Open Access Article
Open Access ArticleGli pathway-targeted Co(III) Schiff base complexes inhibit migration of basal cell carcinoma cells†
Caroline E. Bond a,
Keaton D. Olson‡
a,
Keaton D. Olson‡
 a,
Metehan Punar‡
a,
Metehan Punar‡ a,
Lillian B. Friedman
a,
Lillian B. Friedman a,
Jian-Hong Tang
a,
Jian-Hong Tang ac,
Minrui Luoa,
Matthew D. Baileya,
Robert A. Holmgrenb and
Thomas J. Meade
ac,
Minrui Luoa,
Matthew D. Baileya,
Robert A. Holmgrenb and
Thomas J. Meade *a
*a
aDepartments of Chemistry, Molecular Biosciences, Neurobiology, and Radiology, Northwestern University, Evanston, IL, USA. E-mail: tmeade@northwestern.edu
bDepartment of Molecular Biosciences, Northwestern University, Evanston, IL, USA
cSchool of Future Technology, University of Chinese Academy of Sciences, Beijing 101408, P. R. China
First published on 19th March 2025
Abstract
Basal Cell Carcinoma (BCC) is the most frequently diagnosed cancer globally and affects about one in five Americans. Given the frequency of diagnosis, it is surprising that there are very few therapeutic options. Surgical removal is currently the most common treatment option; however, this can lead to noticeable scarring and cosmetic issues. As a result, there is a compelling interest in developing non-invasive therapeutic approaches to this disease. Here, we introduce a new transition metal–DNA derivative called CoGli–GOPEI that inhibits the migration of murine ASZ BCC cells in laboratory experiments. Notably, this complex significantly outperforms two established hedgehog-pathway inhibitors: GANT-61 (an investigational compound) and vismodegib (an FDA-approved drug). These inhibitors target the hedgehog signaling pathway—specifically the Gli family of transcription factors—to slow cancer progression. By effectively reducing cell migration, CoGli–GOPEI offers a less invasive alternative to traditional treatments like surgical resection and chemotherapy. Our results highlight how targeting the Gli transcription factors within the hedgehog pathway can create a novel therapeutic strategy against BCC. The ultimate goal of these new derivates is to reduce the spread of cancer cells while minimizing the downsides of surgery.
Introduction
The use of metals in medicine has grown impressively in recent years due to a significantly advanced understanding of the structures of biologically active metal complexes and metal-containing proteins.1 One landmark in this area was the introduction of cisplatin and related transition metal derivatives as anticancer drugs.2–9 It is becoming clear that developing new inorganic therapeutic agents that can be specifically coupled to a biologically active site by cooperative redox-binding ligation will have a significant impact.10–16 Previous research has shown Co(III) complexes developed for anticancer, enzyme inhibition, and preventative protein aggregation. With a limited number of available cancer treatments for patients impacted by cancers such as Basal Cell Carcinoma (BCC), cobalt-based drugs may serve as a new therapeutic strategy in targeting prevalent cancers.17Basal Cell Carcinoma (BCC) is a form of skin cancer in the body's basal cells. It is the most frequently diagnosed cancer globally and affects about one in five Americans. BCC of the skin is commonly caused by exposure to ultraviolet radiation, with an increased risk of disease development if exposure occurs during childhood and adolescence.18 Physical characteristics such as red and blonde hair, light eye color, and light complexion are associated with increased BCC risk under ultraviolet exposure.19 BCC lesions are primarily located throughout the head and neck region, with pigmentation levels varying between lesions.20,21
Although the mortality rate of BCC is relatively low, available treatments aren't offered to many patients due to a plethora of health risks, including disfigurements such as scars and hypopigmentation of the skin, high levels of invasiveness, and overall difficulties in treating small areas of skin like the eyelids.17 The variability in current surgical treatments, such as excision margins and depth for removal, make recurrences in BCC lesions especially common. Reoccurrence presents a significant issue for patients requiring repeated surgical attention.22 In addition, a uniquely challenging obstacle is the lack of cellular model systems for which early-stage therapeutics can be adequately evaluated, as no human BCC models exist, only murine.
