Open Access Article
Open Access ArticleInvestigation of simple BODIPY dyes as G-quadruplex recognizing ligands†
Jakub Żubertowskia,
Magdalena Rapp *a,
Jan Dolicherb,
Błażej Rubiś
*a,
Jan Dolicherb,
Błażej Rubiś b and
Anna Dembska
b and
Anna Dembska *a
*a
aFaculty of Chemistry, Adam Mickiewicz University, Uniwersytetu Poznańskiego 8, Poznań 61-614, Poland. E-mail: magdrapp@amu.edu.pl; aniojka@amu.edu.pl
bDepartment of Clinical Chemistry and Molecular Diagnostics, University of Medical Sciences, Rokietnicka 3, 60-806 Poznań, Poland
First published on 17th February 2025
Abstract
An important class of fluorescent dyes used in studying interactions and visualization of vital biomolecules are compounds with a skeleton origin 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene, known as BODIPY. The objects of the presented study are the simple, polar and hydrophobic 3,7-dimethyl-substituted-core BODIPY dyes with the unmodified or modified phenyl aromatic ring at the meso position. Their optical properties as well as binding interactions with different DNA forms (i-motif, parallel G4, antiparallel G4, hybrid G4, dsDNA and ssDNA) were investigated by biophysical methods. The BODIPY derivatives interact more preferably with tetraplexes than other DNA forms. Especially, ligand 1 and 3 exibit tendency to destabilize parallel c-MYC G-quadruplex. The experiments with peroxidase-mimicking DNAzymes manifest that the main interaction between these BODIPY ligands and parallel G-quadruplex occurs via end-stacking mode. Moreover, their biological activity was evaluated by MTT assay.
1. Introduction
Fluorescent dyes are promising tools in studying the interactions and visualization of important biomolecules (cellular components).1 Among them, the important class is origin 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene, known as BODIPY (BODIPY). Due to their attractive properties such as intense absorption peak with high molar absorption coefficient in visible region, high fluorescence quantum yield that is insensitive to pH and solvent polarity as well as its narrow long-wave emission,2 these dyes have been found useful in bio-imaging,3–9 including tumor imaging in vivo,7,10–12 bio-labeling,13,14 photodynamic therapy15–17 and drug delivery.18–22 In comparison with commercially available phenyl BODIPY derivatives, the application of strongly electron withdrawing pentafluorophenyl substituent at the meso-position of BODIPY not only influences on the absorption and emission energies of the boron-dipyrromethene core but also made it an attractive platform for functionalization of BODIPY dyes.23–25 Additionally, some fluorinated probes were described as laser dyes,26 or pH sensors,27 as well as tools for surface analysis.28 In comparison, the azide introduction to BODIPY core offers the opportunity for further convenient transformation,29 application in azide–alkyne cycloaddition “click” reaction30–35 or Staudinger-Bertozzi ligation.36 Consequently, this group of dyes has been used as viscosity or H2S sensors,37,38 for biolabeling purposes,35,36 as well as in light-promoted surfaces patterning.39 Moreover, the BODIPY functionalized with sulfonate moieties or quaternary ammonium groups were applied as devices imaging microviscosimetry in appropriate compartment of living cells.40,41 Other water-soluble BODIPY compounds were also used as biolabeling probes.42,43 Thereby the BODIPY-based dyes have been used particularly for specific DNA or RNA detection.44,45 Recently, the results concerning intramolecular energy transfer and fluorescence quenching of BODIPY-bounded to double-stranded DNA were presented,46 while in case of cis-Pt-BODIPY complex bounding with DNA through groove and/or by an intercalative mode was reported.47 Further strategy involving application of 5′-BODIPY labeled 15-mer thrombin binding aptamer gave the possibility to monitor duplex-antiparallel G-quadruplex conversion.48 Moreover, the linear 19-mer tagged at 5′ end with BODIPY fluorophore and DABCYL at 3′ end as its quencher were applied to visualize the i-motif formation in DNA device.49 Alternatively, the BODIPY linked to nucleoside were described as molecular rotor DNA and has been applied as in vitro and in vivo microenvironment sensor,50 while the introduction of BODIPY to the DNA strand assembler on gold surface has allowed to develop the reversible DNA-based switchable device responding to pH changes.51 The BODIPY dyes were also applied as fluorescent nucleotide terminators for DNA sequencing by synthesis (SBS).52,53 Furthermore, the series of BODIPY dyes bearing two hydrophilic PEG chains and one hydrophobic BODIPY tail included in dendrimer-based lipid nanoparticles as mRNA delivery system and near-infrared (NIR) imaging were investigated in vitro and in vivo.11Moreover, a conjugates of the fluorophore BODIPY and the macrocyclic heptaozaxole54 or Phen-DC3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55
55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) were used to visualize G-quadruplexes (G4) in cell-free and cell-based assays systems. Lately, a few reports have been published studying the binding ability of BODIPY derivatives towards G4 DNA.56–59
were used to visualize G-quadruplexes (G4) in cell-free and cell-based assays systems. Lately, a few reports have been published studying the binding ability of BODIPY derivatives towards G4 DNA.56–59
G-quadruplex are DNA secondary structures formed in guanine-rich sequences60 and have been found to play an important role in regulating different biological processes such as replication, transcription, translation, and genomic stability. Their biological significance is a consequence of fact that guanine-rich sequences forming G-quadruplexes are present in different regions in the human genome, such as telomeres and the promoter region of different genes, including oncogene promoters.61–65 Therefore, the rational design of small molecules able to recognize, bind with high affinity and stabilize these non-canonical DNA become one of the key approaches in the development of new therapeutic and diagnostic strategies in cancer diseases.66 So far, a large number of G4-targeting ligands have been designed and synthesized, most of which exhibit common structural elements, such as a polycyclic aromatic core responsible for π-stacking interactions with the external G4 quartet and the presence of positively charged substituents that can interact electrostatically with the grooves of G4 DNA.67,68 Moreover, taking into account structural diversity of G-quadruplexes, an effort had been made to design and develop both stabilizing and destabilizing ligands which acquire specificity and selectivity without compromising affinity.69,70 For example, studies by Li et al. revealed high selectivity of BODIPY derivatives towards parallel c-MYC G4.59
Encouraged by these findings we decided to examine the potential of a few small BODIPY derivatives 1–6 to serve as a G4 ligands. The chosen BODIPY derivatives possess phenyl ring at meso position and 3,7-dimethyl-substituted or 3,7-dimethyl-2,8-disulfo-substituted-core. Our molecular target was the parallel G-quadruplex formed by the 5′-TGAGGGTGGGTAGGGT GGGTAA-3′ DNA sequence derived from the nuclease hypersensitive region of the c-MYC promoter (c-MYC G4).61 This choice was dictated not only by the fact that the c-MYC G4 is the most extensively studied and the best-characterized of all proto-oncogenes, due to its involvement in cellular proliferation and cell growth,71,72 but also by high selectivity of BODIPY derivatives towards c-MYC G4.59 All the ligands were tested against a parallel c-MYC G4 by means of UV-Vis, fluorescence and circular dichroism spectroscopy (CD). The CD technique is an essential tool providing information about the structure formation, stability and topology of G-quadruplexes, it is also commonly used to study the interaction between ligands and G-quadruplex structures.73–76 For example, the presence of induced exciton bands can be used as a method to distinguish between different binding modes G4 with ligand.73 The steady-state fluorescence measurements and DNAzyme assay were applied to explore binding properties between tested BODIPY dyes and c-MYC G4.
The BODIPY derivatives with best binding affinity were also tested against different DNA forms (i-motif, parallel G4, antiparallel G4, hybrid G4, dsDNA, and ssDNA) as well as their biological activity was evaluated.
2. Results and discussion
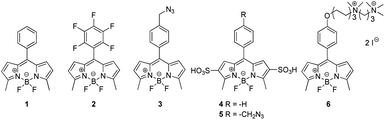
For our studies, we synthesized BODIPY derivatives 1–6, chemical structures of which are shown in Fig. 1. Compound 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77 possesses the phenyl aromatic ring, which in the case of compound 2
77 possesses the phenyl aromatic ring, which in the case of compound 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 is penta-fluorinated. Fluorine is inductively electron-withdrawing but electron-donating by resonance.78
23 is penta-fluorinated. Fluorine is inductively electron-withdrawing but electron-donating by resonance.78
Fluorine substituents diminish the π electron density of phenyl, enhancing the change in the quadrupole moment in aromatic ring facilitating participation in stacking79,80 and increasing the lipophilicity of compound. The ability of fluorine to participate in the formation of hydrogen bonds is crucial to biological activity of fluorochemicals.81 Therefore, we assumed that penta-fluorination of aromatic ring in 2 will increase the expected π–π interactions with G4 tetrad. The planar azide moieties in BODIPY dyes 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41 and 5
41 and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41 offer the opportunity to interact with G-quadruplex structures acting as supramolecular template directing bioorthogonal macrocyclization.82 Moreover, the compounds 4 and 5 contain polar sulfonic groups, and were supposed to serve as a comparison to nonpolar compounds 1 and 2. Finally, the derivative 6
41 offer the opportunity to interact with G-quadruplex structures acting as supramolecular template directing bioorthogonal macrocyclization.82 Moreover, the compounds 4 and 5 contain polar sulfonic groups, and were supposed to serve as a comparison to nonpolar compounds 1 and 2. Finally, the derivative 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41 with aliphatic moiety containing positively charged (cationic) quaternary ammonium groups should presumably interact with the phosphate backbone to anchor the molecule in the DNA and to enhance their solubility. The role of “tails” substituents in better and more efficiently tuned selectivity with respect to G-tetrad stacking-only focused species was evidenced by many research.83 The boron-dipyrromethene (BODIPY) derivatives 1–6 were prepared accordingly to previously reported methods.23,41,77 Moreover, the single crystal of compound 2 was obtained and its structure confirms the presence of a pentafluorinated phenyl ring twisted in the relationship to BODIPY core containing boron centre (see: ESI†).
41 with aliphatic moiety containing positively charged (cationic) quaternary ammonium groups should presumably interact with the phosphate backbone to anchor the molecule in the DNA and to enhance their solubility. The role of “tails” substituents in better and more efficiently tuned selectivity with respect to G-tetrad stacking-only focused species was evidenced by many research.83 The boron-dipyrromethene (BODIPY) derivatives 1–6 were prepared accordingly to previously reported methods.23,41,77 Moreover, the single crystal of compound 2 was obtained and its structure confirms the presence of a pentafluorinated phenyl ring twisted in the relationship to BODIPY core containing boron centre (see: ESI†).
The compounds 1–3 are hardly soluble in aqueous solution, thus these compounds were dissolved in DMSO directly before spectroscopic measurements. Stock solutions of BODIPY derivatives 4–6 were prepared in high-purity water. The values of molar absorption coefficients for tested ligand (in Tris buffer) have been determined – both in presence and absence of potassium ions and are collected in Table S1.†
2.1. Optical properties of BODIPY 1–6
Although meso-phenyl-modified BODIPY derivatives are a group of recognizable and widely applied fluorophores, the possible impact of experimental conditions on their spectral properties tends to be omitted in applicational studies. In our case, the tested compounds 1–6 are fluorescent and their emission bands are located between 500 to 600 nm with maximum around 520 nm for most ligands (Fig. 2); except of ligand 2 possessing red-shifted maximum (536 nm) due to presence of fluorine in the phenyl ring. Two phenomena seem to be of particular importance for tested compounds, especially for their emission efficiency, that are: twisted intramolecular charge transfer (TICT) and aggregation.2,3,84–89 To verify if TICT could alter emissive properties of any tested compound we have examined their fluorescence in THF/water mixtures of different ratios (fw) (Fig. S2†). Anticipated decrease of fluorescence intensity has been initially observed for all non-water-soluble compounds 1–3, until to the certain point (fw > 30%, fw > 60% and fw > 60% for 1, 2 and 3, respectively) at which the fluorescence is revitalized to drop once again when fw reaches almost 100%. For water soluble compounds 4 and 5 the emission intensity increases gradually with the increase of a mixture's polarity – with the emission maximum being blue-shifted, while for 6 after the initial enhancement, emission intensity drops gradually. Gradual fluorescence intensification of 4 and 5 upon the addition of water is intuitive for compounds which are water-soluble (due to the presence of sulfonic groups) and observed hypsochromism (shifts from 536 nm to 517 nm for 4 and 539 nm to 518 nm for 5) are caused by a solvatochromic effect. However, to understand the profiles obtained for remaining four compounds and changes observed, one should refer to another process – aggregation. Obtained absorption (Fig. 3 and S3†) and emission spectra (Fig. S4†) for all tested compounds during gradual increase of their concentration from 1 to 10 μM indicates the tendency of 1–3 and 6 to form aggregates, which was seen as the broadening of a spectrum.84 | ||
| Fig. 2 Fluorescence spectra of BODIPY derivatives 1–6 (1 μM). All spectra were measured in Tris–HCl buffer (10 mM, pH 7.2) containing 100 mM KCl. | ||
 | ||
| Fig. 3 Normalized absorption spectra of 1 (A) and 3 (B) upon gradual concentration increase. All spectra were measured in Tris–HCl buffer (10 mM, pH 7.2) containing 100 mM KCl. | ||
From red-shifted absorption maximum of 1–3 and 6 the type of aggregates can be determined as J-aggregates – end-to-end stacking.87,88 Results obtained shows no evidence for aggregation of 4 and 5, as expected due to the repulsion between negatively charged sulfonic groups present in their structure (Fig. 1).
To verify the aggregation induced emission (AIE) properties90 of tested compounds we have examined their fluorescence in DMSO/water mixtures of different ratios (fw) (Fig. S5†). The significant increase in the fluorescence intensity of 1 and 2 was observed upon enhancing fw from 0 to 60% and 80%, respectively. With a further increase of fw to 99%, the AIE effect diminished which can be attributed to their precipitation.90 In contrast, the fluorescence intensity of 3 and 6 decreased gradually with increase of the water fraction. The latter changes in the fluorescence intensity suggested a significant aggregation quenched emission (ACQ) effect of 3 and 6 (Fig. S5C and F†).
At this point formation of aggregates can be compared to structural modification of fluorophores, introduced during molecules design, which hampers the rotation between donor and acceptor units and disables the charge transfer.84,86 Therefore, when 1–3 and 6 are introduced to the THF solutions with increasing water fractions the TICT is promoted by the solvent polarity, seen as initial decrease of emission intensity, until the point at which polarity of the mixture induces fluorophores aggregation. Disabled rotation reduces TICT process and aggregates revitalize their emission, until the water fraction reaches almost 100% and fluorescence intensity drops, which could be seen as the result of the formation of greater, non-emissive aggregates. Interestingly, despite being perfectly water-soluble, compound 6 displays properties similar to those of insoluble 1–3 rather than to 4 and 5. Introduction of quaternary ammonium groups guarantees water-solubility, however their location in long aliphatic chain causes 6 to be amphiphilic, which explains its partial similarity to behavior of 1–3 when introduced to mixtures of varying polarities.
Temperature is another variable known to impact emissive properties of systems with coexisting TICT and aggregation processes. It has been observed that cooling can cause the profound increase in emission intensity of many AIE systems by suppression of intramolecular rotation.91,92 In contrary, aggregation quenched emission (ACQ) systems should exhibit opposite behavior upon decrease of temperature. We observed the drop of fluorescence intensity when the temperature was increased from 0 to 30 °C for aqueous solutions of 1 and 4–6, while for 2 and 3 the opposite appeared and emission intensity increased (Fig. S6 and S7†). There are limited reports of BODIPY derivatives exhibiting AIE phenomenon which boost their emission intensity with the temperature increase and most of those concern measurements taken in nonpolar, organic solvents – like in series of benzodithiophene BODIPY derivatives synthesized by Sengupta et al.93 Searching for the cause of distinct and surprising behavior of 2, one should consider that introduction of fluorine atoms to the phenyl ring of 2 diminishes its electron density and could impact both TICT and AIE processes of this molecule. As fluorine is an EWG (electron-withdrawing group), phenyl part of a BODIPY becomes a weaker donor for charge transfer. Tendency for aggregates formation or their self-assembly's patterns could also be affected as electrons withdrawal would disfavor π–π interactions of aromatic rings. On the other hand, the presence of –CH2N3 moiety in 3 introduced another rotating junction – it has been noticed that additional sites for intramolecular rotations can result in less emissive fluorophores.84 On the other hand, rotation at the junction with azide moiety could also disturb overall molecule's planarity and as such alter the aggregation pattern or its dynamics. Such subtle changes are known to greatly impact both arrangement and spectral properties of molecular rotors.94,95
During the preparation of abovementioned experiments, we have also noticed that intensities of both absorption and fluorescence decreases in time if the sample solutions are vigorously mixed, but only for derivatives able to form aggregates – 1–3 and 6. Absorption spectra of all derivatives were measured for one hour, in 10 minutes intervals, with sample solutions being mixed on vortex mixer for 10 second after each measurement. Analogical measurement was taken for non-mixed samples. Obtained results (Fig. S8 and S9†) clearly indicates that mixing of fluorophore's solution decrease absorption intensity but only for aggregating ones, while there is no effect on absorption of non-aggregating compounds 4 and 5. Additionally, when used quartz cuvettes were emptied and field with DMSO as a wash, the absorption spectra characteristic for given BODIPY derivative still could be obtained. Once again, such observation was made only for aggregative compounds 1–3 and 6, while for 4 and 5 there were no traces of absorption after DMSO wash (Fig. S8 and S9†). We assume that nanosized aggregates are formed on the walls of a cuvette even thou they are macroscopically clear and transparent and the addition of DMSO causes them to come apart, which results in observed sharp spectra. Fluctuations of absorbance and fluorescence intensities while mixing are commonly observed among molecular rotors, as mixing can promote their rotation.96 Additionally, while vigorous mixing, the solution in the cuvette gets aerated. As the concentration of gasses such introduced to the solution increase, it decreases the density of a water-based solution.97 Less-dens media are also known to enhance molecular rotation and as such – favor TICT.
The variety of conditions and variables can influence on optical properties of BODIPY dyes. Consequently, data analysis could be challenging for systems in which multiple variables coexist.
2.2. Investigation upon interaction between BODIPY and c-MYC G4
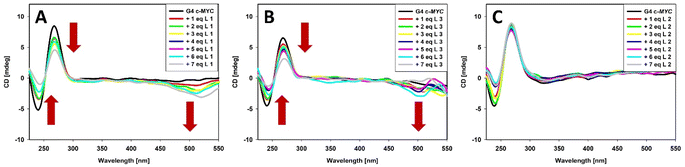
As mentioned, studies by Li et al. revealed high selectivity of BODIPY derivatives towards parallel c-MYC G4.59 Therefore, we decided to investigate ability of tested ligands to recognize c-MYC G-quadruplex structure. First, we performed circular dichroism (CD) titration experiment by adding incremental BODIPY to pre-folded c-MYC G4 (Fig. 4 and S10†). Before titration, the pre-folded G-quadruplex structure was confirmed by circular dichroism measurement (CD). According to the literature, we observed a strong positive band at 264 nm and a smaller negative at 245 nm characteristic for the parallel G-quadruplex structure formed by c-MYC.73,76 Next, we added the increasing amounts of each ligand (L) to c-MYC G4 solutions and in all cases the CD signals at 264 nm decreased (Fig. 4 and S10†). For example, ligand 3 induced a strong decrease (57.21%, at 7 mol equiv.) of the CD signal of the G4 collected at 264 nm and a smaller negative band at 245 nm (Fig. 4B).Interestingly, we have also noticed that CD signal of c-MYC G4 at 264 nm did not decrease gradually after addition of BODIPY ligands. Therefore, we decided to investigate interaction between BODIPY ligands 1–6 and G4 by adding small amounts (0.25 equiv., 12 times) of BODIPY ligands to 5 μM solution of folded c-MYC G4. As shown in Fig. S11 and Table S2,† we can observe the fluctuations in intensity of the positive band peak at 265 nm as well as the negative band at range from 505–520 nm for ligands 1–5. In case of ligand 6 we rather noticed gradual decrease of the positive CD band. The UV-Vis spectra measured parallel with CD spectra also indicate the similar tendency for absorbance at 260 nm upon addition of ligands 1–6 (Fig. S11†), which also imply interactions between tested BODIPY ligands and folded c-MYC G4. The observed fluctuations and final decrease in CD signal indicated that BODIPY molecules 1–6 can be classified as G-quadruplex destabilizing compounds. However, caution should be exercised when making such conclusions based only on CD titrations used to study DNA/small molecule interactions because of possible ICD contributions and other possible artefacts (e.g., aggregation) that cannot be easily unravelled.98
What is important, none of the studied BODIPY-derived ligands did affect the position of the bands of the c-MYC G4, which indicate that the G4 retains the parallel topology. Observed fluctuations in the intensity of the CD signals after addition of BODIPY ligands suggests end-stacking interactions with G4 tetrad rather than process of disturbing the perfect arrangement of tetrads in the G-quadruplex by compounds 1–6.99,100 Moreover, in case of compounds 1 and 3 the induced negative signals (ICD) were clearly observed in the long-wavelength region where BODIPY-based ligands exhibit absorption band (around 500 nm) (Fig. 4). This observation rather excludes the groove binding to G-quadruplex structures, the binding mode with characteristic positive ICD signals in the long-wavelength absorption region of the ligand.76,101–106 The presence of ICD suggests close proximity between the BODIPY ring of ligand 1 or 3 and G-tetrad(s) and strong π–π interactions between them.107 Using CD, we also checked if ligands 1–6 were able to induce G-quadruplex structure formation. We found that the addition up to 7 equivalents of tested ligands did not result in the formation of the G-quadruplex structure (Fig. S12†). Based on our experiments we supposed that the level of fluctuations in CD spectra observed both for G4 and single-stranded DNA (Fig. S13†) after binding with BODIPY ligands comes from each ligand ability to form aggregates due to binding to DNA and complexity equilibria between free ligands and DNA/BODIPY complexes. As it has been proposed for porphyrins, there exist a few possibilities of ligand/G4 interactions, which are in equilibrium.108 Thus, different species are present in solution, among them there are free ligand monomers, monomers bound to DNA and ligand aggregates formed on the surface of DNA molecules. We supposed that this scenario is possible in case of studied BODIPY ligands and it is responsible for fluctuations observed in their CD and UV spectra.
To verify if the main way of interaction between BODIPY derivatives and G4 is through end-stacking interaction, we decided to use a G4/hemin peroxidase inhibition assay.57,109 G4-hemin complex behaves as DNAzyme and exhibits peroxidase activity, i.e. it is able to oxidize ABTS in the presence of H2O2.110 The hemin molecule binds to the DNA oligonucleotide through an end-stacking interaction between the pyrrole rings of porphyrin and DNA nucleobases. This interaction mode requires the easy access of hemin to the G-quartet; thus, the G4s forming parallel topologies are known to produce DNAzymes exhibiting higher peroxidase activity.111 One should remember that the examined c-MYC G4 structure has a sidewise-type of loops, which facilitates ligand/G-tetrad π–π stacking interaction. We assumed that our ligands would compete with hemin to bind on the G-tetrad surface, that should cause the decrease in the catalytic oxidation ability of DNAzyme. Thus, for our purpose we used two parallel G-quadruplexes: c-MYC G4 as well as Cat G4. The latter one possesses the sequence 5′-TGG GTA GGG CGG GTT GGG AAA-3′and it also adopts the parallel topology.112 Among tested ligands only ligands 1 and 3 inhibit the activity of both complexes: c-MYC G4/hemin as well as Cat G4/hemin (compare Fig. 5, S14 and S15†). The fact that such results were obtained for those two ligands is in a good agreement with previously discussed CD spectra, in which we observed ICD bands only for these ligands. However, the clear inhibition activity of c-MYC G4/hemin or Cat G4/hemin complexes was observed only in presence of BODIPY derivative 1 as indicated by their initial reaction rates 5.0 × 10−4 [1 s−1] and 10.0 × 10−4 [1 s−1] compared with values 8.0 × 10−4 [1 s−1] and 17.0 × 10−4 [1 s−1] for c-MYC G4/hemin or Cat G4/hemin, respectively. These results support the conclusion that binding process of tested BODIPY-based compounds on the parallel G-quadruplexes DNA can occur via end-stacking interaction with external G-tetrads of G-quadruplexes. According to literature61 the 5′-end G-tetrad is more accessible, providing a larger hydrophobic surface, where the ligand could more easily stack and interact stronger compared to the 3′-end G-tetrad. Taking into account the results of DNAzyme assay, we can assume that our BODIPY ligands are rather stacked on G-tetrad at 3′end as hemin has been demonstrated to bind selectively to the 3′-terminal G-tetrad of parallel G4s.113
We may also speculate the BODIPY molecules may induce the changes in conformation of the flanking bases at 3′ termini as was demonstrated for quindoline/c-MYC114 or 9CI/c-MYC G-quadruplex interactions.115 Moreover, such structural arrangement could also be explanation of the ICD band observed in case of ligand 1 and 3. We realize that such hypothesis should be further verified by molecular dynamic simulations or NMR studies.
To assess the affinity of BODIPY ligands 1–6 to G-quadruplex structure, we performed fluorescence titration experiment by adding incremental c-MYC G4 to solution of BODIPY and vice versa (reverse titration). Addition of c-MYC G4 to 1 μmolar solution of ligands causes quenching of their fluorescence except of BODIPY 4 and 5 (Fig. 6A). In case of 2 and 6 a dramatic decrease was observed in fluorescence intensity (around 90%) with no shifts for the maximum peaks. However, the reference experiments reveled that emission of ligands also decreases upon addition of the same amount (in ml) of KCl/Tris buffer (Fig. 6B). This effect we attributed to the aggregation of these ligands, which we demonstrated earlier (see part 2.2). As previously, the fluorescence emission of ligands 4 and 5 (with sulfonic group in BODIPY core) was stable and almost unaffected by addition of salt. Taking into account that observed changes in fluorescence of ligands 1–3 and 6 are not only caused by G4 but also by salt (or only mixing) it became clear that these data could not be used to obtain the reliable values Kb of binding constants. Thus, we decided to perform reverse fluorescence titrations to estimate the binding affinities of the tested BODIPY compounds. Such approach was used by other researchers studying ligands/G4 complexes,116,117 although it seems to be controversial due to fact that the fluorescence molecule is added to G4 solution and the inner filter effect (IFE) can contribute to level of final emission.
Anyway, we believe that obtained results at least can help to distinguish which of tested ligands binds with highest affinity to G-quadruplex. In our case, the ligands were added to 2 μM G4 solution in stepwise mode to reach the final concentration 10 or 14 μM (Fig. S16†). Interestingly, for emission of ligand 2 we observed the increase of weak fluorescence band located near 505 nm, which we attributed to aggregates formation. In the case of hydrophilic ligands 4–6, no plateau was observed for titration plots. Next, such obtained, reverse fluorescence titration data were fitted to the non-competitive DNA-binding model.118 The calculated binding constants, Kb values may be ranked as follows: 7.80 ± 0.03 × 106 for 3, 5.40 ± 2.10 × 106 for 1, 2.51 ± 0.80 × 105 for 6, 1.92 ± 0.10 × 105 for 5, 1.48 ± 0.03 × 105 for 4 and 1.30 ± 0.03 × 105 for 2. The obtained results indicated that BODIPY 1 and 3 bind with high affinity to G4 c-MYC, that is in good agreement with observed DNAzyme inhibition, as only ligands 1 and 3 bind to parallel G4 with affinity sufficient to compete with hemin molecule.
To investigate the binding stoichiometries of tested BODIPY derivatives with the G-quadruplex we applied a fluorescence-based Job plot. In this method, the total molar concentration of BODIPY ligand and c-MYC G4 were kept constant, but their mole fractions were varied.119 The valuable plots were obtained only for ligand 1 and water-soluble ligands 4–6. As shown in Fig. S17,† inflection points in the graphs are observed at the DNA fraction of 0.5 for all complexes. These results indicated that BODIPY ligands bind to c-MYC G4 DNA by 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry. One should remember, that Job plot stoichiometry represents only the major binding event and the higher binding stoichiometries are difficult to determine precisely in case of ligands able to bind by multiple modes such as end-stacking, electrostatic interactions, groove binding.120 We cannot exclude such scenario, especially for ligands 2 and 3 due to their tendency to aggregation, and thus possibility of self-association on the DNA backbone, analogously as proposed by Gandini et al. for porphyrins.108 Nevertheless, the main interaction of our BODIPY ligands with G4 seems to be end-stacking process with 1
1 binding stoichiometry. One should remember, that Job plot stoichiometry represents only the major binding event and the higher binding stoichiometries are difficult to determine precisely in case of ligands able to bind by multiple modes such as end-stacking, electrostatic interactions, groove binding.120 We cannot exclude such scenario, especially for ligands 2 and 3 due to their tendency to aggregation, and thus possibility of self-association on the DNA backbone, analogously as proposed by Gandini et al. for porphyrins.108 Nevertheless, the main interaction of our BODIPY ligands with G4 seems to be end-stacking process with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio.
1 ratio.
2.3. The interaction between BODIPY and different DNA structures
The DNA melting studies are often used as fast technique to screening the selectivity of ligands towards different DNA. Among tested ligands, 1 and 3 exhibited considerable selectivity for parallel G-quadruplexes as indicted by binding studies (Kd in micromolar range).Thus, we decided to check their ability to discrimination between parallel G-quadruplex topology c-MYC G4 and other structural DNA form. For our studies we used hybrid-type G-quadruplex 22HT/K+, antiparallel G-quadruplex 22HT/Na+, double-stranded DNA ds26, single-stranded DNA and i-motif (iM) c-MYC (Table S3†).60 Therefore, we acquired UV melting curves for different DNA tetraplexes with and w/o BODIPY ligands 1 and 3 at 295 nm (Table 1 and Fig. S18†). The most pronounced effect can be seen for ligand 3/c-MYC G4 complex with Tm 70.0 °C, which is 6.4 °C lower than for free c-MYC G4. Interestingly, changes in Tm indicate that ligand 1 and 3 exhibit destabilizing effect for G4 with different topologies, but in contrary exhibits stabilizing effect for iM. It is worth to mention that the rest among tested ligands seems to stabilize c-MYC G4 (Fig. S19†).
| System | Tm ± SD [°C] | ΔTm [°C] | ΔH [kcal mol−1] | ΔS [cal mol−1 K−1] | ΔG (310 K) [kcal mol−1] | |
|---|---|---|---|---|---|---|
| c-MYC G4 (parallel) | DNA | 76.4 ± 0.5 | −52 | −149 | −5.8 | |
| +Ligand 1 | 76.2 ± 0.5 | −0.2 | −38 | −108 | −4.5 | |
| +Ligand 3 | 70.0 ± 0.3 | −6.4 | −28 | −83 | −2.3 | |
| 22HT/Na+ (antiparallel) | DNA | 57.5 ± 0.5 | −31 | −93 | −2.2 | |
| +Ligand 1 | 56.1 ± 0.4 | −1.4 | −37 | −111 | −2.6 | |
| Ligand 3 | 54.0 ± 0.2 | −3.5 | −32 | −99 | −6.3 | |
| 22HT/K+ (hybrid) | DNA | 63.2 ± 0.5 | −35 | −104 | −2.8 | |
| +Ligand 1 | 61.3 ± 0.3 | −1.9 | −32 | −96 | −2.2 | |
| +Ligand 3 | 62.5 ± 0.1 | −0.7 | −30 | −91 | −1.8 | |
| iM c-MYC (i-motif) | DNA | 53.9 ± 0.5 | −60 | −184 | −3.0 | |
| +Ligand 1 | 54.3 ± 0.2 | + 0.4 | −57 | −174 | −3.1 | |
| +Ligand 3 | 55.2 ± 0.1 | + 1.3 | −56 | −170 | −3.3 | |
A thermodynamic analysis of the denaturation profiles (shown in Fig. S18†) was performed according to two-state model, where the folded G-quadruplex is in equilibrium with the unfolded strand. The enthalpy and entropy were calculated from classical relation as follows: ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (K) = ΔH − TΔS. The equilibrium constant K can be written as K = f/(1 − f), where for intramolecular equilibrium f is the fraction of folded oligonucleotide. In all cases G-quadruplex or i-motif formation is enthalpically favorable and entropically unfavorable as observed previously.121 The thermodynamic data collected in Table 1 have shown that ligand/G4 binding events are the exotermic reactions. For all oligonucleotides the ΔG of tetraplex formation was negative in the appropriate buffer at physiological temperature (37 °C), indicating that the predominant species was the folded form in this temperature (Table 1). The interaction with ligands is associated with a net free energy change (ΔG). For example, ligand 3 binds to c-MYC G4 causing ΔG change from −5.8 to −2.3 kcal mol−1, whereas to 22HT/Na+ causing ΔG change from −2.2 to −6.3 kcal mol−1. Thus, ligand 3 pronounces destabilizing effect on c-MYC G4 and opposite on 22HT/Na+.
(K) = ΔH − TΔS. The equilibrium constant K can be written as K = f/(1 − f), where for intramolecular equilibrium f is the fraction of folded oligonucleotide. In all cases G-quadruplex or i-motif formation is enthalpically favorable and entropically unfavorable as observed previously.121 The thermodynamic data collected in Table 1 have shown that ligand/G4 binding events are the exotermic reactions. For all oligonucleotides the ΔG of tetraplex formation was negative in the appropriate buffer at physiological temperature (37 °C), indicating that the predominant species was the folded form in this temperature (Table 1). The interaction with ligands is associated with a net free energy change (ΔG). For example, ligand 3 binds to c-MYC G4 causing ΔG change from −5.8 to −2.3 kcal mol−1, whereas to 22HT/Na+ causing ΔG change from −2.2 to −6.3 kcal mol−1. Thus, ligand 3 pronounces destabilizing effect on c-MYC G4 and opposite on 22HT/Na+.
The CD spectra of pre-annealed solutions of G-quadruplexes of different topologies as well as i-motif and single-stranded and double-stranded DNA were recorded prior addition of BODIPY ligands in stepwise mode as previously (Fig. 7, S12A, C and S20†). In general, none of ligands (1 and 3) affect the positions of the CD signals, suggesting that the DNA topology is unchanged. Moreover, no significant changes in the long-wave part of CD spectra were observed even after the addition of 7 equivalents of ligand 3 to antiparallel G4 (22HT/Na+) or hybrid G4 (22HT/K+). The latter results suggest that binding mode of 3 to these G4s can be different than in complexes with parallel c-MYC G4. However, the ligand 1 induced CD band around 510 nm in case of antiparallel G4 (22HT/Na+) or hybrid G4 (22HT/K+), analogously to parallel G4 (c-MYC G4) (Fig. S20A and B†). Summarizing, the performed CD titration experiments of the different DNA structure with 1 and 3 also showed that these BODIPY ligands interact more preferably with tetraplexes than ds26 or ssDNA. The interaction of BODIPY derivatives 1–6 with iM will be explored deeply in the future.
2.4. The cytotoxicity of BODIPY derivatives
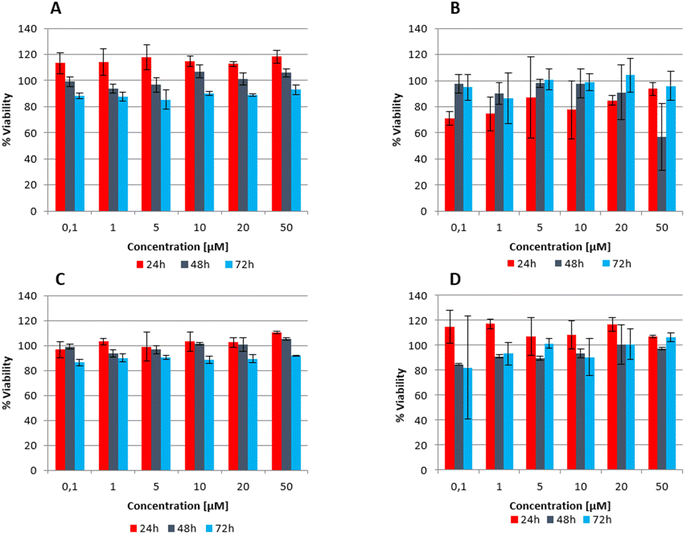
Taking into account the potential applications of BODIPY dyes to serve as DNA G-quadruplex interacting compounds or bio-imaging agents, their biological activity was investigated. For this purpose, the chosen BODIPY derivatives (ligand 1 and ligand 3) were tested for their contribution to cellular metabolic activity and the viability and cytotoxicity in the T-47D human breast carcinoma cell line according the MTT assay protocol. In addition, the human fetal lung fibroblast MRC-5 cell line was used to assess the effects of the compounds on normal cells. The cells were treated with varying concentrations of the synthesized compounds, ranging from 0.1 μM to 50 μM. The graphs of percentage on viability of T-47D and MRC-5 cells against concentrations of the tested ligand 1 and ligand 3 (after 24 h, 48 h and 72 h of incubation) are presented in Fig. 8.Evaluation of dose-response to tested ligands 1 and 3 measured at rising concentrations indicated their non-cytotoxic effect. Both tested ligands slightly affected cell viability in a dose-dependent, but not significant manner. It was observed that ligand 1 after 24 h treatment increased T-47D viability from 12% to 18% for 5 μM or 50 μM, while viability reduction from 5–25% was observed for MRC-5 cells. To compare, the slight cytotoxic activity against T-47D and for MRC-5 cells after 48 h (viability >94% and >90%, respectively) and 72 h (viability >85%) was detected. Only in the case of the concentration 50 μM of BODIPY 1, the highest effect on MRC-5 cell after 48 h of incubation (viability 57%) was observed. Interestingly, the opposite tendency occurred in case of both cell lines ligand 3 treatment. Thus, the higher viability (from 107% to 117% for 1 μM ligand) of non-tumorigenic MRC-5 cells was observed after 24 h of incubation compared to 102% (1 μM) for T-47D cells. The results for T-47D and MRC-5 cells after 48 h treatment with 0.1 μM ligand 3 were 85%, compared to viability 82% after 72 h of incubation (Fig. 8).
Summarizing, in the ligand 1 and 3 concentration range of 0.1–50 μM, the chosen BODIPY derivatives indicated non-cytotoxicity against non-tumorigenic lung fibroblast cells, nor breast cancer cells. The interactions of ligands 1 and 3 with G4 structures demonstrated during our studies in vitro seems to be not associated with their anticancer activity, as it was shown by in cellulo evaluations. On the other hand, the non-cytotoxic properties of the BODIPY dyes 1 and 3 would be the assets in their potential applications in neurological disorders and age-related diseases investigations, as reported for other small molecules that destabilize G-quadruplex.122
However, more in-depth studies in the context of neurodegenerative diseases should be performed. G4 and i-motif-binding compounds are known for their ability to bind promoters and affect the metabolism of mitochondria that are critical for age-related and neurodegenerative diseases.123 Thus, further studies should involve evaluation of cortical neurons in the context of ROS levels assessment followed by evaluation of autophagy, inflammation and gene expression analysis. Similarly, such studies should be accompanied by 3D cell culture models and animal studies evaluation. Moreover, the evaluated compounds 1 and 3 seem to be the good candidates for the bio-imaging as the BODIPY-based compounds have already been found as fluorescent probes in Alzheimer disease and other related processes.124
3. Materials and methods
3.1. Materials
DNA oligonucleotides were purchased from Genomed (Warsaw, Poland) and were used without further purification. The strand concentrations were determined at 260 nm at 85 °C using extinction coefficients of 254![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1 (c-MYC G4), 221
600 M−1 cm−1 (c-MYC G4), 221![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 013 M−1 cm−1 (Cat G4), 282
013 M−1 cm−1 (Cat G4), 282![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1 (c-MYC i-motif), 251
700 M−1 cm−1 (c-MYC i-motif), 251![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 M−1 cm−1 (22HT G4), 282
800 M−1 cm−1 (22HT G4), 282![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 M−1 cm−1 (ds26) as calculated taking the literature values of molar absorptivity of nucleotides.125 Tris base (CAS number 77-86-1), Tris–HCl (CAS number 1185-53-1) and dimethylarsinic acid sodium salt trihydrate (CAS number 6131-99-3), hemin, H2O2, ABTS (2,2′-azinobis(3-ethylbenzothiozoline)-6-sulfonic acid) were purchased from Merck Life Science (Poznań, Poland) and used as received. A stock solution of hemin was prepared in DMSO at a concentration of 0.01 M, and was stored in a freezer for up to one month. RPMI1640 medium for culture the epithelial T-47D human breast carcinoma cells was purchased from (Biochrom GmbH, Merck Millipore, Germany), MTT solution (5 mg mL−1) from Sigma-Aldrich, St. Louis, MO, USA and used as received.
100 M−1 cm−1 (ds26) as calculated taking the literature values of molar absorptivity of nucleotides.125 Tris base (CAS number 77-86-1), Tris–HCl (CAS number 1185-53-1) and dimethylarsinic acid sodium salt trihydrate (CAS number 6131-99-3), hemin, H2O2, ABTS (2,2′-azinobis(3-ethylbenzothiozoline)-6-sulfonic acid) were purchased from Merck Life Science (Poznań, Poland) and used as received. A stock solution of hemin was prepared in DMSO at a concentration of 0.01 M, and was stored in a freezer for up to one month. RPMI1640 medium for culture the epithelial T-47D human breast carcinoma cells was purchased from (Biochrom GmbH, Merck Millipore, Germany), MTT solution (5 mg mL−1) from Sigma-Aldrich, St. Louis, MO, USA and used as received.
3.2. Synthesis of BODIPY derivatives and preparation of the stock solutions of 1–6
The details of preparation of compounds 1–6 are included in ESI.† Compound 1 is also commercially available. Compound 4 is newly synthesized by us. The purity of each ligand was examined on the basis of their NMR spectra. The details of crystal structure determination and crystal data of compound 2 (No. CCDC: 2241316) are included in ESI†.Ligands 1–3 were dissolved in DMSO, whereas ligands 4–6 were dissolved in H2O for obtaining stock solutions of ∼1.5 mM concentration and were stored at 4 °C. The ligands 2 and 3 were stocked no longer than 48 h.
3.3. Preparation DNA samples for spectroscopic measurements
A day before measurements, the DNA solutions were pre-annealed in appropriate buffer at 90 °C for 5 min followed by slow cooling, and storing at 4 °C overnight. Concentration of DNA samples were 5 μM/strand or 2 μM/strand. DNA oligonucleotides were dissolved in a 10 mM Tris–HCl buffer (pH 7.2) w/o salt (ss c-MYC) or with 100 mM KCl (c-MYC G4), with 100 mM NaCl (22HT/Na), 100 mM KCl (22HT/K), 100 mM NaCl (ds26) or in cacodylate buffer (10 mM, pH 5.5) with 100 mM KCl (iM c-MYC).3.4. Apparatus
Circular dichroism (CD) spectra were recorded on a Jasco J-810 spectropolarimeter (Jasco, Tokyo, Japan). Fluorescence spectra were acquired using a Jasco Spectrofluorometer (Tokyo, Japan) or Cary Eclipse (Agilent Technologies). UV-Vis spectra were obtained using a Jasco spectrophotometer (Jasco, Tokyo, Japan) equipped with a temperature controller. Cell viability was quantified spectrophotometrically using a Labsystems Multiscan RC plate reader (Thermo Fisher Scientific, Helsinki, Finland).3.5. Circular dichroism (CD) measurements
Circular dichroism (CD) spectropolarimeter was equipped with a temperature controller, in the spectral range from 210 to 700 nm with 1000 nm min−1 scan speed and bandwidth of 1 nm. Each spectrum was recorded in quartz cuvettes of 10 mm path length and averaged from 3 scans. Concentration of DNA samples were 5 μM/strand or 2 μM per strand. Each ligand was added to DNA solutions in stepwise manner starting from 0.25 to 5 molar or 0.5 to 10 molar equivalents, unless otherwise noted.3.6. Fluorescence spectroscopy
The emission spectra were recorded in the wavelength range of 490–650 nm at 25 °C using an excitation wavelength (λex) of 475 nm for all the samples. A quartz cuvette with a 10 mm path length in the excitation direction and a 4 mm path length in the emission direction were used. All emission spectra were uncorrected.3.7. Melting temperature (Tm) determination by UV spectroscopy
The melting curves of the samples were obtained using a spectrophotometer equipped with a Jasco temperature controller. A day before measurements, the 2 μM oligonucleotide solutions were preannealed in appropriate buffers as indicated in 3.3 and in buffer with low concentrations of stabilizing cation (10 mM Tris–HCl buffer (pH 7.2) containing 90 mM LiCl and 10 mM KCl for c-MYC G4). The melting profiles were recorded in the absence and presence of 3 equiv. of ligands (1 or 3) in 10–90 °C range with a 1 °C min−1 temperature gradient and data were collected at 295 nm. All experiments were carried out using quartz cuvettes with a 10 mm optical path. The melting temperatures (Tm) were determined as the maximum of the first derivative of the heating curves. Each Tm value was an average of three independent measurements.3.8. Peroxidase activity measurements
Samples containing 1 μM DNA, 10 mM Tris–HCl buffer (pH 7.2), and 100 mM KCl were heated for 5 min at 95 °C, and allowed to cool down on the ice bath for 15 min. Then, the samples were divided into two equal parts, that were treated as follows. To the first part hemin was added to obtain 1 μM solution and the obtained mixture was incubated at RT for 30 min. Next, tested ligand was added to get 1 μM concentration, and the mixture was incubated at lab bench for additional 30 min. Parallelly, to the second part of the samples, only hemin was added to get 1 μM solution, followed by the incubation at lab bench for 30 min. After addition of the peroxidase substrate (ABTS), 140 μl of samples were transferred into microplate wells. The time profiles of absorbance were recorded using a M200 microplate reader (Tecan, Mannedorf, Switzerland), after initialization of the reaction with the addition of H2O2 (final sample volume = 150 μl). Absorbance changes for ABTS oxidation were monitored at 414 nm in 10 s intervals for 15 min.3.9. MTT assay
The epithelial T-47D human breast carcinoma cells, were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum and insulin (8 μg mL−1; Gibco). A non-tumorigenic human fetal lung fibroblast MRC-5 cells were cultured in EMEM medium supplemented with 10% fetal bovine serum. Before reaching confluence, cells were counted and passaged into 96-well plates (5000 cells per well). Cells were cultured for 24, 48 or 72 h with or without ligand at 0.1–50 μM. The solvent, DMSO in a concentration of 0.10%, was also applied as a control. After this time 10 μL of MTT solution (5 mg mL−1) was added to each well. The plate was incubated at 37 °C for 2 h followed by 100 μL of solubilization buffer (10% SDS in 0.01 M HCl) addition. Cell viability was quantified spectrophotometrically. Cytotoxicity rate was expressed as the absorbance of a sample comparing to control cells, obtained values were calculated using CalcuSyn (Biosoft, Cambridge, UK) and Excel software (Microsoft, Redmond, WA, USA). The obtained results were statistically analyzed using the Welsh test and online programs: Statology and GraphPad. Each experiment was repeated at least three times. All experiments were performed in minimal exposure to light.4. Conclusions
We have studied the ability of three hydrophobic as well as three hydrophilic BODIPY dyes to recognize, stabilize or induce G-quadruplex (G4) formation. The spectroscopic studies performed for BODIPY dyes have revealed tendency of 1–3 and 6 to form aggregates, which have influence on their fluorescence properties and later were found to impact their interaction's mode with studied DNA tetraplexes. Therefore, the complexity of fluorescence signal of tested compounds forced us to use reverse fluorescence titration approach to evaluate their binding affinity.Among tested BODIPY derivatives, ligands 1 and 3 indicated the highest affinity to parallel c-MYC G4 and are able to compete with hemin to bind on the G-tetrad surface. The main mode of those interactions occurs via end-stacking and binding stoichiometry 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The end-stacking binding mode with external G-tetrads was also confirmed by circular dichroism spectroscopy titration. The strong negative ICD in the long wavelength absorption region of achiral BODIPY-based ligands 1 and 3 around 500 nm indicates that π–π end-stacking interactions may be the dominant interaction mode between compounds 1 or 3 and c-MYC G4. The CD spectra titrations also revealed that none of tested ligands with BODIPY skeleton were able to induce formation of a stable G4. Moreover, CD studies upon interactions of 1 and 3 with different DNA forms indicated that these ligands interact more preferably with tetraplexes (both G4s and i-motifs) than ssDNA and dsDNA. However, the ligand 1 induced CD band around 510 nm in case of antiparallel G4 (22HT/Na+) or hybrid G4 (22HT/K+), which suggest that binding mode of 1 to these G4s can be similar to complexes with parallel c-MYC G4. A decrease in CD signal intensity (the position of signals has not changed) recorded upon the addition of up to five equivalents of ligands, suggesting the presence of ligand-dependent perturbations in the studied G4 structure. However, this may be the result of the stacking interaction and not the destabilization of G4. To verify this hypothesis we performed a thermodynamic analysis of the melting profiles of different topology G4s with and w/o BODIPY derivatives. The latter results confirmed that the ligand 1 and 3 interacted the strongest with c-MYC G4 as changes in enthalpy and entropy are the most pronounced and a net free energy changes confirm the destabilizing effect of these ligands on c-MYC G4.
1. The end-stacking binding mode with external G-tetrads was also confirmed by circular dichroism spectroscopy titration. The strong negative ICD in the long wavelength absorption region of achiral BODIPY-based ligands 1 and 3 around 500 nm indicates that π–π end-stacking interactions may be the dominant interaction mode between compounds 1 or 3 and c-MYC G4. The CD spectra titrations also revealed that none of tested ligands with BODIPY skeleton were able to induce formation of a stable G4. Moreover, CD studies upon interactions of 1 and 3 with different DNA forms indicated that these ligands interact more preferably with tetraplexes (both G4s and i-motifs) than ssDNA and dsDNA. However, the ligand 1 induced CD band around 510 nm in case of antiparallel G4 (22HT/Na+) or hybrid G4 (22HT/K+), which suggest that binding mode of 1 to these G4s can be similar to complexes with parallel c-MYC G4. A decrease in CD signal intensity (the position of signals has not changed) recorded upon the addition of up to five equivalents of ligands, suggesting the presence of ligand-dependent perturbations in the studied G4 structure. However, this may be the result of the stacking interaction and not the destabilization of G4. To verify this hypothesis we performed a thermodynamic analysis of the melting profiles of different topology G4s with and w/o BODIPY derivatives. The latter results confirmed that the ligand 1 and 3 interacted the strongest with c-MYC G4 as changes in enthalpy and entropy are the most pronounced and a net free energy changes confirm the destabilizing effect of these ligands on c-MYC G4.
Summarizing, we showed that the simple BODIPY dyes with the unmodified or modified phenyl aromatic ring at meso position may serve as novel G4-recognizing ligands. In case of two ligands, we observed the distinct tendency toward disrupting the G4 structure and for this reason these ligands are good candidates for bio-imaging or to study neurodegenerative as well as age-related diseases.
Data availability statement
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization: AD and MR; synthesis, structure determination: MR; CD, UV and fluorescence data collection: AD and JŻ; peroxidase activity measurements: JŻ; biological experiments: JD and BR; data analysis AD, MR and JŻ; interpretation of results: AD, MR, JŻ and BR; draft manuscript preparation: AD and MR, editing AD, MR, JŻ; revision: AD, MR, and BR. All authors reviewed the results and approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Adam Mickiewicz University, Poznań, Poland. Authors thank prof. Bernard Juskowiak for valuable discussions; prof. Maciej Kubicki and Grzegorz Dutkiewicz for crystal structure determination of 2, Joanna Kosman for support during experiments with DNAzymes.References
- R. P. Haugland, Handbook of Fluorescent Probes and Research Chemicals, 6th edn, Molecular Probes: Eugene, OR, 1996; ISBN 0965224007 Search PubMed.
- A. Loudet and K. Burgess, Chem. Rev., 2007, 107, 4891 CrossRef CAS PubMed.
- N. Boens, V. Leen and W. Dehaen, Chem. Soc. Rev., 2012, 41, 1130 RSC.
- S. Adhikari, J. Moscatelli, E. M. Smith, C. Banerjee and E. M. Puchner, Nat. Commun., 2019, 10, 3400 CrossRef PubMed.
- M. Okamoto, S. Kobayashi, H. Ikeuchi, S. Yamada, K. Yamanouchi, K. Nagasawa, S. Maekawa, T. Kato and I. Shimizu, Steroids, 2012, 77, 845 CrossRef CAS PubMed.
- J. Karolin, L. B.-A. Johansson, L. Strandberg and T. Ny, J. Am. Chem. Soc., 1994, 116, 7801 CrossRef CAS.
- F. An, J. Xin, C. Deng, X. Tan, O. Aras, N. Chen, X. Zhang and R. Ting, J. Mater. Chem. B, 2021, 9, 9308 RSC.
- B. Sui, S. Tang, A. W. Woodward, B. Kim and K. D. Belfield, Eur. J. Org Chem., 2016, 2851 CrossRef CAS.
- R. D. Moriarty, A. Martin, K. Adamson, E. O'Reilly, P. Mollard, R. J. Forster and T. E. Keyes, J. Microsc., 2014, 253, 204 CrossRef CAS PubMed.
- X. He, H. Li, S. Liu, Y. Li, X. Lin, H. Zheng, Z. Zhou and D. Zeng, ChemistrySelect, 2022, 7(4), e202103821 CrossRef CAS.
- H. Xiong, S. Liu, T. Wei, Q. Cheng and D. J. Siegwart, J. Controlled Release, 2020, 325, 198 CrossRef CAS PubMed.
- H. Xiong, H. Zuo, Y. Yan, G. Occhialini, K. Zhou, Y. Wan and D. J. Siegwart, Adv. Mater., 2017, 29, 1700131 CrossRef PubMed.
- Q. Zheng, G. Xu and P. N. Prasad, Chemistry, 2008, 14, 5812 CrossRef CAS PubMed.
- M.-C. Yee, S. C. Fas, M. M. Stohlmeyer, T. J. Wandless and K. A. Cimprich, J. Biol. Chem., 2005, 280, 29053 CrossRef CAS PubMed.
- A. Kamkaew, S. H. Lim, H. B. Lee, L. V. Kiew, L. Y. Chung and K. Burgess, Chem. Soc. Rev., 2013, 42, 77 Search PubMed.
- J. Zou, Z. Yin, K. Ding, Q. Tang, J. Li, W. Si, J. Shao, Q. Zhang, W. Huang and X. Dong, ACS Appl. Mater. Interfaces, 2017, 9, 32475 CrossRef CAS PubMed.
- C. Li, W. Lin, S. Liu, W. Zhang and Z. Xie, J. Mater. Chem. B, 2019, 7, 4655 RSC.
- E. Antina, N. Bumagina, Y. Marfin, G. Guseva, L. Nikitina, D. Sbytov and F. Telegin, Molecules, 2022, 27, 1396 CrossRef CAS PubMed.
- Y. Luo and G. D. Prestwich, Bioconjugate Chem., 1999, 10, 755 CrossRef CAS PubMed.
- J. Li, I. H. Kim, E. D. Roche, D. Beeman, A. S. Lynch, C. Z. Ding and Z. Ma, Bioorg. Med. Chem. Lett., 2006, 16, 794 CrossRef CAS PubMed.
- Y. Jang, T.-I. Kim, H. Kim, Y. Choi and Y. Kim, ACS Appl. Bio Mater., 2019, 2, 2567 CrossRef CAS PubMed.
- A. Sharma, A. Khatchadourian, K. Khanna, R. Sharma, A. Kakkar and D. Maysinger, Biomaterials, 2011, 32, 1419 CrossRef CAS PubMed.
- A. May, J. Mack, T. Nyokong and J. Porphyr, Phthalocyanines, 2020, 24, 1129 CrossRef CAS.
- A. K. May, C. Chiyumba, J. Harris, J. Mack, T. Nyokong and J. Porphyr, Phthalocyanines, 2022, 26(11), 691 CrossRef CAS.
- M. De Vetta and I. Corral, Comput. Theor. Chem., 2019, 1150, 110 CrossRef CAS.
- J. Bañuelos, V. Martín, C. F. A. Gómez-Durán, I. J. Arroyo Córdoba, E. Peña-Cabrera, I. García-Moreno, Á. Costela, M. E. Pérez-Ojeda, T. Arbeloa and I. López Arbeloa, Chemistry, 2011, 17, 7261 CrossRef PubMed.
- O. Galangau, C. Dumas-Verdes, R. Méallet-Renault and G. Clavier, Org. Biomol. Chem., 2010, 8, 4546 RSC.
- M. Hecht, T. Fischer, P. Dietrich, W. Kraus, A. B. Descalzo, W. E. S. Unger and K. Rurack, ChemistryOpen, 2013, 2, 25 CrossRef CAS PubMed.
- A. Oliden-Sánchez, R. Sola-Llano, J. Bañuelos, I. García-Moreno, C. Uriel, J. C. López and A. M. Gómez, Front. Chem., 2019, 7, 801 CrossRef PubMed.
- A. M. Hansen, A. L. Sewell, R. H. Pedersen, D.-L. Long, N. Gadegaard and R. Marquez, Tetrahedron, 2013, 69(39), 8527 CrossRef CAS.
- M. Ceulemans, K. Nuyts, W. De Borggraeve and T. Parac-Vogt, Inorganics, 2015, 3(4), 516 CrossRef CAS.
- C. Felion, R. Lopez-Gonzalez, A. L. Sewell, R. Marquez and C. Gauchotte-Lindsay, ACS Omega, 2022, 7(45), 41284 CrossRef CAS PubMed.
- D. Khuong Mai, B. Kang, T. Pegarro Vales, I. W. Badon, S. Cho, J. Lee, E. Kim and H.-J. Kim, Molecules, 2020, 25, 3340 CrossRef PubMed.
- C. Wang, F. Xie, N. Suthiwangcharoen, J. Sun and Q. Wang, Sci. China:Chem., 2011, 55, 125 CrossRef.
- J.-J. Shie, Y.-C. Liu, Y.-M. Lee, C. Lim, J.-M. Fang and C.-H. Wong, J. Am. Chem. Soc., 2014, 136, 9953 CrossRef CAS PubMed.
- M. Verdoes, B. I. Florea, U. Hillaert, L. I. Willems, W. A. van derLinden, M. SaeHeng, D. V. Filippov, A. F. Kisselev, G. A. van der Marel and H. S. Overkleeft, ChemBioChem, 2008, 9, 1735 CrossRef CAS PubMed.
- T. Saha, D. Kand and P. Talukdar, Org. Biomol. Chem., 2013, 11, 8166 RSC.
- Q. Zhao, C. Yin, J. Kang, Y. Wen and F. Huo, Dyes Pigm., 2018, 159, 166 CrossRef CAS.
- G. Raffy, R. Bofinger, A. Tron, A. D. Guerzo, N. D. McClenaghan and J.-M. Vincent, Nanoscale, 2017, 9, 16908 RSC.
- I. López-Duarte, T. T. Vu, M. A. Izquierdo, J. A Bull and M. K. Kuimova, Chem. Commun., 2014, 50, 5282 RSC.
- L. Michels, V. Gorelova, Y. Harnvanichvech, J. W. Borst, B. Albada, D. Weijers and J. Sprakel, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18110 CrossRef CAS PubMed.
- G. Fan, L. Yang and Z. Chen, Front. Chem. Sci. Eng., 2014, 8, 405 CrossRef CAS.
- O. Dilek and S. L. Bane, Bioorg. Med. Chem. Lett., 2009, 19, 6911 Search PubMed.
- S. Kurata, T. Kanagawa, K. Yamada, M. Torimura, T. Yokomaku, Y. Kamagata and R. Kurane, Nucleic Acids Res., 2001, 29(6), e34 CrossRef CAS PubMed.
- H.-A. Wagenknecht and T. Ehrenschwender, Synthesis, 2008, 2008, 3657 CrossRef.
- J. P. Rostron, G. Ulrich, P. Retailleau, A. Harriman and R. Ziessel, New J. Chem., 2005, 29, 1241 RSC.
- V. Ramu, P. Kundu, P. Kondaiah and A. R. Chakravarty, Inorg. Chem., 2021, 60, 6410 Search PubMed.
- P. S. Deore, D. V. Soldatov and R. A. Manderville, Sci. Rep., 2018, 8, 16874 CrossRef PubMed.
- Y. Wang, X. Li, X. Liu and T. Li, Chem. Commun., 2007, 42, 4369 Search PubMed.
- D. Dziuba, P. Jurkiewicz, M. Cebecauer, M. Hof and M. Hocek, Angew Chem. Int. Ed. Engl., 2016, 55, 174 Search PubMed.
- S. Wang, H. Liu, D. Liu, X. Ma, X. Fang and L. Jiang, Angew Chem. Int. Ed. Engl., 2007, 46, 3915 CrossRef CAS PubMed.
- Q. Meng, D. H. Kim, X. Bai, L. Bi, N. J. Turro and J. Ju, J. Org. Chem., 2006, 71, 3248 CrossRef CAS PubMed.
- J. Ju, D. H. Kim, L. Bi, Q. Meng, X. Bai, Z. Li, X. Li, M. S. Marma, S. Shi and J. Wu, et al., Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19635 CrossRef CAS PubMed.
- M. Tera, K. Iida, K. Ikebukuro, H. Seimiya, K. Shin-Ya and K. Nagasawa, Org. Biomol. Chem., 2010, 8, 2749 RSC.
- B. Prasad, M. Doimo, M. Andréasson, V. L’Hôte, E. Chorell and S. Wanrooij, Chem. Sci., 2022, 13, 2347 RSC.
- M.-Q. Wang, J.-J. Gao, Q.-Q. Yu and H.-B. Liu, New J. Chem., 2020, 44, 13557 RSC.
- L. Zhang, J. C. Er, K. K. Ghosh, W. J. Chung, J. Yoo, W. Xu, W. Zhao, A. T. Phan and Y.-T. Chang, Sci. Rep., 2014, 4, 3776 CrossRef PubMed.
- M. Deiana, K. Chand, E. Chorell and N. Sabouri, J. Phys. Chem. Lett., 2023, 14, 1862 CrossRef CAS PubMed.
- H. Y. Li, H.-W. Cao, X.-X. Lang, Y.-S. Chen and M.-Q. Wang, J. Mater. Chem. B, 2022, 10, 7772 RSC.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402 CrossRef CAS PubMed.
- J. Dai, M. Carver, L. H. Hurley and D. Yang, J. Am. Chem. Soc., 2011, 133, 17673 CrossRef CAS PubMed.
- G. N. Parkinson, M. P. H. Lee and S. Neidle, Nature, 2002, 417, 876 CrossRef CAS PubMed.
- R. I. Mathad, E. Hatzakis, J. X. Dai and D. Z. Yang, Nucleic Acids Res., 2011, 39, 9023 Search PubMed.
- T. S. Dexheimer, D. Sun and L. H. Hurley, J. Am. Chem. Soc., 2006, 128, 5404 CrossRef CAS PubMed.
- S. Rankin, A. P. Reszka, J. Huppert, M. Zloh, G. N. Parkinson, A. K. Todd, S. Ladame, S. Balasubramanian and S. Neidle, J. Am. Chem. Soc., 2005, 127, 10584 CrossRef CAS PubMed.
- T. M. Ou, Y. J. Lu, J. H. Tan, Z. S. Huang, K. Y. Wong and L. Q. Gu, ChemMedChem, 2008, 3, 690 CrossRef CAS PubMed.
- S. M. Haider, S. Neidle and G. N. Parkinson, Biochimie, 2011, 93, 1239 Search PubMed.
- S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261 CrossRef CAS PubMed.
- J. Figueiredo, J.-L. Mergny and C. Cruz, Life Sci., 2024, 340, 122481 Search PubMed.
- S. Asamitsu, T. Bando and H. Sugiyama, Chem.–Eur. J., 2019, 25, 417 CrossRef CAS PubMed.
- M. Eilers and R. N. Eisenman, Genes Dev., 2008, 22, 2755 CrossRef CAS PubMed.
- N. Meyer and L. Z. Penn, Nat. Rev. Cancer, 2008, 8, 976 Search PubMed.
- S. Paramasivan, I. Rujan and P. H. Bolton, Methods, 2007, 43, 324 Search PubMed.
- J. Kypr, I. Kejnovská, D. Renciuk and M. Vorlícková, Nucleic Acids Res., 2009, 37, 1713 CrossRef CAS PubMed.
- E. W. White, F. Tanious, M. A. Ismail, A. P. Reszka, S. Neidle, D. W. Boykin and W. D. Wilson, Biophys. Chem., 2007, 126, 140 Search PubMed.
- M. Vorlíčková, I. Kejnovská, J. Sagi, D. Renčiuk, K. Bednářová, J. Motlová and J. Kypr, Methods, 2012, 57, 64 CrossRef PubMed.
- W. Qin, M. Baruah, M. Van der Auweraer, F. C. De Schryver and N. Boens, J. Phys. Chem. A, 2005, 109, 7371 Search PubMed.
- T. Siodła, W. P. Ozimiński, M. Hoffmann, H. Koroniak and T. M. Krygowski, J. Org. Chem., 2014, 79, 7321 Search PubMed.
- C. Y. Kim, P. P. Chandra, A. Jain and D. W. Christianson, J. Am. Chem. Soc., 2001, 123, 9620 Search PubMed.
- K. E. Riley and K. M. Merz Jr, J. Phys. Chem. B, 2005, 109, 17752 CrossRef CAS PubMed.
- P. Kirsch, Modern Fluoroorganic Chemistry: Synthesis, Reactivity, Applications, Wiley, 2013, ISBN 9783527331666 Search PubMed.
- R. Chaudhuri, T. Prasanth and J. Dash, Angew. Chem., 2023, 62(7), e202215245 CrossRef CAS PubMed.
- T. Biver, Molecules, 2022, 27, 4165 CrossRef CAS PubMed.
- R. Hu, E. Lager, A. Aguilar-Aguilar, J. Liu, J. W. Y. Lam, H. H. Y. Sung, I. D. Williams, Y. Zhong, K. S. Wong, E. Peña-Cabrera and B. Z. Tang, J. Phys. Chem. C Nanomater. Interfaces, 2009, 113, 15845 CrossRef CAS.
- Z. R. Grabowski, K. Rotkiewicz and W. Rettig, Chem. Rev., 2003, 103, 3899 CrossRef PubMed.
- C. Wang, W. Chi, Q. Qiao, D. Tan, Z. Xu and X. Liu, Chem. Soc. Rev., 2021, 50, 12656 RSC.
- Y. S. Marfin, E. A. Banakova, D. A. Merkushev, S. D. Usoltsev and A. V. Churakov, J. Fluoresc., 2020, 30, 1611 CrossRef CAS PubMed.
- S. Choi, J. Bouffard and Y. Kim, Chem. Sci., 2014, 5, 751 RSC.
- X. Wang, X. Lin, R. Li, Z. Wang, W. Liu, L. Chen, N. Chen, T. Dai, S. Sun and Z. Li, et al., Molecules, 2022, 27, 193 CrossRef CAS PubMed.
- F. H. Stootman, D. M. Fisher, A. Rodger and J. R. Aldrich-Wright, Analyst, 2006, 131(10), 1145 RSC.
- J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718 CrossRef CAS PubMed.
- M. M. Ogle, A. D. Smith McWilliams, M. J. Ware, S. A. Curley, S. J. Corr and A. A. Martí, J. Phys. Chem. B, 2019, 123, 7282 CrossRef CAS PubMed.
- P. R. Aswathy, S. Sharma, N. P. Tripathi and S. Sengupta, Chemistry, 2019, 25, 14870 CrossRef CAS PubMed.
- X. Ma, J. Cheng, J. Liu, X. Zhou and H. Xiang, New J. Chem., 2015, 39, 492 RSC.
- X. Ma, R. Sun, J. Cheng, J. Liu, F. Gou, H. Xiang and X. Zhou, J. Chem. Educ., 2016, 93, 345 CrossRef CAS.
- M. A. Haidekker and E. A. Theodorakis, Org. Biomol. Chem., 2007, 5, 1669 RSC.
- H. Watanabe and K. Iizuka, Metrologia, 1985, 21, 19 CrossRef CAS.
- J. Mitteaux, P. Lejault, F. Wojciechowski, A. Joubert, J. Boudon, N. Desbois, C. P. Gros, R. H. E. Hudson, J.-B. Boule, A. Granzhan and D. Monchaud, J. Am. Chem. Soc., 2021, 143(32), 12567 CrossRef CAS PubMed.
- B. Dumat, G. Bordeau, E. Faurel-Paul, F. Mahuteau-Betzer, N. Saettel, M. Bombled, G. Metgé, F. Charra, C. Fiorini-Debuisschert and M.-P. Teulade-Fichou, Biochimie, 2011, 93, 1209 CrossRef CAS PubMed.
- K. Bhadra and G. S. Kumar, Biochim. Biophys. Acta, 2011, 1810, 485 CrossRef CAS PubMed.
- M. Eriksson and B. Nordén, Methods Enzymol., 2001, 340, 68 CAS.
- M. Hranjec, I. Piantanida, M. Kralj, L. Suman, K. Pavelić and G. Karminski-Zamola, J. Med. Chem., 2008, 51, 4899 CrossRef CAS PubMed.
- S. Catoen-Chackal, M. Facompré, R. Houssin, N. Pommery, J.-F. Goossens, P. Colson, C. Bailly and J.-P. Hénichart, J. Med. Chem., 2004, 47, 3665 CrossRef CAS PubMed.
- T. Yamashita, T. Uno and Y. Ishikawa, Bioorg. Med. Chem., 2005, 13, 2423 CrossRef CAS PubMed.
- H. Sun, Y. Tang, J. Xiang, G. Xu, Y. Zhang, H. Zhang and L. Xu, Bioorg. Med. Chem. Lett., 2006, 16, 3586 CrossRef CAS PubMed.
- J. Dash, P. S. Shirude, S.-T. D. Hsu and S. Balasubramanian, J. Am. Chem. Soc., 2008, 130, 15950 CrossRef CAS PubMed.
- A. Głuszyńska, J. Kosman, S. S. Chuah, M. Hoffmann and S. Haider, Pharmaceuticals, 2024, 17, 912 CrossRef PubMed.
- S. C. M. Gandini, I. E. Borissevitch, J. R. Perussi, H. Imasato and M. Tabak, J. Lumin., 1998, 78, 199853 CrossRef.
- X. Cheng, X. Liu, T. Bing, Z. Cao and D. Shangguan, Biochemistry, 2009, 48, 7817 CrossRef CAS PubMed.
- J. Kosman and B. Juskowiak, Adv. Biochem. Eng./Biotechnol., 2020, 170, 59 CrossRef CAS PubMed.
- D.-M. Kong, J. Wu, N. Wang, W. Yang and H.-X. Shen, Talanta, 2009, 80, 459 CrossRef CAS PubMed.
- J. Kosman, K. Żukowski and B. Juskowiak, Open Chem., 2019, 17, 1157 CAS.
- K. Saito, H. Tai, M. Fukaya, T. Shibata, R. Nishimura, S. Neya and Y. Yamamoto, J. Biol. Inorg. Chem., 2012, 17, 437 CrossRef CAS PubMed.
- M. A. S. Abdelhamid, A. J. Gates and Z. A. E. Waller, Biochemistry, 2019, 58(4), 245 CrossRef CAS PubMed.
- Q. Zhai, C. Gao, J. Ding, Y. Zhang, B. Islam, W. Lan, H. Hou, H. Deng, J. Li and Z. Hu, et al., Nucleic Acids Res., 2019, 47, 2190 Search PubMed.
- V. Gabelica, R. Maeda, T. Fujimoto, H. Yaku, T. Murashima, M. Sugimoto and D. Miyoshi, Biochemistry, 2013, 52, 5620 Search PubMed.
- R. Gargallo, A. Aviñó, R. Eritja, P. Jarosova, S. Mazzini, L. Scaglioni and P. Taborsky, Spectrochim. Acta, Part A, 2021, 248, 119185 CrossRef CAS PubMed.
- F. H. Stootman, D. M. Fisher, A. Rodger and J. R. Aldrich-Wright, Analyst, 2006, 131(10), 1145 RSC.
- P. Job, Ann Chim., 1928, 9, 113 CAS.
- N. C. Sabharwal, J. Chen, J. H. Lee, C. M. A. Gangemi, A. D'Urso and L. A. Yatsunyk, Int. J. Mol. Sci., 2018, 19, 3686 Search PubMed.
- J. L. Mergny, A. T. Phan and L. Lacroix, FEBS Lett., 1998, 435(1), 74 CrossRef CAS PubMed.
- P. Lejault, J. Mitteaux, F. R. Sperti and D. Monchaud, Cell Chem. Biol., 2021, 28(4), 436 CrossRef CAS PubMed.
- N. Sanghai and G. K. Tranmer, Cells, 2023, 12(18), 2318 CrossRef CAS PubMed.
- D. Abramchuk, A. Voskresenskaya, I. Kuzmichev, A. Erofeev, P. Gorelkin, M. Abakumov, E. Beloglazkina and O. Krasnovskaya, Eur. J. Med. Chem., 2024, 276, 116682 CrossRef CAS PubMed.
- A. V. Tataurov, Y. You and R. Owczarzy, Biophys. Chem., 2008, 133, 66 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, compounds characterization (NMR spectra, X-ray structure), spectroscopy measurements (CD, UV, FL), DNAzyme and MTT assays. CCDC 2241316. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ra08464k |
| This journal is © The Royal Society of Chemistry 2025 |