 Open Access Article
Open Access ArticleNew quinazolone–sulfonate conjugates with an acetohydrazide linker as potential antimicrobial agents: design, synthesis and molecular docking simulations†
Asmaa F. Kassemab,
Sherif S. Ragab c,
Mohamed A. Omarb,
Najla A. Altwaijry
c,
Mohamed A. Omarb,
Najla A. Altwaijry d,
Mohamed Abdelraofe,
Ahmed Temirakb,
Asmaa Salehd and
Aladdin M. Srour
d,
Mohamed Abdelraofe,
Ahmed Temirakb,
Asmaa Salehd and
Aladdin M. Srour *f
*f
aDepartment of Chemistry, College of Science and Humanities in Al-Kharj, Prince Sattam Bin Abdulaziz University, Al-Kharj, 11942, Saudi Arabia
bChemistry of Natural and Microbial Products Department, Pharmaceutical and Drug Industries Research Institute, National Research Centre, Dokki, Giza 12622, Egypt
cPhotochemistry Department, Chemical Industries Research Institute, National Research Centre (NRC), 33 El-Behouth St., P.O. 12622, Dokki, Giza, Egypt
dDepartment of Pharmaceutical Sciences, College of Pharmacy, Princess Nourah Bint Abdulrahman University, P.O. Box 84428, Riyadh 11671, Saudi Arabia
eMicrobial Chemistry Department, National Research Centre, Biotechnology Research Institute, Giza, Egypt
fDepartment of Therapeutic Chemistry, Pharmaceutical and Drug Industries Research Institute, National Research Centre, Dokki, Giza 12622, Egypt. E-mail: aladdinsrour@gmail.com
First published on 13th January 2025
Abstract
A novel molecular design based on a quinazolinone scaffold was developed via the attachment of aryl alkanesulfonates to the quinazolinone core through a thioacetohydrazide azomethine linker, leading to a new series of quinazolinone–alkanesulfonates 5a–r. The antimicrobial properties of the newly synthesized quinazolinone derivatives 5a–r were investigated to examine their bactericidal and fungicidal activities against bacterial pathogens like Bacillus subtilis, Staphylococcus aureus (Gram-positive), Pseudomonas aeruginosa, Klebsiella pneumonia, Sallmonella Typhimurium (Gram-negative), in addition to Candida albicans (unicellular fungal). The tested compounds demonstrated reasonable bactericidal activities compared to standard drugs. Notably, derivatives 5g and 5k exhibited the greatest MIC values against Candida albicans, while 5g was the best against Staphylococcus aureus with MIC of 11.3 ± 2.38 μg mL−1, two-fold efficacy more than that was recorded with sulfadiazine. Furthermore, 5k significantly prevented biofilm formation for all bacterial pathogens, with a percentage ratio reaching 63.9%, surpassing the standard drug Ciprofloxacin. Additionally, 5k caused elevated lipid peroxidation (LPO) when added to the tested microbial pathogens. Confocal Laser Scanning Microscopy (CLSM) visualization revealed fewer live cells after treatment. Molecular docking studies showed that the quinazolinone derivatives bind strongly to the DNA gyrase enzyme, with the acid hydrazide core interacting effectively with key residues GLU50, ASN46, GLY77, and ASP136, consistent with their antimicrobial activity. Additionally, these compounds exhibited promising physicochemical properties, paving the way for discovering new antimicrobial drugs.
1. Introduction
Increasing multidrug-resistant (MDR) microorganisms have become one of the most crucial threats to healthcare in the current era.1 The World Health Organization describes antimicrobial resistance as a natural phenomenon caused by bacteria that can no longer respond to treatments to which they were previously sensitive.2,3 Despite recent advances in resistance mechanisms, drug-resistant microorganisms continue to pose a threat to our ability to heal common diseases and are becoming increasingly deadly.4–6 In the past, antibiotic-resistant bacteria were rare and mainly associated with nosocomial infections, but today, they have become very common. Especially alarming is the pace at which multi- and pan-resistant bacteria spread around the globe and infect people with diseases that are not treatable with antibiotics or other antimicrobials.2 Moreover, the coronavirus 2 pandemic (COVID-19)7–9 also worsened the MDR issue as a result of the widespread exposure of patients to antibiotics.9 The most common pathogens include Enterococcus faecalis, Klebsiella pneumonia, Staphylococcus species involving Staphylococcus aureus and Staphylococcus epidermidis, in addition to the predominant pathogens involving Enterobacter, Pseudomonas aeruginosa, and Acinetobacter baumannii.10–12 The new versions of antibiotics that are currently approved have a similar structure close to the previous drugs, thus they are expected to confront the same pathways of bacterial resistance.13,14 As a result, there is a pressing requirement to create and develop new antibiotics using innovative methods to effectively combat multidrug-resistant organisms.15 In their continuous quest to find lead compounds capable of combating bacterial resistance, researchers have identified a range of compounds, and the potential to develop antimicrobial agents is ongoing in all directions. In this context, molecular hybridization emerged as a promising methodology effectively utilized to exert synergistic effects against MDR diseases by encompassing multiple pharmacophores in a single molecule.16,17Quinazolinones, a bicyclic system of pyrimidinone and benzene rings, dominate the field of medicinal chemistry and represent the winning horse in drug discovery.18,19 In terms of synthetic as well as biological implications, quinazolinones are quite intriguing and could provide effective treatment options for public health issues. Although quinazolinones were reported for a long time, their synthesis and applications in diverse fields are still topical and have captivated organic and medicinal chemists for decades.20 Numerous quinazolinone-based drugs have been approved and spread in the markets such as idelalisib, arofuto, afloqualone, mecloqualone, ispinesib, balaglitazone, nolatrexed, raltiltrexed, halofuginone.21 Albaconazole22 and Fluquinconazole23 represent the more interesting quinazolinone-based drugs in our study where they are widely used as antimicrobial agents (Fig. 1). Recently, gained momentum has been directed to the utilization of quinazolinone as a core for the formulation of new antibacterial drug leads.24
 | ||
| Fig. 1 Marketed drugs containing quinazolinone, hydrazide–hydrazone, or sulfonate motifs along with our molecular design. | ||
On the other hand, hydrazide–hydrazone derivatives, hydrazide molecules with azomethine functionality (–CONHN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), were shown to exhibit prominent pharmacological and biological properties, such as antimicrobial, anti-inflammatory, analgesic, anticancer, anticonvulsant, antiviral, antiprotozoal, antiplatelet, antimalarial, antimycobacterial, vasodilator, and antischistosomiasis activities.25,26 Various studies reported that hydrazide–hydrazone functionalities in biological systems can easily undergo hydrolysis, which can be advantageous in treating a variety of life-threatening diseases.27,28 The antimicrobial activity of this class of compounds represents the most frequently encountered in scientific literature among the biological properties.27 Indeed, nitrofurazone, furazolidone, and nitrofurantoin (Fig. 1) are widely used antimicrobial drugs and are considered typical examples of chemotherapeutic agents containing hydrazide–hydrazone moiety.29
CH–), were shown to exhibit prominent pharmacological and biological properties, such as antimicrobial, anti-inflammatory, analgesic, anticancer, anticonvulsant, antiviral, antiprotozoal, antiplatelet, antimalarial, antimycobacterial, vasodilator, and antischistosomiasis activities.25,26 Various studies reported that hydrazide–hydrazone functionalities in biological systems can easily undergo hydrolysis, which can be advantageous in treating a variety of life-threatening diseases.27,28 The antimicrobial activity of this class of compounds represents the most frequently encountered in scientific literature among the biological properties.27 Indeed, nitrofurazone, furazolidone, and nitrofurantoin (Fig. 1) are widely used antimicrobial drugs and are considered typical examples of chemotherapeutic agents containing hydrazide–hydrazone moiety.29
Sulfonates have found widespread use in medical research and have significant pharmacological applications. The distinctive physicochemical properties of sulfonate-based compounds support their affinity for lipid phases causing a facile penetration through cell membranes to interact with the target sites.30 Fenson, Chlorfenson, and Genite are aryl sulfonates that have long been marketed as insecticides,31,32 while busulfan is an alkyl sulfonate drug that was approved as a chemotherapy for the treatment of chronic myeloid leukemia (CML)33 (Fig. 1). Aryl alkanesulfonates are a class of sulfonate-containing compounds of both alkyl and aryl substituents and have been screened as antifungal34 anticancer,35 acaricidal agents,24 in addition to their use as inhibitors of carbonic anhydrase.36 Using these results in conjunction with our previous research on pyrimidine antibacterial agents.1 Our efforts are focused on developing novel molecular designs to get new quinazolinone-based scaffolds with anticipated antimicrobial activity. To achieve this, 2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetohydrazide 3 has been employed as a promising precursor for synthesizing new antimicrobial candidates. The design of this new series incorporates quinazolinone and sulfonate moieties, strategically positioned to bind with ARG136 and ARG76, respectively. Furthermore, the amide NH group of the acetohydrazide linker is designed to form hydrogen bonds with GLY77. This approach is compared to the known inhibitor 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide (CWW), to improve binding affinity and antimicrobial efficacy,37,38 Fig. 1. The antimicrobial properties of the newly synthesized quinazolinone acetohydrazide sulfonates derivatives were investigated to examine their bactericidal and fungicidal activities against some clinical microbial pathogens.
