free-energy basins across both AChE and BACE-1 highlight its promise as a dual inhibitor, potentially offering more reliable and potent enzyme modulation in neurodegenerative disease therapies.
Electrostatic analysis of AChE and BACE-1 complexes
Using the Adaptive Poisson–Boltzmann Solver (APBS) approach,90 we evaluated the electrostatic complementarity of tripeptides within the binding pockets of AChE and BACE-1 (Fig. 4). Among the tested tripeptides, WHM exhibited the highest electrostatic compatibility with AChE. The enzyme's binding pocket presented a strong, localized negative potential that aligns well with WHM's positively charged residues, fostering robust ionic interactions and conferring high binding specificity (Fig. 4A). In contrast, HMW showed a more balanced mix of negative and positive potentials within the active site, suggesting diverse interaction modes but somewhat reduced electrostatic focus compared to WHM. WMH and HWM both displayed dispersed or smooth electrostatic profiles with weaker negative potentials, indicating a diminished reliance on ionic interactions and a likely dependence on non-ionic forces (e.g., vdW or hydrophobic) for binding.A similar trend emerged in BACE-1 complexes, where WHM again demonstrated the most favorable electrostatic complementarity (Fig. 4B). The enzyme's pronounced negative potential complemented WHM's positive charge distribution, resulting in strong ionic interactions and high affinity. HMW exhibited moderate electrostatic complementarity, enabling multiple interaction modes but at a slightly reduced overall affinity compared to WHM. Conversely, WMH and HWM displayed weaker negative potentials that were either dispersed or smoothly graded, suggesting lower specificity and a greater need for non-ionic interactions to stabilize binding. Collectively, these electrostatic profiles identify WHM as the most promising dual inhibitor of AChE and BACE-1, owing to its consistently strong and focused electrostatic complementarity that promotes stable binding. HMW's balanced profile offers adaptable binding modes, though with lower specificity than WHM. In contrast, WMH and HWM rely more on non-ionic interactions due to dispersed or weak electrostatic potentials, making them less suitable when robust ionic contacts are crucial for inhibition.
FEP calculation
FEP analyses for both AChE and BACE-1 revealed a consistent hierarchy among the four tripeptides (WHM, HMW, WMH, and HWM), with WHM displaying the highest affinity in both systems (Table 3 and Fig. S4†). WHM's ΔGFEP values (−33.31 ± 8.31 kcal mol−1 for AChE and −38.30 ± 1.96 kcal mol−1 for BACE-1) underscored its robust electrostatic and vdW interactions that align with its robust docking scores and pronounced structural perturbations observed in MD simulations. In contrast, HMW displayed the least favorable binding free energies for both AChE (−3.72 ± 2.23 kcal mol−1) and BACE-1 (−12.31 ± 6.47 kcal mol−1), while WMH and HWM exhibited intermediate affinity profiles in each system.| Enzyme | Ligand | ΔGcou | ΔGvdw | ΔGFEP |
|---|---|---|---|---|
| a Error is the standard error of mean. The unit is of kcal mol−1. | ||||
| AChE | WHM | −20.25 | −13.06 | −33.31 ± 8.31 |
| HMW | 5.78 | −9.51 | −3.72 ± 2.23 | |
| WMH | −16.53 | −6.90 | −23.42 ± 9.28 | |
| HWM | −20.40 | −8.26 | −28.67 ± 9.79 | |
| BACE-1 | WHM | −31.78 | −6.52 | −38.30 ± 1.96 |
| HMW | −6.65 | −5.67 | −12.31 ± 6.47 | |
| WMH | −21.50 | −5.69 | −27.19 ± 10.77 | |
| HWM | −18.93 | −6.76 | −25.69 ± 8.30 | |
The larger negative binding free energies obtained from FEP, relative to ML and docking predictions, arise from FEP's rigorous thermodynamic treatment, which explicitly incorporates both enthalpic and entropic contributions through detailed MD sampling. Unlike docking and ML methods, which rely on approximations or empirical scoring functions, FEP calculations inherently provide greater thermodynamic accuracy. Although absolute binding energy values vary across these methods, the binding free energies derived from FEP closely matched the trends observed in MD simulations and docking studies, validating the use of FEP as a critical refinement step to differentiate closely related ligands initially identified by ML and docking.
WHM's consistently strong performance across all analyses can be attributed to its optimal balance of hydrophobic, electrostatic, and hydrogen-bonding interactions. In AChE, WHM formed multiple π–π stacking interactions with key residues (e.g., Trp286 and Tyr341), while in BACE-1, HBs with Asp228 and π-cation interactions enhanced its binding specificity. These interactions likely contribute to its ability to stabilize altered conformations of catalytic residues, thereby potentially inhibiting enzymatic activity. Overall, these findings reinforce the superior dual-inhibitory potential of WHM, meriting further exploration for multi-target therapeutic strategies against AD disease.
ADMET
The absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis of these tripeptides provides critical insights into their pharmacokinetic profiles. As shown in Table S3,† all peptides are lipophilic (log P > 0), a key physicochemical property influencing solubility, permeability, potency, selectivity, and toxicity. Consistent with typical peptide behavior, all four tripeptides exhibit poor brain penetration, as reflected by their negative logBB values (logarithm of the brain-to-blood concentration ratios) and BBB values (brain-to-blood concentration ratios) less than 1.91 With respect to oral absorption, each tripeptide demonstrates a nearly identical human intestinal absorption (HIA) value (∼54.86%), suggesting moderate gastrointestinal uptake, aligning with common absorption patterns of small peptides. All four peptides also display moderate plasma protein binding (PPB ≈ 50%), and a predicted Caco-2 permeability coefficient of about 20 implies moderate permeability through Caco-2 cells. Furthermore, none of the compounds appear to act as inhibitors or substrates of CYP_2D6—an enzyme critical for the metabolism and elimination of many clinical drugs.Toxicity was assessed through mutagenicity, carcinogenicity (Ames_test), and hERG inhibition values. While all four tripeptides proved mutagenic, they were non-carcinogenic in mouse models. Nevertheless, the high-risk profiles for hERG_inhibition point to potential cardiotoxicity stemming from disruption of potassium ion channel function. Consequently, further optimization via structural modifications, alternative delivery strategies, and in vitro experimentation is warranted to mitigate these risks.
Ultimately, our research contributes to the ongoing efforts in developing effective and safe therapeutic agents for AD by exploring the potential of natural tripeptides as dual inhibitors of AChE and BACE-1. By integrating ML models, molecular docking, MD simulations, FEP calculation and pharmacokinetic predictions, we provide a comprehensive understanding of the molecular mechanisms underlying enzyme inhibition by these tripeptides. This approach facilitates the rational design of novel compounds with improved efficacy and pharmacological profiles, potentially accelerating the discovery of multi-targeted therapeutics for AD.
Conclusion
In this study, we conducted a comprehensive computational screening of 8000 tripeptides to identify potential dual inhibitors of AChE and BACE-1, key enzymes implicated in AD pathology. Initial screening using ML models based on the XGBoost algorithm enabled rapid and accurate estimation of binding free energies, narrowing the candidates to four top leads: WHM, HMW, WMH, and HWM. Further refinement using mVina docking analysis confirmed favorable interactions between these tripeptides and the active sites of AChE and BACE-1, mediated by hydrogen bonds and π–π stacking interactions. MD simulations demonstrated that tripeptide binding effectively disrupted the catalytic triad of AChE and the catalytic dyad of BACE-1, indicating potential inhibition of their enzymatic activity. FEL analyses further supported the stability of the AChE/BACE-1-tripeptide complexes. Electrostatic analyses highlighted the strong complementarity between the tripeptides and enzyme active sites, providing a structural basis for their inhibitory potential. Finally, FEP calculations provided quantitative confirmation of the robust binding affinities exhibited by the promising tripeptides. Among the candidates, WHM emerged as the most promising dual inhibitor, exhibiting strong binding affinity, and stability. However, potential cardiotoxicity risks associated with the tripeptide must be addressed to fully realize its therapeutic potential. This study underscores the effectiveness of an integrated computational workflow in identifying WHM as promising lead for dual inhibitors targeting AD and highlights the need for further experimental validation.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
All authors contribute to preparation of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Ho Chi Minh City Department of Science and Technology under project code 115/QĐ-SKHCN.References
- K. Blennow, M. J. de Leon and H. Zetterberg, Lancet, 2006, 368, 387–403 CrossRef CAS PubMed.
- P. Scheltens, K. Blennow, M. M. Breteler, B. De Strooper, G. B. Frisoni, S. Salloway and W. M. Van der Flier, Lancet, 2016, 388, 505–517 CrossRef CAS PubMed.
- P. Scheltens, B. De Strooper, M. Kivipelto, H. Holstege, G. Chételat, C. E. Teunissen, J. Cummings and W. M. van der Flier, Lancet, 2021, 397, 1577–1590 CrossRef CAS PubMed.
- T. Guo, D. Zhang, Y. Zeng, T. Y. Huang, H. Xu and Y. Zhao, Mol. Neurodegener., 2020, 15, 1–37 CrossRef PubMed.
- M. P. Murphy and H. LeVine III, J. Alzheimers Dis., 2010, 19, 311–323 Search PubMed.
- S.-S. Yoon and S. A. Jo, Biol. Ther., 2012, 20, 245 CAS.
- G. S. Bloom, JAMA Neurol., 2014, 71, 505–508 CrossRef PubMed.
- Y. Chen and Y. Yu, J. Neuroinflammation, 2023, 20, 165 CrossRef PubMed.
- T. E. Golde, Neurotherapeutics, 2023, 19, 209–227 CrossRef PubMed.
- K. Anitha, M. K. Singh, K. Kohat, S. Chenchula, R. Padmavathi, L. S. Amerneni, V. Vardhan, M. R. Chavan and S. Bhatt, Mol. Neurobiol., 2024, 1–19 Search PubMed.
- M. C. Tartaglia and M. Ingelsson, Mol. Diagn. Ther., 2024, 1–16 Search PubMed.
- J. Folch, D. Petrov, M. Ettcheto, S. Abad, E. Sánchez-López, M. L. García, J. Olloquequi, C. Beas-Zarate, C. Auladell and A. Camins, Neural Plast., 2016, 2016, 8501693 Search PubMed.
- Y. Peng, H. Jin, Y.-h. Xue, Q. Chen, S.-y. Yao, M.-q. Du and S. Liu, Front. Aging Neurosci., 2023, 15, 1206572 CrossRef CAS PubMed.
- C. A. Lemere and E. Masliah, Nat. Rev. Neurol., 2010, 6, 108–119 CrossRef CAS PubMed.
- C. Song, J. Shi, P. Zhang, Y. Zhang, J. Xu, L. Zhao, R. Zhang, H. Wang and H. Chen, Transl. Neurodegener., 2022, 11, 18 CrossRef CAS PubMed.
- D. L. Castillo-Carranza, M. J. Guerrero-Muñoz and R. Kayed, ImmunoTargets Ther., 2013, 19–28 Search PubMed.
- A. Esquer, F. Blanc and N. Collongues, Neurol. Ther., 2023, 12, 1883–1907 CrossRef PubMed.
- M. V. F. Silva, C. d. M. G. Loures, L. C. V. Alves, L. C. De Souza, K. B. G. Borges and M. d. G. Carvalho, J. Biomed. Sci., 2019, 26, 1–11 CrossRef PubMed.
- H. Soreq and S. Seidman, Nat. Rev. Neurosci., 2001, 2, 294–302 CrossRef CAS PubMed.
- V. N. Talesa, Mech. Ageing Dev., 2001, 122, 1961–1969 CrossRef CAS PubMed.
- G. Marucci, M. Buccioni, D. Dal Ben, C. Lambertucci, R. Volpini and F. Amenta, Neuropharmacology, 2021, 190, 108352 CrossRef CAS PubMed.
- T. C. d. Santos, T. M. Gomes, B. A. S. Pinto, A. L. Camara and A. M. D. A. Paes, Front. Pharmacol., 2018, 9, 1192 CrossRef PubMed.
- S. L. Cole and R. Vassar, Mol. Neurodegener., 2007, 2, 1–25 CrossRef PubMed.
- A. K. Ghosh and H. L. Osswald, Chem. Soc. Rev., 2014, 43, 6765–6813 RSC.
- H. Hampel, R. Vassar, B. De Strooper, J. Hardy, M. Willem, N. Singh, J. Zhou, R. Yan, E. Vanmechelen and A. De Vos, Biol. Psychiatry, 2021, 89, 745–756 CrossRef CAS PubMed.
- N. M. Moussa-Pacha, S. M. Abdin, H. A. Omar, H. Alniss and T. H. Al-Tel, Med. Res. Rev., 2020, 40, 339–384 CrossRef PubMed.
- E. McDade, I. Voytyuk, P. Aisen, R. J. Bateman, M. C. Carrillo, B. De Strooper, C. Haass, E. M. Reiman, R. Sperling and P. N. Tariot, Nat. Rev. Neurol., 2021, 17, 703–714 CrossRef PubMed.
- M. Muttenthaler, G. F. King, D. J. Adams and P. F. Alewood, Nat. Rev. Drug Discov., 2021, 20, 309–325 CrossRef CAS PubMed.
- A. C.-L. Lee, J. L. Harris, K. K. Khanna and J.-H. Hong, Int. J. Mol. Sci., 2019, 20, 2383 CrossRef PubMed.
- Z. Yu, S. Wu, W. Zhao, L. Ding, Y. Fan, D. Shiuan, J. Liu and F. Chen, Food Funct., 2018, 9, 1173–1178 RSC.
- K. Singh, A. Kaur, B. Goyal and D. Goyal, ACS Chem. Neurosci., 2024, 15, 2545–2564 CrossRef CAS PubMed.
- R. Dai, Y. Sun, R. Su and H. Gao, Biomed. Pharmacother., 2022, 154, 113576 CrossRef CAS PubMed.
- P. Ung and D. A. Winkler, J. Med. Chem., 2011, 54, 1111–1125 CrossRef CAS PubMed.
- M. J. Humphrey and P. S. Ringrose, Drug Metab. Rev., 1986, 17, 283–310 CrossRef CAS PubMed.
- S. Santos, I. Torcato and M. A. Castanho, Pept. Sci., 2012, 98, 288–293 CrossRef CAS PubMed.
- L. Zhao, D. Li, X. Qi, K. Guan, H. Chen, R. Wang and Y. Ma, Food Funct., 2022, 13, 10851–10869 RSC.
- Z. Yu, H. Ji, J. Shen, R. Kan, W. Zhao, J. Li, L. Ding and J. Liu, Food Funct., 2020, 11, 6643–6651 RSC.
- Z. Yu, W. Dong, S. Wu, J. Shen, W. Zhao, L. Ding, J. Liu and F. Zheng, J. Sci. Food Agric., 2020, 100, 2648–2655 CrossRef CAS PubMed.
- B. Kaur, R. Kaur, Vivesh, S. Rani, R. Bhatti and P. Singh, ACS Omega, 2024, 9, 12896–12913 CAS.
- X. Zhao, Q. Hu, X. Wang, C. Li, X. Chen, D. Zhao, Y. Qiu, H. Xu, J. Wang and L. Ren, Eur. J. Med. Chem., 2024, 116810 CrossRef CAS PubMed.
- Y. Zhu, K. Xiao, L. Ma, B. Xiong, Y. Fu, H. Yu, W. Wang, X. Wang, D. Hu and H. Peng, Bioorg. Med. Chem., 2009, 17, 1600–1613 CrossRef CAS PubMed.
- S. J. Y. Macalino, V. Gosu, S. Hong and S. Choi, Arch. Pharmacal Res., 2015, 38, 1686–1701 CrossRef CAS PubMed.
- C. M. Song, S. J. Lim and J. C. Tong, Briefings Bioinf., 2009, 10, 579–591 CrossRef CAS PubMed.
- Q. M. Thai, H. T. T. Phung, N. T. Tung, L. H. Tran and S. T. Ngo, Chem. Phys. Lett., 2025, 141899 CrossRef CAS.
- J. Fan, A. Fu and L. Zhang, Quant. Biol., 2019, 7, 83–89 CrossRef CAS.
- L. Pinzi and G. Rastelli, Int. J. Mol. Sci., 2019, 20, 4331 CrossRef CAS PubMed.
- B. J. Bender, S. Gahbauer, A. Luttens, J. Lyu, C. M. Webb, R. M. Stein, E. A. Fink, T. E. Balius, J. Carlsson and J. J. Irwin, Nat. Protoc., 2021, 16, 4799–4832 CrossRef CAS PubMed.
- N. M. Tam, T. H. Nguyen, M. Q. Pham, N. D. Hong, N. T. Tung, V. V. Vu, D. T. Quang and S. T. Ngo, J. Mol. Graph. Model., 2023, 124, 108535 CrossRef CAS PubMed.
- J. D. Durrant and J. A. McCammon, BMC Biol., 2011, 9, 1–9 CrossRef PubMed.
- H. Alonso, A. A. Bliznyuk and J. E. Gready, Med. Res. Rev., 2006, 26, 531–568 CrossRef CAS PubMed.
- X. Liu, D. Shi, S. Zhou, H. Liu, H. Liu and X. Yao, Expet Opin. Drug Discov., 2018, 13, 23–37 CrossRef CAS PubMed.
- T. H. Nguyen, N. Q. Anh Pham, Q. M. Thai, V. V. Vu, S. T. Ngo and J.-T. Horng, ACS Omega, 2024, 9, 48505–48511 CrossRef CAS PubMed.
- Q. M. Thai, T. H. Nguyen, H. T. T. Phung, M. Q. Pham, N. K. T. Pham, J.-T. Horng and S. T. Ngo, RSC Adv., 2024, 14, 18950–18956 RSC.
- Q. M. Thai, M. Q. Pham, P.-T. Tran, T. H. Nguyen and S. T. Ngo, R. Soc. Open Sci., 2024, 11, 240546 CrossRef CAS PubMed.
- T. H. Nguyen, Q. M. Thai, M. Q. Pham, P. T. H. Minh and H. T. T. Phung, Mol. Divers., 2024, 28, 553–561 CrossRef CAS PubMed.
- J. Cheung, E. N. Gary, K. Shiomi and T. L. Rosenberry, ACS Med. Chem. Lett., 2013, 4, 1091–1096 CrossRef CAS PubMed.
- U. Neumann, M. Ufer, L. H. Jacobson, M. L. Rouzade-Dominguez, G. Huledal, C. Kolly, R. M. Lüönd, R. Machauer, S. J. Veenstra and K. Hurth, EMBO Mol. Med., 2018, 10, e9316 CrossRef PubMed.
- T. H. Nguyen, P.-T. Tran, N. Q. A. Pham, V.-H. Hoang, D. M. Hiep and S. T. Ngo, ACS Omega, 2022, 7, 20673–20682 CrossRef CAS PubMed.
- Q. M. Thai, T. H. Nguyen, G. B. Lenon, H. T. Thu Phung, J.-T. Horng, P.-T. Tran and S. T. Ngo, J. Mol. Graph. Model., 2025, 134, 108906 CrossRef CAS PubMed.
- M. K. Gilson, T. Liu, M. Baitaluk, G. Nicola, L. Hwang and J. Chong, Nucleic Acids Res., 2016, 44, D1045–D1053 CrossRef CAS PubMed.
- T. Liu, Y. Lin, X. Wen, R. N. Jorissen and M. K. Gilson, Nucleic Acids Res., 2007, 35, D198–D201 CrossRef CAS PubMed.
- T. Chen and C. Guestrin, presented in part at the Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, California, USA, 2016 Search PubMed.
- D. K. Duvenaud, D. Maclaurin, J. Iparraguirre, R. Bombarell, T. Hirzel, A. Aspuru-Guzik and R. P. Adams, presented in part at the Advances in Neural Information Processing Systems, 2015, 2015 Search PubMed.
- B. Ramsundar, P. Eastman, P. Walters and V. Pande, Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More, O'Reilly, 2019 Search PubMed.
- T. D. Quang, D. T. M. Dung, Q. M. Thai, P.-T. Tran, S. T. Ngo and T. H. Nguyen, ChemRxiv, Cambridge Open Engage, Cambridge, 2025, preprint, DOI:10.26434/chemrxiv-2025-xccmh.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- T. N. H. Pham, T. H. Nguyen, N. M. Tam, T. Y. Vu, N. T. Pham, N. T. Huy, B. K. Mai, N. T. Tung, M. Q. Pham, V. V. Vu and S. T. Ngo, J. Comput. Chem., 2021, 43, 160–169 CrossRef PubMed.
- J. Cheung, E. N. Gary, K. Shiomi and T. L. Rosenberry, ACS Med. Chem. Lett., 2013, 4, 1091–1096 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- A. E. Aliev, M. Kulke, H. S. Khaneja, V. Chudasama, T. D. Sheppard and R. M. Lanigan, Proteins: Struct., Funct., Bioinf., 2014, 82, 195–215 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, J. Comb. Chem., 1997, 18, 1463–1472 CAS.
- D. Hamelberg and J. A. McCammon, J. Am. Chem. Soc., 2004, 126, 7683–7689 CrossRef CAS PubMed.
- R. W. Zwanzig, J. Chem. Phys., 1954, 22, 1420–1426 CrossRef CAS.
- S. T. Ngo, Q. M. Thai, T. H. Nguyen, N. N. Tuan, T. N. H. Pham, H. T. T. Phung and D. T. Quang, RSC Adv., 2024, 14, 14875–14885 RSC.
- C. H. Bennett, J. Comput. Phys., 1976, 22, 245–268 CrossRef.
- A. Amadei, A. B. M. Linssen and H. J. C. Berendsen, Proteins: Struct., Funct., Genet., 1993, 17, 412–425 CrossRef CAS PubMed.
- X. Daura, K. Gademann, B. Jaun, D. Seebach, W. F. van Gunsteren and A. Mark, Angew. Chem., Int. Ed., 1999, 38, 236–240 CrossRef CAS.
- E. Papaleo, P. Mereghetti, P. Fantucci, R. Grandori and L. De Gioia, J. Mol. Graph. Model., 2009, 27, 889–899 CrossRef CAS PubMed.
- S. Lee, I. Lee, H. Kim, G. Chang, J. Chung and K. No, EuroQSAR 2002 Designing Drugs and Crop Protectants: Processes, Problems and Solutions, 2003, pp. 418–420 Search PubMed.
- P. Schrödinger LLC, Schrödinger Release 2020-4: Maestro, 2020 Search PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- C. A. Caffalette, R. A. Corey, M. S. Sansom, P. J. Stansfeld and J. Zimmer, Nat. Commun., 2019, 10, 1–11 CrossRef PubMed.
- W. R. Grither and G. D. Longmore, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E7786–E7794 CrossRef CAS PubMed.
- M. Noike, T. Matsui, K. Ooya, I. Sasaki, S. Ohtaki, Y. Hamano, C. Maruyama, J. Ishikawa, Y. Satoh and H. Ito, Nat. Chem. Biol., 2015, 11, 71–76 CrossRef CAS PubMed.
- L. Hong, G. Koelsch, X. Lin, S. Wu, S. Terzyan, A. K. Ghosh, X. C. Zhang and J. Tang, Science, 2000, 290, 150–153 CrossRef CAS PubMed.
- R. A. Friesner, J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis and P. S. Shenkin, J. Med. Chem., 2004, 47, 1739–1749 CrossRef CAS PubMed.
- Q. M. Thai, H. T. T. Phung, N. Q. A. Pham, J.-T. Horng, P.-T. Tran, N. T. Tung and S. T. Ngo, J. Biomol. Struct. Dyn., 2024, 1–9 CrossRef PubMed.
- E. Jurrus, D. Engel, K. Star, K. Monson, J. Brandi, L. E. Felberg, D. H. Brookes, L. Wilson, J. Chen, K. Liles, M. Chun, P. Li, D. W. Gohara, T. Dolinsky, R. Konecny, D. R. Koes, J. E. Nielsen, T. Head-Gordon, W. Geng, R. Krasny, G.-W. Wei, M. J. Holst, J. A. McCammon and N. A. Baker, Pro. Sci., 2018, 27, 112–128 CrossRef CAS PubMed.
- A. Ajay, G. W. Bemis and M. A. Murcko, J. Med. Chem., 1999, 42, 4942–4951 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: All-atom RMSD of simulated AChE/BACE-1 complexes; time dependence of energies for complexes; the coordinate of the minima corresponds to the most stable configuration; and ADMET profiles of the four tripeptides. See DOI: https://doi.org/10.1039/d5ra00709g |
| This journal is © The Royal Society of Chemistry 2025 |