 Open Access Article
Open Access ArticleFiber ring laser biosensor based on no-core fibers for label-free DNA biomolecule measurements
Lina Wang *ab and
Chong Lia
*ab and
Chong Lia
aCollege of Electronic Engineering, Huainan Normal University, Huainan 232038, China. E-mail: wanglina2017@email.szu.edu.cn
bShenzhen Key Laboratory of Sensor Technology, Shenzhen University, Shenzhen 518060, China
First published on 21st January 2025
Abstract
An erbium-doped fiber ring laser based on a single-mode fiber–no-core fiber–single-mode fiber (SMF-NCF-SMF) structure was constructed and experimentally demonstrated for label-free DNA hybridization measurement. The SMF-NCF-SMF structure acts as a sensing element and a filter to select the laser wavelength. The proposed fiber ring laser sensor exhibits a high optical signal-to-noise ratio (SNR, >50 dB) and narrow full width at half maximum (FWHM, <0.05 nm). Its refractive index sensitivity is 116.8 nm per RIU in the range of 1.3406–1.3705, and its detection limit is 1.79 × 10−4 RIU. By continuously monitoring the laser wavelength, we successfully achieved label-free measurement of complementary DNA (cDNA) at concentrations as low as 1 μM. Subsequently, the specificity of the sensor was detected by non-complementary DNA (N-cDNA). Experimental results show that the fiber ring laser biosensor has the advantages of simple operation, label-free measurement, and high specificity. Furthermore, it shows a broad application prospect in several fields, especially in key areas such as medical diagnosis and cancer screening.
1. Introduction
Among various biosensors, DNA biosensors are important tools for detecting DNA variation and gene expression, which are of great significance in the fields of genetics, pathology, pharmacogenetics, food safety and criminology.1,2 Currently, the polymerase chain reaction (PCR),3 loop-mediated isothermal amplification (LAMP),4 denaturing high-performance liquid chromatography (DHPLC)5 and fluorescent labelling6 have been widely used for DNA detection. However, these techniques have disadvantages such as high reagent consumption and dependence on highly specialized operations, which leads to high cost, increased complexity and significant time-consumption. In contrast, fiber-optic label-free DNA sensors have unique advantages in biomedical detection owing to their compact structure, high sensitivity, anti-electromagnetic interference, in situ detection, multi-parameter sensing and low cost.7 Various optical fiber sensing schemes have been proposed for label-free DNA detection. On the basis of their sensing mechanisms, they can be classified as optical fiber grating (OFG),8,9 surface plasmon resonance (SPR),10 Mach-Zehnder interferometry (MZI),11,12 Michelson interferometry (MI),13,14 Fabry-Perot interferometry (FPI)15 and Sagnac interferometry (SI).16However, sensing systems with broadband light sources (BLSs) or amplified spontaneous emission (ASE) may have poor resolution and low detection accuracy. To solve the problems of low output spectral intensity and a large full width at half maximum of traditional sensing systems, tunable erbium-doped fiber (EDF) ring lasers have been widely studied.17–19 In 2017, Fen Yu et al. fabricated a fiber ring laser based on the SMF-TCF-SMF structure for refractive index measurement.20 Its refractive index sensitivity was 44.88 nm per RIU. A high optical signal-to-noise ratio of >50 dB and a narrow full width half maximum of <0.11 nm were obtained. In the same year, Lu Cai et al. designed a fiber ring laser based on a core-offset Mach-Zehnder interferometer.21 Its refractive index sensitivity was 38.1 nm per RIU and detection limit was 4.54 × 10−4. However, to date, there have been fewer studies on fiber ring laser sensors for the detection of DNA hybridization, which forms the purpose of this paper.
In this paper, a fiber ring laser biosensor based on an SMF-NCF-SMF structure for label-free DNA detection is proposed. The sensing head is fabricated by splicing a section of no-core fibers between two single-mode fibers, which has the advantages of offering a simple structure and easy fabrication. Compared with the traditional sensing system, the output spectral line width of the fiber ring laser is much narrower, thereby improving spectral resolution. In addition, the SMF-NCF-SMF structure can be used not only as a band-pass filter for the laser, but also as a sensing head. The functionalized sensing head enables specific recognition of low concentrations of cDNA.