2. Results and discussion
2.1. Chemistry
Our molecular design involves 2-((4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetohydrazide 3 as a promising precursor for synthesizing new quinazolinone candidates. The latter conger was installed via attachment of aryl alkanesulfonates to the quinazolinone core via thioacetohydrazide azomethine linker, leading to a new series of quinazolinone-alkanesulfonates 5a–r. The synthetic route toward this series was depicted in Scheme 1. Starting with the reaction of 2-amino-3-methylbenzoic acid or 2-amino-5-chlorobenzoic acid with phenyl isocyanate in ethanol in the presence of Et3N yielded the corresponding 2-mercapto-3-phenylquinazolin-4(3H)-ones 1a,b. Esterification of 1a,b was then carried out using methyl bromoacetate to afford the corresponding esters 2a,b which were then converted to the corresponding acetohydrazide analogs after heating under reflux in ethanol with hydrazine hydrate to give 3a,b. Derivatives 3a,b were the precursors for the final quinazoline-alkanesulfonate targets 5a–r when heated in ethanol under reflux with various para-substituted aromatic aldehydes (bearing different alkane sulfonate moieties namely: methane, ethane or propane sulfonate moieties) 4a–i. IR, 1H & 13C-NMR, and elemental analyses have been exploited to reveal the chemical structure of the newly synthesized conjugates 5a–r. Specifically, the IR spectra of 5a–r revealed a strong stretching band around 3170 cm−1 indicating the NH group, while the characteristic absorption bands of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N appeared at 1669–1683 cm−1 and 1618–1636 cm−1, respectively. Moreover, the SO2 band was observed between 1340 and 1388 cm−1. The 1H NMR spectra of 5a–r confirmed the existence of alkanesulfonate moieties where they appeared within the upfield region as singlet signals, triplet–quartet signals, or triplet–sextet–triplet signals for methyl, ethyl, or propyl chains, respectively. Furthermore, the methylene protons were exhibited as singlets at δH = 4.46–4.51 ppm. Furthermore, 1H NMR spectra of 5a–r also showed two singlet signals in the range of δH = 3.88–4.00 ppm and δH = 4.00–4.51 ppm with the integration of 2H each corresponding to SCH2C
N appeared at 1669–1683 cm−1 and 1618–1636 cm−1, respectively. Moreover, the SO2 band was observed between 1340 and 1388 cm−1. The 1H NMR spectra of 5a–r confirmed the existence of alkanesulfonate moieties where they appeared within the upfield region as singlet signals, triplet–quartet signals, or triplet–sextet–triplet signals for methyl, ethyl, or propyl chains, respectively. Furthermore, the methylene protons were exhibited as singlets at δH = 4.46–4.51 ppm. Furthermore, 1H NMR spectra of 5a–r also showed two singlet signals in the range of δH = 3.88–4.00 ppm and δH = 4.00–4.51 ppm with the integration of 2H each corresponding to SCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O protons that validate the presence of iminol and amide tautomers, respectively. The existence of such tautomerism was also supported by the presence of two singlet signals with total integration of 1H due to the azomethine protons (HC
O protons that validate the presence of iminol and amide tautomers, respectively. The existence of such tautomerism was also supported by the presence of two singlet signals with total integration of 1H due to the azomethine protons (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) of both tautomers within the ranges of δH = 7.98–8.06 and δH = 8.14–8.28 ppm, and two peaks with only 1H integration appeared in the ranges δH = 11.66–11.80 ppm and δH = 11.70–11.83 ppm, due to OH proton in case of iminol and NH proton in amide tautomer, respectively. On the other hand, the characteristic peaks that represent the carbons of alkanesulfonate groups were depicted at the aliphatic zone of the 13C NMR spectra in the range of δC = 8.00–52.46 ppm, while the peaks observed between δC = 34.26 ppm and δC = 35.01 ppm assumed to be for the methylene carbons. The characteristic azomethine carbons (HC
N) of both tautomers within the ranges of δH = 7.98–8.06 and δH = 8.14–8.28 ppm, and two peaks with only 1H integration appeared in the ranges δH = 11.66–11.80 ppm and δH = 11.70–11.83 ppm, due to OH proton in case of iminol and NH proton in amide tautomer, respectively. On the other hand, the characteristic peaks that represent the carbons of alkanesulfonate groups were depicted at the aliphatic zone of the 13C NMR spectra in the range of δC = 8.00–52.46 ppm, while the peaks observed between δC = 34.26 ppm and δC = 35.01 ppm assumed to be for the methylene carbons. The characteristic azomethine carbons (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) appeared between δC = 145.52 ppm and δC = 145.93 ppm, whereas the carbonyl carbon of the acetohydrazide moiety was detected in the range of δC = 167.32–168.75 ppm. Spectral charts of the new chemical entities 5a–r are included in in ESI file (Fig. S2–S37),† The purity of the final compounds was determined by LC-MS using the area percentage method on the UV trace recorded at a wavelength of 254 nm and found to be >95%, representative charts of HPLC purity are displayed in ESI file (Fig. S38–S40).†
N) appeared between δC = 145.52 ppm and δC = 145.93 ppm, whereas the carbonyl carbon of the acetohydrazide moiety was detected in the range of δC = 167.32–168.75 ppm. Spectral charts of the new chemical entities 5a–r are included in in ESI file (Fig. S2–S37),† The purity of the final compounds was determined by LC-MS using the area percentage method on the UV trace recorded at a wavelength of 254 nm and found to be >95%, representative charts of HPLC purity are displayed in ESI file (Fig. S38–S40).†
2.2. Antimicrobial activity
The newly synthesized molecules 5a–r have been evaluated for their in vitro antibacterial properties, on Bacillus subtilis, Staphylococcus aureus (Gram-positive), Pseudomonas aeruginosa, Klebsiella pneumonia, Salmonella typhimurium (Gram-negative), and Candida albicans (unicellular fungal) (Fig. S1†). The preliminary results displayed in Table S1† revealed that derivatives 5a–c,e–h,k,l,n,p were found to be potent inhibitors against some of the tested pathogens, while not able to inhibit the proliferation of other microbial strains. The high resistance of some pathogens reflects their virulence and rapid proliferation ability even upon being treated with some standard antibiotics such as Cephradine. Accordingly, most of the tested molecules failed to show any activity on MDR pathogens such as Staphylococcus aureus MRSA, Salmonella typhimurium, and Pseudomonas aeruginosa, which the inhibitory activity ranged from inactive to weak (not exceeding 3 mm of the inhibition zone). Derivatives 5f, 5g, and 5k demonstrated promising inhibitory activity against all MDR pathogens, particularly the methyl-substituted quinazoline's phenyl derivatives 5f and 5g rather than the corresponding chloro analog 5k. Moreover, compounds 5a, 5f, 5g and 5h displayed plausible inhibitory effects as anti-candida, which could represent good antifungal candidates among the tested series. On the other hand, no promising inhibition response was obtained by the tested series 5a–r towards the MDR; Salmonella typhimurium, and Pseudomonas aeruginosa except derivatives 5a, 5f, and 5g. Bacillus subtilis also exhibited a potent resistance towards most of the tested molecules, except for derivatives 5a, 5f, and 5g (methyl-substituted quinazoline's phenyl derivatives) and 5k, 5n, and 5o (chloro-substituted quinazoline's phenyl derivatives). Nonetheless, a reasonable inhibitory activity towards Klebsiella pneumonia was noticed for all derivatives except for derivatives 5e and 5n, Table S1.† Consequently, the minimum inhibition concentration (MIC) for the most potent molecules 5f, 5g and 5k has been determined to provide the lowest microbial growth following the standard procedure.39 As shown in Table 2, brilliant inhibitory activity at lower concentrations has been recorded by derivatives 5g and 5k against Candida albicans (9 ± 3.17 and 10 ± 2.55 μg mL−1, respectively) which exceeds by 7-fold the activity of the standard antifungal agent Sulfadiazine (72 ± 3.44 μg mL−1). Likewise, derivative 5g also exhibited a potent inhibitory effect towards Staphylococcus aureus at 11.3 ± 2.38 μg mL−1, reflecting a 2-fold efficacy more than the standard antibacterial agent Sulfadiazine (24 ± 3.92 μg mL−1) Table 1. Furthermore, compound 5f provided a significant MIC value against both Gram-positive pathogens and Gram-negative bacteria, the MIC values of compound 5f against Bacillus subtilis, Klebsiella pneumonia, Salmonella typhimurium and Pseudomonas aeruginosa were exceeding to that calculated for the corresponding reference antibiotic agents.| Compd. ID | (MIC, μg mL−1) | |||||
|---|---|---|---|---|---|---|
| Unicellular fungal | Gram-positive bacteria | Gram-negative bacteria | ||||
| Candida albicans | Bacillus subtilis | Staphylococcus aureus | Salmonella typhimurium | Klebsiella pneumonia | Pseudomonas aeruginosa | |
| a Bacillus subtilis, Staphylococcus aureus (Gram-positive), Pseudomonas aaerginousea, Klebsiella pneumonia, Sallmonella Typhemerium (Gram-negative), and Candida albicans (unicellular fungal) Dimethyl sulfoxide (DMSO) was used as a negative control. | ||||||
| 5f | 48 ± 4.18 | 23 ± 2.21 | 50 ± 0.88 | 49 ± 1.28 | 22 ± 4.72 | 45 ± 6.11 |
| 5g | 9 ± 3.17 | 48 ± 2.25 | 11.3 ± 2.38 | 95 ± 2.05 | 99 ± 5.02 | 96 ± 4.83 |
| 5k | 10 ± 2.55 | 49 ± 2.77 | 52 ± 0.28 | 21 ± 5.35 | 44 ± 1.88 | 97 ± 3.88 |
| Sulfadiazine | 72 ± 3.44 | 46 ± 4.18 | 24 ± 3.92 | 94 ± 2.29 | 47 ± 3.18 | 70 ± 4.99 |
2.3. Antibiofilm of the targeted molecules
Inhibiting biofilm formation is one of the most effective ways to eradicate bacterial proliferation under stress conditions. Most bacterial pathogens protect themselves from stress by forming biofilms. Furthermore, biofilm formation is an important mechanism that makes bacterial pathogens more resistant to many traditional antibiotics. Biofilms primarily form in hostile environments surrounding the bacterial medium. This biofilm mechanism is commonly associated with the high resistance of bacterial pathogens and prevents the complete inhibition of bacterial cells when treated with antibacterial agents. Therefore, biofilm inhibitors are considered highly important compounds for enhancing the efficacy of antibacterial agents. Consequently, discovering new molecules that prevent or inhibit biofilm formation by bacterial pathogens has become increasingly desirable.To investigate the biofilm inhibitory activity of our potent molecules, we utilized the crystal violet method to assess their effectiveness in eradicating established biofilms of bacterial pathogens. As indicated in Table 2, derivative 5k demonstrated a significant ability to prevent biofilm formation by bacterial pathogens, particularly against Bacillus subtilis and Salmonella typhimurium. Similarly, compound 5f showed considerable biofilm inhibition of Staphylococcus aureus, exhibiting antibiofilm activity that exceeded that of the standard drug, Ciprofloxacin. A slightly lower activity was observed for compound 5g, particularly against Gram-negative bacteria. However, despite the promising activity of 5k, a remarkably high resistance to biofilm formation was detected in Pseudomonas aeruginosa.
| Sample no. | Biofilm inhibition (%) | ||||
|---|---|---|---|---|---|
| Gram-positive bacteria | Gram-negative bacteria | ||||
| Bacillus subtilis | Staphylococcus aureus | Klebsiella pneumonia | Salmonella typhimurium | Pseudomonas aeruginosa | |
| a Dimethyl sulfoxide (DMSO) was used as a negative control. | |||||
| 5f | 56.4 ± 3.12 | 45.1 ± 1.62 | 59.6 ± 2.22 | 37.4 ± 1.82 | 34.8 ± 4.07 |
| 5g | 45.1 ± 2.72 | 34.7 ± 4.02 | 32.8 ± 1.92 | 33.9 ± 2.94 | 20.1 ± 2.32 |
| 5k | 63.7 ± 0.92 | 42.9 ± 2.62 | 53.1 ± 2.12 | 61.9 ± 1.72 | 38.7 ± 3.66 |
| Ciprofloxacin | 49.2 ± 1.16 | 30.9 ± 4.52 | 40.8 ± 3.88 | 58.3 ± 4.02 | 45.2 ± 1.18 |
2.4. Effect of the targeted molecules on the bacterial lipid peroxidation (LPO)
A difficult penetration of the bacterial cytoplasmic membrane is considered an important tool to evaluate the inhibition by the tested treatments, which demonstrates the abilities of these treatments to make differences in the inhibition process, oxidative stress on the bacterial cell wall usually triggers oxidation of the cell wall fatty acid content leading to easily penetration of the tested compound inside the bacterial cell. The level of the obtained lipid peroxidation refers to the success of the treatment in overcoming the lysis cell wall problem. Therefore, an LPO test took place to evaluate the oxidation potential of bacterial cell membrane fatty acids by targeted molecules. Accordingly, treatments of each tested bacterial strain using the potent molecules at the MIC level were investigated (Table 3).| Sample no. | Lipid peroxidation efficiency (%) | ||||
|---|---|---|---|---|---|
| Gram-positive bacteria | Gram-negative bacteria | ||||
| Bacillus subtilis | Staphylococcus aureus | Klebsiella pneumonia | Salmonella typhimurium | Pseudomonas aeruginosa | |
| 5f | 212.5 ± 5.52 | 151.2 ± 1.92 | 230.4 ± 2.12 | 273.1 ± 4.33 | 140.4 ± 5.02 |
| 5g | 219.7 ± 2.71 | 118.4 ± 2.26 | 285.2 ± 1.44 | 210.4 ± 4.66 | 175.2 ± 3.29 |
| 5k | 377.6 ± 3.44 | 202.2 ± 2.17 | 295.2 ± 1.89 | 278.3 ± 5.07 | 181.3 ± 6.06 |
| Ciprofloxacin | 417.7 ± 2.83 | 319.8 ± 3.26 | 327.4 ± 4.42 | 294.8 ± 1.99 | 346.3 ± 2.62 |
In this respect, there were significant variations in lipid peroxidation activity between the targeted molecules. There is a significant LP activity of 5k when added to Bacillus subtilis, Salmonella typhimurium, and Klebsiella pneumonia. The LP result for these pathogens proved to be somewhat close to that obtained by the standard antibacterial agents. Moreover, 5k also caused an elevated LP in the case of Staphylococcus aureus (202.2 ± 2.17%), however a lower LP activity in the case of Pseudomonas aeruginosa was indicated when compared to the standard drug. Overall, the positive response of the bacterial pathogens, Bacillus subtilis, Salmonella typhimurium, and Klebsiella pneumonia to the LP activity was demonstrated for the tested compounds. In addition, the strong resistance of both Pseudomonas aeruginosa and Staphylococcus aureus toward the LP activity reflected their virulence.
2.5. Confocal laser scanning microscope
The different observations of the potent compounds (i.e. 5f and 5k) effectiveness on the microbial pathogens were detected by a Confocal Laser Scanning Microscope (CLSM). As shown in (Fig. 2), a clear effect of the potent molecules on the biofilm and morphological observations was indicated. Since, a high concentration of the dead cells (red color) was found to be more prevalent, while a lower number of live cells were observed after treatment. The activity of the potent molecules was sharply indicated in the live cell concentration, particularly in the case of Bacillus subtilis, Salmonella typhimurium and Klebsiella pneumonia.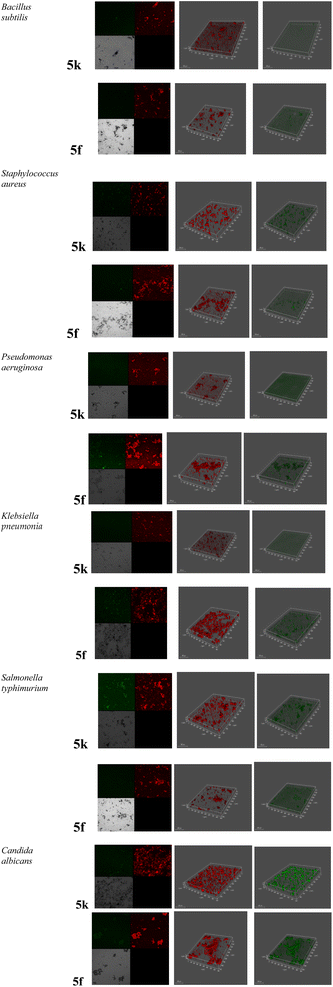 | ||
| Fig. 2 The effect of the promising compounds 5f and 5k on the proliferation of the microbial pathogens via Confocal Laser Scanning Microscope (CLSM). | ||
The right set of panels displayed the strains were allowed to form on glass-bottomed chambers for 24 h with acridine dye before being treated with the indicated antimicrobials (green color). The medium set of panels displayed the concentration of dead cells after treatment with target molecules (red color). The left set of panels displayed the negative control (black), untreated cells (off-white), live cells (green) and dead cells (red).
2.6. Cytotoxic properties
The safety of the powerful antimicrobial compounds, 5f, 5g, and 5k, on the BJ1 cell line was assessed using the MTT assay. Notably, the study's findings showed that these substances did not cause cytotoxicity to the tested cells; however, their safety profile should be assumed after the necessary in vivo investigations are completed (Table S3†).2.7. Docking studies on DNA gyrase enzyme
DNA gyrase is an essential enzyme in bacteria, responsible for introducing negative supercoils into DNA, which is crucial for DNA replication, transcription, and overall genomic stability. This enzyme inhibition is particularly effective against a broad spectrum of Gram-negative and some Gram-positive bacteria, making it a prime target for antibacterial agents. In our study, we investigated the antimicrobial properties of newly synthesized quinazolinone derivatives 5a–r against various Gram-negative bacteria (Klebsiella pneumoniae, Salmonella typhimurium, Pseudomonas aeruginosa) and Gram-positive bacteria (Bacillus subtilis, Staphylococcus aureus). We performed detailed molecular docking studies of these compounds with the DNA gyrase enzyme to elucidate the molecular basis of their antimicrobial effects. The E. coli Gyrase B enzyme in complex with the known inhibitor 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide (CWW) was obtained from the Protein Data Bank (PDB code: 6F86).40,41 Biovia Discovery Studio was employed to prepare the DNA gyrase enzyme for docking studies.41,42 Extraneous chains and water molecules were removed, retaining HOH616 due to its role in forming a hydrogen-bonding network with the pyridyl ring's nitrogen atom and the amino acid residues ASP73, THR165, and GLY77.41 Polar hydrogens were added. Partial charges were adjusted to accurately represent electrostatic interactions between the ligand and the binding site, thereby influencing the predicted binding pose and interaction quality. Docking simulations were then performed using Autodock 4 to predict the interactions of the quinazolinone derivatives with the DNA gyrase enzyme.43 Autodock 4 was then used to predict the interactions between the synthesized quinazolinone derivatives (5a–r) and the DNA gyrase enzyme. This allowed us to calculate binding energies and visualize ligand positioning within the enzyme's active site, providing deeper insights into the binding mechanisms. Moreover, to validate our docking procedure, we re-docked the co-crystallized native ligand (CWW) into the DNA gyrase active site. The resulting pose closely matched the original binding conformation, with binding affinities of −11.72 kcal mol−1. It also demonstrated a RMSD (root mean square deviation) of 0.65. This close alignment indicates a successful reproduction of the native ligand's binding mode, thus confirming the validity of our docking protocol, Table S3.†Detailed interaction analysis revealed that the terminal pyridyl moiety of CWW forms a hydrogen bond with ARG136 and engages in hydrophobic interactions with the ARG76 residue. The amide NH group exhibits hydrogen bonding with the GLY77 residue (Fig. 3). The pyridyl core of CWW is stabilized in the active site by a direct hydrogen bond with THR165 and an additional water-mediated hydrogen bond involving HOH616. The urea side chain also forms significant hydrogen bonds with ASP73 and ASN46. Additionally, the native ligand participates in several hydrophobic interactions (van der Waals, alkyl, and carbon–hydrogen bonds) with key residues such as VAL167, ALA47, VAL43, ILE78, ILE94, and PRO79 (Fig. 3). Molecular docking studies were meticulously conducted to investigate the interactions of quinazolinone derivatives 5f, 5g, and 5k within the active site of the DNA gyrase enzyme. These studies aimed to understand how these compounds could potentially inhibit DNA gyrase, providing insights into their antimicrobial properties. In general, the sulfonate group in the quinazolinone derivatives was found to engage in hydrogen bonding, similar to the carbonyl group of the urea side chain in the native ligand, interacting with several amino acids such as ASN43, VAL120, SER121, ILE78, and ARG136 (Fig. 3–6). This indicates that the sulfonate group may significantly stabilize the compounds within the enzyme active site.
 | ||
| Fig. 3 (A) Superimposition of the native co-crystallized ligand CWW and the docked ligand within the DNA gyrase enzyme's binding site (PDB ID: 6F86). The native ligand's carbon atoms are depicted in violet, while the docked ligand's carbon atoms are shown in orange. (B) 2D interaction diagram of the docked CWW with the DNA gyrase enzyme. | ||
Moreover, the acid hydrazide core of the quinazolinone derivative exhibited similar binding interactions to the amide linker of the native ligand. This core formed interactions with residues GLU50, ASN46, GLY77, and ASP136, suggesting that the acid hydrazide core is crucial for the binding affinity and proper positioning of the derivatives within the active site. The carbonyl group in the acid hydrazide core of compound 5f exhibited a notable hydrogen bonding interaction with the water molecule HOH616 as depicted in Fig. 4. This interaction is similar to that exhibited by the core pyridyl ring in the native ligand, as illustrated in Fig. 3.
Besides, the nitrogen atom of the quinazolinone moiety demonstrated a binding pattern analogous to the nitrogen of the terminal pyridyl moiety in the native ligand, particularly with the ASP136 residue. This interaction was notably observed with compound 5k, highlighting the importance of this moiety in the binding process (Fig. 3 and 6). Comparing the molecular interactions of our compounds with the native ligand showed that the sulfonyl group increased the binding interactions with several H-bonds. Also, we noticed that changing the position of the sulfonyl group from the m- to the p- position of the terminal phenyl ring did not affect the binding interactions. Also, we noticed that the ester group near the sulfonyl group increased the binding interactions of the quinazolinone derivatives as observed in compounds 5f and 5g, which could be attributed to the presence of several H-bond acceptor groups and their tendency to establish several H-bonding within the DNA gyrase binding pocket or maybe also some intramolecular H-bonds. Finally, we think his comprehensive analysis of the molecular interactions highlights the critical binding features that contribute to the antimicrobial activity of the quinazolinone derivatives, thereby providing valuable insights for future design and optimization of potent antimicrobial agents.
2.8. In silico pharmacokinetic analysis
The predicted pharmacokinetic characteristics and drug-like properties of the newly synthesized quinazolinone derivatives 5a–r were assessed through the SwissADME server, emphasizing key pharmacokinetic considerations. These compounds exhibit metabolic stability and are not substrates for the various isoforms of CYP450 enzymes, which is significant for minimizing drug–drug interactions and enhancing metabolic predictability. Additionally, they are not substrates for P-glycoprotein (P-gp), indicating that they are likely to remain within cells and exert their therapeutic action without being effluxed by P-gp, which is a common resistance mechanism in many cancers and bacterial infections.These derivatives display acceptable aqueous solubility, with a calculated topological polar surface area (TPSA) of approximately 150 Å2, which is beneficial for their absorption and permeability. The compounds also exhibit suitable lipophilicity, with a consensus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value around 4.1, suggesting a balanced hydrophilic–lipophilic profile that is crucial for bioavailability and cell membrane permeability (Table 4). Regarding Lipinski's rule of five, the compounds show only one violation: their molecular weight exceeds 500 daltons, averaging around 550 daltons. While this might typically raise concerns about permeability and absorption, the other favorable properties mitigate this issue to some extent.
P value around 4.1, suggesting a balanced hydrophilic–lipophilic profile that is crucial for bioavailability and cell membrane permeability (Table 4). Regarding Lipinski's rule of five, the compounds show only one violation: their molecular weight exceeds 500 daltons, averaging around 550 daltons. While this might typically raise concerns about permeability and absorption, the other favorable properties mitigate this issue to some extent.
| No. | MW (g mol−1) | #H-bond acceptor | #H-bond donors | TPSA | Consensus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
P-gp substrate | Synthetic accessibility | Lipinski #violations | Pains #alerts |
|---|---|---|---|---|---|---|---|---|---|
| 5a | 522.60 | 7 | 1 | 153.40 | 3.68 | No | 3.82 | 1 | 0 |
| 5b | 536.62 | 8 | 1 | 156.89 | 4.51 | No | 4.08 | 1 | 0 |
| 5c | 550.65 | 8 | 1 | 156.89 | 4.90 | No | 4.22 | 1 | 0 |
| 5d | 580.68 | 9 | 1 | 166.12 | 4.94 | No | 4.36 | 1 | 0 |
| 5e | 566.65 | 9 | 1 | 166.12 | 4.61 | No | 4.23 | 1 | 0 |
| 5f | 580.68 | 9 | 1 | 166.12 | 4.94 | No | 4.36 | 1 | 0 |
| 5g | 552.62 | 9 | 1 | 166.12 | 4.19 | No | 4.07 | 1 | 0 |
| 5h | 566.65 | 9 | 1 | 166.12 | 4.57 | No | 4.22 | 1 | 0 |
| 5i | 580.68 | 9 | 1 | 166.12 | 4.84 | No | 4.35 | 1 | 0 |
| 5j | 543.01 | 8 | 1 | 156.89 | 4.41 | No | 3.79 | 1 | 0 |
| 5k | 557.04 | 8 | 1 | 156.89 | 4.74 | No | 3.94 | 1 | 0 |
| 5l | 571.07 | 8 | 1 | 156.89 | 5.18 | No | 4.07 | 2 | 0 |
| 5m | 573.04 | 9 | 1 | 166.12 | 4.44 | No | 3.93 | 1 | 0 |
| 5n | 587.07 | 9 | 1 | 166.12 | 4.79 | No | 4.08 | 1 | 0 |
| 5o | 601.09 | 9 | 1 | 166.12 | 5.15 | No | 4.22 | 1 | 0 |
| 5p | 573.04 | 9 | 1 | 166.12 | 4.41 | No | 3.92 | 1 | 0 |
| 5q | 587.07 | 9 | 1 | 166.12 | 4.79 | No | 4.08 | 1 | 0 |
| 5r | 601.09 | 9 | 1 | 166.12 | 5.13 | No | 4.21 | 1 | 0 |
Moreover, these compounds demonstrate good synthetic accessibility, with an average score of 4 on the SwissADME scale, where 1 indicates an easy synthesis and 10 indicates a hard synthesis. This score reflects a moderate ease of synthesis, which is advantageous for further development and large-scale production. Overall, the quinazolinone derivatives exhibit promising pharmacokinetic properties. Future modifications could focus on reducing the molecular weight and enhancing aqueous solubility to optimize these compounds for further drug development. These adjustments could improve their pharmacodynamic profiles and therapeutic efficacy.
3. Materials and methods
The details about material and methods used in this study are provided in ESI file.†3.1. Chemistry
3.1.1.1. 4-((2-(2-((8-Methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)phenyl methanesulfonate (5a). Yield: 84%; mp 250–252 °C; IR (KBr) cm−1, ν: 3320 (OH, tautomer), 3174 (NH), 1676 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1618 (C
O), 1618 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1363 (SO2), 1207 (C–O, C–N); 1H NMR δ (ppm): 2.46 (s, 3H, CH3), 3.42 (s, 3H, SO2CH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.51 (s, 2H, CH2, amide tautomer), 7.34–7.37 (m, 1H, H-6′), 7.41–7.42 (d, 2H, J = 6.0 Hz, H-2′′ and H-6′′), 7.48 (brs, 2H, H-2 and H-6), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.65 (brs, 1H, H-7′), 7.81 (brs, 2H, H-3 and H-5), 7.90–7.92 (m, 1H, H-5′), 8.05 (s, 1H, CH
N), 1363 (SO2), 1207 (C–O, C–N); 1H NMR δ (ppm): 2.46 (s, 3H, CH3), 3.42 (s, 3H, SO2CH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.51 (s, 2H, CH2, amide tautomer), 7.34–7.37 (m, 1H, H-6′), 7.41–7.42 (d, 2H, J = 6.0 Hz, H-2′′ and H-6′′), 7.48 (brs, 2H, H-2 and H-6), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.65 (brs, 1H, H-7′), 7.81 (brs, 2H, H-3 and H-5), 7.90–7.92 (m, 1H, H-5′), 8.05 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.24 (s, 1H, CH
N, iminol tautomer), 8.24 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.76 (s, 1H, OH/NH tautomer), 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 16.62 (CH3), 34.86 (CH2), 37.52 (SO2CH3), 122.63 (C-2 and C-6), 124.12 (C-4′a), 125.41 (C-5′), 128.29 (C-2′′ and C-6′′), 128.61 (C-4), 129.32 (C-3′′ and C-5′′), 129.46 (C-3 and C-5), 129.86 (C-4′′), 133.21 (C-6′), 133.96 (C-1′′), 135.08 (C-8′), 135.84 (C-7′), 141.77 (C-8′a), 145.53 (HC
N, amide tautomer), 11.76 (s, 1H, OH/NH tautomer), 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 16.62 (CH3), 34.86 (CH2), 37.52 (SO2CH3), 122.63 (C-2 and C-6), 124.12 (C-4′a), 125.41 (C-5′), 128.29 (C-2′′ and C-6′′), 128.61 (C-4), 129.32 (C-3′′ and C-5′′), 129.46 (C-3 and C-5), 129.86 (C-4′′), 133.21 (C-6′), 133.96 (C-1′′), 135.08 (C-8′), 135.84 (C-7′), 141.77 (C-8′a), 145.53 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.80 (C-1), 155.90 (C-2′), 160.87 (CO-quinazolinone), 168.57 (CONH); anal. calc. for C25H22N4O5S2 (522.59) C, 57.46; H, 4.24; N, 10.72. Found: C, 57.31; H, 4.38; N, 10.64.
N), 149.80 (C-1), 155.90 (C-2′), 160.87 (CO-quinazolinone), 168.57 (CONH); anal. calc. for C25H22N4O5S2 (522.59) C, 57.46; H, 4.24; N, 10.72. Found: C, 57.31; H, 4.38; N, 10.64.
3.1.1.2. 4-((2-(2-((8-Methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)phenyl ethanesulfonate (5b). Yield: 87%; mp 229–321 °C; IR (KBr) cm−1, ν: 3317 (OH, tautomer), 3177 (NH), 1677 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1619 (C
O), 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1388 (SO2), 1219 (C–O, C–N); 1H NMR δ (ppm): 1.38 (t, 3H, J = 5.5 Hz, CH3CH2), 2.45 (s, 3H, CH3), 3.56 (q, 2H, J = 4.5 Hz, CH3CH2), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.34 (t, 1H, J = 7.5 Hz, H-6′), 7.37–7.40 (m, 2H, H-2′′ and H-6′′), 7.47 (d, 2H, J = 6.0 Hz, H-2 and H-6), 7.59 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.65 (d, 1H, J = 6.5 Hz, H-7′), 7.80 (d, 2H, J = 8.5 Hz, H-3 and H-5), 7.91 (d, 1H, J = 7.5 Hz, H-5′), 8.05 (s, 1H, C
N), 1388 (SO2), 1219 (C–O, C–N); 1H NMR δ (ppm): 1.38 (t, 3H, J = 5.5 Hz, CH3CH2), 2.45 (s, 3H, CH3), 3.56 (q, 2H, J = 4.5 Hz, CH3CH2), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.34 (t, 1H, J = 7.5 Hz, H-6′), 7.37–7.40 (m, 2H, H-2′′ and H-6′′), 7.47 (d, 2H, J = 6.0 Hz, H-2 and H-6), 7.59 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.65 (d, 1H, J = 6.5 Hz, H-7′), 7.80 (d, 2H, J = 8.5 Hz, H-3 and H-5), 7.91 (d, 1H, J = 7.5 Hz, H-5′), 8.05 (s, 1H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.24 (s, 1H, CH
N, iminol tautomer), 8.24 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.76 (s, 1H, OH/NH, tautomer) 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.01 (CH3CH2), 16.65 (CH3), 34.88 (CH2), 44.82 (CH3CH2), 119.25 (C-4′a), 122.53 (C-2 and C-6), 124.13 (C-5′), 125.43 (C-4), 128.31 (C-2′′ and C-6′′), 128.61 (C-4′′), 129.33 (C-3′′ and C-5′′), 129.49 (C-3 and C-5), 133.13 (C-6′), 134.00 (C-1′′), 135.10 (C-8′), 135.86 (C-7′), 141.82 (C-8′a), 145.55 (HC
N, amide tautomer), 11.76 (s, 1H, OH/NH, tautomer) 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.01 (CH3CH2), 16.65 (CH3), 34.88 (CH2), 44.82 (CH3CH2), 119.25 (C-4′a), 122.53 (C-2 and C-6), 124.13 (C-5′), 125.43 (C-4), 128.31 (C-2′′ and C-6′′), 128.61 (C-4′′), 129.33 (C-3′′ and C-5′′), 129.49 (C-3 and C-5), 133.13 (C-6′), 134.00 (C-1′′), 135.10 (C-8′), 135.86 (C-7′), 141.82 (C-8′a), 145.55 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.67 (C-1), 155.91 (C-2′), 160.91 (CO-quinazolinone), 168.60 (CONH); anal. calc. for C26H24N4O5S2 (536.62): C, 58.19; H, 4.51; N, 10.44. Found: C, 58.22; H, 4.46; N, 10.28.
N), 149.67 (C-1), 155.91 (C-2′), 160.91 (CO-quinazolinone), 168.60 (CONH); anal. calc. for C26H24N4O5S2 (536.62): C, 58.19; H, 4.51; N, 10.44. Found: C, 58.22; H, 4.46; N, 10.28.
3.1.1.3. 4-((2-(2-((8-Methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)phenyl propane-1-sulfonate (5c). Yield: 84%; mp 231–233 °C; IR (KBr) cm−1, ν: 3315 (OH, tautomer), 3178 (NH), 1676 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1619 (C
O), 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1370 (SO2), 1210 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 8.0 Hz, CH3CH2CH2), 1.85 (sextet, 2H, J = 7.5 Hz, CH3CH2CH2), 2.45 (s, 3H, CH3), 3.54 (t, 2H, J = 8.0 Hz, CH3CH2CH2), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.34 (t, 1H, J = 7.5 Hz, H-6′), 7.37–7.40 (m, 2H, H-2′′ and H-6′′), 7.47 (dd, 2H, J = 7.5, 1.5 Hz, H-2 and H-6), 7.59–7.60 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.65 (d, 1H, J = 7.0 Hz, H-7′), 7.80 (d, 2H, J = 7.5 Hz, H-3 and H-5), 7.91 (d, 1H, J = 8.0 Hz, H-5′), 8.05 (s, 1H, CH
N), 1370 (SO2), 1210 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 8.0 Hz, CH3CH2CH2), 1.85 (sextet, 2H, J = 7.5 Hz, CH3CH2CH2), 2.45 (s, 3H, CH3), 3.54 (t, 2H, J = 8.0 Hz, CH3CH2CH2), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.34 (t, 1H, J = 7.5 Hz, H-6′), 7.37–7.40 (m, 2H, H-2′′ and H-6′′), 7.47 (dd, 2H, J = 7.5, 1.5 Hz, H-2 and H-6), 7.59–7.60 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.65 (d, 1H, J = 7.0 Hz, H-7′), 7.80 (d, 2H, J = 7.5 Hz, H-3 and H-5), 7.91 (d, 1H, J = 8.0 Hz, H-5′), 8.05 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.24 (s, 1H, CH
N, iminol tautomer), 8.24 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.76 (s, 1H, OH/NH, tautomer), 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.31 (CH3CH2CH2), 16.64 (CH3), 16.93 (CH3CH2CH2), 34.88 (CH2), 51.49 (CH3CH2CH2), 119.25 (C-4′a), 122.53 (C-2 and C-6), 124.12 (C-5′), 125.41 (C-4), 128.29 (C-2′′ and C-6′′), 128.62 (C-4′′), 129.32 (C-3′′ and C-5′′), 129.49 (C-3 and C-5), 133.10 (C-6′), 133.97 (C-1′′), 135.08 (C-8′), 135.85 (C-7′), 141.80 (C-8′a), 145.52 (HC
N, amide tautomer), 11.76 (s, 1H, OH/NH, tautomer), 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.31 (CH3CH2CH2), 16.64 (CH3), 16.93 (CH3CH2CH2), 34.88 (CH2), 51.49 (CH3CH2CH2), 119.25 (C-4′a), 122.53 (C-2 and C-6), 124.12 (C-5′), 125.41 (C-4), 128.29 (C-2′′ and C-6′′), 128.62 (C-4′′), 129.32 (C-3′′ and C-5′′), 129.49 (C-3 and C-5), 133.10 (C-6′), 133.97 (C-1′′), 135.08 (C-8′), 135.85 (C-7′), 141.80 (C-8′a), 145.52 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.63 (C-1), 155.88 (C-2′), 160.88 (CO-quinazolinone), 168.58 (CONH); anal. calc. for C27H26N4O5S2 (550.65): C, 58.89; H, 4.76; N, 10.17. Found: C, 58.74; H, 4.59; N, 10.29.
N), 149.63 (C-1), 155.88 (C-2′), 160.88 (CO-quinazolinone), 168.58 (CONH); anal. calc. for C27H26N4O5S2 (550.65): C, 58.89; H, 4.76; N, 10.17. Found: C, 58.74; H, 4.59; N, 10.29.
3.1.1.4. 2-Methoxy-4-((2-(2-((8-methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)-acetyl)hydrazono)methyl)phenyl methanesulfonate (5d). Yield: 80%; mp 242–244 °C; IR (KBr) cm−1, ν: 3321 (OH, tautomer), 3170 (NH), 1671 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1630 (C
O), 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1340 (SO2), 1217 (C–O, C–N); 1H NMR δ (ppm): 2.46 (s, 3H, CH3), 3.38 (s, 3H, SO2CH3), 3.88 (s, 3H, OCH3), 4.00 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.32–7.37 (m, 3H, H-6′, H-2′′ and H-6′′), 7.48 (brs, 2H, H-3 and H-7′), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.64–7.66 (m, 2H, H-5 and H-6), 7.90–7.92 (m, 1H, H-5′), 8.02 (s, 1H, CH
N), 1340 (SO2), 1217 (C–O, C–N); 1H NMR δ (ppm): 2.46 (s, 3H, CH3), 3.38 (s, 3H, SO2CH3), 3.88 (s, 3H, OCH3), 4.00 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.32–7.37 (m, 3H, H-6′, H-2′′ and H-6′′), 7.48 (brs, 2H, H-3 and H-7′), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.64–7.66 (m, 2H, H-5 and H-6), 7.90–7.92 (m, 1H, H-5′), 8.02 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.22 (s, 1H, CH
N, iminol tautomer), 8.22 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 16.71 (CH3), 34.85 (CH2), 38.43 (SO2CH3), 56.05 (OCH3), 111.17 (C-3), 119.26 (C-4′a), 119.36 (C-6), 124.15 (C-5), 125.21 (C-5′), 125.45 (C-4), 129.34 (C-2′′ and C-6′′), 129.50 (C-3′′ and C-5′′), 129.90 (C-4′′), 134.01 (C-6′), 134.24 (C-1′′), 135.13 (C-8′), 135.87 (C-7′), 138.79 (C-8′a), 142.16 (C-1), 145.56 (HC
N, amide tautomer), 11.80 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 16.71 (CH3), 34.85 (CH2), 38.43 (SO2CH3), 56.05 (OCH3), 111.17 (C-3), 119.26 (C-4′a), 119.36 (C-6), 124.15 (C-5), 125.21 (C-5′), 125.45 (C-4), 129.34 (C-2′′ and C-6′′), 129.50 (C-3′′ and C-5′′), 129.90 (C-4′′), 134.01 (C-6′), 134.24 (C-1′′), 135.13 (C-8′), 135.87 (C-7′), 138.79 (C-8′a), 142.16 (C-1), 145.56 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.72 (C-2), 155.91 (C-2′), 160.92 (CO-quinazolinone), 168.60 (CONH); anal. calc. for C26H24N4O6S2 (552.62): C, 56.51; H, 4.38; N, 10.14. Found: C, 56.38; H, 4.20; N, 10.09.
N), 151.72 (C-2), 155.91 (C-2′), 160.92 (CO-quinazolinone), 168.60 (CONH); anal. calc. for C26H24N4O6S2 (552.62): C, 56.51; H, 4.38; N, 10.14. Found: C, 56.38; H, 4.20; N, 10.09.
3.1.1.5. 2-Methoxy-4-((2-(2-((8-methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)-acetyl)hydrazono)methyl)phenyl ethanesulfonate (5e). Yield: 85%; mp 216–218 °C; IR (KBr) cm−1, ν: 3322 (OH), 3172 (NH), 1675 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1636 (C
O), 1636 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1359 (SO2), 1219 (C–O, C–N); 1H NMR δ (ppm): 1.39 (t, 3H, J = 6.0 Hz, CH2CH3), 2.45 (s, 3H, CH3), 3.51 (q, 2H, J = 5.5 Hz, CH2CH3), 3.88 (s, 3H, OCH3), 4.00 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.33–7.34 (m, 3H, H-6′, H-2′′ and H-6′′), 7.47 (brs, 2H, H-3 and H-7′), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.64–7.66 (m, 2H, H-5 and H-6), 7.90–7.92 (m, 1H, H-5′), 8.02 (s, 1H, C
N), 1359 (SO2), 1219 (C–O, C–N); 1H NMR δ (ppm): 1.39 (t, 3H, J = 6.0 Hz, CH2CH3), 2.45 (s, 3H, CH3), 3.51 (q, 2H, J = 5.5 Hz, CH2CH3), 3.88 (s, 3H, OCH3), 4.00 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.33–7.34 (m, 3H, H-6′, H-2′′ and H-6′′), 7.47 (brs, 2H, H-3 and H-7′), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.64–7.66 (m, 2H, H-5 and H-6), 7.90–7.92 (m, 1H, H-5′), 8.02 (s, 1H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.22 (s, 1H, CH
N, iminol tautomer), 8.22 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.79 (s, 1H, OH/NH, tautomer), 11.81 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.00 (CH2CH3), 16.66 (CH3), 34.85 (CH2), 45.67 (CH2CH3), 56.01 (OCH3), 111.08 (C-3), 119.24 (C-4′a), 119.30 (C-6), 124.12 (C-5), 124.13 (C-5′), 125.41 (C-4), 129.31 (C-2′′ and C-6′′), 129.45 (C-3′′ and C-5′′), 129.89 (C-4′′), 134.06 (C-6′), 134.12 (C-1′′), 135.08 (C-8′), 135.09 (C-7′), 138.68 (C-8′a), 142.09 (C-1), 145.52 (HC
N, amide tautomer), 11.79 (s, 1H, OH/NH, tautomer), 11.81 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.00 (CH2CH3), 16.66 (CH3), 34.85 (CH2), 45.67 (CH2CH3), 56.01 (OCH3), 111.08 (C-3), 119.24 (C-4′a), 119.30 (C-6), 124.12 (C-5), 124.13 (C-5′), 125.41 (C-4), 129.31 (C-2′′ and C-6′′), 129.45 (C-3′′ and C-5′′), 129.89 (C-4′′), 134.06 (C-6′), 134.12 (C-1′′), 135.08 (C-8′), 135.09 (C-7′), 138.68 (C-8′a), 142.09 (C-1), 145.52 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.62 (C-2), 155.89 (C-2′), 160.87 (CO-quinazolinone), 167.32 (CONH); anal. calc. for C27H26N4O6S2 (566.65): C, 57.23; H, 4.63; N, 9.89. Found: C, 57.11; H, 4.50; N, 9.72.
N), 151.62 (C-2), 155.89 (C-2′), 160.87 (CO-quinazolinone), 167.32 (CONH); anal. calc. for C27H26N4O6S2 (566.65): C, 57.23; H, 4.63; N, 9.89. Found: C, 57.11; H, 4.50; N, 9.72.
3.1.1.6. 2-Methoxy-4-((2-(2-((8-methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)-acetyl)hydrazono)methyl)phenyl propane-1-sulfonate (5f). Yield: 78.5%; mp 206–208 °C; IR (KBr) cm−1, ν: 3311 (OH, tautomer), 3173 (NH), 1679 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1620 (C
O), 1620 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1350 (SO2), 1221 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 7.0 Hz, CH3CH2CH2), 1.90 (sextet, 2H, J = 7.0 Hz, CH3CH2CH2), 2.45 (s, 3H, CH3), 3.49 (t, 2H, J = 7.0 Hz, CH3CH2CH2), 3.88 (s, 3H, OCH3), 4.00 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.31–7.35 (m, 3H, H-6′, H-2′′ and H-6′′), 7.47–7.48 (m, 2H, H-3 and H-7′), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.64–7.66 (m, 2H, H-5 and H-6), 7.91 (d, 1H, J = 8.0 Hz, H-6), 8.02 (s, 1H, CH
N), 1350 (SO2), 1221 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 7.0 Hz, CH3CH2CH2), 1.90 (sextet, 2H, J = 7.0 Hz, CH3CH2CH2), 2.45 (s, 3H, CH3), 3.49 (t, 2H, J = 7.0 Hz, CH3CH2CH2), 3.88 (s, 3H, OCH3), 4.00 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.31–7.35 (m, 3H, H-6′, H-2′′ and H-6′′), 7.47–7.48 (m, 2H, H-3 and H-7′), 7.60 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.64–7.66 (m, 2H, H-5 and H-6), 7.91 (d, 1H, J = 8.0 Hz, H-6), 8.02 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.22 (s, 1H, CH
N, iminol tautomer), 8.22 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.79 (s, 1H, OH/NH, tautomer), 11.82 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.37 (CH3CH2CH2), 16.68 (CH3), 17.01 (CH3CH2CH2), 34.82 (CH2), 52.42 (CH3CH2CH2), 56.03 (OCH3), 111.07 (C-3), 119.24 (C-4′a), 119.33 (C-6), 124.13 (C-5), 124.19 (C-5′), 125.42 (C-4), 129.32 (C-2′′ and C-6′′), 129.47 (C-3′′ and C-5′′), 129.87 (C-4′′), 133.98 (C-6′), 134.09 (C-1′′), 135.08 (C-8′), 135.85 (C-7′), 138.69 (C-8′a), 142.12 (C-1), 145.54 (HC
N, amide tautomer), 11.79 (s, 1H, OH/NH, tautomer), 11.82 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.37 (CH3CH2CH2), 16.68 (CH3), 17.01 (CH3CH2CH2), 34.82 (CH2), 52.42 (CH3CH2CH2), 56.03 (OCH3), 111.07 (C-3), 119.24 (C-4′a), 119.33 (C-6), 124.13 (C-5), 124.19 (C-5′), 125.42 (C-4), 129.32 (C-2′′ and C-6′′), 129.47 (C-3′′ and C-5′′), 129.87 (C-4′′), 133.98 (C-6′), 134.09 (C-1′′), 135.08 (C-8′), 135.85 (C-7′), 138.69 (C-8′a), 142.12 (C-1), 145.54 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.65 (C-2), 155.89 (C-2′), 160.89 (CO-quinazolinone), 168.56 (CONH); anal. calc. for C28H28N4O6S2 (580.67): C, 57.92; H, 4.86; N, 9.65. Found: C, 57.80; H, 4.71; N, 9.50.
N), 151.65 (C-2), 155.89 (C-2′), 160.89 (CO-quinazolinone), 168.56 (CONH); anal. calc. for C28H28N4O6S2 (580.67): C, 57.92; H, 4.86; N, 9.65. Found: C, 57.80; H, 4.71; N, 9.50.
3.1.1.7. 2-Methoxy-5-((2-(2-((8-methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)-acetyl)hydrazono)methyl)phenyl methanesulfonate (5g). Yield: 86%; mp 229–231 °C; IR (KBr) cm−1, ν: 3316 (OH, tautomer), 3180 (NH), 1669 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1628 (C
O), 1628 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1344 (SO2), 1229 (C–O, C–N); 1H NMR δ (ppm): 2.46 (s, 3H, CH3), 3.38 (s, 3H, SO2CH3), 3.90 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.25–7.29 (m, 1H, H-3), 7.33–7.36 (m, 1H, H-6′), 7.48 (d, 2H, J = 7.0 Hz, H-2′′ and H-6′′), 7.60 (d, 2H, J = 6.5 Hz, H-3′′ and H-5′′), 7.63–7.65 (m, 3H, H-4, H-7′ and H-4′′), 7.90–7.92 (m, 2H, H-6 and H-5′), 7.99 (s, 1H, CH
N), 1344 (SO2), 1229 (C–O, C–N); 1H NMR δ (ppm): 2.46 (s, 3H, CH3), 3.38 (s, 3H, SO2CH3), 3.90 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.25–7.29 (m, 1H, H-3), 7.33–7.36 (m, 1H, H-6′), 7.48 (d, 2H, J = 7.0 Hz, H-2′′ and H-6′′), 7.60 (d, 2H, J = 6.5 Hz, H-3′′ and H-5′′), 7.63–7.65 (m, 3H, H-4, H-7′ and H-4′′), 7.90–7.92 (m, 2H, H-6 and H-5′), 7.99 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.16 (s, 1H, CH
N, iminol tautomer), 8.16 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.68 (s, 1H, OH/NH, tautomer), 11.71 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 16.69 (CH3), 35.01 (CH2), 38.41 (SO2CH3), 56.21 (OCH3), 113.67 (C-3), 119.25 (C-4′a), 121.47 (C-6), 121.74 (C-4), 124.11 (C-5′), 125.39 (C-6), 127.21 (C-5), 129.33 (C-2′′ and C-6′′), 129.47 (C-3′′ and C-5′′), 129.85 (C-4′′), 134.05 (C-1′′), 135.06 (C-8′), 135.88 (C-7′), 138.10 (C-8′a), 141.97 (C-1), 145.54 (HC
N, amide tautomer), 11.68 (s, 1H, OH/NH, tautomer), 11.71 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 16.69 (CH3), 35.01 (CH2), 38.41 (SO2CH3), 56.21 (OCH3), 113.67 (C-3), 119.25 (C-4′a), 121.47 (C-6), 121.74 (C-4), 124.11 (C-5′), 125.39 (C-6), 127.21 (C-5), 129.33 (C-2′′ and C-6′′), 129.47 (C-3′′ and C-5′′), 129.85 (C-4′′), 134.05 (C-1′′), 135.06 (C-8′), 135.88 (C-7′), 138.10 (C-8′a), 141.97 (C-1), 145.54 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 152.65 (C-2), 155.94 (C-2′), 160.90 (CO-quinazolinone), 168.39 (CONH); anal. calc. for C26H24N4O6S2 (552.62): C, 56.51; H, 4.38; N, 10.14. Found: C, 56.63; H, 4.51; N, 10.28.
