Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A new natural Cyperol A together with five known compounds from Cyperus rotundus L.: isolation, structure elucidation, DFT analysis, insecticidal and enzyme-inhibition activities and in silico study†
Saqib Hussain Bangash ab,
Muhammad Ibrahim*b,
Akbar Ali
ab,
Muhammad Ibrahim*b,
Akbar Ali *c,
Chen-Yang Weia,
Amjad Hussaind,
Moazama Riazb,
Muhammad Fayyaz Ur Rehman
*c,
Chen-Yang Weia,
Amjad Hussaind,
Moazama Riazb,
Muhammad Fayyaz Ur Rehman e,
Faiz Ahmed
e,
Faiz Ahmed c,
Rashad Al-Salahi
c,
Rashad Al-Salahi f and
Wen-Wei Tang*a
f and
Wen-Wei Tang*a
aGuangxi Key Laboratory of Agro-Environment and Agric-Product Safety, National Demonstration Center for Experimental Plant Science Education, College of Agriculture, Guangxi University, Nanning, Guangxi, People's Replublic of China. E-mail: wenweitg@163.com
bDepartment of Applied Chemistry, Government College University Faisalabad, Pakistan. E-mail: ibrahimchem@gmail.com
cDepartment of Chemistry, Government College University Faisalabad, Pakistan. E-mail: akbarali@gcuf.edu.pk; akbarchm@gmail.com
dInstitute of Chemistry, University of Okara, Okara-56300, Punjab, Pakistan
eInstitute of Chemistry, University of Sargodha, Sargodha, Pakistan
fDepartment of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, Riyadh, 11451, Saudi Arabia
First published on 11th April 2025
Abstract
One new natural benzaldehyde derivative (1), together with five known compounds, was isolated from the methanolic extract of the whole plant of Cyperus rotundus L., which is a globally distributed noxious weed. The structure of compound (1) (named Cyperol A) was determined using various NMR methods, including 1H, 13C, COSY, HMBC, HSQC and NOESY, and mass spectrometric techniques, including EIMS. The newly isolated compound (1) was subjected to optimization using computer-assisted calculation via DFT methods for natural bond orbital (NBO) and frontier molecular orbital (FMO) analyses and compared with carbofuran, which is used to control the pest brown planthopper. The in vitro insecticidal efficacy of compounds 1–6 was evaluated against Nilaparvata lugens. Compound 1 demonstrated exceptional lethal and notable enzyme inhibitory effects. Furthermore, compound 1 was investigated in silico for its anti-pesticidal activities targeting the BPH (Nilaparvata lugens (Stål)) key enzymes, such as glutathione S-transferase (GST) and acetylcholinesterase (AChE). Compound 1 showed good docking scores of −9.75 kcal mol−1 against GST, forming hydrogen bonds with its active site, and −10.56 kcal mol−1 with AChE owing to its high potential for hydrogen bonding.
1. Introduction
Purple nutsedge (Cyperus rotundus L.) is a member of the Cyperaceae family and is considered one of the most noxious weeds in the world.1 It is widely distributed in the tropical and subtropical regions, infesting fields that include sugarcane, maize, and even rice. In the rice fields, brown planthoppers (Nilaparvata lugens (Stål)) have been reported as one of the most common pests. Interestingly, C. rotundus has never been reported as a food source or host of brown planthoppers.2 Meanwhile, studies have shown that this weed contains many phytochemicals, including steroids, triterpenes, tannins, anthraquinones, and alkaloids, which are known for their potent insecticidal activities.3 Additionally, oils extracted from this weed have been reported to function as natural insecticides, serving as toxic fumigants4 and insect repellents5 with ovicidal potential. Thus, in recent years, there has been a growing interest in using compounds derived from C. rotundus to manage pests, particularly in the stored products.6 Therefore, we speculate that certain natural phytochemicals may be responsible for the brown planthoppers not choosing C. rotundus as a food source or host.Density functional theory (DFT) is an exciting computer-assisted calculation technique used to compute molecular structures, which aids in identifying plant-derived substances and confirming their vibrational frequencies and molecular parameters derived from experimental spectroscopic techniques. Additionally, it plays a key role in facilitating molecular modelling, which has revolutionized our understanding of the fundamental physical and chemical interactions and the prediction of the physiochemical properties of natural compounds extracted from plants.7–9 The toxicity of insecticidal compounds is often linked to their impact on biochemical pathways, particularly through enzyme inhibition activities.10 Organophosphates, carbamates, and certain botanical compounds inhibit acetylcholinesterase (AChE) activity, leading to neuronal overstimulation, muscle tremors, convulsions, and ultimately insect mortality.11,12 Furthermore, molecular docking is an emerging and powerful technique that enables in-depth exploration of ligand–receptor interactions and precise predictions of optimal binding configurations between ligands and targeted proteins.13 These methods efficiently screen large chemical libraries and assist researchers in understanding the extracted natural compounds.14
In the current study, five known compounds and Cyperol A (a novel natural compound) were isolated from the whole plant of C. rotundus. The structure of Cyperol A was elucidated using spectroscopic techniques, including NMR, EIMS, and HRMS. The newly isolated natural aromatic aldehyde was subjected to DFT studies, along with assessments of its lethal activity and inhibitory effects on AChE and GST against N. lugens. Additionally, molecular docking studies were conducted to evaluate its potential as a natural insecticide. Notably, the results from these analyses provide valuable insights into C. rotundus as a promising source of eco-friendly insecticides that can contribute to sustainable pest management solutions in agriculture.
2. Results and discussion
The methanolic extract (300 g) of the whole plant of C. rotundus was fractionated using repeated column chromatography over silica gel, Sephadex (LH-20) and normal phase chromatography, resulting in the isolation of a new natural product (1), along with five known compounds: biochanin A (2), cyperotundol (3), p-coumaric acid (4), chlorogenic acid (5) and quercetin (6)15,16 (Fig. 1).Compound 1 was isolated as a yellowish amorphous solid; m.p: 186–188 °C, IR (CHCl3) ύmax cm−1: 3400 cm−1 (OH), 1720 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); UV λmax nm (MeOH) (log
O); UV λmax nm (MeOH) (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 305 (2.8); EI-MS: m/z [M]+ = 166.13 (calculated 166.13 for C8H6O4). The 1H-NMR spectrum of compound 1 was resolved into four singlets including one signal in the downfield region resonating at δH 10.21 (H-8), characteristic of an aldehydic proton, which indicated the presence of an aldehydic group, and two aromatic methine protons at δH 7.24 (1H, s, H-5) and δH 7.33 (1H, s, H-7) indicated a tetra-substituted aromatic ring in the compound. A doubly integrated singlet, characteristic of methylene protons, resonated at δH 6.20 (2H, s, H-2), while a singlet corresponding to a single OH group was observed at δH 7.52 (Fig. S1†). The 13C-NMR spectrum of compound 1 was resolved into 8 resonances. The DEPT-90 and 135 helped in identifying the presence of one oxy-methylene group, indicated by a downfield signal at δC 104.0 (C-2), and two methine carbons at δC 107.7 (C-5) and δC 105.3 (C-7). The four quaternary carbons in the aromatic region were assigned as δC 128.3 (C-4), δC 152.3 (C-6), δC 151.6 (C-1a), and δC 147.3 (C-3a). A single downfield signal at δC 186.9 (C-8) in the broadband, DEPT-90 and DEPT-135 of the 13C-NMR spectra indicated the presence of an aldehyde function in the compound. The HMBC spectrum (Fig. 2 and S8†) displayed the interactions of the OH proton (δO
ε): 305 (2.8); EI-MS: m/z [M]+ = 166.13 (calculated 166.13 for C8H6O4). The 1H-NMR spectrum of compound 1 was resolved into four singlets including one signal in the downfield region resonating at δH 10.21 (H-8), characteristic of an aldehydic proton, which indicated the presence of an aldehydic group, and two aromatic methine protons at δH 7.24 (1H, s, H-5) and δH 7.33 (1H, s, H-7) indicated a tetra-substituted aromatic ring in the compound. A doubly integrated singlet, characteristic of methylene protons, resonated at δH 6.20 (2H, s, H-2), while a singlet corresponding to a single OH group was observed at δH 7.52 (Fig. S1†). The 13C-NMR spectrum of compound 1 was resolved into 8 resonances. The DEPT-90 and 135 helped in identifying the presence of one oxy-methylene group, indicated by a downfield signal at δC 104.0 (C-2), and two methine carbons at δC 107.7 (C-5) and δC 105.3 (C-7). The four quaternary carbons in the aromatic region were assigned as δC 128.3 (C-4), δC 152.3 (C-6), δC 151.6 (C-1a), and δC 147.3 (C-3a). A single downfield signal at δC 186.9 (C-8) in the broadband, DEPT-90 and DEPT-135 of the 13C-NMR spectra indicated the presence of an aldehyde function in the compound. The HMBC spectrum (Fig. 2 and S8†) displayed the interactions of the OH proton (δO![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 7.52) with C-5 and C-7, which helped in determining the position of the hydroxy group at C-6. Other observed HMBC correlations included H-5 with C-4, C-6, C-7, C-3a and C-8; H-7 with C-5, C-6, C-1a, C-9 on one side; and the HMBC interactions of H-8 with C-4, C-5 and C-3a, which helped in identifying the position of the carboxyl group at C-4 on the aromatic ring. The important HMBC interactions of δ 2H at C-2 with C-1a and C-3a confirmed the presence of a dioxole group and its position at C-1a and C-3a on the aromatic ring. These data closely aligned with those of 6-hydroxy-2H-1,3-benzodioxole-5-carbaldehyde,17 confirming the position of the aldehyde group at C-4 in compound 1. Additionally, the NOSEY and COSY NMR spectra showed no correlations (Fig. S5 and S6,† respectively). Consequently, the structure of compound 1 was elucidated as 6-hydroxy-2H-1,3-benzodioxole-4-carbaldehyde, as shown in Fig. 1. Thus, the compound was named Cyperol A.
7.52) with C-5 and C-7, which helped in determining the position of the hydroxy group at C-6. Other observed HMBC correlations included H-5 with C-4, C-6, C-7, C-3a and C-8; H-7 with C-5, C-6, C-1a, C-9 on one side; and the HMBC interactions of H-8 with C-4, C-5 and C-3a, which helped in identifying the position of the carboxyl group at C-4 on the aromatic ring. The important HMBC interactions of δ 2H at C-2 with C-1a and C-3a confirmed the presence of a dioxole group and its position at C-1a and C-3a on the aromatic ring. These data closely aligned with those of 6-hydroxy-2H-1,3-benzodioxole-5-carbaldehyde,17 confirming the position of the aldehyde group at C-4 in compound 1. Additionally, the NOSEY and COSY NMR spectra showed no correlations (Fig. S5 and S6,† respectively). Consequently, the structure of compound 1 was elucidated as 6-hydroxy-2H-1,3-benzodioxole-4-carbaldehyde, as shown in Fig. 1. Thus, the compound was named Cyperol A.
The spectroscopic data of compounds 2–6 were compared with those of previously reported compounds and were found to be identical to biochanin A (2), cyperotundol (3), p-coumaric acid (4), chlorogenic acid (5) and quercetin (6).15,16
2.1. Computational procedures
 | (1) |
In Cyperol A, the highest stabilization energy (E2) is reported (Table S1†) due to the conjugate interaction of the filled molecular orbital lone pair of (O12), (O17) and (O18) with unfilled (π* C11![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16), (π* C1
C16), (π* C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2) and (π* C1
C2) and (π* C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2) molecular orbital, respectively. The E2 of these interactions are 20.39, 28.14 and 27.07 kJ mol−1, respectively, which is due to intramolecular charge transfer (ICT), resulting in the high stabilization of the investigated compound. The strong electron donor interactions, π(C1
C2) molecular orbital, respectively. The E2 of these interactions are 20.39, 28.14 and 27.07 kJ mol−1, respectively, which is due to intramolecular charge transfer (ICT), resulting in the high stabilization of the investigated compound. The strong electron donor interactions, π(C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2) → π*(C3
C2) → π*(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4) (18.50 kJ mol−1), π(C1
C4) (18.50 kJ mol−1), π(C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2) → π*(C5
C2) → π*(C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16) (18.73 kJ mol−1), π(C3
C16) (18.73 kJ mol−1), π(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4) → π*(C1
C4) → π*(C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2) (17.87 kJ mol−1), π(C3
C2) (17.87 kJ mol−1), π(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4) → π*(C5
C4) → π*(C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16) (15.20 kJ mol−1), π(C5
C16) (15.20 kJ mol−1), π(C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16) → π*(C1
C16) → π*(C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2) (18.35 kJ mol−1), π(C5
C2) (18.35 kJ mol−1), π(C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16) → π*(C3
C16) → π*(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4) (15.29 kJ mol−1), and π(C5
C4) (15.29 kJ mol−1), and π(C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C16) → π*(C11 = O12) (16.59 kJ mol−1) in the aromatic ring are identified. Another prominent intramolecular hyperconjugative interaction occurs between the lone pair on (O12) and the antibonding σ* orbitals of (C11) and σ*(C11–H13) having the E2 of 11.52 and 19.07 kJ mol−1, respectively. These intramolecular charge transfers (ICT) contribute to the overall stabilization of the investigated compound.
C16) → π*(C11 = O12) (16.59 kJ mol−1) in the aromatic ring are identified. Another prominent intramolecular hyperconjugative interaction occurs between the lone pair on (O12) and the antibonding σ* orbitals of (C11) and σ*(C11–H13) having the E2 of 11.52 and 19.07 kJ mol−1, respectively. These intramolecular charge transfers (ICT) contribute to the overall stabilization of the investigated compound.
 | ||
| Fig. 4 Frontier molecular orbitals of Cyperol A and carbofuran at the B3LYP/6-311G (d,p) level; red and green color represent positive and negative phases, respectively. | ||
Meanwhile, the charges were transferred to the fused carbon atom and the adjacent carbon atom of the benzodioxole nucleus. In FMO analysis, higher reactivity is estimated by a low energy gap, while lower reactivity is estimated by a larger energy gap.23 Besides the FMOs analysis, the energies of the HOMO and LUMO are valuable for calculating reactivity descriptors such as ionization potential (I), electron affinity (A), chemical hardness (η), global softness (ω), and chemical potential (μ), as defined by Koopman's theorem.24,25 The electron affinity (A) and ionization potential (I) can be calculated using eqn (2) and (3) (Table S3†).26 Meanwhile, global hardness (η), electronegativity (X), and chemical potential (μ) were calculated using eqn (4)–(6) (Table S3†).27 Electrophilicity (ω) was calculated to determine the charge transfer using eqn (7) (Table S3†), while global softness (σ) was calculated using eqn (8) (Table S3†).28,29
In Table 1, the computed values for ionization potential (I), electron affinity (A), global hardness (η), electronegativity (X), chemical potential (μ), electrophilicity (ω), and global softness are shown. The electron donor and acceptor behavior of materials can be described using the ionization potential (IP) and electron affinity (EA), respectively. The calculated values of IP and EA were 5.60 eV and 1.78 eV. The predicted value of electronegativity (3.69 eV) was significantly high. The stability and reactivity of the material can be predicted by global hardness (η), and the value of η for Cyperol A was 1.91 eV. Furthermore, the predicted values of chemical potential (μ), chemical softness (σ), and global electrophilicity index (ω) were −1.91, 0.52, and 0.95, respectively. All these molecular descriptors indicate that Cyperol A has greater biological potential compared to the BPH insecticide carbofuran.
| Molecular descriptors | Cyperol A | Carbofuran |
|---|---|---|
| EHOMO (eV) | −5.60 | −8.35 |
| ELUMO (eV) | −1.78 | −4.27 |
| ΔEHOMO–LUMO (eV) | 3.82 | 4.08 |
| Ionization potential, IP (eV) | 5.60 | 8.35 |
| Electron affinity, EA (eV) | 1.78 | 4.27 |
| Electronegativity, χ (eV) | 3.69 | 6.31 |
| Global hardness, η (eV) | 1.91 | 2.04 |
| Chemical potential, μ (eV) | −1.91 | −2.04 |
| Chemical softness, σ (eV−1) | 0.52 | 0.49 |
| Global electrophilicity index, ω (eV) | 0.95 | 1.02 |
| Dipole moment (debye) | 5.41 | 6.09 |
| Polarizability α (a.u) | 125.05 | 137.78 |
2.2. Vibrational analysis
The theoretical FTIR bands and relative intensities of Cyperol A were computed at the DFT/B3LYP/6-311G (d,p) level to predict the vibrational modes (Table S2†).30,31![[double bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e001.gif) C stretching vibration. The experimental FTIR stretching vibration of the C
C stretching vibration. The experimental FTIR stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond is generally located between 1660 and 1600 cm−1 in FTIR spectra, while for the aromatic system, it is found between 1600 and 1475 cm−1.34 The stretching C
C bond is generally located between 1660 and 1600 cm−1 in FTIR spectra, while for the aromatic system, it is found between 1600 and 1475 cm−1.34 The stretching C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibration for the aromatic region was observed at 1693–1460 and 1322–1230 cm−1 for 1. The bending C
C vibration for the aromatic region was observed at 1693–1460 and 1322–1230 cm−1 for 1. The bending C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibrations were found at 395, 482, 494, 513, 608, 618, 702, 735, 760, 818, 832, 953 and 395 cm−1 for compound 1. The rotational vibrations were found at 1131, 1021, 833, 688, 612, 505, 408, 338, 245, 210, 196, 164, 158, and 101 cm−1 for Cyperol A.
C vibrations were found at 395, 482, 494, 513, 608, 618, 702, 735, 760, 818, 832, 953 and 395 cm−1 for compound 1. The rotational vibrations were found at 1131, 1021, 833, 688, 612, 505, 408, 338, 245, 210, 196, 164, 158, and 101 cm−1 for Cyperol A.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration is typically found between 1850 and 1600 cm−1.35 For Cyperol A, the C
O stretching vibration is typically found between 1850 and 1600 cm−1.35 For Cyperol A, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of the aldehyde group was located at 1768 cm−1. The C–H bending vibrations of the aldehyde group were located at 1443, 1432, 1021, 505, 408, and 196 cm−1. The rotational vibrations were found at 93, 152, 156, 199, 267, 290, 313, 354 and 395 cm−1.
O stretching vibration of the aldehyde group was located at 1768 cm−1. The C–H bending vibrations of the aldehyde group were located at 1443, 1432, 1021, 505, 408, and 196 cm−1. The rotational vibrations were found at 93, 152, 156, 199, 267, 290, 313, 354 and 395 cm−1.2.3. Time- and dose-dependent toxicity of compound 1
Previously, for carbofuran, the highest mortality of 73.3% was recorded at a dosage of 0.5 kg ai ha−1, which was effective in controlling the brown planthopper (BPH) at 72 hours after treatment (HAT).36 Similarly, carbofuran exhibited significant dose- and time-dependent mortality against the brown planthopper (Nilaparvata lugens), with 90.0% mortality observed 24 hours after treatment (HAT), increasing to 100% at 48 HAT and remaining consistent at 72 HAT.37 The primary challenge in controlling the brown planthopper (Nilaparvata lugens) is the development of insecticide resistance. Different studies have reported varying LC50 values for insecticides against N. lugens. In one study, the LD50 value for carbofuran was reported to be 20.3 μg g−1, with a 95% confidence interval ranging from 13.4 to 91.1 μg g−1.38A diverse group of isolated natural compounds, present in numerous plant species, exhibits a wide range of biological activities. Many natural compounds have gained attention as promising botanical insecticides due to their potent antifeedant and insecticidal properties against various insect pests.39 Only a few rare natural compounds isolated from plants have been demonstrated to possess anti-complement and antifeedant activities.40 However, this study represents the first report on the contact toxicity of naturally isolated Cyperol A against N. lugens.
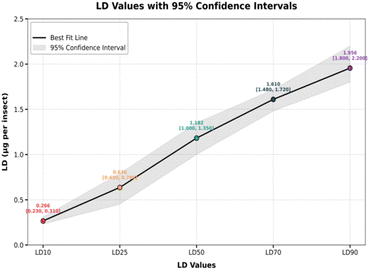
In the current study, compound 1 exhibited potent insecticidal activity, characterized by both time- and dose-dependent toxicity, making it a highly effective candidate for pest control. The LD50 value of 1.18 μg per insect (95% CI: 0.89–1.38) underlines its potency, achieving 50% mortality at relatively low doses. Even minimal concentrations are toxic, as evidenced by the LC10 and LC25 values of 0.26 μg per insect and 0.63 μg per insect, respectively. At higher doses, the efficacy is enhanced, with LC70 and LC90 values of 1.61 μg per insect and 1.95 μg per insect, respectively. The steep slope of the Hill (1.158) signifies a sharp increase in mortality with increasing doses, which is characteristic of potent insecticides. Moreover, the high R-squared value of 0.9368 validates the reliability of the data, confirming the compound's robust dose–response relationship. These findings highlight the efficiency of compound 1 in achieving significant insect mortality, providing flexibility in dose application based on pest severity (Fig. 6).
There is also a marked time dependency, with mortality rates progressively increasing over time. At lower time points, such as 8 hours, mortality is modest, even at higher doses. However, with prolonged exposure at 24, 48, and 72 hours, the mortality rates rise sharply, demonstrating the prolonged efficacy of the compound. For instance, at 1 μg per insect, mortality increases from approximately 20% at 8 hours to almost 70% at 72 hours (Fig. 7).
This comparison highlights the superior efficacy of compound 1 as an insecticide against N. lugens and underlines its potential for effective pest control applications in agriculture.
In contrast, compounds 2 to 6 exhibited significantly lower mean mortality percentages across all dose levels. When the LD50 value exceeds 2 μg per insect, it is categorized as non-toxic.41 In our study, compounds 2–6 are classified as non-toxic based on their mortality results, as their LD50 values exceed 2 μg per insect.
For instance, at the highest tested dose (2.4 μg per insect), compound 2 achieved only 16% mean mortality, while compound 6 achieved 21.8%. This comparison highlights the superior efficacy of compound 1 as an insecticide against N. lugens and underlines its potential for effective pest control applications in agriculture.
2.4. Time- and dose-dependent AChE inhibition by compound 1
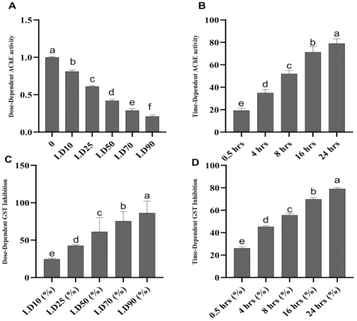
The brown planthopper (Nilaparvata lugens) exhibits significant insecticide resistance due to reduced AChE sensitivity. The IC50 values previously reported for carbofuran are 1.5 × 10−2 mM and 8.4 × 10−2 mM in the resistant Rc-30 and Rf-30 strains, respectively, compared to 7.8 × 10−4 mM in the susceptible (S) strain. These values correspond to 19.2- and 107.7-fold insensitivity ratios, indicating a marked reduction in carbofuran efficacy against resistant populations.38In our study, compound 1 demonstrated the most potent acetylcholinesterase (AChE) inhibition, exhibiting strong dose- and time-dependent effects compared to other compounds. At the LD50 dose (1.182 μg per insect), the inhibition rate increases progressively over time, starting at 19.3% at 0.5 hours and peaking at 79% at 24 hours. This trend highlights the prolonged effect and significant inhibitory potential of compound 1, indicating that it maintains high efficacy over extended periods. Its superior performance at all time intervals reflects its ability to efficiently disrupt AChE activity, making it highly effective as a pesticide.
In contrast, compounds 2–6 exhibit considerably weaker AChE inhibition. For instance, at 0.5 hours, compound 2 inhibits AChE by only 9%, while compound 6 achieves only 7%. At 24 hours, the inhibition rates for these compounds remain significantly lower, with compound 2 reaching 26% and compound 6 achieving 25%, both substantially below 79% of compound 1. Similarly, compounds 3–5 display lower inhibition rates and do not match the effectiveness of compound 1 across all time intervals. This comparative analysis clearly identifies compound 1 as the most promising candidate for pest control due to its robust and sustained AChE inhibition over time, which is critical for the effective disruption of the insect nervous system function.
2.5. Time and dose-dependent inhibition of glutathione S-transferase (GST)
Detoxification enzymes, including carboxylesterases (CarE), acid and alkaline phosphatases (ACP and ALP), cytochrome P450 monooxygenases (P450s), and glutathione S-transferases (GSTs), are crucial for neutralizing toxic compounds.10 A reduction in the activity of these enzymes is closely linked to a decreased ability of insects to metabolize and eliminate insecticides.42 Previous research indicates that exposure to certain compounds, such as kaempferol-3-O-glu-rha-glu, quercetin-3-O-glu-rha-glu,43 and pectolinarigenin,44 can significantly inhibit the activities of P450s and GSTs, which are phase I detoxification enzymes. Such inhibition weakens the detoxification capacity and reduces the metabolic ability of N. lugens. These effects correlate with the increased mortality in insects exposed to insecticides.45In the dose-dependent assay, compound 1 exhibited the strongest inhibition at all doses tested, with inhibition reaching 86.3% at LD90, demonstrating its high potency and efficacy. The inhibition was consistent across doses, and its variability was low (±1.6% SD). Compound 2 showed moderate inhibition, with a peak of 48.5% at LD90, while compound 3 displayed the weakest inhibition, reaching only 36.9% at the same dose. Compounds 4–6 showed intermediate inhibition, with values ranging from 43.2% (compound 4) to 41.7% (compound 6) at LD90 (Fig. 8).
In the time-dependent assay, compound 1 again exhibited the strongest inhibition, achieving 79.1% at 24 hours with low variability (±1.3% SD). The other compounds showed a gradual increase in inhibition over time. Compound 2 reached 43.2% at 24 hours, while compounds 3–6 exhibited relatively weaker inhibition, with 28.1% (compound 3) to 38.8% (compound 4) at the same time point.
The exceptional performance of compound 1 in both assays highlights its potential as a highly effective insecticide. Its potent and consistent inhibition of GST makes it a promising candidate for insecticide development, offering rapid and sustained activity. The potent insecticidal effects of compound 1 could be pivotal in developing more efficient and eco-friendly pest management strategies, potentially reducing reliance on traditional chemical insecticides and contributing to sustainable agricultural practices. Further research into optimizing the formulation and application of compound 1 could enhance its utility in integrated pest management (IPM) systems, ensuring long-term control of pests like the brown plant hopper (BPH).
3. In silico studies
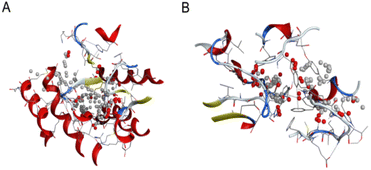
Plant-derived substances are eco-friendly and effective against insects, making them potential alternatives to synthetic insecticides. Finding new substances that bind molecular targets could be made easier and faster through molecular docking. Insects have important detoxifying enzymes such as glutathione S-transferase (GST), acetylcholinesterase (AChE), and cytochrome P450. This enzyme system (CYP450) contributes to their resistance to several insecticides and environmental stresses; thus, targeting these enzymes is a promising approach for the development of new insecticides (Fig. 9).46,47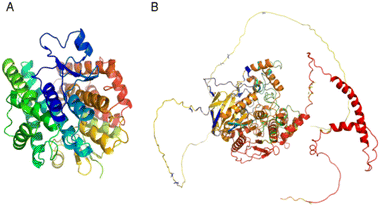 | ||
| Fig. 9 Three-dimensional (3D) structures of N. lugens enzymes: (A) glutathione S-transferase (GST) from the protein data bank (PDB), (B) acetylcholinesterase (AChE) from AlphaFold. | ||
The glutathione S-transferase of N. lugens structures was retrieved from the Protein Data Bank (PDB) database (PDB code: 3WYW) and Acetylcholinesterases (AChE) with accession number AF-G9BJC1-F1. The structural model was obtained from the AlphaFold database (https://www.AlphaFold.org).46 Polar hydrogen atoms were added to the protein and ligand by MOE (Molecular Operating Environment version 2019.0102), and energy minimization was applied to prepare them for docking, followed by 3D protonation.47
3.1. Validation of the modelled structures
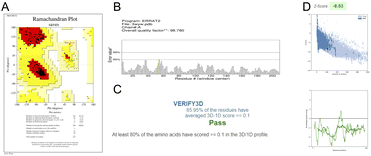
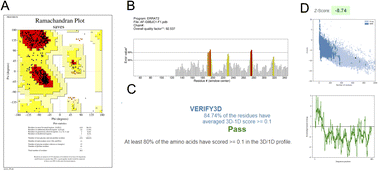
Different types of stereochemical parameters of the protein structure were predicted online using PROCHECK, ERRAT, and Verify3D (https://www.saves.mbi.ucla.edu). PROCHECK analyzes the overall model geometry by generating the Ramachandran plot with favorable and non-favorable residue regions. ERRAT is a database of highly refined protein structures that uses a nine-residue sliding window to assess the residue versus error relationship. Verify3D validates a 3D structure in terms of loops, sheets, and alpha helices.48,49The Ramachandran plots for GST and AChE are given in Fig. 10A and 11A. In ERRAT, the GST and AChE structures show overall quality factors of 98.78 and 92.53, respectively (Fig. 10B and 11B). In Verify3D model verification, where the minimum passing score was 80%, the 3D models received scores of 85.95 and 84.74% for GST and AChE (Fig. 10C and 11C). The ProSA-web server was utilized to compare the protein structural models to the PDB's protein structures based on Z-score.50
A plot of the residual energy and the input structure's Z-score was provided by the application SWISS-MODEL. The Z-score showed Z-scores of −8.53 and −8.74 for GST and AChE (Fig. 10D and 11D), which represent excellent models.
3.2. Active site prediction
Docking efficiency is significantly increased when the location of the active sites or binding sites is known before the docking procedures. Cavity detection tools can be used to find probable active sites within proteins. Blind docking is the process of docking without making any assumptions about the binding location.51 The type of interactions between protein molecules dictates their biological roles, which involve a few residues and binding sites where the interaction occurs. One of the most important steps in finding new chemical entities in molecular docking for structure-based drug design is identifying the binding site, which is accomplished by examining the physicochemical and shape properties of the protein area. The target proteins and binding sites with little to no ability to bind ligands can be excluded from the binding site identification process.52,53Twenty-six high-potential binding sites were found in GST and the one that has the following amino acids was chosen: VAL8, PRO9, GLY10, SER11, ALA12, PRO13, THR33, ASP34, LEU35, LYS36, HIS40, GLN51, HIS52, ASN53, VAL54, PRO55, ASN65, GLU66, SER67, ARG68, MET103, TYR107, GLN108, GLY111, ASP112, TYR115, PRO116, PHE119, LYS128, GLU201, GLY202, GLY205, PHE206, GLN208, MET209, TYR100, GLY104, THR105 GLN108, SER109, ASP112, PRO116, ASP127, LYS128, LYS131, ASP134, ALA135 and PHE138.
Furthermore, 30 binding sites in AChE were analysed, and focusing on the binding sites with amino acids ILE199, ASP201, SER210, TRP213, PHE245, GLY246, GLY247, GLY248, TYR250, SER251, GLY252, THR253, LEU256, TYR259, ALA280, GLU327, SER328, ALA329, GLY330, ALA331, TRP409, GLY410, THR411, LEU412, GLY413, ILE414, CYS415, GLU416, PHE417, TYR457, PHE458, TYR461, HIS462, HIS568, GLY569 and ILE572 were selected. Previously, azadirachtin was found to interact with acetylcholinesterase of Tribolium castaneum.54,55
Moreover, an activity prediction test was also employed, following the method described by Empereur-Mot et al., to confirm the modelled protein's active site.56 Finally, the new isolated compound was accurately docked into the proposed active sites, as shown in Fig. 12 (Table 2).
3.3. ADME (absorption, distribution, metabolism, and excretion) analysis
The Swiss ADMET online tool was utilized to predict the absorption, distribution, metabolism, elimination and pharmacokinetic properties of molecules to evaluate the Lipinski five rules (MW, iLOGP, HBAs, and HBDs).57 MW was recorded as 166.13 g mol−1, and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P was 1.33. Cyperol A presented no violations, and according to Lipinski's rule of five, it is the best drug candidate.
P was 1.33. Cyperol A presented no violations, and according to Lipinski's rule of five, it is the best drug candidate.
3.4. Molecular docking
Molecular docking stimulates interactions between the ligand and the protein at the atomic level, clarifies basic biochemical processes, and characterizes the behavior of small molecules at the target protein binding sites. The two fundamental steps in the docking process are predicting the ligand's orientation and position within these sites, also known as pose, and evaluating the binding affinity.58 The most crucial element of the structure-based drug design is the scoring function. The scoring functions are mathematical functions used to roughly forecast their binding affinities. In the ligand–receptor interactions, the lowest scores are ideal and are frequently utilized in virtual screening and drug discovery.59 The outcomes of our docking study demonstrated that the chosen inhibitor docked within the pockets of N. lugens AChE and GST target proteins has potential interactions with AChE and GST. Following the in silico docking, the lowest S score of Cyperol A with GST was assessed, which established the strongest interaction with a minimum S score of −9.75, followed by −9.42 (Table 2). AChE shows strong and the best interactions with Cyperol A, with a docking score of −10.83, followed by −9.97, as shown in (Table 3). This indicates the best docking score and shows the capability of Cyperol A in insecticide development.3.5. Protein–ligand interaction
PLIP (https://plip-tool.biotec.tu-dresden.de) was used to map the interaction of GST and AChE with Cyperol A. Cyperol A formed hydrogen bonds with two specific amino acids (PHE119 and GLY120) in its interaction with GST. The bond distances were measured to be 2.39 and 3.21 Å. Cyperol A also formed one hydrophobic interaction with the amino acid PRO116 with GST (Fig. 13A).Nine hydrogen bonds and two hydrophobic interactions were also found between Cyperol A and AChE. The observed hydrogen bonds with amino acids GLY246A, GLY247, TYR259, TYR259, SER328, ALA329, GLY330, ALA331, and HIS568, with bond distances measuring 3.07, 4.0, 2.61, 2.61, 2.18, 2.93, 2.37, 2.36 and 1.91 Å, respectively. Hydrophobic interactions of Cyperol A were also found with amino acids GLU327 and SER328 (Fig. 13B).
4. Method and materials
4.1. General experimental
The JASCO A-302 and Shimadzu UV-240 spectrophotometers were used to record the IR and UV spectra, respectively, while the JASCO P-2000 Polarimeter was used to record the specific rotations of all the isolated compounds. 1H and 13C-NMR spectra were taken on Bruker Avance NMR spectrometers (300, 400, and 500 MHz). Finnigan MAT 95 XP, Finnigan MAT 311, and Jeol JMS HX 110 mass spectrometers were used for low-resolution EI, HREI-MS, HRMS Q-Exactive MS-US Thermo Fisher Scientific, and FABMS investigations, respectively. Silica gel (70–230 mesh, Kieselgel 60) was used for the column chromatography, and TLCs were performed on F254 aluminium sheets (0.25 mm).4.2. Plant material
The C. rotundus plant material was collected from the Okara region, Punjab, Pakistan, and a voucher specimen (A.R., no. 112) was submitted to the Department of Botany's Herbarium of Government College University Faisalabad, Pakistan.4.3. Extraction and isolation
The plant was dried under the shade and powdered. The dried powdered material was then soaked in 95% methanol at room temperature (3 × 5 days × 25 L) to obtain 300 g of crude extracts of Cyperus rotundus.4.3.1.1 Vacuum liquid chromatography. The crude extract of C. rotundus, weighing 300 g, was subjected to vacuum liquid chromatography using an ethyl acetate/n-hexane solvent system and 1.0 kg of silica gel to obtain fractions (I–V): (Fr. I, 0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10), (Fr. II, 1
10), (Fr. II, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9), (Fr. III, 2
9), (Fr. III, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8), (Fr. IV, 4
8), (Fr. IV, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6), and (Fr. V, 6
6), and (Fr. V, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4).
4).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9), (Fr. V2, 2
9), (Fr. V2, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8), (Fr. V3, 3
8), (Fr. V3, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7), and (Fr. V4, 5
7), and (Fr. V4, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5).
5).Fraction V3 (5.3 g) was further subjected to column chromatography to get three fractions using pure methanol as the solvent system in a Sephadex (LH-20) column (500 g, 3 × 60 cm): (Fr. V3A, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9), (Fr. V3B, 3
9), (Fr. V3B, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7), and (Fr. V3C, 2
7), and (Fr. V3C, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). A new compound 1 (178 mg) was purified from Fr. V3A (965 mg) through normal phase column chromatography in an acetone/n-hexane 3
3). A new compound 1 (178 mg) was purified from Fr. V3A (965 mg) through normal phase column chromatography in an acetone/n-hexane 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 solvent system using 20 g of normal phase silica gel in a column (1 cm × 10 cm). Compound 2 (57 mg) and compound 3 (20.5 mg) were purified from Fr. V3C (744 mg) by normal phase column chromatography using 100 g silica gel in a column (1 cm × 40 cm) with an acetone/n-hexane 5
7 solvent system using 20 g of normal phase silica gel in a column (1 cm × 10 cm). Compound 2 (57 mg) and compound 3 (20.5 mg) were purified from Fr. V3C (744 mg) by normal phase column chromatography using 100 g silica gel in a column (1 cm × 40 cm) with an acetone/n-hexane 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 solvent system.
5 solvent system.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), (IIB, 1
4), (IIB, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), (IIC, 2.5
4), (IIC, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5), (IID, 2.5
3.5), (IID, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5), and (IIE, 3.5
2.5), and (IIE, 3.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5). Fr. IIB (7.4 g) was fractionated using 300 g of silica gel in a column (3 cm × 40 cm) to get three minor fractions: (IIB1, 0
1.5). Fr. IIB (7.4 g) was fractionated using 300 g of silica gel in a column (3 cm × 40 cm) to get three minor fractions: (IIB1, 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), (IIB2, 1
4), (IIB2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), (IIB3, 1.5
4), (IIB3, 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5), and (Fr. IIB4, 2
3.5), and (Fr. IIB4, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). Fr. IIB3 (1.6 g) was fractionated into five minor fractions using 200 g of silica gel with column: (2 cm × 40 cm) (IIB3A, 0
3). Fr. IIB3 (1.6 g) was fractionated into five minor fractions using 200 g of silica gel with column: (2 cm × 40 cm) (IIB3A, 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), (IIB3B, 1
4), (IIB3B, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), (IIB3C, 1.5
4), (IIB3C, 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5), (IIB3D, 2
3.5), (IIB3D, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), and (IIB3E, 2.5
3), and (IIB3E, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5).