2. Materials and methods
2.1 Sensor structure and fabrication
Fig. 1 shows the schematic of the multimode interference sensor based on SMF-NCF-SMF. The effective refractive index, diameter and length of the NCF are 1.457, 125 μm and 59 mm, respectively. This sensor head is simple to fabricate by fusing a section of the NCF between two SMFs. The optical fiber waveguide presents different propagation constants and mode field distributions due to variations in the RI of the external environment. From the MMI principle, the output interference spectrum is shifted. Therefore, the measurement of the external environment RI can be achieved by calculating the relationship between the RI and the characteristic wavelength.222.2 Theoretical simulation
Fig. 2(a) shows beam propagation along the NCF at 1550 nm, and the focusing will occur periodically at specific positions of the NCF. The simulations are performed based on a beam propagation method (BPM). The simulated transmission spectrum based on the SMF-NCF-SMF structure with a wide bandwidth is shown in Fig. 2(b). The transmission peak wavelength λk can be expressed as follows:
 | (1) |
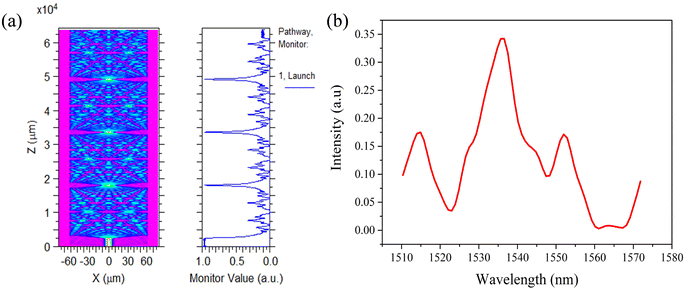 | ||
| Fig. 2 (a) Simulated beam propagation along the NCF at 1550 nm. (b) Simulated transmission spectrum based on the SMF-NCF-SMF structure. | ||
2.3 Experimental materials
Poly-L-lysine (PLL), phosphate buffer solution (1 × PBS, pH = 7.2–7.4) and DNA synthesis sequences for the experiment were purchased from Sangon Biotech engineering Co., Ltd (Shanghai, China). The DNA sequences were kept frozen in a refrigerator, and the test experiments were performed at room temperature (26 °C ± 0.1 °C). All the DNA sequences are shown in Table 1.| Biomolecules | Sequence (5′–3′) |
|---|---|
| Probe DNA (pDNA) | 5′-AGGAGGAGACTTAAGTAAAA-3′ |
| Complementary DNA (cDNA) | 5′-TTTTACTTAAGTCTCCTCCT-3′ |
| Non-complementary DNA (N-cDNA) | 5′-CTCACGTTAATGCATTTTGGTC-3′ |
| FAM fluorescent labeled pDNA | 5′-FAM-AGGAGGAGACTTAAGTAAAA-3′ |
N-cDNA is used to explore the specificity of DNA molecular hybridization. The FAM fluorophore is a fluorescein derivative, also known as carboxy-fluorescein. With the help of FAM fluorescent-labeled DNA, the binding of DNA molecules to the fiber surface can be verified. The solution required for the experiment is configured as follows:
(a) PLL solution: 25 mg of PLL powder is added to 2.5 mL of ultrapure water and shaken well. The configured PLL stock solution with a concentration of 10 mg mL−1 is divided into centrifuge tubes and stored in the refrigerator. In the experiment, the stock solution is diluted to a coating solution with a concentration of 0.1 mg mL−1.
(b) DNA solution: pDNA solution (10 μM), cDNA solution (1 μM, 3 μM, 5 μM, and 10 μM) and N-cDNA solution (5 μM) are prepared by pipetting PBS into centrifuge tubes containing the DNA sequence. All the DNA solutions are stored in a refrigerator at 4 °C and protected from light.
2.4 Measurement system
The sensing system of the fiber ring laser based on the NCF is shown in Fig. 3. The fiber ring cavity laser consists of a 980 nm pump, a wavelength division multiplexer (1550 nm/980 nm, WDM), a section of erbium-doped fibers (EDFs) with a length of 2 m, an isolator (ISO), a sensing head, a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 coupler and an optical spectrum analyzer. The EDF is used as the gain medium, and the ISO is used to maintain the unidirectionality of the laser. When transmitted light reaches the sensing head, the formed interference spectrum acts as a wavelength selector. Only one peak is amplified into a laser line, and the laser is finally outputted from the 10% port of the coupler to the OSA after multiple cycles in the cavity. As the interference spectrum of the sensing head varies with external environmental parameters (e.g., temperature, strain and refractive index), the laser wavelength also changes. Therefore, the detection of DNA hybridization can be achieved using OSA to monitor the laser wavelength.
90 coupler and an optical spectrum analyzer. The EDF is used as the gain medium, and the ISO is used to maintain the unidirectionality of the laser. When transmitted light reaches the sensing head, the formed interference spectrum acts as a wavelength selector. Only one peak is amplified into a laser line, and the laser is finally outputted from the 10% port of the coupler to the OSA after multiple cycles in the cavity. As the interference spectrum of the sensing head varies with external environmental parameters (e.g., temperature, strain and refractive index), the laser wavelength also changes. Therefore, the detection of DNA hybridization can be achieved using OSA to monitor the laser wavelength.
2.5 Functionalization of the biosensor
Fig. 4 shows the fiber surface functionalization and biological bonding process. The details of the process are as follows:(a) The fiber sensing head is cleaned with ultrapure water and dried in air for 5 minutes. To prevent damage, the sensing head is fixed on a slide.
(b) The cleaned fiber is placed in a plasma cleaner for surface treatment. This method is less time-consuming and more efficient than chemical etching and avoids the harmful effects of chemicals.
(c) The prepared PLL solution is injected into the flow cell and left to stand for 1 hour. The sensing element is then dried in air for 5 minutes. PLL attached to the fiber surface forms a thin molecular layer with uniform thickness and enhanced stability.
(d) The pDNA strand has a negatively charged phosphate group that binds to the +NH2 groups on PLL via electrostatic attraction. Using this method, pDNA can be immobilized on the surface of the optical fiber.
(e) cDNA hybridization experiments were performed to evaluate the sensing performance of the prepared sensor. When cDNA molecules are present in the solution, pDNA and cDNA combine to become a double helix structure at room temperature. The attachment of a larger number of biomolecules leads to an increase in RI on the fiber surface, resulting in a shift in the laser wavelength.
3. Results and discussion
3.1 RI sensing performance
Firstly, the response sensitivity of the sensor to different concentrations of sodium chloride (NaCl) solution was experimentally investigated, and the results are shown in Fig. 5(a and b). The RI of NaCl solution ranges from 1.3406 to 1.3705, which was determined using a digital Abbe refractometer. The variation in the laser wavelength with the RI is shown in Fig. 5(a). As the RI increases, the peak shifts towards longer wavelengths. The peak intensity remains almost constant, and the SNR is higher than 50 dB. As shown in Fig. 5(b), the RI sensitivity of the fiber ring laser based on the SMF-NCF-SMF structure is 116.8 nm per RIU. There is a good linear relationship between the wavelength shift and RI, and the R2 value is 0.991. The detection limit (DL) of the experimental system can be expressed as follows:
 | (2) |
 | (3) |
| σampl-noise ≈ (FWHM)/(4.5 × (SNR)0.25) | (4) |
 | (5) |
 | ||
| Fig. 5 The shift in the laser wavelength with the RI. (b) The relationship between wavelength shift and the RI. | ||
Table 2 lists the performance of our proposed sensor compared with that of other sensors. As shown in Table 2, although the proposed sensor does not stand out in terms of RI sensitivity, its detection accuracy is much more superior and improves by an order of magnitude compared to other that of types of sensors.
| Sensing configuration | SNR (dB) | FWHM (nm) | Sensitivity | DL | Ref. |
|---|---|---|---|---|---|
| Tapered photonic crystal fiber interferometer | 15 | 15 | 750 nm per RIU | 4.52 × 10−3 RIU | 23 |
| Cladding-etched thin-core fiber | 15 | 18 | 493.9 nm per RIU | 4.14 × 10−3 RIU | 24 |
| Single mode tapered fiber-optic interferometer | 10 | 30 | 1.5 × 10−3 nm per RIU | 6.24 × 10−3 RIU | 25 |
| Fiber ring laser sensor | >50 | <0.05 | 116.8 nm per RIU | 1.79 × 10−4 RIU | This work |
3.2 Immobilization of PLL and pDNA
The wavelength acquisition function is used to monitor the functionalization and biomolecule binding on the surface of the optical fiber in real time, as shown in Fig. 6(a and b). The wavelength response induced by PLL binding on the fiber surface is shown in Fig. 6(a). It can be seen that the peak shifts towards the longer wavelength and stabilizes at about 60 minutes, with a maximum shift of 50 pm. This is mainly due to the fact that PLL is deposited on the surface, which increases the RI of the fiber surface, resulting in the shift of the laser wavelength.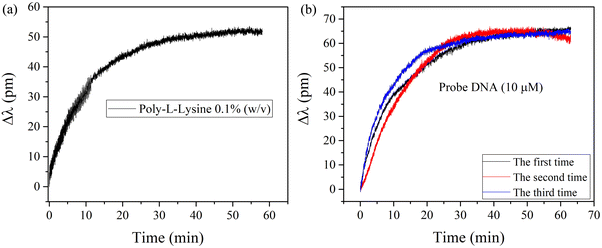 | ||
| Fig. 6 (a) Wavelength shift of the sensor after PLL functional modification. (b) Wavelength response of 10 μM pDNA. | ||
The results of the continuous wavelength acquisition of 10 μM pDNA by the functionalized fiber ring laser are shown in Fig. 6(b). The experiment was repeated at least three times to ensure the accuracy of detection results. The laser wavelength shifts towards a longer wavelength and tends to be stable at around 30 minutes. The total wavelength shifts are 65 pm, 62 pm and 64 pm, respectively. The maximum difference in wavelength shift is as low as 3 pm. The good consistency of the three tests shows that the sensor has good repeatability.
Using a FAM fluorescent labeled pDNA molecule, we verified that the laser wavelength shift is due to the binding of DNA biomolecules on the fiber surface. The excitation and emission wavelengths of FAM fluorescent dye are close to those of FITC fluorescein at 492 nm and 518 nm, respectively. The obtained images using the FITC module of a fluorescence microscope are shown in Fig. 7(a and b). As shown in Fig. 7(a), no fluorescence appeared on the sensor surface of unlabeled pDNA. In contrast, the surface of the sensor fluorescent labeled with FAM is excited with a very bright green fluorescence, as shown in Fig. 7(b). This result confirms that the functionalization process described above can successfully immobilize DNA on the optical fiber, thus inducing a wavelength shift in the spectrum.
 | ||
| Fig. 7 (a) Fluorescence microscopy images of the sensor with the pDNA molecule. (b) Microscopy images with the FAM fluorescent labeled pDNA molecule. | ||
3.3 Label free detection of cDNA
To explore the application of the fiber ring laser sensor in biochemical detection, cDNA samples at concentrations of 1 μM, 3 μM, and 5 μM are tested. Each concentration is tested at least three times, and the results are shown in Fig. 8(a–c). The peak wavelength tends to be stable at around 30 minutes, and the maximum wavelength shifts for each concentration are 25 pm, 50 pm and 85 pm, respectively. The proposed sensor can be reused by simply cleaning the fiber with a plasma cleaner to remove the DNA adsorbed on the fiber surface. The wavelength shift difference between the three tests is small, which indicates that the sensor has good repeatability. | ||
| Fig. 8 Response curves for specific binding of different concentrations of target solutions: (a) 1 μM cDNA; (b) 3 μM cDNA; (c) 5 μM cDNA. (d) Wavelength fluctuation test of the sensor. | ||
The fluctuation of the sensor in solution greatly affects its biosensing results, especially for low concentration detection. The sensor is immersed in PBS buffer solution, and the laser peak wavelength was collected continuously for 60 min, and the results are shown in Fig. 8(d). It can be seen that the sensor has good wavelength stability. The total wavelength shift throughout the test is 7.75 pm with a standard deviation of 1.19 pm.
The relationship between wavelength shift and cDNA concentration is shown in Fig. 9(a). When the detection concentration increases from 1 μM to 10 μM, the sensor gradually reaches saturation. As shown in the illustration, the linear sensitivity is 15.33 pm μM−1 in the concentration range of 1 μM to 5 μM. The small relative standard deviation (RSD) indicates that the measurement error of the sensor is small and its repeatability is good. Considering the triple standard deviation, the LOD of the sensor can be calculated as 0.23 μM.
 | ||
| Fig. 9 (a) Wavelength shift and error of different concentrations of cDNA hybridization. (b) Comparison of the wavelength shift of complementary and non-complementary cDNA. | ||
To evaluate the specificity of the fiber ring laser sensor for DNA hybridization, we performed controlled experiments using 1 μM, 3 μM, 5 μM and 10 μM cDNA and N-cDNA, and the results are shown in Fig. 9(b). The sensor exhibited different wavelength shifts for the same concentration of cDNA and N-cDNA. For sample solutions with concentrations of 1 μM, 3 μM, 5 μM and 10 μM cDNA, the wavelength shifts observed within 60 min are 24.5 pm, 50.9 pm, 85.6 pm and 89.8 pm, respectively. In contrast, for 1 μM, 3 μM, 5 μM and 10 μM N-cDNA, the sensor showed wavelength shifts of 3.5 pm, 11 pm, 23.6 pm and 23.8 pm, which are mainly due to the electrostatic adsorption of N-cDNA by a very small number of PLL molecules that were not bound to the probe. Within 60 min, the wavelength shift of 1 μM cDNA (24.5 pm) is seven times that of 1 μM N-cDNA (3.5 pm). Even for high concentration of 10 μM biomolecules, the wavelength shift of cDNA (89.8 pm) is 3.77 times that of N-cDNA (23.8 pm). These results confirm that the biosensor exhibits good specificity for cDNA detection even in the presence of high concentrations of non-specific biomolecules, ensuring high detection accuracy.
4. Conclusion
In summary, a novel method for label-free DNA hybridization detection based on the SMF-NCF-SMF structure of an erbium-doped fiber ring laser is proposed. Experimental and simulation results show that the fiber ring laser exhibits a high optical SNR and narrow FWHM, which significantly improves the detection accuracy of the sensing system. In the RI range of 1.3406–1.3705, the sensitivity reaches 116.8 nm per RIU, and the detection limit is 1.7 × 10−4 RIU. The gradual modification and functionalization process of the sensing fiber is monitored in real time by continuously detecting the laser wavelength, and label-free measurements of cDNA with concentration as low as 1 μM are achieved. In addition, the specificity and repeatability of the fiber ring laser functionalized by PLL and pDNA are tested. The sensing system demonstrates high specificity and good repeatability, which opens up a promising platform for label-free DNA detection.Data availability
All data supporting the findings of this study are included within the manuscript.Author contributions
L. W.: conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, writing-original draft, visualization, writing-reviewing and editing, and funding acquisition. C. L: data curation, visualization, formal analysis, software, and resources.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by Natural Science Foundation of Shenzhen University (No. 860-000002110620), Stabilization Support Program for Higher Education Institutions of Shenzhen (No. 20200811232156001), Shenzhen Science and Technology Innovation Commission (No. JCYJ20190808120801661), Anhui Province College Student Innovation Training Program Project (No. S201410381003), and Scientific Research Initiation Grant Project of Huainan Normal University (No. 823064).References
- H. M. R. Goncalves, L. Moreira and L. Pereira, Biosens. Bioelectron., 2015, 84, 30–36 CrossRef PubMed.
- Y. Kuo, Y. Chen and P. A. Huang, IEEE Open J. Nanotechnol., 2020, 1, 157–162 Search PubMed.
- S. A. Sugiarti and N. Nurhayati, Abinawanto, J. Phys.:Conf. Ser., 2021, 1725, 012046 CrossRef CAS.
- W. S. Kao, L. S. Yu and C. H. Lin, IEEE Sens. J., 2023, 23, 13876–13881 CAS.
- L. Jean-Christophe, L. G. Marie-Suzanne, L. M. Cedric, F. Claude and M. Remi, Int. J. Pediatr. Otorhinolaryngol., 2017, 102, 80–85 CrossRef.
- A. Niammusik, C. Thammakhet-Buranachai and P. Thavarungkul, Siam Physics Congress (SPC 2019): Physics beyond Disruption Society, 2019, 6–7 Search PubMed.
- A. Aray, F. Chiavaioli and M. Arjmand, Biophoton, 2016, 9, 1077–1084 CrossRef CAS.
- A. Bekmurzayeva and K. Dukenbayev, Sensors, 2018, 18, 4298 CrossRef.
- S. Schulze, M. Wehrhold and C. Hille, Sensors, 2018, 18, 2844 CrossRef PubMed.
- S. Kumar, R. Sing, B. K. Kaushik, N. Chen, Q. Yang and X. Zhang, IEEE Sens. J., 2019, 19, 7399–7406 CAS.
- Y. Huang, Z. Tian, L. Sun, D. Sun, J. Li, Y. Ran and B. Guan, Opt. Express, 2015, 23, 26962 CrossRef CAS.
- Y. Wen, L. Ting and B. Ji, Sens. Actuators, B, 2016, 228, 322–329 CrossRef.
- R. Gao, D. Lu, J. Cheng, Y. Jiang, L. Jiang, J. Xu and Z. Qi, Biosens. Bioelectron., 2016, 86, 321–329 CrossRef CAS PubMed.
- X. Hu, Y. Zhao, Y. Peng, R. Tong, H. Zheng, J. Zhao and S. Hu, Sens. Actuators, B, 2022, 370, 132467 CrossRef CAS.
- F. Li, X. Li, X. Zhou, P. Gong, Y. Zhang, Y. Zhao and L. V. Nguyen, Sens. Actuators, B, 2022, 354, 131212 CrossRef CAS.
- S. Gao, L. Sun, J. Li, L. Jin, Y. Ran, Y. Huang and B. Guan, Opt. Express, 2017, 25, 13305 CrossRef CAS.
- L. Xue, Opt. Express, 2005, 13, 142–147 CrossRef PubMed.
- W. L. Yong and B. Lee, IEEE Photonics Technol. Lett., 2003, 15, 795–797 Search PubMed.
- Z. Fu and D. Yang, Opt Laser. Technol., 2009, 41, 392–396 CrossRef CAS.
- Y. Fen, Opt. Eng., 2017, 56, 126105 Search PubMed.
- L. Cai and Y. Zhao, Sens. Actuators, B, 2017, 242, 673–678 CrossRef CAS.
- C. Sun, M. Wang and J. Liu, IEEE Photonics Technol. Lett., 2016, 28, 1 Search PubMed.
- S. J. Qiu, Y. Chen, J. L. Kou, F. Xu and Y. Q. Lu, Appl. Opt., 2011, 50, 4328–4332 CrossRef PubMed.
- B. Xu, Y. Li, X. Dong and S. Jin, Chin. Phys. Lett., 2012, 29, 094203 CrossRef.
- T. K. Yadav, R. Narayanaswamy, M. H. A. Bakar, Y. M. Kamil and M. A. Mahdi, Opt. Express, 2014, 22, 22802–22807 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |