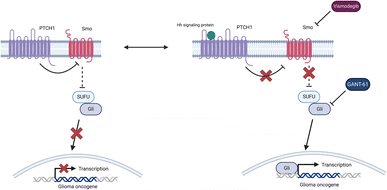
Specific cell signaling pathways have been found to play an intrinsic role in BCC's oncogenesis and disease progression, such as the hedgehog (Hh) signaling pathway.23 Mutations of Patched1 (PTCH1), a BCC tumor suppressant protein, are often linked with BCC lesions within this pathway. This mutation is prominent in patients with Gorlin syndrome, characterized by many basal cell carcinomas affecting organs throughout the body due to loss-of-function mutations in PTCH1.24 The Hh signaling pathway is activated when the Hh signaling protein binds to PTCH1. Without the Hh protein present in this signaling cascade, PTCH1 inhibits the activity of Smoothened (SMO), a transmembrane protein localized to the cell surface. When SMO is inhibited, the fused (SUFU) suppressor is attached to glioma-associated oncogene (Gli) transcription factors and blocks their translocation to the nucleus. When the Hh protein binds to PTCH1, the disinhibition of SMO occurs, and SUFU releases Gli transcription factors that can go on to activate genes that contribute to the formation of BCC lesions.25
Vismodegib is one of eleven FDA-approved chemotherapeutic treatments for BCCs, of which six are generic versions of the other five. This complex is an SMO inhibitor that directly binds SMO and blocks downstream hedgehog signaling. It is recommended for patients not approved for surgical removal of BCCs or radiation therapy treatments (Fig. 1).26 Vismodegib is taken orally and can reduce the size of lesions to allow patients to become approved candidates for surgery. However, this process typically requires several months, and with off-target effects such as loss of taste, alopecia, and muscle cramps, alternative approaches are needed.27,28 Finally, chemoresistance is a significant concern because secondary BCC lesions may grow and inhibit therapeutic function.17,20,21
The Gli family of transcription factors has been shown to play a role in malignant growth and maintenance of multiple forms of cancer, including BCC, when abnormal pathway activation occurs.29 Additionally, the highly conserved DNA binding domain of Gli proteins underscores the importance of finding effective, non-invasive treatments for this family, as the treatment could be utilized for various cancers that utilize Gli transcription factors.30
Currently, there are no examples of agents that target the Gli family of zinc finger transcription factors (ZnFTFs) due to the absence of targeting. ZnFTFs are transcription factors which contain a zinc-finger binding domain that binds DNA to aid in gene regulation.30 This difficulty stems from these transcription factors do not have well-defined binding pockets.31 Nonetheless, efforts have been made to identify molecules that can achieve binding. One of the more promising molecules that could inhibit Gli transcription factors is GANT-61, a derivative of Gli antagonists (GANT). GANT-61 has been shown to disrupt many Hh signaling pathways associated with various cell carcinomas.32,33 GANT-61 can achieve this feat by binding directly with Gli and inhibiting DNA interactions, thus preventing oncogenic transcription. However, GANT-61 exhibits insufficient bioavailability and solubility, indicating that this molecule is not an effective treatment for BCC.34
Previous research regarding zinc finger domain disruption has revealed the potential of cobalt(III) Schiff base conjugates (CoSB) as inhibitors, providing a potential avenue for use in Gli inhibition.35,36 These conjugates preferentially bind to histidine residues in Cys2His2 zinc fingers of Gli transcription factors.35 This action displaces the zinc and changes the zinc finger's tetrahedral configuration to an octahedral arrangement. The change in the zinc fingers' molecular geometry – via disruption of the ββα structural motif needed for gene regulation essential for Gli binding and transcription capabilities – is thus irreversibly inhibited.37 Using a Gli-specific oligonucleotide chemically attached to CoSB (CoGli), CoGli can favorably bind to Gli's zinc finger domain, limiting off-target effects.38
However, complications with delivering negatively charged DNA molecules into cells (a result of the negatively charged backbone of the Gli DNA sequence) arise due to the hydrophobic interior of cell membranes, which repel the entry of said molecules.39,40 Thus, an effective delivery method must be employed to ensure the transfection and subsequent delivery of DNA-conjugated compounds.
Previous drug delivery research has shown that liposomal transfection agents and positively charged polymers such as branched polyethyleneimine (PEI) have been proven as promising agents to combat this issue.41 PEI is especially applicable due to its high endosomal and lysosomal escape rates from the cell. PEI has also been shown to have increased permeabilization capability when attached to a relevant complex-delivery system, such as graphene oxide (GO) nanoparticles.39 The use of GO nanoparticles has shown promising results for the intracellular delivery of chemotherapeutics because of rich surface chemistry and rapid levels of chemical uptake.40,42,43
The modification of GO's surface with polyethyleneimine (PEI) electrostatically binds DNA-based molecules (such as CoGli) and delivers them within the cell to reduce levels of cytotoxicity.38 GOPEI refers to the graphene oxide with polyethyleneimine on the surface. Previous research has demonstrated GOPEI's higher efficacy over unmodified PEI due to the increased internalization and retention exhibited upon GOPEI. Thus, combining CoGli with GOPEI may form a compound (CoGli–GOPEI) that acts as an Hh pathway inhibitor.38
Therefore, this compound was developed to enable the intracellular delivery of CoGli, the DNA-targeted, Gli-specific CoSB inhibitor (Fig. 2). Herein, we report using CoGli–GOPEI as a new compound that inhibits the migration of the murine ASZ basal cell carcinoma cell line in vitro.
 | ||
| Fig. 2 Structure and DNA sequence of CoGli. The Co(III) Schiff base complex tethered to the Gli transcription factor targeted sequence. The asterisks denote phosphorothioate linkages on the 3′ and 5′ end to prevent degradation by endonucleases. The synthesis was done according to literature with modifications made to enhance yield.38 | ||
Results and discussion
Gli transcription factors play an essential role in the formation of BCC lesions through their ability to activate several genes known to play prominent roles in the formation of lesions.38 The Gli family of ZnFTFs contains Cys2His2 sites, where cobalt(III) Schiff-base DNA conjugates have been shown to change the molecular arrangement for DNA binding.35,44 The challenge for targeting ZnFTFs due to the lack of defined binding pockets and their role in developing lesions led to our previously reported studies using a CoGli–GOPEI conjugate.15,38 The CoSB-Gli DNA annealing was confirmed with circular dichroism (CD) before GOPEI was added (Fig. S2†). High-performance liquid chromatography was used to purify the CoGli conjugates. All the aforementioned derivatives were investigated using a transwell migration assay to determine the percentage of cell migration.To compare the potential inhibition of CoGli–GOPEI against industry standards vismodegib (an FDA-approved chemotherapeutic for BCC lesions) and GANT-61 (a compound in clinical development for Gli transcription factor specific inhibition), the individual inhibition potentials of vismodegib and GANT-61 were tested. Tested concentrations were chosen based on reported values and previous work.45–47 Of the two compounds, decreased concentrations of vismodegib showed similar inhibition effects. Effectiveness was based on the inhibition of cell migration. Although neither vismodegib nor GANT-61 significantly reduced cell migration when assessed with a two-tailed Student's t-test compared to their controls, even at the highest doses, approximately 80% normalized migration was observed (Fig. 3).
Previously, we investigated CoGli–GOPEI regarding cell migration inhibition in 2D models.15,38 Herein, we describe a 3D model that shows that the same compound significantly inhibits cell migration, showing only 16% migration at a dose of 2.5 μM and even less at higher doses (Fig. 4). Significance was determined using a two-tail Student's t-test. This decrease in cell migration suggests inhibiting the Gli transcription factors may disrupt an essential step in BCC's tumor progression. The improved efficacy is partly due to the conjugate's targeted and preferential delivery component (GOPEI), which allows the cell to uptake CoGli (Fig. 4).38
Graphene oxide was chosen as the delivery platform to aid cellular uptake, a limiting factor of previous complexes' therapeutic impact.34 The positively charged nature of PEI provides enhanced electrostatic interaction with DNA-conjugate molecules. GOPEI served as a functional delivery system for cellular uptake while limiting side effects such as decreased cell movement and cellular cytotoxicity (Fig. 5). [Co(acacen)(NH3)2Cl] was used as a control complex to determine the impact of these CoSB conjugates on cell migration (Fig. 5A), as these derivative mimics the CoSB used in CoGli–GOPEI (Fig. 4A). The same concentrations were used for GOPEI and [Co(acacen)(NH3)2Cl] as controls. Increased concentrations of [Co(acacen)(NH3)2Cl] caused a two-fold decrease in cellular migration per tested dose, while at the same concentrations, GOPEI caused no significant reduction in cellular migration (Fig. 5D).
To analyze the raw data, all 8 images taken per well were used. The percent area of the well covered by cells was calculated by dividing the computed area covered by cells by the total area of the well. The percent areas for all 8 images for a given well were averaged to give each well its average percent area. A total of 3 averages per dose were calculated (n = 3), each using 8 images. The control wells' average percent area was also calculated using this procedure. This value was then used to normalize every well's average percent area. This allowed for a 2-tailed paired Student's t-test to calculate p-values between doses to determine statistical significance, with p < 0.05.
These results highlight the success of the CoGli–GOPEI delivery and its ability to inhibit the Hh signaling pathway through the specific targeting of Gli transcription factors. To our knowledge, this is the first report on the use of CoGli–GOPEI in a 3D model, which is a better representation of body conditions. The statistically significant inhibition of ASZ cell migration seen in vitro help guide future in vitro and in vivo studies of the compound's use as a therapeutic for BCC.
Conclusions
CoGli–GOPEI has previously been characterized chemically and biologically in static experiments.15,38 Here, we have shown the characterization and effectiveness of these derivatives in a dynamic biological environment. Cell migration, tumor growth, and progression are influenced by the tumor microenvironment in vivo.48 The microenvironment that expands during tumorigenesis and tumor development provides a protection barrier against the host's immune system by inducing immunosuppressive cytokine production.48,49Inhibiting the growth and spread of the tumor microenvironment by limiting cell migration may result in the ability of complete surgical removal and a lower risk of recurrence. The success of our compound in this novel environment further highlights the possibility for clinical translation in the future. Therefore, the CoGli–GOPEI platform offers an alternative therapeutic approach for inhibiting the Gli family transcription factors activated by the hedgehog signalling pathway. Understanding cellular behavior in the presence of these targeted Gli transcription factor inhibitors will provide mechanistic information about the role of the hedgehog pathway in cellular migration in BCC. This work establishes a framework for future BCC work in limiting tumor progression and provides a platform to build upon for the targeting of Gli family transcription factors in various cell pathways.
Experimental
Cell culture
ASZ cells were cultured in the growth medium [M154CF calcium free base (Gibco, 500 mL) supplemented with fetal bovine serum (GenClone, 10 mL), 0.2 M calcium chloride (Gibco, 125 μL), and penicillin–streptomycin (Gibco, 5 mL)]. The fetal bovine serum was chelexed, as described below, prior to its addition to the growth medium. The sterile media solution was vacuum filtered (0.2 μm aPES membrane, 75 mm diameter) and stored at 4 °C. Cells were grown in an environment of 5% carbon dioxide at 37 °C in T-75 culture flasks (Corning). Cells were split every two days to a new T-75 culture flask with Dulbecco's phosphate-buffered saline (DPBS, GenClone), pre-made media, and trypsin (TrypLE Express (1× Dissociation Reagent), Gibco) that were warmed in a 37 °C water bath for 20 minutes prior to splitting.The cell culture was washed twice (2 mL per wash) with the warmed DPBS and treated with 2 mL of trypsin for cell de-adhesion, including 10 minute incubation periods between each step. After detachment and centrifugation (10 minutes, 300 rcf, 4 °C in tabletop Eppendorf Centrifuge 5810 R, 15 Amp Version), much of the trypsin was aspirated, with the remainder quenched upon the addition of fresh growth medium. 2 mL of the 5 mL cell suspension was grown in a new T-75 culture flask with 8 mL of fresh growth medium.
Chelex fetal bovine serum (FBS)
50 grams of Chelex resin (BioRad Chelex100 NC9085110, Mesh: 200/400) was washed with ultrapure water (500 mL) for 20 minutes. The mixture was vacuum-filtered, with the resin being retained. The washing and recollection of the resin were repeated 4 additional times with the following components in their respective order: ultrapure water, 1 M HCl, 1 M NaOH, ultrapure water, 0.5 M sodium acetate (MilliporeSigma). Following the final filtration of sodium acetate, the resin was equilibrated for 10 minutes in phosphate-buffered saline (PBS (500 mL), Corning) while stirring. After, the mixture was vacuum filtered, with the resin being retained. The resin was equilibrated in fresh FBS for 10 minutes (500 mL).The mixture's pH was adjusted to a pH of 7.4 (Thermo Scientific Orion Star A211 pH meter). The mixture was filtered, with the resin being retained. The resin was washed with ultrapure water (500 mL), and the pH was adjusted to 7.4. The mixture was vacuum filtered, with the resin being retained. The resin was added to FBS and stirred for an hour (500 mL). The mixture was vacuum-filtered, and the resin was discarded. The collected sterile FBS solution was vacuum filtered (0.2 μm aPES membrane, 75 mm diameter) and stored at −80 °C until use.
Serum-free media
The serum-free media [M154CF calcium free base (Gibco, 500 mL) supplemented with 0.2 M calcium chloride (Gibco, 125 μL) and penicillin–streptomycin (Gibco, 5 mL)] is necessary to aid in the initial migration of cells in the insert due to its lack of FBS and associated nutrients.CoGli single strand DNA purification
Co-Gli ssDNA was purified using high-performance liquid chromatography (HPLC). An analytical-scale Agilent 1260 Infinity II system was equipped with a diode-array detector. DNA elution was monitored at 260 nm. Separation using a DNAPac™ PA200 Analytical column (4.0 mm × 250 mm, 8 μM, Thermo Fisher). Mobile phases consisted of A: Tris buffer (pH 8.0) and B: Tris buffer containing 1.25 M NaCl (pH 8.0). The elution followed (t = 0 min, 32% B; t = 17 min, 46.4% B; t = 17.1 min, 75% B; t = 19.1 min, 75% B; t = 19.2 min, 32% B; t = 24 min, 32% B). Collected fractions were investigated by MALDI-TOF MS to identify the fraction containing Co-Gli ssDNA. The product was isolated between 9.7 and 14.5 min. MS (MALDI-TOF) m/z: [M–H] – calculated for 5840; found 5839.165.Based on previous literature, CoGli–GOPEI, GOPEI, and the annealed DNA mixture were combined in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.79 ratio (in grams) of DNA
3.79 ratio (in grams) of DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GOPEI, comparable to a 1
GOPEI, comparable to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio used in literature to ensure complete DNA loading.25
4 ratio used in literature to ensure complete DNA loading.25
Preparation of samples for matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS)
MALDI matrix was prepared by dissolving 2,5-dihydroxyacetophenone (DHAP, 25 mg) in methanol (333 μL), followed by adding ammonium citrate until solution saturation. Solutions of aqueous analyte (0.5 μL) were mixed with matrix (1 μL). The combined solution was then deposited on a MALDI grid and allowed to dry. MALDI-TOF MS data was collected using a Bruker RapiFlex Tissue Typer using negative mode with a mass range of 4–20 kilodaltons 150 kV (Fig. S1†).Desalting of Co-Gli single strand DNA
After purification, the isolated cobalt-conjugated ssDNA was lyophilized and redissolved in type 1 ultrapure water (3 mL). Desalting was performed using a pre-packed NAP-25 G25 Sephadex column (Cytiva) with water as the eluent. Fractions were collected and monitored by UV/vis spectroscopy for the presence of ssDNA. The ssDNA-containing fractions were combined, and ssDNA concentration was determined by UV/vis (ε260 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 770
770![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1).
000 M−1 cm−1).
Complementary strand annealing
The complementary strand of DNA (IDT) was added to the HPLC-purified cobalt single-strand DNA (IDT) at a 2 to 1 ratio. The solution was heated at 95 °C for 3 minutes and left to slowly cool overnight.Circular dichroism of annealed DNA
Circular dichroism was collected on the Circular Dichroism Spec J-1700 in Northwestern University's Keck Biophysics Facility. All experiments were conducted using a 10 mm Quartz cuvette (Aldrich Chemical), 200–400 nm wavelength range, 50 nm min−1 scan in continuous mode, and 4 second digital integration time (D.I.T.). A 180 mM PBS buffer blank was run at a total volume of 700 μL. The complementary single strand of DNA (IDT) was run with a volume of 700 μL consisting of 680 μL of 180 mM PBS buffer and 20 μL DNA sample. All annealed samples had a total volume of 700 μL consisting of 680 μL of 180 mM PBS buffer and 20 μL of respective DNA samples. Data analysis was performed using the Jansco CD analysis software SpectraManager. Baseline correction was performed, and smoothing was applied at a value of 15. Subtraction was performed at a factor of 0.1 with a step of 0.1 (Fig. S2†).Transwell migration
All complex stock solutions can be found in Table S4.† All solutions used during experimentation were homogeneous, and employing sterile techniques dramatically limits the possibility of the seen debris being any other foreign particulates. On day 1, the cells were passaged as normal from the above protocol (Cell Culture), stopping at the post-centrifugation resuspension. Cell count estimation was obtained using a hemacytometer to determine the amount needed to obtain approximately 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well of a 6 well culture plate (Corning) when diluted to a final volume of 1 mL per well. The plated cells were incubated at 37 °C overnight, and the remaining resuspension can be used to passage a new T-75 culture flask stock (Table S1†).
000 cells per well of a 6 well culture plate (Corning) when diluted to a final volume of 1 mL per well. The plated cells were incubated at 37 °C overnight, and the remaining resuspension can be used to passage a new T-75 culture flask stock (Table S1†).
On day 2, the pre-made media was warmed at 37 °C for 20 minutes. All plated wells were media changed and replenished with appropriate complex concentration and fresh growth medium for a final volume of 900 μL per well. The cells were again incubated overnight at 5% carbon dioxide, 37 °C (Table S2†).
On day 3, the DPBS, media, and trypsin were pre-warmed to 37 °C. The cell passaging procedure described above was followed through resuspension, with the only difference being the volumes used for washing (2 mL) and trypsin (400 μL) per well. A total volume of 500 μL per insert (8.0 μm pore size, 12 mm diameter, tissue culture treated; Millicell) was composed of cell resuspension (200 μL), desired complex concentration, and fresh serum-free medium. A total volume of 500 μL in the well was composed of fresh growth medium and the desired complex concentration. Inserts were staged in empty wells and transferred to the well solutions once all components had been added independently. The plate was incubated for 2 days at 5% carbon dioxide at 37 °C (Table S3†).
On day 5, the DPBS was warmed in the 37 °C water bath for 20 minutes. Media solutions were removed from both insert and well for all, and cells were fixed with 4% paraformaldehyde in PBS (600 μL, Thermo Fisher Scientific) at room temperature during the 10 minute incubation period. Paraformaldehyde was removed and replaced with methanol (600 μL, Sigma-Aldrich) in each insert to permeabilize the cells, which were then incubated at room temperature for 10 minutes. Methanol was removed, and crystal violet stain (400 μL, Sigma-Aldrich) was added to each insert, which was incubated at room temperature for 30 minutes. The crystal violet stain was removed. Each insert was washed three times with DPBS (600 μL). The inserts were immediately imaged using Olympus BX53 Widefield microscope 10× magnification at Northwestern University's Biological Imaging Facility (BIF) and Q-capture Pro 7 software. The Q-capture Pro 7 software's resolution was 2560 × 1920 FF, auto white balance applied, and auto adjust exposure applied. Filters were not used when capturing images, and a total of 8 images were captured per insert.
Transwell migration image processing and analysis
Image processing was done using the free image processing package Fiji. The analysis settings were defined by selecting the area and area fraction before image processing occurred. Color deconvolution is applied using the H&E vector, where image 1 is selected for subsequent processing. Of the three images produced from color deconvolution, image 1 displays the best shade separation between the stained cells and the background. A minimum threshold with the red setting was applied prior to the watershed threshold being performed. Each image was analyzed, and the area fraction value was recorded and saved.All data analyses were performed in Microsoft Excel to calculate a well's normalized average percent area migration (average percent area) of ASZ cells. First, all 8 images taken per well were used. The area of the well covered by cells was calculated using Fiji software for each image. The percent area was then calculated by dividing the computed area covered by cells by the total area of the well. The percent areas for all 8 images for a given well were averaged to give each well its average percent area. The process mentioned earlier was used to calculate the control wells' average percent area (i.e., the wells with a 0 μM dose). This value was then used to normalize every well's average percent area, resulting in a normalized average percent area for each well. From these normalized values, a 2-tailed paired Student's t-test was used to calculate p-values between doses to determine statistical significance with p < 0.05.
Data availability
Transwell migration cell imaging data for this article can be found on a GitHub repository named the title of the paper (https://github.com/carolinebond/Gli-Pathway-Targeted-Complexes-Inhibit-Migration-of-Basal-Cell-Carcinoma-Cells.git). Spectral data and experimental calculations can be found in the ESI.†Author contributions
Bond, Caroline E.: conceptualization, methodology, software, investigation, data curation, writing – original draft, writing – review and editing, visualization, supervision. Olson, Keaton D.: formal analysis, investigation, data curation, visualization, writing – original draft, writing – review and editing. Punar, Metehan: investigation, data curation, writing – original draft. Friedman, Lillian B.: investigation, visualization. Tang, Jian-Hong: methodology, investigation. Luo, Minrui: investigation, writing – original draft. Bailey, Matthew D.: methodology, supervision. Holmgren, Robert A.: writing – review and editing, project administration. Meade, Thomas J.: conceptualization, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported through the National Institute of Health R01NS115571. KO and MP were funded by the WCAS-Baker-Faculty-Research-Grant. We thank Professor Tony Oro for providing the ASZ cell line, Naedum DomNwachukwu for assisting in characterizing the GOPEI, Professor Mark Hersam's lab for providing the probe-sonicated 150 nm graphene oxide, and Dr Meghan W. Dukes for her preliminary work that initiated the work done in this paper. Her advice and guidance were greatly appreciated. This work used the Keck Biophysics Facility of Northwestern University, which is partially supported by the NCI Cancer Center Support Grant #P30 CA060553. Microscopy was performed at the Biological Imaging Facility at Northwestern University (RRID:SCR_017767), graciously supported by the Chemistry for Life Processes Institute, the NU Office for Research, the Department of Molecular Biosciences, and the Rice Foundation. The authors thank Dr David Kirchenbuechler for his assistance and guidance in the image analysis procedure.References
- J. Karges, R. W. Stokes and S. M. Cohen, Metal Complexes for Therapeutic Applications, Trends Chem., 2021, 3(7), 523–534, DOI:10.1016/j.trechm.2021.03.006.
- M. Eberl, D. Mangelberger, J. B. Swanson, M. E. Verhaegen, P. W. Harms, M. L. Frohm, A. A. Dlugosz and S. Y. Wong, Tumor Architecture and Notch Signaling Modulate Drug Response in Basal Cell Carcinoma, Cancer Cell, 2018, 33(2), 229–243, DOI:10.1016/j.ccell.2017.12.015.
- P. C. Bruijnincx and P. J. Sadler, New trends for metal complexes with anticancer activity, Curr. Opin. Chem. Biol., 2008, 12(2), 197–206, DOI:10.1016/j.cbpa.2007.11.013.
- E. Meggers, From conventional to unusual enzyme inhibitor scaffolds: the quest for target specificity, Angew Chem. Int. Ed. Engl., 2011, 50(11), 2442–2448, DOI:10.1002/anie.201005673.
- L. Gourdon, K. Cariou and G. Gasser, Phototherapeutic anticancer strategies with first-row transition metal complexes: a critical review, Chem. Soc. Rev., 2022, 51(3), 1167–1195, 10.1039/d1cs00609f.
- Y. Zhong, C. Jia, X. Zhang, X. Liao, B. Yang, Y. Cong, S. Pu and C. Gao, Targeting drug delivery system for platinum(IV)-based antitumor complexes, Eur. J. Med. Chem., 2020, 194, 112229, DOI:10.1016/j.ejmech.2020.112229.
- B. S. Murray and P. J. Dyson, Recent progress in the development of organometallics for the treatment of cancer, Curr. Opin. Chem. Biol., 2020, 56, 28–34, DOI:10.1016/j.cbpa.2019.11.001.
- J. Shao, Q. Zhang, J. Wei, Z. Yuchi, P. Cao, S. Q. Li, S. Wang, J. Y. Xu, S. Yang and Y. Zhang, et al., Synthesis, crystal structures, anticancer activities and molecular docking studies of novel thiazolidinone Cu(II) and Fe(III) complexes targeting lysosomes: special emphasis on their binding to DNA/BSA, Dalton Trans., 2021, 50(38), 13387–13398, 10.1039/d1dt02180j.
- T. Ashraf, B. Ali, H. Qayyum, M. S. Haroone and G. Shabbir, Pharmacological aspects of Schiff base metal complexes: a critical review, Inorg. Chem. Commun., 2023, 150, 110449, DOI:10.1016/j.inoche.2023.110449.
- M. C. Heffern, N. Yamamoto, R. J. Holbrook, A. L. Eckermann and T. J. Meade, Cobalt derivatives as promising therapeutic agents, Curr. Opin. Chem. Biol., 2013, 17(2), 189–196, DOI:10.1016/j.cbpa.2012.11.019.
- C. R. Munteanu and K. Suntharalingam, Advances in cobalt complexes as anticancer agents, Dalton Trans., 2015, 44(31), 13796–13808, 10.1039/c5dt02101d.
- C. M. Hirschbiegel, X. Zhang, R. Huang, Y. A. Cicek, S. Fedeli and V. M. Rotello, Inorganic nanoparticles as scaffolds for bioorthogonal catalysts, Adv. Drug Deliv. Rev., 2023, 195, 114730, DOI:10.1016/j.addr.2023.114730.
- Z. Pei, H. Lei and L. Cheng, Bioactive inorganic nanomaterials for cancer theranostics, Chem. Soc. Rev., 2023, 52(6), 2031–2081, 10.1039/d2cs00352j.
- S. Narayana, B. H. J. Gowda, U. Hani, S. S. Shimu, K. Paul, A. Das, S. Ashique, M. G. Ahmed, M. A. Tarighat and G. Abdi, Inorganic nanoparticle-based treatment approaches for colorectal cancer: recent advancements and challenges, J. Nanobiotechnol., 2024, 22(1), 427, DOI:10.1186/s12951-024-02701-3.
- M. W. Dukes and T. J. Meade, Modulation of Hedgehog Signaling for the Treatment of Basal Cell Carcinoma and the Development of Preclinical Models, Biomedicines, 2022, 10(10), 2376, DOI:10.3390/biomedicines10102376.
- H. Y. Khan, A. Ahmad, M. N. Hassan, Y. H. Khan, F. Arjmand and R. H. Khan, Advances of metallodrug-amyloid β aggregation inhibitors for therapeutic intervention in neurodegenerative diseases: evaluation of their mechanistic insights and neurotoxicity, Coord. Chem. Rev., 2024, 501 DOI:10.1016/j.ccr.2023.215580.
- M. Nakayama, K. Tabuchi, Y. Nakamura and A. Hara, Basal cell carcinoma of the head and neck, J. Skin Cancer, 2011, 2011, 496910, DOI:10.1155/2011/496910.
- R. P. Gallagher, Sunlight exposure, pigmentary factors, and risk of nonmelanocytic skin cancer. I. Basal cell carcinoma, Arch. Dermatol., 1995, 131(2), 157–163, DOI:10.1001/archderm.1995.01690140041006.
- J. T. Lear, B. B. Tan, A. G. Smith, W. Bowers, P. W. Jones, A. H. Heagerty, R. C. Strange and A. A. Fryer, Risk factors for basal cell carcinoma in the UK: case-control study in 806 patients, J. R. Soc. Med., 1997, 90(7), 371–374, DOI:10.1177/014107689709000704.
- A. G. Marzuka and S. E. Book, Basal cell carcinoma: pathogenesis, epidemiology, clinical features, diagnosis, histopathology, and management, Yale J. Biol. Med., 2015, 88(2), 167–179 Search PubMed.
- D. Altamura, S. W. Menzies, G. Argenziano, I. Zalaudek, H. P. Soyer, F. Sera, M. Avramidis, K. DeAmbrosis, M. C. Fargnoli and K. Peris, Dermatoscopy of basal cell carcinoma: morphologic variability of global and local features and accuracy of diagnosis, J. Am. Acad. Dermatol., 2010, 62(1), 67–75, DOI:10.1016/j.jaad.2009.05.035.
- S. S. Patel, S. H. Cliff and P. Ward Booth, Incomplete removal of basal cell carcinoma: what is the value of further surgery?, Oral Maxillofac. Surg., 2013, 17(2), 115–118, DOI:10.1007/s10006-012-0348-3.
- S. Gupta, N. Takebe and P. Lorusso, Targeting the Hedgehog pathway in cancer, Ther. Adv. Med. Oncol., 2010, 2(4), 237–250, DOI:10.1177/1758834010366430.
- H. Hahn, C. Wicking, P. G. Zaphiropoulous, M. R. Gailani, S. Shanley, A. Chidambaram, I. Vorechovsky, E. Holmberg, A. B. Unden and S. Gillies, et al., Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome, Cell, 1996, 85(6), 841–851, DOI:10.1016/s0092-8674(00)81268-4.
- J. Taipale, M. K. Cooper, T. Maiti and P. A. Beachy, Patched acts catalytically to suppress the activity of Smoothened, Nature, 2002, 418(6900), 892–897, DOI:10.1038/nature00989.
- S. Aditya and A. Rattan, Vismodegib: a smoothened inhibitor for the treatment of advanced basal cell carcinoma, Indian Dermatol. Online J., 2013, 4(4), 365–368, DOI:10.4103/2229-5178.120685.
- N. Bertrand, P. Guerreschi, N. Basset-Seguin, P. Saiag, A. Dupuy, S. Dalac-Rat, V. Dziwniel, C. Depoortere, A. Duhamel and L. Mortier, Vismodegib in neoadjuvant treatment of locally advanced basal cell carcinoma: first results of a multicenter, open-label, phase 2 trial (VISMONEO study): Neoadjuvant Vismodegib in Locally Advanced Basal Cell Carcinoma, EClinicalMedicine, 2021, 35, 100844, DOI:10.1016/j.eclinm.2021.100844.
- K. Fife, R. Herd, S. Lalondrelle, R. Plummer, A. Strong, S. Jones and J. T. Lear, Managing adverse events associated with vismodegib in the treatment of basal cell carcinoma, Future Oncol., 2017, 13(2), 175–184, DOI:10.2217/fon-2016-0296.
- M. Kasper, G. Regl, A. M. Frischauf and F. Aberger, GLI transcription factors: mediators of oncogenic Hedgehog signalling, Eur. J. Cancer, 2006, 42(4), 437–445, DOI:10.1016/j.ejca.2005.08.039.
- K. W. Kinzler and B. Vogelstein, The GLI gene encodes a nuclear protein which binds specific sequences in the human genome, Mol. Cell. Biol., 1990, 10(2), 634–642, DOI:10.1128/mcb.10.2.634-642.1990.
- A. Chen and A. N. Koehler, Transcription Factor Inhibition: Lessons Learned and Emerging Targets, Trends Mol. Med., 2020, 26(5), 508–518, DOI:10.1016/j.molmed.2020.01.004.
- T. B. S. de Araujo, L. d. O. S. da Rocha, M. T. A. Vidal, P. L. C. Coelho, M. G. D. Reis, B. S. d. F. Souza, M. B. P. Soares, T. A. Pereira, R. D. Coletta and D. P. Bezerra, et al., GANT61 Reduces Hedgehog Molecule (GLI1) Expression and Promotes Apoptosis in Metastatic Oral Squamous Cell Carcinoma Cells, Int. J. Mol. Sci., 2020, 21(17), 6076, DOI:10.3390/ijms21176076.
- K. Harada, R. Ohashi, K. Naito and K. Kanki, Hedgehog Signal Inhibitor GANT61 Inhibits the Malignant Behavior of Undifferentiated Hepatocellular Carcinoma Cells by Targeting Non-Canonical GLI Signaling, Int. J. Mol. Sci., 2020, 21(9), 3126, DOI:10.3390/ijms21093126.
- A. Borah, V. Palaninathan, A. R. Girija, S. Balasubramanian, A. K. Rochani, T. Maekawa and D. S. Kumar, Poly-Lactic-Co-Glycolic Acid Nanoformulation of Small Molecule Antagonist Gant61 for Cancer Annihilation by Modulating Hedgehog Pathway, NanoWorld J., 2017, 3(1), 1–10, DOI:10.17756/nwj.2017-038.
- A. S. Harney, J. Lee, L. M. Manus, P. Wang, D. M. Ballweg, C. LaBonne and T. J. Meade, Targeted inhibition of Snail family zinc finger transcription factors by oligonucleotide-Co(III) Schiff base conjugate, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(33), 13667–13672, DOI:10.1073/pnas.0906423106.
- A. Y. Louie and T. J. Meade, A cobalt complex that selectively disrupts the structure and function of zinc fingers, Proc. Natl. Acad. Sci. U. S. A., 1998, 95(12), 6663–6668, DOI:10.1073/pnas.95.12.6663.
- M. C. Heffern, J. W. Kurutz and T. J. Meade, Spectroscopic elucidation of the inhibitory mechanism of Cys2His2 zinc finger transcription factors by cobalt(III) Schiff base complexes, Chemistry, 2013, 19(50), 17043–17053, DOI:10.1002/chem.201301659.
- M. W. Dukes, E. A. Bajema, T. J. Whittemore, R. A. Holmgren and T. J. Meade, Delivery of Targeted Co(III)-DNA Inhibitors of Gli Proteins to Disrupt Hedgehog Signaling, Bioconjug. Chem., 2022, 33(4), 643–653, DOI:10.1021/acs.bioconjchem.2c00063.
- Z. Sadeghi, P. Maleki, S. A. Mohammadi Bondarkhilli, M. Mohammadi and J. Raheb, Dataset on cytotoxicity effect of polyethylenimine-functionalized graphene oxide nanoparticles on the human embryonic carcinoma stem cell, NTERA2 cell line, Data Brief, 2019, 26, 104487, DOI:10.1016/j.dib.2019.104487.
- M. Shu, F. Gao, M. Zeng, C. Yu, X. Wang, R. Huang, J. Yang, Y. Su, N. Hu and Z. Zhou, et al., Microwave-Assisted Chitosan-Functionalized Graphene Oxide as Controlled Intracellular Drug Delivery Nanosystem for Synergistic Antitumour Activity, Nanoscale Res. Lett., 2021, 16(1), 75, DOI:10.1186/s11671-021-03525-y.
- J. Casper, S. H. Schenk, E. Parhizkar, P. Detampel, A. Dehshahri and J. Huwyler, Polyethylenimine (PEI) in gene therapy: current status and clinical applications, J. Controlled Release, 2023, 362, 667–691, DOI:10.1016/j.jconrel.2023.09.001.
- B. Dabrowski, A. Zuchowska, A. Kasprzak, G. Z. Zukowska and Z. Brzozka, Cellular uptake of biotransformed graphene oxide into lung cells, Chem. Biol. Interact., 2023, 376, 110444, DOI:10.1016/j.cbi.2023.110444.
- A. K. Geim and K. S. Novoselov, The rise of graphene, Nat. Mater., 2007, 6(3), 183–191, DOI:10.1038/nmat1849.
- A. S. Harney, T. J. Meade and C. LaBonne, Targeted inactivation of Snail family EMT regulatory factors by a Co(III)-Ebox conjugate, PLoS One, 2012, 7(2), e32318, DOI:10.1371/journal.pone.0032318.
- H. Wang, Q. Meng, Y. Ding, M. Xiong, M. Zhu, Y. Yang, H. Su, L. Gu, Y. Xu and L. Shi, et al., USP28 and USP25 are downregulated by Vismodegib in vitro and in colorectal cancer cell lines, FEBS J., 2021, 288(4), 1325–1342, DOI:10.1111/febs.15461.
- A. Ishii, K. Shigemura, K. Kitagawa, S. Y. Sung, K. C. Chen, C. Yi-Te, M. C. Liu and M. Fujisawa, Anti-tumor Effect of Hedgehog Signaling Inhibitor, Vismodegib, on Castration-resistant Prostate Cancer, Anticancer Res., 2020, 40(9), 5107–5114, DOI:10.21873/anticanres.14514.
- T. Ishii, Y. Shimizu, K. Nakashima, S. Kondo, K. Ogawa, S. Sasaki and H. Matsui, Inhibition mechanism exploration of investigational drug TAK-441 as inhibitor against Vismodegib-resistant Smoothened mutant, Eur. J. Pharmacol., 2014, 723, 305–313, DOI:10.1016/j.ejphar.2013.11.014.
- E. Chiang, H. Stafford, J. Buell, U. Ramesh, M. Amit, P. Nagarajan, M. Migden and D. Yaniv, Review of the Tumor Microenvironment in Basal and Squamous Cell Carcinoma, Cancers, 2023, 15(9), 2453, DOI:10.3390/cancers15092453.
- M. Yamamura, R. L. Modlin, J. D. Ohmen and R. L. Moy, Local expression of antiinflammatory cytokines in cancer, J. Clin. Invest., 1993, 91(3), 1005–1010, DOI:10.1172/JCI116256.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra00326a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |