Open Access Article
Open Access ArticleSynthesis, radiolabeling, and biological evaluation of methyl 6-deoxy-6-[18F]fluoro-4-thio-α-D-maltotrioside as a positron emission tomography bacterial imaging agent†
Kiyoko Takemiya *a,
Wonewoo Seobc,
Ronald J. Vollbc,
Sheng Zhaod,
Giji Josepha,
Shelly Wang
*a,
Wonewoo Seobc,
Ronald J. Vollbc,
Sheng Zhaod,
Giji Josepha,
Shelly Wang a,
Fanxing Zengbc,
Jonathon A. Nyebce,
Niren Murthy
a,
Fanxing Zengbc,
Jonathon A. Nyebce,
Niren Murthy d,
W. Robert Taylorafg and
Mark M. Goodman‡
bc
d,
W. Robert Taylorafg and
Mark M. Goodman‡
bc
aDivision of Cardiology, Department of Medicine, Emory University School of Medicine, 1750 Haygood Dr NE, Atlanta, Georgia 30322, USA. E-mail: kiyoko.takemiya@emory.edu
bDepartment of Radiology and Imaging Sciences, School of Medicine, Emory University, 1841 Clifton Road NE, Atlanta, Georgia 30322, USA
cCenter for Systems Imaging, Emory University, 1364 Clifton Rd NE, Atlanta, Georgia 30022, USA
dDepartment of Bioengineering, University of California at Berkeley, Stanley Hall 306, Berkeley, California 94720, USA
eDepartment of Radiology and Radiological Science, Medical University of South Carolina, 261 Calhoun Street, Charleston, South Carolina 29425, USA
fJoseph Maxwell Cleland Atlanta VA Medical Center, 1670 Clairmont Road, Decatur, Georgia 30033, USA
gWallace H. Coulter Department of Biomedical Engineering, School of Medicine, Emory University, 1750 Haygood Dr NE, Atlanta, Georgia 30322, USA
First published on 21st March 2025
Abstract
We developed fluorine-18 ([18F]) labeled methyl 6-deoxy-6-fluoro-4-thio-α-D-maltotrioside ([18F]MFTMT) for bacterial imaging and evaluated its stability and efficacy in vitro and in vivo. We found that Staphylococcus aureus (S. aureus) internalized [18F]MFTMT whereas Escherichia coli (E. coli) and CHO-K1 cells did not, in in vitro. Positron emission tomography imaging with [18F]MFTMT revealed that radioactivity accumulated not only in the S. aureus-infected group but also in the E. coli-infected and non-infectious inflammation groups. Further studies revealed that rat serum digested [18F]MFTMT into [18F]-methyl 6-deoxy-6-fluoro-4-thio-α-D-maltoside ([18F]MFTM), while [18F]MFTMT was stable in human serum for 210 min. [18F]MFTM was identified as the only radioactive metabolite in vivo. Similar to [18F]MFTMT, [18F]MFTM was internalized only by S. aureus. [18F]MFTM was identified as the only radioactive metabolite in vivo. We found that the sodium–glucose co-transporter 1 (SGLT1) is expressed in inflammatory tissue, and SGLT1 overexpressing cells showed increased retention of [18F]MFTMT and [18F]MFTM in vitro. Our study showed that the thio-glycosyl bond is stable against enzymatic digestion, and maltotetraose or a longer maltodextrin backbone is desirable for bacteria-specific imaging to avoid nonspecific uptake by SGLT1.
Introduction
Bacterial infections pose formidable health challenges, often leading to severe tissue damage and necessitating major surgical interventions.1–7 Diseases such as endocarditis and infections associated with medical devices are particularly concerning, underscoring the critical need for accurately localizing bacteria within target organs or tissues to facilitate their diagnosis and treatment.6,8–14 Early detection of bacterial infections using non-invasive methods could enable the timely administration of antibiotic therapy, potentially preventing tissue damage and reducing treatment costs.Currently, no established non-invasive diagnostic methods specifically detect the existence of bacteria in deep organs and/or tissue directly. Non-invasive diagnostic methods, such as computed tomography (CT) and ultrasound, detect the tissue damage induced by bacterial infection, and contrast CT and 2-deoxy-2-[18F]fluoro-D-glucose ([18F]FDG) positron emission tomography (PET) imaging detect the increase of blood flow and glucose uptake induced by inflammation.15 However, there can be false positive results from these tests because of sterile inflammation. Thus, for the specific detection of bacterial infection, new methods to detect the existence of bacteria are required.
We have been focusing on the differences in sugar metabolism between bacteria and mammalian cells. Bacteria utilize maltose and maltodextrins via the maltodextrin transporter and phosphotransferase system (PTS) as both are specific and ubiquitous expressed in bacteria. Mammalian cells do not have maltodextrin transporter nor PTS, and they do not internalize maltose and maltodextrins. Comparing the maltodextrin transporter and PTS, the maltodextrin transporter has tremendous potential for in vivo bacterial imaging16–19 because of its specific expression in bacteria and presence in both Gram-positive and Gram-negative bacteria and because of its capacity to transport a molecule conjugated to maltodextrin.20–24 The maltodextrin transporter actively transports maltodextrins up to maltoheptaose and can recognize imaging molecules conjugated to the maltodextrin backbone.16–23,25 Several studies have validated the efficacy of maltodextrin-based imaging probes in targeting the maltodextrin transporter and visualizing bacteria in vivo.16–19,25–27 For example, probes incorporating a maltohexaose backbone conjugated with imaging molecules at the anomeric carbon demonstrated high sensitivity and specificity in both in vitro and in vivo bacterial imaging scenarios.16–19
Among many imaging modalities, such as PET, CT and magnetic resonance imaging (MRI), the radioactive tracers are known to be more sensitive than other modalities.28–30 Fluorine-18 ([18F]) can replace the hydroxyl group by derivatization of maltodextrins or can be conjugated with a linker molecule.17,18,31 We have shown that [18F] conjugated maltohexaose ([18F]MHF) accumulates only in bacterial infection lesions and does not accumulate in non-infectious inflammation lesions.17,18 In contrast, [18F]FDG accumulates in both bacterial infection lesion and non-infectious inflammation lesion.18 Our previous studies demonstrated the potential of maltohexaose-based imaging probes for the specific detection of bacterial infections.17–19
One limitation of the maltohexaose backbone lies in its susceptibility to degradation via serum amylases and α-glucosidases, generating various sugars that may be internalized by mammalian cells, causing nonspecific uptake of labeled metabolites, such as glucose and maltose.32–36
To enhance the stability of the maltodextrin backbone, we propose incorporating a thio-glycosyl bond to confer resistance against enzymatic digestion to improve stability of the maltodextrin backbone. This study focused on the development of fluorine-18 labeled methyl 6-deoxy-6-fluoro-4-thio-α-D-maltotrioside ([18F]MFTMT) and assessed its serum stability. Additionally, we investigated the uptake of [18F]MFTMT by bacteria in vitro and assessed its efficacy in detecting bacterial infections in vivo using a device infection model.
Results
Chemistry
The synthesis of the cold standard methyl 6-deoxy-6-fluoro-4-thio-α-D-maltotrioside (6) and the synthesis of the [18F]-labeling precursor methyl 6-(4-bromobenzenesulfonyloxy(brosyloxy))-4-thio-α-D-maltotrioside (10) is described in Schemes 1 and 2, respectively. The main advantage for the 4-bromobenzenesulfonyloxy ester is that nucleophilic substitution proceeds more efficiently compared to the use of the corresponding 4-methylbenzenesulfonyloxy ester.37Methyl 6-deoxy-6-fluoro-4-thio-α-D-maltotrioside 6 (MFTMT) was synthesized in a four-step reaction sequence starting from methyl 6-deoxy-6-fluoro-α-D-galactopyranose38 1 (Scheme 1). Reaction of 1 with benzoyl chloride in pyridine afforded the 2,3-dibenzoyl compound 2. Treatment of 2 with triflic anhydride in pyridine gave the triflate 3. Reaction of 3 with 2,3,6-tri-O-acetyl-S-acetyl-(2,3,4,6-tetra-O-acetyl-S-acetyl-1-thio-α-D-glucopyranosyl)-α-D-glucopyranose39 4 according to the literature procedure40 afforded 4-thio-maltotrioside 5. Removal of the acetate and benzoate groups by treatment with NaOCH3–CH3OH gave 6 followed by purification on silica resulting in a 51% yield. Compound 6 was prepared in 9% overall yield from compound 1.
We employed a similar route to synthesize the [18F]-labeling precursor methyl 6-brosyloxy-4-thio-α-D-maltotrioside 10 (Scheme 2). Methyl 2,3-di-O-benzoyl-4-trifluoromethylsulfonyl-6-O-trityl-α-D-galactopyranoside41 7 was treated with 2,3,6-tri-O-acetyl-S-acetyl-(2,3,4,6-tetra-O-acetyl-S-acetyl-1-thio-α-D-glucopyranosyl)-α-D-glucopyranose to afford the trityl ether 8. A sequential reaction of 8 with aqueous acetic acid and brosyl chloride afforded the C-6 brosyloxy precursor 10 resulting in a 65% yield.
We next turned our attention to the synthesis of methyl 6-deoxy-6-fluoro-4-thio-α-D-maltoside. It was anticipated that this compound may be the product of rat serum α-glucosidase metabolism of methyl 6-deoxy-6-fluoro-4-thio-α-D-maltotrioside. Methyl 6-deoxy-6-fluoro-4-thio-α-D-maltoside 13 was synthesized in a two-step reaction sequence starting from methyl 2,3-di-O-benzoyl-4-O-trifluoromethylsulfonyl-6-deoxy-6-fluoro-α-D-galactopyranoside 3 (Scheme 3). Reaction of 3 with 2,3,4,6-tetra-O-acetyl-S-acetyl-1-thio-α-D-glucopyranose42 11 according to the literature procedure40 afforded 4-thio-maltoside 12 in 59% yield. Removal of the acetate and benzoate groups by treatment with NaOCH3–CH3OH gave 13 following purification on silica with a 99% yield.
Radiochemistry
The automated synthesis of [18F]MFTMT ([18F]6) (Scheme 4) was carried out in a cGMP compliant device, the GE TracerLab FX 2N. Compound [18F]6, ([18F]MFTMT) was obtained by sequentially reacting 10 with K18F/Kryptofix-222, complex in CH3CN at 100 °C, followed by the removal of the acetate and benzoate groups of [18F]14 with using sodium ethoxide (NaOEt–)–ethanol (EtOH) at 100 °C. Following the removal of the maltriose-protecting groups, the reaction mixture was neutralized, and [18F]MFTMT was purified by semi-preparative high-performance liquid chromatography (HPLC) using 6% EtOH–water as the eluent. The product was sterilized by passing through a Millex®-GS sterile filter (0.22 μm). A total of 188 mCi (6956 MBq) of [18F]MFTMT in a decay-corrected radiochemical yield (RCY) of 20% was obtained from 1.56 Ci, 57.72 GBq of [18F]-fluoride in a synthesis time of 80 min. The purified final product [18F]MFTMT was diluted in a saline solution (pH = 6) and used for in vitro and positron emission tomography (PET) imaging studies. Radiochemical and chemical purity were >95% and the specific activity = 1.9 Ci (70.3 GBq) per μmol (ESI Fig. S1†).[18F]MFTMT was identified by co-injection with non-radiolabeled MFTMT ([19F]MFTMT (6)) by HPLC analysis using a Waters reverse phase analytical HPLC column (ESI Fig. S2†).
Biological evaluation of [18F]MFTMT
To understand biological characteristics of [18F]MFTMT, we evaluted the uptake of [18F]MFTMT by bacteria and mammlian cells in vitro. The efficacy of [18F]MFTMT to detect bacterial infection was determined by [18F]MFTMT PET imaging.Bacteria uptake assays
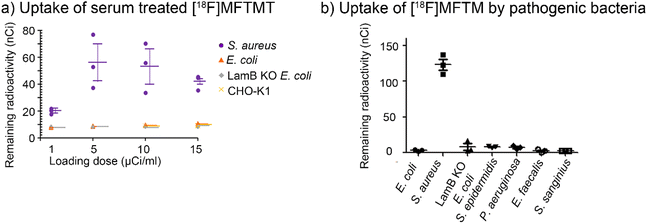
The affinity of [18F]MFTMT was evaluated using bacteria and mammalian cells. Fig. 1a shows that S. aureus internalized [18F]MFTMT, whereas E. coli did not. The numerical values of uptake of radioactivity by bacteria and CHO-K1 cells are shown in ESI Table S1.† The level of radioactivity internalized by 1 × 108 colony-forming units (CFU) of S. aureus after incubation with 5 to 20 μCi mL−1 of [18F]MFTMT was increased with the loading radioactivity. The Km and Vmax of S aureus with [18F]MFTMT was calculated as Km = 151.2 ± 224.7 nCi and Vmax = 18.1 ± 24.3 nCi min−1/108 CFU by using the Prism software. In contrast, the observed radioactivity in E. coli, LamB KO E. coli, and CHO-K1 cells was nearing the detection limitation of radioactivity even after incubation with 20 μCi mL−1 of [18F]MFTMT. These results showed that while S. aureus internalized [18F]MFTMT, E. coli and mammalian cells internalized minimal amounts of this compound.We further evaluated if other pathogenic bacteria for implantable medical device infections internalized [18F]MFTMT.43–45 As shown in Fig. 1b, S. aureus significantly internalized [18F]MFTMT (133.3 ± 10.0 nCi). On the other hand, the retained amounts of [18F]MFTMT in bacteria other than S. aureus were small, and no significant differences were observed. These results showed that [18F]MFTMT specifically accumulates in S. aureus but not in other bacteria. The numerical values of uptake of radioactivity by bacteria are shown in ESI Table S2.† showed that while S. aureus internalized [18F]MFTMT, E. coli and mammalian cells internalized minimal amounts of this compound.
We further evaluated if other pathogenic bacteria for implantable medical device infections internalized [18F]MFTMT.43–45 As shown in Fig. 1b, S. aureus significantly internalized [18F]MFTMT (133.3 ± 10.0 nCi). On the other hand, the retained amounts of [18F]MFTMT in bacteria other than S. aureus were small, and no significant differences were observed. These results showed that [18F]MFTMT specifically accumulates in S. aureus but not in other bacteria. The numerical values of uptake of radioactivity by bacteria are shown in ESI Table S2.†
In the uptake studies with bacteria and CHO-K1 cells, both [18F]MFTMT showed high selectivity for S. aureus compared with mammalian cells and pathogenic bacteria other than S. aureus.
In vivo PET imaging of rat skin pocket infections using [18F]MFTMT
We evaluated the accumulation of [18F]MFTMT in a rat skin-pocket-infection model.18 To examine whether the selectivity of [18F]MFTMT for S. aureus is maintained in vivo, we used four groups, control, E. coli infected, S. aureus infected and non-infectious inflammation groups. At the time of PET imaging, the body weight of rats was 298.0 ± 3.6 g and the amount of injected radioactivity per gram was 0.84 ± 0.02 μCi g−1. The body weight and dosage of radioactivity showed no significant differences between the four groups (ESI Fig. S11†).To understand the basic characteristics of [18F]MFTMT PET imaging, we evaluated the distribution of radioactivity in the control rats. The accumulation of radioactivity in the major organs and tissue was calculated as standardized uptake value (SUV), and the SUV of the organs and tissue were compared. The kidneys showed the highest accumulation which was significantly increased compared with other organs and tissue, suggesting that the radiotracer is excreted via the kidneys. Among the organs and tissue excluding the kidney, the liver showed significantly increased accumulation of radioactivity compared with other organs and tissue (ESI Fig. S12†). These results suggest that the majority of [18F]MFTMT and its metabolite were excreted into urine via the kidneys and feces via the liver. Other than these metabolic organs, the brain, skeletal muscles, and the subcutaneous fat and soft tissue showed significantly reduced accumulation compared with the blood. In our studies, the ratio of free [18F] was less than 4%, thus, although some non-specific accumulation in the bones was observed, it was not significantly increased compared to the radioactivity in the blood. These findings show that the distribution of radioactivity derived from [18F]MFTMT is not evenly distributed in the body and that to evaluate the accumulation of the tracer induced by bacterial infection, the radioactivity in the target lesion should be compared with that in the non-infected area of the same organ/tissue.
Despite demonstrating specificity in in vitro uptake studies with bacteria and mammalian cells, Fig. 2a shows the accumulation of radioactivity not only in the S. aureus-infected group but also in the E. coli-infected and non-infectious inflammation groups. The PET imaging of coronal and sagittal views is shown in the ESI Fig. S12.† The accumulation of radioactivity in mock-up areas were compared using the ratio of SUV of the mock-up area to that of the normal skin area. The average accumulation of radioactivity in the mock-up area (SUV mean ratio) was significantly increased in the E. coli-infected, S. aureus-infected, and non-infectious inflammation groups compared with the control group (Fig. 2b). The highest radioactivity in the region of interest (ROI) (SUV max ratio) in the S. aureus-infected group was significantly increased compared to the control group, representing a 1.49-fold increase. The SUV max ratios of the E. coli-infected and non-infectious inflammation groups did not significantly differ from the S. aureus-infected group. The radioactivity in the normal skin area did not differ among these four groups. The numeric values of PET imaging analysis are shown in ESI Table S3.† These results suggest that both S. aureus and the inflammatory tissue of rats accumulated [18F]MFTMT and/or its metabolite(s).
 | ||
| Fig. 2 [18F]MFTMT PET imaging of device infection model in rats. (a) [18F]MFTMT PET imaging was conducted on a skin-pocket-infection model in rats, utilizing sum images from 30 to 60 min for evaluation. The location of the mock-up was confirmed by merging images of PET and CT. Coronal and sagittal sections of PET images are shown in the ESI Fig. S12.† No radioactivity accumulation was observed in the mock-up area in the control rats. Radioactivity accumulated not only in the mock-up area in S. aureus-infected rats but also in E. coli-infected rats and rats with non-infectious inflammation. Arrows show mock-up areas. (b) Radioactivity accumulation around the mock-up area and normal skin was compared using the ratio of SUV in the mock-up area and normal skin area. The mean SUV ratio in the mock-up area was significantly increased in S. aureus-infected rats, E. coli-infected rats, and non-infectious inflammation rats compared to control rats. SUV max ratio was increased in S. aureus-infected rats. SUVs in the normal skin area were similar across the four groups. The remaining radioactivity in the blood was evaluated by placing the ROI on the left ventricle, indicated as a ratio to the injected radioactivity (% ID per g tissue). * Indicates p < 0.05. | ||
Blood radioactivity was also assessed by placing the ROI within the left ventricle in a coronal view to estimate blood radioactivity retention in rats. The remaining radioactivity in the blood was calculated as a ratio relative to the injection dose per gram (% ID per g tissue). At the time of imaging, the remaining radioactivity in the blood showed no significant difference in the four groups (Fig. 2b).
Our previous study with the same device infection model showed that [18F]FDG significantly accumulated both in the S. aureus infected and non-infectious inflammation lesions compared with control lesions (SUV mean ratios 2.86, 2.62 and 1.54, respectively).18 On the other hand, [18F]MHF accumulated only in the S. aureus infected lesion compared with non-infectious inflammation and control lesions (SUV mean ratio 2.87, 1.26 and 1.61, respectively), and different from [18F]FDG PET imaging, [18F]MHF showed obvious selectivity for bacteria.18 To explore why [18F]MFTMT lost the selectivity to S. aureus in in vivo imaging, we further investigate the in vitro and in vivo metabolites of [18F]MFTMT and possible transporters in mammalian cells.
In vitro and in vivo metabolite(s) of [18F]MFTMT
Uptake of serum treated [18F]MFTMT and [18F]MFTM by bacteria
We investigated whether [18F]MFTM, the metabolite of [18F]FMTMT, was still selective for S. aureus. Fig. 5a shows the uptake of serum treated [18F]MFTMT by bacteria and CHO-K1 cells. The numerical values of uptake of radioactivity by bacteria and CHO-K1 cells are shown in ESI Table S4.† Similar to non-processed [18F]MFTMT, S. aureus internalized serum treated [18F]MFTMT. Because of the large dispersion of data at each concentration, the kinetics could not be calculated. On the other hand, E. coli, LamB KO E. coli, and CHO-K1 cells showed minimal internalization of radioactivity even when incubated with 15 μCi mL−1 of serum-treated [18F]MFTMT. These results show that both [18F]MFTMT and its serum metabolite, [18F]MFTM, are only internalized by S. aureus.We also evaluated whether [18F]MFTM was internalized common pathogens for implantable device infections. The purity of [18F]MFTM was confirmed by HPLC (ESI Fig. S11†). Fig. 5b shows that similar to [18F]MFTMT, S. aureus significantly internalized [18F]MFTM (123.1 ± 7.7 nCi), but bacteria other than S. aureus internalized little of [18F]MFTM with no significant differences in each other. The numeric values of uptake of [18F]MFTM are shown in the ESI Table S5.† These results showed that the selectivity of [18F]MFTM to S. aureus.
Sodium glucose co-transporter (SGLT) 1 is the potential cause of [18F]MFTMT accumulation in inflamed tissue
We explored the underlying reasons for the accumulation of radioactivity derived from [18F]MFTMT in mammalian tissue. [18F]MFTMT and its metabolite, [18F]MFTM, possess glucoside structures. Therefore, we hypothesized that glucose transporters might be involved in the unexpected accumulation of radioactivity observed in E. coli-infected rats and rats with non-infectious inflammation.In mammalian tissue, glucose is transported through glucose transporters (GLUTs) and sodium–glucose co-transporters (SGLTs).46–48 GLUTs are widely expressed and transport monosaccharides, such as glucose and fructose, while they do not transport disaccharides and longer maltodextrins.48–50 GLUTs are also sensitive to the modification on the anomeric carbon and do not transport α-methyl-D-glucoside.46,51 Conversely, SGLTs are expressed in specific tissues and organs, such as the intestine, kidneys, and liver.52–56 As shown in Fig. 3a, radioactivity accumulation was localized in the inflammatory area, liver, and kidneys, suggesting that GLUTs might not contribute to this unexpected radioactivity accumulation. Thus, we evaluated whether SGLTs, the alternative glucose transport system in mammalian tissue, were expressed in the region around the mock-ups, and whether SGLT1 and SGLT2 interact with [18F]MFTMT and [18F]MFTM, the metabolite of [18F]MFTMT.
Expression of SGLT1 and SGLT2 around the mock-up area
We evaluated the expression of SGLT1 and SGLT2 around the mock-ups by immunohistochemistry and quantitative polymerase chain reaction (qPCR). Fig. 6a and b show that both SGLT1 and SGLT2 are highly expressed in fibroblasts (vimentin-positive cells) within the inflammatory area, respectively.In qPCR analysis, the expression of SGLT1 and SGLT2 was evaluated by expression relative to β-actin. As shown in Fig. 7, the messenger ribonucleic acid (mRNA) for SGLT1 and SGLT2 in the inflammatory tissues were significantly increased compared to the tissue from the control animals.
Interaction of [18F]MFTMT and [18F]MFTM with SGLTs
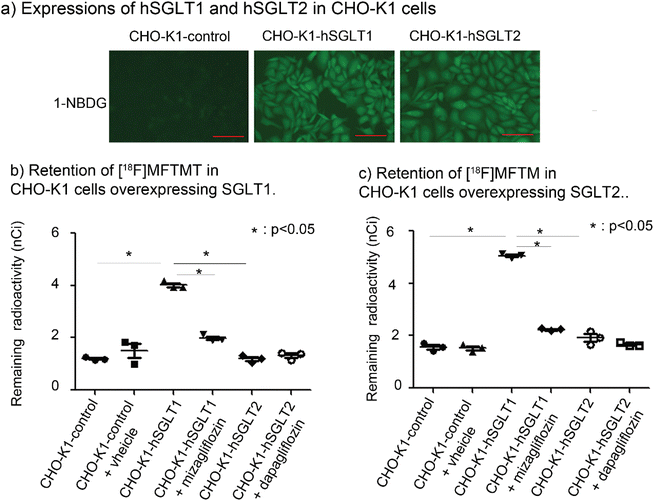
To investigate the potential involvement of SGLTs in the accumulation of radioactivity derived from [18F]MFTMT, we established CHO-K1 cells expressing human SGLT1 (CHO-K1-hSGLT1) and SGLT2 (CHO-K1-hSGLT2).57–59 The functional expression of these genes was confirmed by measuring the uptake of 1-NBDG, a fluorescent substrate specific for SGLT1 and SGLT2 (Fig. 8a).60As shown in Fig. 8b, the remaining radioactivity in CHO-K1-hSGLT1 cells incubated with [18F]MFTMT was 4.0 nCi which was significantly higher than that in control CHO-K1 cells and CHO-hSGLT2 cells. Similar to the cells exposed to [18F]MFTMT, the remaining radioactivity in the CHO-K1 hSGLT1 cells incubated with [18F]MFTM was 5.1 nCi which was significantly increased compared with CHO-K1 control and CHO-K1 hSGLT2 cells (Fig. 8c). The retention of [18F]MFTMT and [18F]MTM in CHO-K1-hSGLT1 cells was approximately 3.3 times and 3.2 times that of control CHO-K1 cells, respectively. Moreover, the retention of radioactivity in CHO-K1 hSGLT1 cells treated with mizagliflozin, a specific SGLT1 inhibitor, was significantly suppressed (Fig. 8b and c). The numeric values of remaining radioactivity in the CHO-K1 cells are shown in ESI Table S6.†
These results demonstrate that SGLT1 is highly expressed in inflammatory tissue and is potentially involved in the accumulation of [18F]MFTMT and [18F]MFTM, likely elucidating the loss of specificity of [18F]MFTMT and [18F]MFTM observed in vivo despite its specificity in vitro.
Discussion
We synthesized [18F]MFTMT and assessed its uptake by bacteria, its stability against enzymatic digestion, and its effectiveness at detecting bacterial infections both in vitro and in vivo.Inflammatory tissue, serum, and the drug-metabolizing organs, the liver and kidneys. Our findings suggest that α-glucosidase can digest the glycosyl bond, while the thio-glycosyl bond demonstrates resistance to enzymatic digestion for at least 1 hour.
Our in vitro uptake experiments revealed that S. aureus internalized both [18F]MFTMT and [18F]MFTM, on the other hand, other bacteria and mammalian cells internalized minimal amounts of [18F]MFTMT and [18F]MFTM. [18F]MFTMT has three characteristic modifications from maltotriose, a (1–4)-thio-glycosyl bond, 6-deoxy-6-fluoro modification, and -O-methyl modification on the anomeric carbon. [18F]MFTM, the metabolite of [18F]MFTMT also has the same modifications. Among the bacteria used for this study, the maltose/maltodextrin transport systems in E. coli have been extensively studied providing insight into how these modifications can affect transport into E. coli.20–22,61–79 In E. coli, the maltoporin, LamB, situated on the outer membrane, initially allows the permeation of maltodextrins into the periplasmic space.67–74 Inside the periplasmic space, the maltose/maltodextrin-binding protein, malE, binds to maltodextrins (forming malE-maltodextrin complex).62–64,66,67 Subsequently, the malE-maltodextrin complex associates with the maltodextrin transporter (malF–malG–malK complex) on the inner membrane, facilitating maltodextrin transport into the cytoplasm.62,63 LamB and malF in the malF–malG–malK complex recognize the non-reducing end of maltodextrins, while malE recognizes its reducing end.62–64,66–74 Studies have shown that modifications on the reducing end of maltodextrin can considerably influence malE binding.62–65 Our first generation [18F]-maltodextrin probe, [18F]MHF, featuring [18F]fluorine attached via a β-glycosidic linkage on the anomeric carbon at the reducing end of maltohexaose, was internalized by both S. aureus and E. coli.16–18 This suggests that malE of E. coli may tolerate only one modification on the anomeric carbon at its reducing end. MalE of E. coli binds to maltose and maltodextrins up to maltoheptaose.67,80,81 Crystallography studies showed that malE of E. coli generates hydrogen bonds with the hydroxyl groups on glucose molecule at the reducing end and the hydroxyl groups on 2, 3, and 6 positions of the glucose molecule next to the reducing end.62,65 When malE of E. coli binds to maltotriose or longer maltodextrins, the number of hydrogen bonds generated between malE of E. coli and maltodextrins increases, while the binding positions malE of E. coli and two glucose molecules from the reducing end are maintained.62,65 This binding to maltose or two glucose molecules from the reducing end induces conformational change of malE of E. coli, making malE of E. coli the “closed” conformation.82,83 MalE of E. coli also known to bind modified maltodextrin analogs such as maltotetraiol. However, the binding conformation remains in the “open” form which is considered as non-functional for transport.64 The carbon–sulfur bond distance in thio-glycosyl bond is longer than the carbon–oxygen bond in the glycosyl bond, resulting in a distinct relative position of the glucose molecule next to the reducing end glucose molecule compared to maltose and maltodextrins.84,85 Consequently, with the 6-deoxy-6-fluoro modification, [18F]MFTMT might exhibit reduced affinity to the malE of E. coli.
PTS of E. coli also transports maltose from the periplasmic space to the cytosol.75–79. [18F]MFTM, the radioactive metabolite of [18F]MFTMT and a maltose analog, was considered as the potential substrate for PTS. However, in PTS, the enzyme IIC component (EIIC) corresponding to maltose is located on the inner membrane, and a crystallography study by McCoy et al. showed that it recognizes the hydroxyl groups on the 6- and 6′-carbons.77–79 Thus, 6-deoxy-6-fluoro-modification in [18F]MFTM, the maltose analog, appears to be critical for binding to ECII for maltose.
The maltose/maltodextrin transport systems, maltodextrin transporter and PTS, in pathogenic bacteria other than E. coli still need to be elucidated. However, our results suggest that the maltodextrin binding protein and ECII for maltose in the evaluated bacteria, Pseudomonas aeruginosa, Staphylococcus epidermidis, Streptococcus sanguinis and Enterococcus faecalis, may also be sensitive to the modifications on the reducing end.
In contrast, S. aureus internalized both [18F]MFTMT and [18F]MFTM. This finding implies that the maltose and maltodextrin transportation systems in S. aureus employ different substrate-binding mechanisms from that of E. coli. Abbott et al. elucidated sugar utilization in Streptococcus pneumoniae and demonstrated that the maltodextrin binding protein of Streptococcus pneumoniae binds to maltotriose and longer maltodextrins. They also showed that PTS transports glucose, maltose and maltotriose, and that the binding conformation of the maltodextrin binding protein with maltoheptaose is different compared to malE in E. coli.23 We recently found that the maltodextrin-binding protein of S. aureus exhibits different binding thermodynamics compared with that of E. coli and like that of S. pneumonia and that it functionally binds to maltotriose and longer maltodextrins.81 Other studies also show that maltodextrin binding proteins of Gram-positive bacteria bind to maltotriose and longer maltodextrins, and PTS of Gram-positive bacteria transports glucose, maltose and maltotriose.86–91 These findings suggest that [18F]MFTMT can be transported via maltodextrin transporter and PTS, and [18F]MFTM can be transported by PTS in S. aureus. We did not specify whether the maltodextrin transporter or PTS in S. aureus internalized [18F]MFTMT and/or its metabolite, [18F]MFTM. However, our results suggest that maltodextrin transporter and/or PTS in S. aureus are tolerant to the modifications on the reducing end and have different binding affinities and mechanisms to substrates compared to those in E. coli. Our results indicated that the tolerance of maltodextrin transporter and PTS to the modifications on maltose/maltodextrins is different in bacterial species. This finding suggests the possibility to target specific bacterial species using modifications of maltodextrin-based probes. Future studies will explore potential differences in maltodextrin transporters between S. aureus, other pathogenic bacteria and E. coli.
[18F]FDG is assumed to be uniformly distributed in the body with increased accumulation of radioactivity indicating increased glucose uptake caused by pathological or physiological changes, such as tumors, inflammation, and muscle activities.92,93 In contrast, the concept of maltodextrin-based imaging probes is that maltodextrin-based imaging probes are only internalized by bacteria and not by mammalian cells.16–18 The injected [18F] labeled maltodextrin-based imaging probes are expected to be rapidly distributed from the blood to interstitial fluid, accumulate in the metabolic organs, and are excreted into urine without being internalized by other tissue and organs. As a result, the remaining radioactivity in the blood and interstitial fluid, which is detected as background is very low, emphasizing the accumulation of radioactivity in bacteria, even if the absolute quantity of radioactivity is small. Therefore, evaluating and understanding whether and how [18F] labeled maltodextrin-based imaging probes and their metabolites accumulate in normal and inflammatory tissue in in vivo is important. To clarify the accumulation of radioactivity in the target area, we used the SUV ratios, the relative the accumulation of radioactivity in the target tissue to the normal tissue, to compare the accumulation of radioactivity between the experimental groups.
We evaluated whether [18F]MFTMT and [18F]MFTM accumulated in mammalian tissue. In [18F]MFTMT PET imaging, we observed radioactivity accumulation not only in the S. aureus-infected group but also in the E. coli-infected and non-infected inflammation groups. Our experiments strongly suggest the involvement of SGLT1, which is highly expressed in inflammatory tissues as the cause of this false positive result. Studies showed that SGLT1 is expressed not only in the inflammatory cells but also in endothelial cells and fibroblasts and that SGLT1 is involved in inflammatory reactions.94–98 In the in vitro uptake studies, when 20 μCi mL−1 of [18F]MFTMT and [18F]MFTM were loaded, the retained radioactivity in 1.2 ×105 of CHO-K1-SGLT1 cells was about 4.0 nCi, and 5.1 nCi, respectively, which was lower than that in 1 × 108 CFUs of S. aureus (124 nCi). The concentration of radioactivity in the in vivo setting is lower than 20 μCi mL−1, and it can be estimated as following. The percentage of extracellular water, including plasma and interstitial fluid, in adult rat is estimated approximately 30%.99 Thus, in our in vivo experiments, when the injected radioactivity was evenly distributed in the extracellular water in the body, the concentration of radioactivity is estimated to be 2.8 μCi mL−1. Assuming that the accumulation of radioactivity in S. aureus and CHO-K1-hSGLT1 cells obey Michaelis–Menten kinetics in which the reaction speed and substrate concentrations interacts linearly in low concentrations, the retention of [18F]MFTMT and [18F]MFTM in 1.2 ×105 of SGLT1 expressing cells is estimated to be 0.56 and 0.71 nCi, respectively, and the accumulation of [18F]MFTMT in 1 × 108 CFUs of S. aureus is estimated to be 20 times higher at 17.6 nCi. However, our study showed that inflammation increases fibroblasts expressing SGLT1 potentially leading to nonspecific accumulation of [18F]MFTMT and/or [18F]MFTM. This is plausible considering the high cell density of the skin at approximately 7 × 108 cells per cm3.100 If 1% of cells in inflammatory tissue expresses SGLT1, the retention of [18F]MFTMT and [18F]MFTM is estimated as 32.7 and 41.7 nCi g−1 tissue, respectively. Our results from immunohistochemistry and qPCR studies show that the increase of the ratio of SGLT1 expressing cells to the total cells in the inflammatory tissue is more than 10% compared to the control tissue, suggesting that the inflammatory tissue retains 327 to 417 nCi g−1 higher radioactivity compared with control tissue. Our skin pocket infection model contains 2.9 × 108 CUF/total infected tissue of S. aureus, and the expected radioactivity in the S. aureus infected tissue is estimated to be about 51 nCi/total infected tissue. Considering the weight of S. aureus infected tissue is more than 1 g and [18F]MFTM has higher retention in SGLT1 overexpressing cells, the increase of radioactivity density in inflammatory tissue caused by SGLT1 might be more than 10 times compared to that by S. aureus infection. As a result, the accumulation of radioactivity in S. aureus infected area did not show a significant difference from that in other inflammation areas induced by oil of turpentine and E. coli infection. We showed that inflammatory tissue expresses SGLT1, and SGLT1 is known to be expressed in many cancers.101–103 Future study needs to ivestigate whether [18F]MFTMT accumulates in cancers.
In our previous studies with [18F]MHF, radioactivity accumulation was only observed in the bacteria-infected area.16–18 In [18F]MHF PET imaging with the same pocket infection model induced by S. aureus and non-infectious inflammation model, the SUV mean ratio in S. aureus infected group was significantly increased compared to control and non-infectious inflammation groups and was 1.8 times to that in control group. In contrast, [18F]FDG PET imaging with the same models, the accumulation of radioactivity in both S. aureus infected and non-infectious inflammation lesions were significantly increased compared with that in control lesion. Different from [18F]MFTMT, [18F]MHF showed efficacy in in vivo bacteria-specific PET imaging compared with the conventional [18F]FDG PET imaging.
There are several notable differences between [18F]MFTMT and [18F]MHF.16–18. [18F]MHF has a maltohexaose backbone and can be digested into [18F]fluoro-maltoside and [18F]fluoro-maltotrioside by serum amylase and further into [18F]fluoro-glucoside by serum α-glucosidase, which is highly active in rodents.16–18,33,35,36 These metabolites, including [18F]fluoro-glucoside, are not transported by GLUTs on mammalian cells because of the modification on the anomeric carbon.46,48,50 While SGLTs can tolerate modifications on the anomeric carbon, the extended maltohexaose backbone of [18F]MHF might prevent its interaction with SGLT1.48,104–108 Despite this, the flexibility and the size of the [18F]MHF linker could potentially allow for [18F]fluoro-glucoside, resulting from [18F]MHF degradation by serum α-glucosidase, to be transported by SGLTs. However, [18F]MHF and its metabolites are swiftly excreted in the urine, leading to a rapid decline in blood radioactivity levels and minimal radioactivity accumulation in non-infectious inflammation areas.17 Conversely, in [18F]MFTMT PET imaging, blood radioactivity is sustained as compared to [18F]MHF, with blood radioactivity reaching approximately 0.15% ID per g, which is more than 2.5-fold higher than that observed with [18F]MHF in our previous study.17 This difference suggests that the thio-glucosyl bond and the modification at the sixth position on the reducing end glucose in [18F]MFTMT might delay its excretion and/or that of its metabolites. Furthermore, differences in the biological half-life of radioactivity might also contribute to increased radioactivity accumulation in inflammatory tissue. These results demonstrate the necessity of employing maltodextrin backbones longer than maltotriose combined with modifications to prevent enzymatic digestion, in the development of bacteria-specific imaging probes. Considering the molecular size, maltodextrin backbones with nitrogen-glycosyl bond(s) might achieve both resistance to enzymatic digestion and increased affinity for bacteria.85
Limitations
One limitation of this study is that we did not determine whether [18F]MFTMT and [18F]MFTM were transported through SGLT1 or whether they were bound to SGLT1 and were internalized by SGLT1 via endocytosis. Further studies will investigate the interactions between [18F]MFTMT and [18F]MFTM, and native maltose and maltodextrins with SGLT1. These studies will help us to optimize the maltodextrin backbone for selective bacterial imaging.Another limitation is the use of a rat model for [18F]MFTMT PET imaging. We showed that [18F]MFTMT was digested into [18F]MFTM by rat serum while it was stable in human serum. We conducted [18F]MFTMT PET imaging to evaluate the specificity for bacteria in vivo because [18F]MFTM was the only radioactive metabolite of [18F]MFTMT in vitro and because [18F]MFTMT and [18F]MFTM had similar selectivity to bacteria in vitro. Rodent models are well-established and cost-effective methods to evaluate the efficacy of drugs, including imaging probes. However, understanding the differences between humans and rodents is important to evaluate the effects of the compounds. Future studyies may use other animal models or α-glucosidase inhibitor treated rats to evaluate the efficacy of maltodextrin-based imaging probes in vivo.
Experimental section
General information
All solvents were purchased from Biosynth, Fisher Scientific, or Sigma Aldrich and dried over 4 Å molecular sieves (8–12 mesh, Sigma Aldrich). Unless otherwise specified, all commercially available reagents and substrates were used as received. Ultrahigh purity dry air was purchased from nexAir LLC. Thin-layer chromatography (TLC) was performed on Merck silica gel plates and visualized under UV light and/or H2SO4–EtOH. 1H, 13C, and 19F NMR spectra were recorded on Varian Mercury 300, Bruker 600, Varian INOVA 600, INOVA 500, and INOVA 400 spectrometers. Residual solvent resonances were considered internal reference signals. 19F spectra were referenced to KF. Product purification was performed via flash chromatography109 unless otherwise specified. The purity and identity of compounds 2, 3, 5, 6, 8, 9, 10, 12, and 13 were determined by analytical and spectroscopical methods such as TLC, NMR, and high-resolution mass spectrometry (HRMS), and the results are shown in the Experimental Section. All the American Chemical Society (ACS) journals accept HRMS analysis of a new compound as a sufficient proof for its characterization and identity. High-resolution mass spectra were obtained from the Emory University Mass Spectroscopy Facility. [18F]fluoride was produced at the Emory University Center for Systems Imaging using an 11 MeV Siemens RDS 111 negative-ion cyclotron (Knoxville, TN) by the 18O(p, n) [18F] reaction using [18O]H2O (95%). Trap/release cartridges model DW-TRC were purchased from D&W, Inc. (Oakdale, TN). Radiometric TLC was performed with the same type of silica plates from Whatman and analyzed with a Raytest system (model Rita Star, Germany). Isolated radiochemical yields were determined using a dose calibrator (Capintec CRC-712M). Analytical HPLC experiments were performed using a Waters Breeze HPLC system equipped with a Bioscan flow count radioactivity detector and an inline UV detector set to monitor wavelengths at 210, 230, and 254 nm (Atlantis T3 column, Waters; mobile phase: 6% EtOH). All animal experiments were conducted under humane conditions and were approved by the Institutional Animal Use and Care Committee and Radiation Safety Committees at Emory University. Experiments involving infectious materials were approved by the Emory University Environmental Health and Safety Office and conducted in Biosafety Level 2 (BSL-2) laboratories. The purity of all reported compounds is at least 95%, as assessed by 1H NMR, radiometric TLC, or radiometric HPLC unless otherwise stated.Chemistry
1H NMR (300 MHz, CDCl3): δ (ppm): 7.98 (d, 4H, J = 7.8 Hz, ArH), 7.55–7.48 (m, 2H, ArH), 7.40–7.35 (m, 4H, ArH), 5.69 (qd, 2H, J = 11.1 and 3.5 Hz), 5.20 (d, 1H, J = 14.6 Hz), 4.77 (d, 1H, J = 5.6 Hz), 4.61 (d, 1H, J = 5.6 Hz), 4.40 (s, 1H), 4.29 (d,t, 1H, J = 14.6 Hz and 5.9 Hz) 3.46 (s, 3H, OCH3), 2.34 (d, 1H, J = 4.2 Hz).
13C NMR (75 MHz, CDCl3): δ (ppm): 166.3, 165.9, 133.7, 133.5, 130.1, 130.0, 129.5, 129.4, 128.7, 128.6, 97.7, 82.6 (d, JC,F = 169.3 Hz), 71.0, 96.0, 68.4 (d, J = 28.8 Hz), 68.3, 55.8.
HRMS (ESI) m/z found: 405.13428, calculated: 405.13441 for C21H22O7F [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford methyl 2,3-di-O-benzoyl-4-O-trifluoromethylsulfonyl-6-deoxy-6-fluoro-α-D-galactopyranoside (3) as an off white solid (0.72 g, 99%).
1) to afford methyl 2,3-di-O-benzoyl-4-O-trifluoromethylsulfonyl-6-deoxy-6-fluoro-α-D-galactopyranoside (3) as an off white solid (0.72 g, 99%).1H NMR (300 MHz, CDCl3): δ (ppm): 8.06–7.95 (m, 4H, ArH), 7.57–7.50 (m, 2H, ArH), 7.43–7.35 (m, 4H, ArH), 5.91 (dd, 1H, J = 3.5 and 10.4 Hz), 5.57 (dd, 1H, J = 3.5 and 10.4 Hz), 5.63 (d, 1H, J = 2.8 Hz), 5.63 (d, 1H, J = 4.2 Hz), 4.73–4.62 (m, 1H), 4.59–4.46 (m, 2H), 3.48 (s, 3H, OCH3).
\13C NMR (75 MHz, CDCl3): δ (ppm): 165.9, 165.8, 134.0, 133.8, 130.3, 130.1, 128.7, 118.5 (d, JC,F = 319.0 Hz), 97.6, 82.6 (d, JC,F = 4.6 Hz), 80.2 (d, JC,F = 172.7 Hz), 68.3, 67.6, 66.4 (d, JC,F = 25.3 Hz), 56.3.
HRMS (ESI) m/z found: 537.08411, calculated: 537.08369 for C22H21O932S [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 5 as a colorless oil (0.28 g, 37%).
1) to afford 5 as a colorless oil (0.28 g, 37%).1H NMR (300 MHz, CDCl3): δ (ppm): 7.96–7.90 (m, 4H, ArH), 7.55–7.46 (m, 2H, ArH), 7.40–7.32 (m, 4H, ArH), 6.06 (t, 1H, J = 10.4 Hz), 5.65 (d, 1H, J = 6.3 Hz), 5.41–5.34 (m, 2H), 5.22–4.93 (m, 5H), 4.87 (dd, 1H, J = 4.2 and 10.5 Hz), 4.79–4.69 (m, 2H), 4.48 (d, 1H, J = 10.2 Hz), 4.29–4.22 (m, 3H), 4.06–3.87 (m, 4H), 3.44 (s, 3H, OCH3), 3.43 (t, 1H, J = 11.1 Hz), 2.18 (s, 3H, CH3 from acetate), 2.10 (s, 3H, CH3 from acetate), 2.04 (s, 3H, CH3 from acetate), 2.03 (s, 3H, CH3 from acetate), 2.01 (s, 3H, CH3 from acetate), 1.92 (s, 3H, CH3 from acetate), 1.69 (s, 3H, CH3 from acetate).
19F NMR (376 MHz, CDCl3): δ (ppm): −253.7(t,d, J = 47.1 Hz and 27.7 Hz).
13C NMR (75 MHz, CDCl3): δ (ppm): 170.8–169.0, 166.0, 165.5, 133.7, 133.5, 130.1, 129.8, 128.7, 128.6, 97.6, 96.0, 83.4, 82.7 (d, JC,F = 175.6 Hz), 73.1–72.9, 72.2, 70.1–69.3, 68.7, 68.0, 62.9, 61.5, 43.4 (d, JC,F = 6.9 Hz), 21.0–20.8, 20.2.
HRMS (ESI) m/z found: 1039.29124, calculated: 1039.29116 for C47H56FNaO23S [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHCl3
CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water; 25
water; 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). The mixture was filtered, and the filtrate evaporated to dryness under the reduced pressure. The residue was purified by flash column chromatography on silica gel (CH3OH
4). The mixture was filtered, and the filtrate evaporated to dryness under the reduced pressure. The residue was purified by flash column chromatography on silica gel (CH3OH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2
CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O; 27/69/4 to 51/46/3) to afford 6 as white solid (65 mg, 51%).
H2O; 27/69/4 to 51/46/3) to afford 6 as white solid (65 mg, 51%).13C NMR (75 MHz, D2O): δ (ppm): 100.4, 100.1, 86.4, 83.8 (d, JC,F = 168.7 Hz), 77.4, 74.4, 73.5, 73.4, 73.3, 72.6, 72.3, 72.0, 71.4, 70.3 (d, JC,F = 17.3 Hz), 69.9, 61.0, 60.8, 55.8, 46.0.
19F NMR (376 MHz, D2O): δ (ppm) −237.4. t,d, J = 47.1 Hz and 28.3 Hz. KF as an internal standard for 19F NMR.
HRMS(ESI) m/z found: 559.14673, calculated: 539.14817 for C19H33O14FNa32S [M + Na]+.
The reaction mixture was neutralized to pH = 7 with 1 N HCl and extracted with EtOAc (50 mL). The EtOAc phase was washed with water (10 mL × 3), dried over Na2SO4, filtered, and evaporated to dryness under reduced pressure. A solution of trityl triflate 7 (0.54, 0.63 mmol) in DMF (20 mL) and NEt3 (0.19 mL) was added to the thiol residue, and the resulting reaction mixture was stirred at 0 °C for 1 h and room temperature for 48 h under argon. EtOAc (100 mL) was added to the reaction mixture, and the EtOAc solution was washed with water (15 mL × 3) and brine (30 mL), dried over Na2SO4, filtered, and evaporated to dryness under reduced pressure. The residue was purified by flash column chromatography on silica gel (EtOAc/hexane = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 8 as a colorless glassy solid (0.13 g, 12%).
1) to afford 8 as a colorless glassy solid (0.13 g, 12%).
1H NMR (300 MHz, CDCl3): δ (ppm): 7.90 (m, 4H, ArH), 7.58–7.29 (m, 21H, ArH), 5.98 (t, 1H, J = 10.4 Hz), 5.50–5.44 (m, 2H), 5.35 (d, 1H, J = 4.2 Hz), 5.24 (d, 1H, J = 3.5 Hz), 5.15–5.03 (m, 3H), 4.90 (dd, 1H, J = 4.2 and 10.4 Hz), 4.66 (dd, 1H, J = 6.2 and 10.4 Hz) 4.27 (dd, 1H, J = 3.5 and 12.5 Hz), 4.04–43.68 (m, 8H), 3.54 (s, 3H, OCH3), 3.51–3.43 (M, 1H), 2.97 (t, 1H, J = 10.4 Hz), 2.11 (s, 9H, CH3 from acetate), 2.03 (s, 3H, CH3 from acetate), 1.98 (s, 3H, CH3 from acetate), 1.90 (s, 3H, CH3 from acetate), 1.64 (s, 3H, CH3 from acetate).
13C NMR (75 MHz, CDCl3): δ (ppm): 170.79, 170.51, 170.06, 169.70, 169.61, 169.29, 166.11, 165.43, 144.16, 133.55, 133.49, 130.09, 129.82, 129.29, 129.22, 128.91, 128.62, 128.55, 128.25, 127.43, 96.86, 95.86, 87.40, 82.65, 73.74, 73.45, 72.41, 72.10, 70.36, 69.91, 69.54, 68.77, 68.48, 68.18, 65.03, 62.46, 61.46, 55.41, 44.79, 20.92, 20.84.
HRMS (ESI) m/z found: 1279.40261, calculated: 1279.40505 for C66H71O24S [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 9 as a white powder (79 mg, 89%).
1) to afford 9 as a white powder (79 mg, 89%).1H NMR (300 MHz, CDCl3): δ (ppm): 7.96–7.90 (m, 4H, ArH), 7.54–7.46 (m, 2H, ArH), 7.40–7.32 (m, 4H, ArH), 6.04 (t, 1H, J = 10.4 Hz), 5.62 (d, 1H, J = 5.6 Hz), 5.40–5.30 (m, 2H), 5.20–5.04 (m, 4H), 4.87 (dd, 1H, J = 4.2 and 10.4 Hz), 4.75 (dd, 1H, J = 5.6 and 9.4 Hz), 4.56 (d, 1H, J = 9.7 Hz), 4.28–3.83 (m, 9H), 3.47 (t, 1H, J = 11.1 Hz), 3.42 (s, 3H, OCH3), 2.33–2.28 (m, 1H), 2.19 (s, 3H, CH3 from acetate), 2.10 (s, 3H, CH3 from acetate), 2.04 (s, 6H, CH3 from acetate), 2.01 (s, 3H, CH3 from acetate), 1.93 (s, 3H, CH3 from acetate), 1.70 (s, 3H, CH3 from acetate).
13C NMR (75 MHz, CDCl3): δ (ppm): 170.80, 170.76, 170.19, 169.65, 169.44, 166.08, 165.48, 133.58, 133.51, 130.06, 129.82, 129.33, 129.17, 128.68, 128.56, 97.50, 96.07, 83.77, 73.37, 73.28, 72.88, 72.03, 70.93, 70.07, 69.87, 69.54, 69.39, 68.67, 68.12, 63.14, 62.20, 61.62, 55.80, 44.71, 21.06, 20.96, 20.89, 20.80, 20.16.
HRMS (ESI) m/z found: 1037.29419, calculated: 1037.29550 for C47H57O24S [M + H]+.
1H NMR (300 MHz, CDCl3): δ (ppm): 7.92–7.87 (m, 6H, ArH), 7.79–7.75 (m, 2H, ArH), 7.53–7.45 (m, 2H, ArH), 7.39–7.30 (m, 4H, ArH), 5.95 (t, 1H, J = 10.4 Hz), 5.59 (d, 1H, J = 5.6 Hz), 5.40–5.33 (m, 2H), 5.17–5.06 (m, 2H), 4.97 (d, 4.87 J = 3.5 Hz), 4.90–4.84 (m, 2H), 4.74 (dd, 1H, J = 5.6 and 9.8 Hz), 4.71–4.55 (m, 3H), 4.08–3.87 (m, 4H), 3.32 (s, 3H, OCH3), 3.16 (t, 1H, J = 11.1 Hz), 2.17 (s, 3H, CH3 from acetate), 2.10 (s, 3H, CH3 from acetate), 2.04 (s, 6H, CH3 from acetate), 2.00 (s, 3H, CH3 from acetate), 1.93 (s, 3H, CH3 from acetate), 1.69 (s, 3H, CH3 from acetate).
13C NMR (75 MHz, CDCl3): δ (ppm): 170.81, 170.76, 170.55, 170.08, 169.70, 169.66, 169.45, 165.92, 165.38, 135.31, 133.71, 133.59, 132.80, 130.05, 129.87, 129.48, 129.11, 128.98, 128.72, 128.58, 97.16, 96.11, 83.45, 73.12, 72.98, 72.48, 71.92, 70.26, 70.00, 69.84, 69.59, 69.55, 68.71, 68.59, 67.98, 62.74, 61.50, 55.99, 44.91, 21.13, 20.92, 20.84, 20.22.
HRMS (ESI) m/z found: 1255.19745, calculated: 1255.19921 for C53H60BrO26S2 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) to afford 12 as a colorless foamy solid (0.2165 g, 59%).
3) to afford 12 as a colorless foamy solid (0.2165 g, 59%).1H NMR (300 MHz, CDCl3): δ (ppm): 7.96–7.88 (m, 4H, ArH), 7.56–7.45 (m, 2H, ArH), 7.41–7.31 (m, 4H, ArH), 6.05 (t, 1H, J = 10.5 Hz), 5.79 (d, 1H, J = 6.3 Hz), 5.26–5.20 (m, 2H), 5.12–4.86 (m, 4H), 4.73 (dd, 1H, J = 9.9 and 48.9 Hz), 4.79–4.69 (m, 2H), 4.41 (dd, 1H, J = 3.8 Hz and 12.3 Hz), 4.30–4.25 (m, 1H), 4.11 (dd, 1H, J = 2.1 and 12.3 Hz), 3.97 (dd, 1H, J = 10.5 and 28.5 Hz), 3.43 (s, 3H, OCH3), 3.43 (t, 1H, J = 11.1 Hz), 2.12 (s, 3H, CH3 from acetate), 2.02 (s, 3H, CH3 from acetate), 1.94 (s, 3H, CH3 from acetate), 1.71 (s, 3H, CH3 from acetate).
19F NMR (376 MHz, CDCl3): δ (ppm) −253.79 (t,d, J = 47.0 and 28.2 Hz).
13C NMR (150 MHz, CDCl3): δ (ppm) 170.7, 169.9, 169.4, 169.0, 165.8, 165.4, 133.5, 133.3, 129.9, 129.0, 128.9, 128.5, 128.4, 97.4, 83.3, 82.4 (d, JC,F = 175.6 Hz), 73.0, 72.8, 70.1, 69.5, 69.4 (d, JC,F = 18.6 Hz), 68.7, 68.0, 61.4, 55.8, 42.7 (d, JC,F = 6.8 Hz), 20.7, 20.6, 20.0.
HRMS (ESI) m/z found: 773.18867, calculated: 773.18859 for C35H39O15F23Na32S [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2
CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 27/69/4 to 51/46/3) to afford 13 as a white solid (98.9 mg, 99%).
H2O = 27/69/4 to 51/46/3) to afford 13 as a white solid (98.9 mg, 99%).13C NMR (150 MHz, D2O): δ (ppm): 102.3, 88.9, 86.1 (d, JC,F = 169.0 Hz), 76.2, 75.7, 75.6, 74.9, 73.9, 72.4 (d, JC,F = 17.6 Hz), 72.2, 63.1, 58.1, 48.4 (d, JC,F = 5.7 Hz).
19F NMR (376 MHz, D2O): δ (ppm): −237.3 (t,d, J = 47.1 and 28.4 Hz).
HRMS (ESI) m/z found: 397.09437, calculated: 397.0939 for C13H23O9FNa32S [M + Na]+.
Radiochemistry
Biological analyses
CHO-K1 cells were cultured in Ham's F-12 Nutrient Mix medium (Thermo-Fisher Scientific) containing 10% fetal bovine serum (Sigma-Aldrich) and penicillin/streptomycin (Sigma-Aldrich). Cells were seeded onto 6-well plates at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well one day before the experiment. The cells were then loaded with [18F]MFTMT at final concentrations of 5, 10, 15, and 20 μCi mL−1 and incubated at 37 °C for 1 h (N = 3 for each concentration). Subsequently, the cells were washed three times with 2 mL of PBS per well. After that, the cells were detached from the plates using cell scrapers and suspended in 0.5 mL of PBS. The resulting cell suspension was transferred to 1.5 mL tubes for their radioactivity assessment.
000 cells per well one day before the experiment. The cells were then loaded with [18F]MFTMT at final concentrations of 5, 10, 15, and 20 μCi mL−1 and incubated at 37 °C for 1 h (N = 3 for each concentration). Subsequently, the cells were washed three times with 2 mL of PBS per well. After that, the cells were detached from the plates using cell scrapers and suspended in 0.5 mL of PBS. The resulting cell suspension was transferred to 1.5 mL tubes for their radioactivity assessment.
To evaluate the uptake of serum-treated [18F]MFTMT, [18F]MFTMT was incubated with a 1/10 volume of rat serum at 37 °C for 1 h, followed by filtration with a molecular filter (PALL 0.45 μm PTFE ACRODISC CR 13). The resulting serum-treated [18F]MFTMT solution was then analyzed for radioactivity. Bacteria and CHO-K1 cells were prepared as previously described, and the serum-treated [18F]MFTMT was added at final concentrations of 1, 5, 10, and 15 μCi mL−1 and incubated at 37 °C for 1 h (N = 3 for each concentration). After incubation, bacteria and cells were processed as described earlier.
In addition to S. aureus and E. coli, uptake of [18F]MFTMT and [18F]MFTM by Pseudomonas aeruginosa, Staphylococcus epidermidis, Enterococcus faecalis, Streptococcus sanguinis were also evaluated. Enterococcus faecalis and Streptococcus sanguinis were cultured in Brain Heart Infusion Broth (BHI broth, Research Products International) and in aerobic condition. Other bacteria were cultured in LB broth and aerobic condition. [18F]MFTM was generated by digesting 1 mCi mL−1 of [18F]MFTMT by 50 units per mL of α-glucosidase (G5003, Sigma-Aldrich) at 3737 °C for 1 h, then, the enzyme, α-glucosidase was removed using 10 kD molecular filters (Amicon Ultra-0.5 Centrifugal Filter Unit, Millipore). As described above, the bacteria were adjusted to the OD600 of 0.3 (concentration of 1 × 108 CFU mL−1), and incubated with 20 μCi mL−1 of [18F]MFTMT or [18F]MFTM at 37 °C for 1 h (N = 3). Then, 200 μL of bacterial suspension was aliquoted onto the 0.22 μm centrifuge filter cartridges and washed with PBS. The bacterial pellets on the filter cartridges were used to evaluate the radioactivity.
The remaining radioactivity associated with bacteria or CHO-K1 cells was measured using a gamma counter (Cobra II gamma counter, Packard). Radioactivity levels were quantified using standards ranging from 0.0005 to 0.01 μCi (N = 2 for each standard concentration).
In vivo PET imaging
Metabolite(s) of [18F]MFTMT
Following the designated incubation period, 200 μL of the ice-cold methanol was added to each tube and mixed. Subsequently, the samples were centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 relative centrifugal force for 10 min. The supernatants were removed and injected into a Waters Breeze HPLC system equipped with a 5 μL stainless steel injection coil installed and a Waters Atlantis T3, 3 μm, 4.6 mm × 150 mm HPLC column. Elution was achieved using 2% EtOH–water as the eluent over 25 min at a flow rate of 1 mL min−1.
000 relative centrifugal force for 10 min. The supernatants were removed and injected into a Waters Breeze HPLC system equipped with a 5 μL stainless steel injection coil installed and a Waters Atlantis T3, 3 μm, 4.6 mm × 150 mm HPLC column. Elution was achieved using 2% EtOH–water as the eluent over 25 min at a flow rate of 1 mL min−1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) or rabbit anti-SGLT2 antibody (GTX59872, GeneTex, 1
100) or rabbit anti-SGLT2 antibody (GTX59872, GeneTex, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). Concurrently, to identify fibroblasts, a mouse anti-vimentin antibody (NBP1-97672, Novus, 1
100). Concurrently, to identify fibroblasts, a mouse anti-vimentin antibody (NBP1-97672, Novus, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) was used. Following PBS washing, the sections were stained with secondary antibodies: donkey anti-rabbit IgG conjugated with Alexa Fluor 568 (A10042, Invitrogen, 1
100) was used. Following PBS washing, the sections were stained with secondary antibodies: donkey anti-rabbit IgG conjugated with Alexa Fluor 568 (A10042, Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) and donkey anti-mouse IgG conjugated with Alexa 488 (A-21202, Invitrogen, 1
500) and donkey anti-mouse IgG conjugated with Alexa 488 (A-21202, Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500). Finally, the sections were sealed with a mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI, VECTASHIELD with DAPI, H-1800). The expression of SGLT1, SGLT2, and vimentin were observed using a confocal microscope (Leica TCS SP5 II).
500). Finally, the sections were sealed with a mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI, VECTASHIELD with DAPI, H-1800). The expression of SGLT1, SGLT2, and vimentin were observed using a confocal microscope (Leica TCS SP5 II).Interaction of [18F]MFTMT with SGLTs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, following the manufacturer's instructions. The medium was changed one day after the transfection, and the expression of transfected genes was confirmed by observing mCherry expression using a fluorescent microscope (Olympus IX71). Cells were selected with puromycin (10 μg mL−1, Santa Cruz) for 3 weeks and seeded in a 96-well plate at a density of 1 cell per well. The stable expression of hSGLT1 or hSGLT2 was confirmed by assessing the uptake of 1-NBDG (0.1 mM, V41383, InvivoChem), a specific substrate for SGLT1 and SGLT2.60 CHO-K1 cells with the highest uptake of 1-NBDG were designated as CHO-K1-hSGLT1 and CHO-K1-hSGLT2. Among the cells transfected with pCVM6-mCherry, those exhibiting the highest mCherry expression were selected as CHO-K1-control.
4, following the manufacturer's instructions. The medium was changed one day after the transfection, and the expression of transfected genes was confirmed by observing mCherry expression using a fluorescent microscope (Olympus IX71). Cells were selected with puromycin (10 μg mL−1, Santa Cruz) for 3 weeks and seeded in a 96-well plate at a density of 1 cell per well. The stable expression of hSGLT1 or hSGLT2 was confirmed by assessing the uptake of 1-NBDG (0.1 mM, V41383, InvivoChem), a specific substrate for SGLT1 and SGLT2.60 CHO-K1 cells with the highest uptake of 1-NBDG were designated as CHO-K1-hSGLT1 and CHO-K1-hSGLT2. Among the cells transfected with pCVM6-mCherry, those exhibiting the highest mCherry expression were selected as CHO-K1-control.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well one day before the experiment. The cells were then loaded with [18F]MFTMT and [18F]MFTM at a final concentration of 20 μCi mL−1 and incubated at 37 °C for 1 h (N = 3). Subsequently, the cells were washed, and the remaining radioactivity in the cells was evaluated using the method described in the in vitro uptake study. To confirm the specificity of the interactions with SGLT1 and/or SGLT2, some CHO-K1 hSGLT1 and COH-K1 SGLT2 cells were treated with mizagliflozin (MedChemExpress), a specific SGJT1 inhibitor and dapagliflozin (Combi-Blocks), a specific SGLT2 inhibitor at final concentration of 1 μM for 30 min.104,105,115–117 Then, the cells were loaded with [18F]MFTMT and [18F]MFTM at a final concentration of 20 μCi mL−1 (N = 3).
000 cells per well one day before the experiment. The cells were then loaded with [18F]MFTMT and [18F]MFTM at a final concentration of 20 μCi mL−1 and incubated at 37 °C for 1 h (N = 3). Subsequently, the cells were washed, and the remaining radioactivity in the cells was evaluated using the method described in the in vitro uptake study. To confirm the specificity of the interactions with SGLT1 and/or SGLT2, some CHO-K1 hSGLT1 and COH-K1 SGLT2 cells were treated with mizagliflozin (MedChemExpress), a specific SGJT1 inhibitor and dapagliflozin (Combi-Blocks), a specific SGLT2 inhibitor at final concentration of 1 μM for 30 min.104,105,115–117 Then, the cells were loaded with [18F]MFTMT and [18F]MFTM at a final concentration of 20 μCi mL−1 (N = 3).Statistical evaluation
Statistical analysis was conducted using Prism statistical software (Version 5.01). The Student's t-test was utilized to compare two groups, while a one-way analysis of variance was employed with Tukey's multiple comparison tests. All data are presented as mean ± standard error, with a significance level of p < 0.05 considered statistically significant.Conclusions
In this study, we synthesized [18F]MFTMT and assessed its serum stability, in vitro uptake, and in vivo distribution and metabolism. While the thio-glycosyl bond resisted enzymatic digestion, [18F]MFTMT and its metabolite, [18F]MFTM, reduced the affinity for E. coli and other pathogenic bacteria other than S. aureus, and thio-glycosyl bond and/or 6-deoxy-6-fluoro-modification prolonged blood retention of radioactivity. In vivo imaging revealed that radioactivity derived from [18F]MFTMT accumulated not only in S. aureus-infected lesions but also in inflammatory tissue. We further found that [18F]MFTM is the only radioactive metabolite in vivo, and increased SGLT1 expression in the inflammatory tissue likely contributed to non-specific [18F]MFTMT and [18F]MFTM accumulation in PET imaging. In addition, delayed excretion of [18F]MFTMT and/or its metabolites might further increase radioactivity accumulation in inflammatory tissue.Our findings suggest that for bacteria-specific probes for PET imaging, maltotetraose and/or a longer maltodextrin backbone may reduce accumulation in mammalian cells caused by SGLT1 and may be a key for specificity to bacteria. Furthermore, a thio-glycosyl bond may reduce the affinity to maltodextrin transporter and PTS and other approaches such as employing a nitrogen-glycosyl bond may satisfy both stability against enzymatic digestion and specificity for bacteria. Moreover, the optimal position to derivatize the imaging molecule might be different depending on the species of bacteria. Based on these findings, future studies will need to investigate the structure and characteristics of maltodextrin transporters of pathogenic bacteria. These studies will lead to the development of next generation maltodextrin-based imaging probes with nitrogen-glycosyl bond making them resistant to enzymatic digestion while maintaining bacterial selectivity and even bacterial species specificity.
Abbreviations
| 1-NBDG | 1(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-1-deoxy-D-glucose |
| % ID per g tissue | Ratio relative to the injection dose per gram |
| BHI broth | Brain heart infusion broth |
| CFU | colony-forming units |
| CT | Computed tomography |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| E. coli | Escherichia coli |
| E. faecalis | Enterococcus faecalis |
| EIIC | Enzyme IIC component |
| EtOH | Ethanol |
| [18F]FDG | 2-Deoxy-2-[18F]fluoro-D-glucose |
| GLUT | Glucose transporter |
| HPLC | High-performance liquid chromatography |
| LB broth | Luria–Bertani broth |
| MFTM | Methyl 6-deoxy-6-fluoro-4-thio-α-D-maltoside |
| MFTMT | Methyl 6-deoxy-6-fluoro-4-thio-α-D-maltotrioside |
| MHF | Fluoro-maltohexaose |
| MRI | Magnetic resonance imaging |
| NaOEt | Sodium ethoxide |
| NEt3 | Triethylamine |
| NMR | Nuclear magnetic resonance |
| P. aeruginosa | Pseudomonas aeruginosa |
| PBS | Phosphate-buffered saline |
| PET | Positron emission tomography |
| POD | Postoperative day |
| PTS | Phosphotransferase system |
| RCY | Decay-corrected radiochemical yield |
| ROI | Region of interest |
| S. aureus | Staphylococcus aureus |
| S. epidermidis | Staphylococcus epidermis |
| SDS-PAGE | Sodium dodecyl-sulfate polyacrylamide gel electrophoresis |
| SGLT | Sodium glucose co-transporter |
| S. sanguinis | Streptococcus sanguinis |
| SUV | Standardized uptake value |
| TLC | Thin-layer chromatography |
Data availability
The data supporting this article has been included as part of the ESI.†Author contributions
Kiyoko Takemiya: investigation, formal analysis, methodology, visualization, writing – original draft, writing – review & editing, and graphical abstract. Wonewoo Seo: investigation, formal analysis, methodology, and writing – original draft. Ronald J. Voll: investigation, formal analysis. Sheng Zhao: investigation. Giji Joseph: investigation. Shelly Wang: investigation. Fanxing Zeng: investigation. Jonathon A Nye: investigation. Niren Murthy: conceptualization, writing – review & editing, funding acquisition. W. Robert Taylor: conceptualization, writing – review & editing, funding acquisition. Mark M. Goodman: conceptualization, methodology, visualization, writing – original draft, writing – review & editing, funding acquisition, and graphical abstract.Conflicts of interest
Mark Goodman is the principal investigator, a sponsor of this study. The terms of this arrangement have been reviewed and approved by Emory University in accordance with Emory University Policy 7.7, Policy for Investigators Holding a Financial Interest in Research. Kiyoko Takemiya, W. Robert Taylor, Niren Murthy, and Mark M. Goodman have equity interest in Microbial Medical, Inc., which is related to this study. Wonewoo Seo, Ronald J. Voll, Sheng Zhao, Giji Joseph, Shelly Wang, Fanxing Zeng, and Jonathon A. Nye have no conflict of interests.Acknowledgements
This study has been funded in part with Federal funds from the National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Department of Health and Human Services, under Grants No. NIH R01 EB029320 and NIH R01 EB020008. This study is supported by gifts from the True Vine Foundation and an anonymous donor. The authors thank Dr Jaekeun Park for his contribution in the micro-PET studies and Ron Crowe, BCNP, and the radiopharmacy at the Emory University Center for Systems Imaging for production of [18F]fluoride.Notes and references
- A. J. Greenspon, J. D. Patel, E. Lau, J. A. Ochoa, D. R. Frisch, R. T. Ho, B. B. Pavri and S. M. Kurtz, J. Am. Coll. Cardiol., 2011, 58, 1001–1006 CrossRef PubMed.
- B. L. Wilkoff, G. Boriani, S. Mittal, J. E. Poole, C. Kennergren, G. R. Corey, J. C. Love, R. Augostini, S. Faerestrand, S. S. Wiggins, J. S. Healey, R. Holbrook, J. D. Lande, D. R. Lexcen, S. Willey, K. G. Tarakji and W.-I. Investigators, Circ.: Arrhythmia Electrophysiol., 2020, 13, e008280 Search PubMed.
- R. Sanghavi, N. Ravikumar, V. Sarodaya, M. Haq, M. Sherif and A. Harky, Future Cardiol., 2022, 18(11), 891–899 CrossRef CAS PubMed.
- K. G. Tarakji and B. L. Wilkoff, Expet Rev. Cardiovasc. Ther., 2013, 11, 607–616 CrossRef CAS PubMed.
- A. Roque, J. Francisco-Pascual, J. F. Andres-Cordon, N. Fernandez-Hidalgo, J. R. Herance, H. Cuellar-Calabria, S. Aguade-Bruix and M. N. Pizzi, J. Nucl. Cardiol., 2023, 30, 2846–2849 CrossRef PubMed.
- D. C. DeSimone and M. R. Sohail, J. Clin. Microbiol., 2018, 56(7), e01683 CrossRef CAS PubMed.
- B. L. Wilkoff, G. Boriani, S. Mittal, J. E. Poole, C. Kennergren, G. R. Corey, A. D. Krahn, E. J. Schloss, J. L. Gallastegui, R. A. Pickett, R. F. Evonich, S. F. Roark, D. M. Sorrentino, D. P. Sholevar, E. M. Cronin, B. J. Berman, D. W. Riggio, H. H. Khan, M. T. Silver, J. Collier, Z. Eldadah, R. Holbrook, J. D. Lande, D. R. Lexcen, S. Seshadri and K. G. Tarakji, Circ.:Arrhythmia Electrophysiol., 2020, 13, e008503 Search PubMed.
- T. W. van der Vaart, P. M. M. Bossuyt, D. T. Durack, L. M. Baddour, A. S. Bayer, E. Durante-Mangoni, T. L. Holland, A. W. Karchmer, J. M. Miro, P. Moreillon, M. Rasmussen, C. Selton-Suty, V. G. Fowler Jr and J. T. M. van der Meer, Clin. Infect. Dis., 2024, 78(4), 922–929 CrossRef PubMed.
- V. G. Fowler, D. T. Durack, C. Selton-Suty, E. Athan, A. S. Bayer, A. L. Chamis, A. Dahl, L. DiBernardo, E. Durante-Mangoni, X. Duval, C. Q. Fortes, E. Fosbol, M. M. Hannan, B. Hasse, B. Hoen, A. W. Karchmer, C. A. Mestres, C. A. Petti, M. N. Pizzi, S. D. Preston, A. Roque, F. Vandenesch, J. T. M. van der Meer, T. W. van der Vaart and J. M. Miro, Clin. Infect. Dis., 2023, 77, 518–526 CrossRef PubMed.
- A. S. Dababneh and M. R. Sohail, Cleve. Clin. J. Med., 2011, 78, 529–537 CrossRef PubMed.
- K. G. Tarakji, E. J. Chan, D. J. Cantillon, A. L. Doonan, T. Hu, S. Schmitt, T. G. Fraser, A. Kim, S. M. Gordon and B. L. Wilkoff, Heart Rhythm, 2010, 7, 1043–1047 Search PubMed.
- M. Nataloni, M. Pergolini, G. Rescigno and R. Mocchegiani, J. Cardiovasc. Med., 2010, 11, 869–883 CrossRef PubMed.
- R. Margey, H. McCann, G. Blake, E. Keelan, J. Galvin, M. Lynch, N. Mahon, D. Sugrue and J. O'Neill, Europace, 2010, 12, 64–70 Search PubMed.
- B. L. Wilkoff, Heart Rhythm, 2007, 4, 1467–1470 Search PubMed.
- M. Unberath and A. Maier, Medical imaging modalities, in Medical Image Analysis, ed. A. F. Frangi, J. L. Prince and M. Sonka, The MICCAI Society book Series, Academic Press, New York, 2024, ch. 1, pp. 3–19 Search PubMed.
- X. Ning, S. Lee, Z. Wang, D. Kim, B. Stubblefield, E. Gilbert and N. Murthy, Nat. Mater., 2011, 10, 602–607 CAS.
- X. Ning, W. Seo, S. Lee, K. Takemiya, M. Rafi, X. Feng, D. Weiss, X. Wang, L. Williams, V. M. Camp, M. Eugene, W. R. Taylor, M. Goodman and N. Murthy, Angew. Chem., 2014, 53, 14096–14101 CAS.
- K. Takemiya, X. Ning, W. Seo, X. Wang, R. Mohammad, G. Joseph, J. S. Titterington, C. S. Kraft, J. A. Nye, N. Murthy, M. M. Goodman and W. R. Taylor, JACC Cardiovasc. Imaging, 2019, 12(5), 875–886 Search PubMed.
- K. Takemiya, J. J. Roise, M. He, C. Taing, A. G. Rodriguez, N. Murthy, M. M. Goodman and W. R. Taylor, PLoS One, 2021, 16, e0247673 CAS.
- R. Dippel, T. Bergmiller, A. Bohm and W. Boos, J. Bacteriol., 2005, 187, 8332–8339 CAS.
- R. Dippel and W. Boos, J. Bacteriol., 2005, 187, 8322–8331 CAS.
- E. Bordignon, M. Grote and E. Schneider, Mol. Microbiol., 2010, 77, 1354–1366 CAS.
- D. W. Abbott, M. A. Higgins, S. Hyrnuik, B. Pluvinage, A. Lammerts van Bueren and A. B. Boraston, Mol. Microbiol., 2010, 77, 183–199 CrossRef CAS PubMed.
- L. Clermont, A. Macha, L. M. Muller, S. M. Derya, P. von Zaluskowski, A. Eck, B. J. Eikmanns and G. M. Seibold, J. Bacteriol., 2015, 197, 1394–1407 CrossRef CAS PubMed.
- A. Axer, S. Hermann, G. Kehr, D. Clases, U. Karst, L. Fischer-Riepe, J. Roth, M. Fobker, M. Schafers, R. Gilmour and A. Faust, ChemMedChem, 2018, 13, 241–250 CrossRef CAS PubMed.
- G. Gowrishankar, M. Namavari, E. B. Jouannot, A. Hoehne, R. Reeves, J. Hardy and S. S. Gambhir, PLoS One, 2014, 9, e107951 Search PubMed.
- A. Zlitni, G. Gowrishankar, I. Steinberg, T. Haywood and S. Sam Gambhir, Nat. Commun., 2020, 11, 1250 CAS.
- A. L. Bernstein, A. Dhanantwari, M. Jurcova, R. Cheheltani, P. C. Naha, T. Ivanc, E. Shefer and D. P. Cormode, Sci. Rep., 2016, 6, 26177 CrossRef CAS PubMed.
- M. L. James and S. S. Gambhir, Physiol. Rev., 2012, 92, 897–965 CAS.
- M. W. Galper, M. T. Saung, V. Fuster, E. Roessl, A. Thran, R. Proksa, Z. A. Fayad and D. P. Cormode, Investig. Radiol., 2012, 47, 475–481 CAS.
- G. Gowrishankar, J. Hardy, M. Wardak, M. Namavari, R. E. Reeves, E. Neofytou, A. Srinivasan, J. C. Wu, C. H. Contag and S. S. Gambhir, J. Nucl. Med., 2017, 58, 1679–1684 CAS.
- T. Takeuchi, T. Kozu, S. Watanabe, M. Morita, K. Shiratori and I. Shibata, Gastroenterol. Jpn., 1978, 13, 395–400 CrossRef CAS PubMed.
- J. C. Murat, C. Castilla and H. Paris, Comp. Biochem. Physiol. B, 1978, 61, 145–149 CAS.
- P. Coma, L. Gomez-Chacon, B. Garcia-Serrano, E. Fernandez and M. A. Ortiz-Apodaca, Clin. Chem., 1992, 38, 223–226 CAS.
- A. Compton, Biochem. J., 1921, 15, 681–686 CAS.
- H. Bittencourt, M. H. Sleisenger and E. Weser, Gastroenterology, 1969, 57, 410–414 CAS.
- N. Jarkas, R. J. Voll and M. M. Goodman, J. Labelled Compd. Radiopharm., 2013, 56, 539–543 CAS.
- J. Kihlberg, T. Frejd, K. Jansson, A. Sundin and G. Magnusson, Carbohydr. Res., 1988, 176, 271–286 CAS.
- L. Greffe, M. T. Jensen, F. Chang-Pi-Hin, S. Fruchard, M. J. O'Donohue, B. Svensson and H. Driguez, Chemistry, 2002, 8, 5447–5455 CAS.
- M. Blanc-Muesser and H. D. Jacques Defaye, Carbohydr. Res., 1978, 67, 305–328 CAS.
- H. Fujino, T. Fukuda, M. Nagatomo and M. Inoue, J. Am. Chem. Soc., 2020, 142, 13227–13234 CAS.
- M. Blanc-Muesser and H. D. Jacques Defaye, J. Chem. Soc. Perkin Trans. 1, 1982, 15–18 CAS.
- M. R. Sohail, D. Z. Uslan, A. H. Khan, P. A. Friedman, D. L. Hayes, W. R. Wilson, J. M. Steckelberg, S. Stoner and L. M. Baddour, J. Am. Coll. Cardiol., 2007, 49, 1851–1859 CrossRef PubMed.
- S. K. Mulpuru, V. G. Pretorius and U. M. Birgersdotter-Green, Circulation, 2013, 128, 1031–1038 CrossRef PubMed.
- G. M. Viola, L. L. Awan and R. O. Darouiche, Circulation, 2010, 121, 2085–2091 CrossRef PubMed.
- B. Thorens and M. Mueckler, Am. J. Physiol. Endocrinol. Metab., 2010, 298, E141–E145 CrossRef CAS PubMed.
- L. Han, Q. Qu, D. Aydin, O. Panova, M. J. Robertson, Y. Xu, R. O. Dror, G. Skiniotis and L. Feng, Nature, 2022, 601, 274–279 CrossRef CAS PubMed.
- D. Deng and N. Yan, Protein Sci., 2016, 25, 546–558 CrossRef CAS PubMed.
- G. D. Holman, Biochem. J., 2018, 475, 3511–3534 CrossRef CAS PubMed.
- D. Deng, P. Sun, C. Yan, M. Ke, X. Jiang, L. Xiong, W. Ren, K. Hirata, M. Yamamoto, S. Fan and N. Yan, Nature, 2015, 526, 391–396 CrossRef CAS PubMed.
- H. Koepsell, Pfluegers Arch., 2020, 472, 1299–1343 CrossRef CAS PubMed.
- I. Vrhovac, D. Balen Eror, D. Klessen, C. Burger, D. Breljak, O. Kraus, N. Radovic, S. Jadrijevic, I. Aleksic, T. Walles, C. Sauvant, I. Sabolic and H. Koepsell, Pfluegers Arch., 2015, 467, 1881–1898 CrossRef CAS PubMed.
- I. Sabolic, I. Vrhovac, D. B. Eror, M. Gerasimova, M. Rose, D. Breljak, M. Ljubojevic, H. Brzica, A. Sebastiani, S. C. Thal, C. Sauvant, H. Kipp, V. Vallon and H. Koepsell, Am. J. Physiol. Cell Physiol., 2012, 302, C1174–C1188 CAS.
- E. M. Wright, D. D. Loo and B. A. Hirayama, Physiol. Rev., 2011, 91, 733–794 CAS.
- P. Song, A. Onishi, H. Koepsell and V. Vallon, Expert Opin. Ther. Targets, 2016, 20, 1109–1125 CrossRef CAS PubMed.
- L. Ferte, A. Marino, S. Battault, L. Bultot, A. Van Steenbergen, A. Bol, J. Cumps, A. Ginion, H. Koepsell, L. Dumoutier, L. Hue, S. Horman, L. Bertrand and C. Beauloye, Am. J. Physiol. Heart Circ. Physiol., 2021, 320, H838–H853 CAS.
- M. A. Hediger, E. Turk, A. M. Pajor and E. M. Wright, Klin. Wochenschr., 1989, 67, 843–846 CAS.
- M. A. Hediger, E. Turk and E. M. Wright, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 5748–5752 CAS.
- R. G. Wells, A. M. Pajor, Y. Kanai, E. Turk, E. M. Wright and M. A. Hediger, Am. J. Physiol., 1992, 263, F459–F465 CAS.
- H. C. Chang, S. F. Yang, C. C. Huang, T. S. Lin, P. H. Liang, C. J. Lin and L. C. Hsu, Mol. Biosyst., 2013, 9, 2010–2020 CAS.
- W. Boos and H. Shuman, Microbiol. Mol. Biol. Rev., 1998, 62, 204–229 CrossRef CAS PubMed.
- M. L. Oldham, D. Khare, F. A. Quiocho, A. L. Davidson and J. Chen, Nature, 2007, 450, 515–521 CrossRef CAS PubMed.
- M. L. Oldham, S. Chen and J. Chen, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18132–18137 CrossRef CAS PubMed.
- X. Duan, J. A. Hall, H. Nikaido and F. A. Quiocho, J. Mol. Biol., 2001, 306, 1115–1126 CrossRef CAS PubMed.
- F. A. Quiocho, J. C. Spurlino and L. E. Rodseth, Structure, 1997, 5, 997–1015 CrossRef CAS PubMed.
- T. Ferenci, Eur. J. Biochem., 1980, 108, 631–636 CrossRef CAS PubMed.
- T. Ferenci, M. Muir, K. S. Lee and D. Maris, Biochim. Biophys. Acta, 1986, 860, 44–50 CrossRef CAS PubMed.
- R. Benz, A. Schmid, T. Nakae and G. H. Vos-Scheperkeuter, J. Bacteriol., 1986, 165, 978–986 CrossRef CAS PubMed.
- A. Ranquin and P. Van Gelder, Res. Microbiol., 2004, 155, 611–616 CrossRef CAS PubMed.
- R. Benz, A. Schmid and G. H. Vos-Scheperkeuter, J. Membr. Biol., 1987, 100, 21–29 CrossRef CAS PubMed.
- C. Andersen, M. Jordy and R. Benz, J. Gen. Physiol., 1995, 105, 385–401 CrossRef CAS PubMed.
- E. Berkane, F. Orlik, A. Charbit, C. Danelon, D. Fournier, R. Benz and M. Winterhalter, J. Nanobiotechnol., 2005, 3, 3 CrossRef CAS PubMed.
- A. Charbit, Front. Biosci., 2003, 8, s265–s274 CrossRef CAS PubMed.
- K. Denker, F. Orlik, B. Schiffler and R. Benz, J. Mol. Biol., 2005, 352, 534–550 CrossRef CAS PubMed.
- R. Somavanshi, B. Ghosh and V. Sourjik, PLoS Biol., 2016, 14, e2000074 CrossRef PubMed.
- C. Jardin, A. H. Horn, G. Schurer and H. Sticht, J. Phys. Chem. B, 2008, 112, 13391–13400 CrossRef CAS PubMed.
- J. G. McCoy, E. J. Levin and M. Zhou, Biochim. Biophys. Acta, 2015, 1850, 577–585 CrossRef CAS PubMed.
- J. G. McCoy, Z. Ren, V. Stanevich, J. Lee, S. Mitra, E. J. Levin, S. Poget, M. Quick, W. Im and M. Zhou, Structure, 2016, 24, 956–964 CrossRef CAS PubMed.
- Z. Ren, J. Lee, M. M. Moosa, Y. Nian, L. Hu, Z. Xu, J. G. McCoy, A. C. M. Ferreon, W. Im and M. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5962–5967 CrossRef CAS PubMed.
- J. Thomson, Y. Liu, J. M. Sturtevant and F. A. Quiocho, Biophys. Chem., 1998, 70, 101–108 CrossRef CAS PubMed.
- K. Takemiya, S. Wang, Y. Liu, N. Murthy, M. M. Goodman and W. R. Taylor, Biochem. Biophys. Res. Commun., 2024, 695, 149467 CrossRef CAS PubMed.
- F. J. Alvarez, C. Orelle, Y. Huang, R. Bajaj, R. M. Everly, C. S. Klug and A. L. Davidson, Mol. Microbiol., 2015, 98, 878–894 CrossRef CAS PubMed.
- J. C. Spurlino, G. Y. Lu and F. A. Quiocho, J. Biol. Chem., 1991, 266, 5202–5219 CrossRef CAS PubMed.
- J. E. Fleming and H. Lynton, Can. J. Chem., 1967, 45, 353 CrossRef.
- F. H. Allen, O. Kennard, D. G. Watson, L. Brammer, A. G. Orpen and R. Taylor, J. Chem. Soc., Perkin Trans. 2, 1987, S1–S19 RSC.
- N. Sauvageot, A. Mokhtari, P. Joyet, A. Budin-Verneuil, V. S. Blancato, G. D. Repizo, C. Henry, A. Pikis, J. Thompson, C. Magni, A. Hartke and J. Deutscher, J. Bacteriol., 2017, 199(9), e00878 CrossRef PubMed.
- Y. Le Breton, V. Pichereau, N. Sauvageot, Y. Auffray and A. Rince, J. Appl. Microbiol., 2005, 98, 806–813 CrossRef CAS PubMed.
- A. Mokhtari, V. S. Blancato, G. D. Repizo, C. Henry, A. Pikis, A. Bourand, M. de Fatima Alvarez, S. Immel, A. Mechakra-Maza, A. Hartke, J. Thompson, C. Magni and J. Deutscher, Mol. Microbiol., 2013, 88, 234–253 CrossRef CAS PubMed.
- P. Joyet, A. Mokhtari, E. Riboulet-Bisson, V. S. Blancato, M. Espariz, C. Magni, A. Hartke, J. Deutscher and N. Sauvageot, Appl. Environ. Microbiol., 2017, 83(13), e00038 CrossRef PubMed.
- S. A. Shelburne III, H. Fang, N. Okorafor, P. Sumby, I. Sitkiewicz, D. Keith, P. Patel, C. Austin, E. A. Graviss, J. M. Musser and D. C. Chow, J. Bacteriol., 2007, 189, 2610–2617 CrossRef PubMed.
- S. A. Shelburne III, D. B. Keith, M. T. Davenport, N. Horstmann, R. G. Brennan and J. M. Musser, Mol. Microbiol., 2008, 69, 436–452 CrossRef PubMed.
- A. Rahmim, M. A. Lodge, N. A. Karakatsanis, V. Y. Panin, Y. Zhou, A. McMillan, S. Cho, H. Zaidi, M. E. Casey and R. L. Wahl, Eur. J. Nucl. Med. Mol. Imag., 2019, 46, 501–518 CrossRef PubMed.
- G. Tomasi and L. Rosso, Curr. Opin. Pharmacol., 2012, 12, 569–575 CrossRef CAS PubMed.
- R. Kekuda, P. Saha and U. Sundaram, Am. J. Physiol. Gastrointest. Liver Physiol., 2008, 294, G1354–G1361 CrossRef CAS PubMed.
- Y. Li, K. M. Thelen, K. M. Fernandez, R. Nelli, M. Fardisi, M. Rajput, N. L. Trottier, G. A. Contreras and A. J. Moeser, Am. J. Physiol. Gastrointest. Liver Physiol., 2022, 322, G346–G359 CrossRef PubMed.
- J. Park, I. S. Lee, K. H. Kim, Y. Kim, E. J. An and H. J. Jang, Int. J. Mol. Sci., 2019, 20(10), 2537 CrossRef CAS PubMed.
- P. Sharma, V. Khairnar, I. V. Madunic, Y. Singh, A. Pandyra, M. S. Salker, H. Koepsell, I. Sabolic, F. Lang, P. A. Lang and K. S. Lang, Cell. Physiol. Biochem., 2017, 42, 1358–1365 CrossRef CAS PubMed.
- M. Zhao, N. Li and H. Zhou, Drug Des., Dev. Ther., 2023, 17, 2011–2023 CrossRef CAS PubMed.
- P. L. Altman and K. Dittmer, Dorothy., Blood and Other Body Fluids/Analysis and Compilation, Federation of American Societies for Experimental Biology, Washington, 1961 Search PubMed.
- I. A. Hatton, E. D. Galbraith, N. S. C. Merleau, T. P. Miettinen, B. M. Smith and J. A. Shander, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2303077120 CAS.
- C. Scafoglio, B. A. Hirayama, V. Kepe, J. Liu, C. Ghezzi, N. Satyamurthy, N. A. Moatamed, J. Huang, H. Koepsell, J. R. Barrio and E. M. Wright, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E4111–E4119 CrossRef CAS PubMed.
- H. Koepsell, Pharmacol. Ther., 2017, 170, 148–165 CAS.
- S. Tsunokake, E. Iwabuchi, Y. Miki, A. Kanai, Y. Onodera, H. Sasano, T. Ishida and T. Suzuki, Breast Cancer Res. Treat., 2023, 201, 499–513 CrossRef CAS PubMed.
- T. Inoue, M. Takemura, N. Fushimi, Y. Fujimori, T. Onozato, T. Kurooka, T. Asari, H. Takeda, M. Kobayashi, H. Nishibe and M. Isaji, Eur. J. Pharmacol., 2017, 806, 25–31 CAS.
- H. Ohno, Y. Kojima, H. Harada, Y. Abe, T. Endo and M. Kobayashi, Xenobiotica, 2019, 49, 463–473 CrossRef CAS PubMed.
- C. S. Hummel, C. Lu, J. Liu, C. Ghezzi, B. A. Hirayama, D. D. Loo, V. Kepe, J. R. Barrio and E. M. Wright, Am. J. Physiol. Cell Physiol., 2012, 302, C373–C382 CAS.
- M. P. Lostao, B. A. Hirayama, D. D. Loo and E. M. Wright, J. Membr. Biol., 1994, 142, 161–170 CAS.
- Y. Niu, R. Liu, C. Guan, Y. Zhang, Z. Chen, S. Hoerer, H. Nar and L. Chen, Nature, 2022, 601, 280–284 CAS.
- M. Namavari, G. Gowrishankar, A. Srinivasan, S. S. Gambhir, T. Haywood and C. Beinat, J. Labelled Compd. Radiopharm., 2018, 61, 408–414 CrossRef CAS PubMed.
- R. Boellaard, R. Delgado-Bolton, W. J. Oyen, F. Giammarile, K. Tatsch, W. Eschner, F. J. Verzijlbergen, S. F. Barrington, L. C. Pike, W. A. Weber, S. Stroobants, D. Delbeke, K. J. Donohoe, S. Holbrook, M. M. Graham, G. Testanera, O. S. Hoekstra, J. Zijlstra, E. Visser, C. J. Hoekstra, J. Pruim, A. Willemsen, B. Arends, J. Kotzerke, A. Bockisch, T. Beyer, A. Chiti, B. J. Krause and M. European Association of Nuclear, Eur. J. Nucl. Med. Mol. Imag., 2015, 42, 328–354 CrossRef CAS PubMed.
- K. Johnsrud, K. Skagen, T. Seierstad, M. Skjelland, D. Russell and M. E. Revheim, J. Nucl. Cardiol., 2019, 26, 883–893 CrossRef PubMed.
- W. S. Lee, Y. Kanai, R. G. Wells and M. A. Hediger, J. Biol. Chem., 1994, 269, 12032–12039 CrossRef CAS PubMed.
- M. W. Song, W. Cui, C. G. Lee, R. Cui, Y. H. Son, Y. H. Kim, Y. Kim, H. J. Kim, S. E. Choi, Y. Kang, T. H. Kim, J. Y. Jeon and K. W. Lee, Front. Pharmacol, 2023, 14, 1228646 CrossRef CAS PubMed.
- H. Y. Kueh, A. Champhekar, S. L. Nutt, M. B. Elowitz and E. V. Rothenberg, Science, 2013, 341, 670–673 CrossRef CAS PubMed.
- S. Han, D. L. Hagan, J. R. Taylor, L. Xin, W. Meng, S. A. Biller, J. R. Wetterau, W. N. Washburn and J. M. Whaley, Diabetes, 2008, 57, 1723–1729 CAS.
- W. Meng, B. A. Ellsworth, A. A. Nirschl, P. J. McCann, M. Patel, R. N. Girotra, G. Wu, P. M. Sher, E. P. Morrison, S. A. Biller, R. Zahler, P. P. Deshpande, A. Pullockaran, D. L. Hagan, N. Morgan, J. R. Taylor, M. T. Obermeier, W. G. Humphreys, A. Khanna, L. Discenza, J. G. Robertson, A. Wang, S. Han, J. R. Wetterau, E. B. Janovitz, O. P. Flint, J. M. Whaley and W. N. Washburn, J. Med. Chem., 2008, 51, 1145–1149 CrossRef CAS PubMed.
- M. Obermeier, M. Yao, A. Khanna, B. Koplowitz, M. Zhu, W. Li, B. Komoroski, S. Kasichayanula, L. Discenza, W. Washburn, W. Meng, B. A. Ellsworth, J. M. Whaley and W. G. Humphreys, Drug Metab. Dispos., 2010, 38, 405–414 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: 1H, 13C, and 19F NMR spectra for MFTMT (6) and MFTM (13), radiometric HPLC data for [18F]13, timelines of biological experiments, radiometric HPLC data for enzymatically digested [18F]MFTMT, body weights of animals, intensity of injected radioactivity for PET imaging, the numeric values of bacteria/cell uptake studies and PET imaging analysis data, and molecular formula strings. See DOI: https://doi.org/10.1039/d5ra00693g |
| ‡ Co-corresponding author. |
| This journal is © The Royal Society of Chemistry 2025 |