2.5).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 solvent system (Fig. 1).
1 solvent system (Fig. 1).
4.3.4.1 Cyperol A (1). Yellowish amorphous solid; m.p.: 186–188 °C, IR (CHCl3) λmax cm−1: 3400.1 cm−1 (OH), 1733 cm−1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); UV λmax nm (MeOH) (log
O); UV λmax nm (MeOH) (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 305 (2.8); EI-MS: m/z [M]+ = 166.13 (calculated 166.13 for C8H6O4). The calculated HRMS of the compound, C8H6O4, is 166.02606 while the negative ion mode, high-resolution mass spectrometry (HR-ESIMS) performed on a Q-Exactive Mass Spectrometer (Thermo Fisher Scientific, USA) shows a prominent peak at 165.01755.
ε): 305 (2.8); EI-MS: m/z [M]+ = 166.13 (calculated 166.13 for C8H6O4). The calculated HRMS of the compound, C8H6O4, is 166.02606 while the negative ion mode, high-resolution mass spectrometry (HR-ESIMS) performed on a Q-Exactive Mass Spectrometer (Thermo Fisher Scientific, USA) shows a prominent peak at 165.01755.1H NMR (400 MHz, CDCl3): δH 6.20 (2H, s, H-2), 7.24 (1H, s, H-5), 7.33 (1H, s, H-7), δC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) O 10.21 (C
O 10.21 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) O, s, H-8), δO
O, s, H-8), δO![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 7.52 (O
7.52 (O![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) -6, s, HO-8) and 13C-NMR (125 MHz, CDCl3): δC 151.6 (C-1a), 104.0 (C-2), 147.3 (C-3a), 128.3 (C-4), 107.7 (C-5), 152.3 (C-6), 105.3 (C-7), 186.9 (C-8).
-6, s, HO-8) and 13C-NMR (125 MHz, CDCl3): δC 151.6 (C-1a), 104.0 (C-2), 147.3 (C-3a), 128.3 (C-4), 107.7 (C-5), 152.3 (C-6), 105.3 (C-7), 186.9 (C-8).
4.4. DFT calculation
The quantum chemistry computations were performed using the Gaussian 16W software suite on a Dell PC with a 12th Gen Intel® Core™ i7-1255U and 24 GB of RAM.60 The molecular geometry of compound 1 was fully optimized. The B3LYP/6-311G (d,p) method was used to compute the theoretical vibrational spectra. The NBO 3.1 program,61 at the B3LYP/6-311G (d,p) level, was used to construct the natural bond orbital (NBO) and to calculate the intermolecular delocalization or hyperconjugation through second-order interactions between the occupied and unoccupied orbitals of the system. The TD-DFT approach was employed to calculate the electronic characteristics, such as the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO).5. Insect mortality and enzyme inhibition assay
N. lugens was originally obtained from the Integrated Pest Management Laboratory, Department of Entomology, University of Agriculture, Faisalabad, Pakistan. The N. lugens was reared continuously on rice seedlings at 25 ± 1 °C and 70% ± 5% relative humidity with a photoperiod of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 h (light
8 h (light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dark). Insecticidal activities of the six isolated compounds from C. rotundus against N. lugens were determined by a micro-topical toxicity bioassay using a hand microapplicator (Burkard Manufacturing Co., Rickmansworth, England).62,63 Each test material was diluted into a series of concentrations with acetone and applied to 3 to 5 day-old adult insects. Thirty insects were lightly anaesthetized with carbon dioxide. Subsequently, a droplet (0.30, 0.60, 1.20, and 2.40 μg per insect) of each sample solution was applied topically with a hand microapplicator to the mid-abdomen of each insect. Control planthoppers (N. lugens adults) were treated with acetone only. The treated N. lugens were maintained with rice seedlings in a glass tube at 25 ± 1 °C, 65–75% relative humidity, and a light
dark). Insecticidal activities of the six isolated compounds from C. rotundus against N. lugens were determined by a micro-topical toxicity bioassay using a hand microapplicator (Burkard Manufacturing Co., Rickmansworth, England).62,63 Each test material was diluted into a series of concentrations with acetone and applied to 3 to 5 day-old adult insects. Thirty insects were lightly anaesthetized with carbon dioxide. Subsequently, a droplet (0.30, 0.60, 1.20, and 2.40 μg per insect) of each sample solution was applied topically with a hand microapplicator to the mid-abdomen of each insect. Control planthoppers (N. lugens adults) were treated with acetone only. The treated N. lugens were maintained with rice seedlings in a glass tube at 25 ± 1 °C, 65–75% relative humidity, and a light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dark photoperiod of 16
dark photoperiod of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 h. The mortalities were recorded after treatment at 8, 16, 24, 48, and 72 h, and all treatments were replicated three times.
8 h. The mortalities were recorded after treatment at 8, 16, 24, 48, and 72 h, and all treatments were replicated three times.
5.1. Enzyme extraction
The insects were homogenized on ice in a cold homogenization buffer specific for AChE activity. For the AChE assay, a 0.1 M sodium phosphate buffer (pH 7.6) was used. After homogenization, the samples were centrifuged at 3500 rpm for 10 minutes at 4 °C to obtain the supernatant using a TGL-16 M freezing centrifuge (Hunan Xiangyi, China). The clear supernatants were collected and stored at −80 °C. These supernatants were used as crude enzyme extracts for AChE activity analysis.5.2. AChE activity assay
The AChE activity was determined using acetylthiocholine iodide (ATCI) as the substrate.64 In a 96-well microplate, 50 μL of crude enzyme extract was mixed with 100 μL of 0.1 M sodium phosphate buffer (pH 7.6) and 50 μL of ATCI solution (1 mM final concentration). The reaction was initiated by adding the substrate, and the plate was incubated at 25 °C for 10–15 minutes.5.3. Measurement
The reaction was stopped by adding DTNB (5,5′-dithiobis(2-nitrobenzoic acid)) to a final concentration of 0.5 mM. The DTNB reacts with the thiocholine produced by AChE activity to form a yellow product. The absorbance was measured at 412 nm using a Shimadzu UV-2650 spectrophotometer. The six test compounds were dissolved in acetone (final concentration ≤1%) and tested at varying concentrations. The inhibitory effects of these compounds on AChE activity were evaluated by comparing the absorbance in the presence of the test compounds with the control (acetone).The percentage of enzyme inhibition was calculated using the formula:
5.4. Measurement of cellular glutathione (GSH/GSSG) levels
The cellular glutathione content was quantified using a commercial assay kit obtained from Nanjing Jiancheng Bioengineering Institute, China. Insects were homogenized to a 20% (w/v) concentration on ice in the provided reagent IV from the kit. The homogenates were then subjected to centrifugation at 3500 rpm for 10 minutes at 4 °C to obtain the supernatant, which was clear and used for subsequent analysis. The glutathione concentrations (GSH and GSSG) were determined by measuring the absorbance, which was then converted to micromolar (μM) values using a standard curve prepared with known glutathione concentrations. Each experimental treatment was performed with three replicates to ensure statistical reliability.5.5. Statistical analysis
All absorbance measurements were performed in triplicate for each treatment. The data were analyzed using GraphPad Prism to generate dose–response curves and calculate IC50 values. A two-factor ANOVA was performed to evaluate the time- and dose-dependent mortality percentages for compounds 1–6, followed by Tukey's test for post-hoc comparisons, with a significance level set at p < 0.05. Figures were prepared using GraphPad Prism (Premium Version 10). Enzyme activity values for each compound at different time points and concentrations were expressed as mean ± standard deviations (SD).6. In silico study
6.1. Validation of the modeled structures
To optimize the 3D model structures, PROCHECK was used to create a Ramachandran plot analysing the phi and psi distribution of non-glycine and non-proline residues. The ERRAT services provided a quality factor, and Verify3D was also used to check the effectiveness of the structure. A plot of the residual energy and the input structure's Z-score using the ProSAweb server was provided by the application SWISS-MODEL's Z-score to compare the protein structures with the PDB.6.2. Prediction of the active site
In molecular docking studies, identifying binding sites in the modelled protein structures is crucial for predicting potential receptor binding. The MOE site finder tool was employed to discover viable binding sites within the chosen proteins.6.3. Lipinski's rule of five for drug-likeness or ADME (absorption, distribution, metabolism, and excretion) analysis
Lipinski's rule of five directs new drug development, ensuring optimal interaction with the target proteins or enzymes.65 Drug metabolism and pharmacokinetics (DMPK) research is pivotal but intricate, frequently resulting in elevated clinical trial failures.66 SwissADME was used to evaluate the absorption, distribution, metabolism, and excretion to assess drug suitability.676.4. Molecular docking
MOE (Version 2019.0102) was selected for molecular docking due to its intuitive graphical interface, which visualizes the binding residue locations for ligands and receptors. MOE organizes potential binding geometries based on the S-score, a numerical parameter offering a graphical data representation. The S-score is influenced by interactions with solvents, cations, and sulfur lone pairs.68 MOE is used to screen the binding of Cyperol A with GST and AChE. The generalized Born solvation model (GBVI) score function was applied in conjunction with the docking outcomes. A force field-based scoring method, GBVI/WSA dG, calculates the ligand's binding free energy from specific orientations. The S score was used to analyze the interaction outcomes.7. Conclusion
The current research was aimed at the photochemical investigation of C. rotundus plant, resulting in the isolation of one new natural compound, together with five known compounds. The structure of the new compound was determined as Cyperol A by spectroscopic methods, including NMR (1H, 13C, COSY, HMBC, HSQC, NOESY) and EIMS. The newly isolated Cyperol A exhibits a stable structure, allowing further investigation. Additionally, the newly isolated Cyperol A was studied using DFT to determine its electronic features. The in vitro assessment of the insecticidal potential of compounds 1–6 against Nilaparvata lugens revealed that compound 1 exhibited remarkable toxicity and significant inhibition of key enzymatic activities. Moreover, the in silico study of a new natural compound with GST and AchE have also been carried out to determine the efficacy of Cyperol A as a biological pesticide targeting GST and AChE. The findings emphasize the intrinsic insecticidal properties of this new natural compound and its potential as an eco-friendly alternative to synthetic insecticides. Given the environmental concerns associated with conventional insecticides and the emergence of insecticide resistance in pests, natural compounds from plants like C. rotundus L. offer a sustainable and environmentally conscious approach to pest management. Additionally, exploring the molecular properties of these compounds may unveil novel applications in insecticide discovery and allied domains.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Data curation, Saqib Hussain Bangash, Chen-Yang Wei and Wen-Wei Tang; formal analysis, Saqib Hussain Bangash, Akbar Ali and Wen-Wei Tang; investigation, Saqib Hussain Bangash, Amjad Hussain, Rashad Al-Salahi and Wen-Wei Tang; methodology, Saqib Hussain Bangash, Moazama Riaz and Wen-Wei Tang; resources, Saqib Hussain Bangash, Rashad Al-Salahi, Wen-Wei Tang; software, Saqib Hussain Bangash, Muhammad Fayyaz ur Rehman, Faiz Ahmed; supervision, Muhammad Ibrahim and Wen-Wei Tang; validation, Saqib Hussain Bangash, Akbar Ali; visualization, Saqib Hussain Bangash and Chen-Yang Wei; writing – original draft, Saqib Hussain Bangash and Wen-Wei Tang; writing – review & editing, Chen-Yang Wei, Rashad Al-Salahi, Faiz Ahmed, Akbar Ali and Wen-Wei Tang.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project, King Saud University, Riyadh, Saudi Arabia, for funding this work through grant No. RSP2025R353. The authors express their appreciation to the Guangxi University for this work.References
- A. M. Peerzada, Acta Physiol. Plant., 2017, 39, 270 CrossRef.
- X. C. Liu, X. N. Lu, Q. Z. Liu and Z. L. Liu, J. Essent. Oil Bear. Plants, 2016, 19, 640–647 CrossRef CAS.
- R. P. C. Costa, A. L. A. Guimarães and A. C. M. Vieira, Rev. Cienc. Farm. Basica Apl., 2014, 35, 425–433 Search PubMed.
- N. Zandi-Sohani and L. Ramezani, Int. Biodeterior. Biodegrad., 2015, 98, 101–106 CrossRef CAS.
- B. E. Bejnordi, M. Veta, P. J. Van Diest, B. Van Ginneken, N. Karssemeijer, G. Litjens, J. A. Van Der Laak, M. Hermsen, Q. F. Manson and M. Balkenhol, JAMA, 2017, 318, 2199–2210 CrossRef PubMed.
- M. B. Isman, Crop Prot., 2000, 19, 603–608 CrossRef CAS.
- P. A. Leggat, S. Chowanadisai, B. Kukiattrakoon, B. Yapong and U. Kedjarune, Int. Dent. J., 2001, 51, 11–16 CrossRef CAS PubMed.
- J. Velasques, M. H. Cardoso, G. Abrantes, B. E. Frihling, O. L. Franco and L. Migliolo, Pestic. Biochem. Physiol., 2017, 143, 14–25 CrossRef CAS PubMed.
- A. Lakshmanan, A. McBrien, J. Zhang and V. Dhole, in Computer Aided Chemical Engineering, Elsevier, 2014, vol. 34, pp. 186–195 Search PubMed.
- S. Senthil-Nathan, Front. Physiol., 2013, 4, 359 Search PubMed.
- M. López and M. Pascual-Villalobos, Ind. Crops Prod., 2010, 31, 284–288 CrossRef.
- S. Karami-Mohajeri and M. Abdollahi, Hum. Exp. Toxicol., 2011, 30, 1119–1140 CrossRef CAS PubMed.
- M. F. Zaini, S. Arshad, K. Thanigaimani, N. C. Khalib, D. A. Zainuri, M. Abdullah and I. A. Razak, J. Mol. Struct., 2019, 1195, 606–619 CrossRef CAS.
- J. C. Alvarez, Curr. Opin. Chem. Biol., 2004, 8, 365–370 CrossRef CAS PubMed.
- Z. Zhou and W. Yin, Molecules, 2012, 17, 12636–12641 CrossRef CAS PubMed.
- J. J. L. Bezerra, T. G. d. Nascimento, R. U. Kamiya, A. P. d. N. Prata, P. M. d. Medeiros, S. A. S. d. Silva and N. E. d. Melo, Braz. J. Pharm. Sci., 2022, 58, e20205 CrossRef CAS.
- M. I. Momin, N. Koorbanally, D. Ramjugernath and M. D. Bala, Acta Crystallogr., Sect. E:Struct. Rep. Online, 2011, 67, o2681 CrossRef CAS PubMed.
- F. Weinhold, C. Landis and E. Glendening, Int. Rev. Phys. Chem., 2016, 35, 399–440 Search PubMed.
- S. Muthu and G. Ramachandran, Spectrochim. Acta, Part A, 2014, 121, 394–403 CrossRef CAS PubMed.
- J.-n. Liu, Z.-R. Chen and S.-F. Yuan, J. Zhejiang Univ., Sci., B, 2005, 6, 584–589 CrossRef PubMed.
- A. Ali, M. Khalid, M. F. U. Rehman, S. Haq, A. Ali, M. N. Tahir, M. Ashfaq, F. Rasool and A. A. C. Braga, ACS Omega, 2020, 5, 15115–15128 CrossRef CAS PubMed.
- A. M. Asiri, M. Karabacak, M. Kurt and K. A. Alamry, Spectrochim. Acta, Part A, 2011, 82, 444–455 CrossRef CAS PubMed.
- H. Abou-Rachid, Y. Song, A. Hu, S. Dudiy, S. Zybin and W. Goddard III, J. Phys. Chem. A, 2008, 112, 11914–11920 CrossRef CAS PubMed.
- G. Venkatesh, Y. Sixto-López, P. Vennila, Y. S. Mary, J. Correa-Basurto, Y. S. Mary and A. Manikandan, J. Mol. Struct., 2022, 1258, 132678 CrossRef CAS.
- K. Periyasamy, P. Sakthivel, P. Vennila, P. Anbarasan, G. Venkatesh and Y. S. Mary, J. Photochem. Photobiol., A, 2021, 413, 113269 CrossRef CAS.
- K. Fukui, Science, 1982, 218, 747–754 CrossRef CAS PubMed.
- R. G. Parr, R. A. Donnelly, M. Levy and W. E. Palke, J. Chem. Phys., 1978, 68, 3801–3807 CrossRef CAS.
- P. K. Chattaraj and D. R. Roy, Chem. Rev., 2007, 107, PR46–PR74 CrossRef CAS.
- H. Tandon, T. Chakraborty and V. Suhag, J. Math. Chem., 2020, 58, 2188–2196 CrossRef CAS.
- S. Akhter, O. Concepcion, A. F. de la Torre, A. Ali, A. R. Raza, R. Eman, M. Khalid, M. F. ur Rehman, M. S. Akram and H. M. Ali, Arabian J. Chem., 2023, 16, 104570 CrossRef CAS.
- S. Andrade, L. C. Gonçalves and F. E. Jorge, J. Mol. Struct.:THEOCHEM, 2008, 864, 20–25 CrossRef CAS.
- M. Muthukkumar, T. Bhuvaneswari, G. Venkatesh, C. Kamal, P. Vennila, S. Armaković, S. J. Armaković, Y. S. Mary and C. Y. Panicker, J. Mol. Liq., 2018, 272, 481–495 CrossRef CAS.
- Y. Chen, M. Mastalerz and A. Schimmelmann, Int. J. Coal Geol., 2012, 104, 22–33 CrossRef CAS.
- D. L. Pavia, G. M. Lampman, G. S. Kriz and J. A. Vyvyan, Introduction to Spectroscopy, Cengage Learning, 2014 Search PubMed.
- D. Lin-Vien, N. B. Colthup, W. G. Fateley and J. G. Grasselli, The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules, Elsevier, 1991 Search PubMed.
- T. Biswas, M. Jahan, M. Rahman, M. Khan and M. Rahaman, SAARC Journal of Agriculture, 2006, 4, 99–112 Search PubMed.
- M. Sardar, R. Khatun, K. Shariful Islam, T. Haque and M. Jahan, International Journal of Entomology Research, 2019, 4, 54–57 Search PubMed.
- J.-K. Yoo, S.-W. Lee, Y.-J. Ahn, T. Nagata and T. Shono, Appl. Entomol. Zool., 2002, 37, 37–41 CrossRef CAS.
- A. Gonzalez-Coloma, M. Reina, R. Cabrera, P. Castañera and C. Gutierrez, J. Chem. Ecol., 1995, 21, 1255–1270 CrossRef CAS PubMed.
- T. Nohara, Y. Kashiwada, T. Tomimatsu, M. Kido, N. Tokubuchi and I. Nishioka, Tetrahedron Lett., 1980, 21, 2647–2648 CrossRef CAS.
- W. Tang, X. Wei, H. Xu, D. Zeng and L. Long, Fitoterapia, 2009, 80, 286–289 CrossRef CAS PubMed.
- X. Zhang, X. Liao, K. Mao, P. Yang, D. Li, E. Alia, H. Wan and J. Li, Crop Prot., 2017, 94, 106–114 CrossRef CAS.
- X. G. Wang, Q. Li, S. R. Jiang, P. Li and J. Z. Yang, Acta Trop., 2017, 170, 70–78 CrossRef CAS PubMed.
- K. Baskar, C. Muthu and S. Ignacimuthu, Phytoparasitica, 2014, 42, 323–331 CrossRef CAS.
- R. A. Khan, J. Y. Liu, M. Rashid, D. Wang and Y. L. Zhang, Int. J. Mol. Sci., 2013, 14, 5482–5500 CrossRef CAS PubMed.
- R. Pearce and Y. Zhang, Curr. Opin. Struct. Biol., 2021, 68, 194–207 CrossRef CAS PubMed.
- C. Scholz, S. Knorr, K. Hamacher and B. Schmidt, J. Chem. Inf. Model., 2015, 55, 398–406 CrossRef CAS PubMed.
- C. Colovos and T. Yeates, Protein Sci., 1993, 2, 1511–1519 CrossRef CAS PubMed.
- D. Eisenberg, R. Lüthy and J. U. Bowie, in Methods in Enzymology, Elsevier, 1997, vol. 277, pp. 396–404 Search PubMed.
- M. Wiederstein and M. J. Sippl, Nucleic Acids Res., 2007, 35, W407–W410 CrossRef PubMed.
- P. J. Goodford, J. Med. Chem., 1985, 28, 849–857 CrossRef CAS PubMed.
- S. Kalyaanamoorthy and Y.-P. P. Chen, Drug discovery today, 2011, 16, 831–839 CrossRef CAS PubMed.
- B. Nisius, F. Sha and H. Gohlke, J. Biotechnol., 2012, 159, 123–134 CrossRef CAS PubMed.
- A. J. Sami, S. Bilal, M. Khalid, F. R. Shakoori and A. Shakoori, Pak. J. Zool., 2016, 48, 881–886 CAS.
- S. Bilal, M. Mudassir Hassan, M. Fayyaz ur Rehman, M. Nasir, A. Jamil Sami and A. Hayat, Food Chem., 2021, 346, 128894 CrossRef CAS PubMed.
- C. Empereur-Mot, H. Guillemain, A. Latouche, J.-F. Zagury, V. Viallon and M. Montes, J. Cheminf., 2015, 7, 1–17 Search PubMed.
- S. Mendis, P. Puska, B. e. Norrving and W. H. Organization, Global Atlas on Cardiovascular Disease Prevention and Control, World Health Organization, 2011 Search PubMed.
- B. J. McConkey, V. Sobolev and M. Edelman, Curr. Sci., 2002, 845–856 CAS.
- D. Joseph-McCarthy, J. C. Baber, E. Feyfant, D. C. Thompson and C. Humblet, Curr. Opin. Drug Discovery Dev., 2007, 10, 264–274 CAS.
- H. G. Roth, N. A. Romero and D. A. Nicewicz, Synlett, 2016, 27, 714–723 CAS.
- E. D. Glendening, A. E. Reed, J. E. Carpenter and F. Weinhold, NBO Version 3.1, Pittsburgh, PA, 2003 Search PubMed.
- C.-H. Lee, J.-H. Jeon, S.-G. Lee and H.-S. Lee, J. Korean Soc. Appl. Biol. Chem., 2010, 53, 464–469 CrossRef CAS.
- T. Nagata, T. Masuda and S. Moriya, Appl. Entomol. Zool., 1979, 14, 264–269 CrossRef CAS.
- V. Gorun, I. Proinov, V. Băltescu, G. Balaban and O. Bârzu, Anal. Biochem., 1978, 86, 324–326 CrossRef CAS PubMed.
- C. A. Lipinski, Drug Discovery Today:Technol., 2004, 1, 337–341 CrossRef CAS PubMed.
- D. B. Singh, M. K. Gupta, D. V. Singh, S. K. Singh and K. Misra, Interdiscip. Sci.:Comput. Life Sci., 2013, 5, 1–12 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- A. M. Clark and P. Labute, J. Chem. Inf. Model., 2007, 47, 1933–1944 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra00505a |
| This journal is © The Royal Society of Chemistry 2025 |