N), 152.65 (C-2), 155.94 (C-2′), 160.90 (CO-quinazolinone), 168.39 (CONH); anal. calc. for C26H24N4O6S2 (552.62): C, 56.51; H, 4.38; N, 10.14. Found: C, 56.63; H, 4.51; N, 10.28.
3.1.1.8. 2-Methoxy-5-((2-(2-((8-methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)-acetyl)hydrazono)methyl)phenyl ethanesulfonate (5h). Yield: 80.5%; mp 234–236 °C; IR (KBr) cm−1, ν: 3320 (OH, tautomer), 3165 (NH), 1677 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1618 (C
O), 1618 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1350 (SO2), 1211 (C–O, C–N); 1H NMR δ (ppm): 1.39 (t, 3H, J = 7.0 Hz, CH2CH3), 2.46 (s, 3H, CH3), 3.51 (q, 2H, J = 7.5 Hz, CH2CH3), 3.89 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.25–7.28 (m, 1H, H-3), 7.33–7.36 (m, 1H, H-6′), 7.47 (brs, 2H, H-2′′ and H-6′′), 7.60 (brs, 2H, H-3′′ and H-5′′), 7.61–7.64 (m, 3H, H-4, H-7′ and H-4′′), 7.91–7.92 (m, 2H, H-6 and H-5′), 7.99 (s, 1H, CH
N), 1350 (SO2), 1211 (C–O, C–N); 1H NMR δ (ppm): 1.39 (t, 3H, J = 7.0 Hz, CH2CH3), 2.46 (s, 3H, CH3), 3.51 (q, 2H, J = 7.5 Hz, CH2CH3), 3.89 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.25–7.28 (m, 1H, H-3), 7.33–7.36 (m, 1H, H-6′), 7.47 (brs, 2H, H-2′′ and H-6′′), 7.60 (brs, 2H, H-3′′ and H-5′′), 7.61–7.64 (m, 3H, H-4, H-7′ and H-4′′), 7.91–7.92 (m, 2H, H-6 and H-5′), 7.99 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.16 (s, 1H, CH
N, iminol tautomer), 8.16 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.67 (s, 1H, OH/NH, tautomer), 11.70 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.02 (CH2CH3), 16.71 (CH3), 34.97 (CH2), 45.73 (CH2CH3), 56.22 (OCH3), 113.63 (C-3), 119.27 (C-4′a), 121.42 (C-6), 121.59 (C-4), 124.13 (C-5′), 125.42 (C-6), 127.15 (C-5), 129.34 (C-2′′ and C-6′′), 129.49 (C-3′′ and C-5′′), 129.89 (C-4′′), 134.08 (C-1′′), 135.08 (C-8′), 135.88 (C-7′), 138.02 (C-8′a), 142.02 (C-1), 145.56 (HC
N, amide tautomer), 11.67 (s, 1H, OH/NH, tautomer), 11.70 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.02 (CH2CH3), 16.71 (CH3), 34.97 (CH2), 45.73 (CH2CH3), 56.22 (OCH3), 113.63 (C-3), 119.27 (C-4′a), 121.42 (C-6), 121.59 (C-4), 124.13 (C-5′), 125.42 (C-6), 127.15 (C-5), 129.34 (C-2′′ and C-6′′), 129.49 (C-3′′ and C-5′′), 129.89 (C-4′′), 134.08 (C-1′′), 135.08 (C-8′), 135.88 (C-7′), 138.02 (C-8′a), 142.02 (C-1), 145.56 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 152.61 (C-2), 155.95 (C-2′), 160.93 (CO-quinazolinone), 168.40 (CONH); anal. calc. for C27H26N4O6S2 (566.65): C, 57.23; H, 4.63; N, 9.89. Found: C, 57.38; H, 4.51; N, 9.75.
N), 152.61 (C-2), 155.95 (C-2′), 160.93 (CO-quinazolinone), 168.40 (CONH); anal. calc. for C27H26N4O6S2 (566.65): C, 57.23; H, 4.63; N, 9.89. Found: C, 57.38; H, 4.51; N, 9.75.
3.1.1.9. 2-Methoxy-5-((2-(2-((8-methyl-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)-acetyl)hydrazono)methyl)phenyl propane-1-sulfonate (5i). Yield: 90.5%; mp 256–258 °C; IR (KBr) cm−1, ν: 3315 (OH, tautomer), 3172 (NH), 1673 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1631 (C
O), 1631 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1349 (SO2), 1221 (C–O, C–N); 1H NMR δ (ppm): 1.03 (t, 3H, J = 7.0 Hz, CH3CH2CH2), 1.88 (sextet, 2H, J = 7.0 Hz, CH3CH2CH2), 2.46 (s, 3H, CH3), 3.47 (t, 2H, J = 6.0 Hz, CH3CH2CH2), 3.89 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.24–7.28 (m, 1H, H-3), 7.32–7.35 (m, 1H, H-6′), 7.48 (brs., 2H, H-2′′ and H-6′′), 7.60 (brs., 2H, H-3′′ and H-5′′), 7.61–7.64 (m, 3H, H-4, H-7′ and H-4′′), 7.90–7.92 (m, 2H, H-6 and H-5′), 7.98 (s, 1H, CH
N), 1349 (SO2), 1221 (C–O, C–N); 1H NMR δ (ppm): 1.03 (t, 3H, J = 7.0 Hz, CH3CH2CH2), 1.88 (sextet, 2H, J = 7.0 Hz, CH3CH2CH2), 2.46 (s, 3H, CH3), 3.47 (t, 2H, J = 6.0 Hz, CH3CH2CH2), 3.89 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.50 (s, 2H, CH2, amide tautomer), 7.24–7.28 (m, 1H, H-3), 7.32–7.35 (m, 1H, H-6′), 7.48 (brs., 2H, H-2′′ and H-6′′), 7.60 (brs., 2H, H-3′′ and H-5′′), 7.61–7.64 (m, 3H, H-4, H-7′ and H-4′′), 7.90–7.92 (m, 2H, H-6 and H-5′), 7.98 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.17 (s, 1H, CH
N, iminol tautomer), 8.17 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.68 (s, 1H, OH/NH, tautomer), 11.72 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.37 (CH3CH2CH2), 16.68 (CH3), 17.01 (CH3CH2CH2), 34.82 (CH2), 52.42 (CH3CH2CH2), 56.03 (OCH3), 111.67 (C-3), 119.24 (C-4′a), 119.33 (C-6), 124.13 (C-4), 124.19 (C-5′), 125.42 (C-6), 129.32 (C-2′′ and C-6′′), 129.47 (C-3′′ and C-5′′), 129.87 (C-4′′), 133.98 (C-5), 134.09 (C-1′′), 135.08 (C-8′), 135.85 (C-7′), 138.69 (C-8′a), 142.12 (C-1), 145.54 (HC
N, amide tautomer), 11.68 (s, 1H, OH/NH, tautomer), 11.72 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.37 (CH3CH2CH2), 16.68 (CH3), 17.01 (CH3CH2CH2), 34.82 (CH2), 52.42 (CH3CH2CH2), 56.03 (OCH3), 111.67 (C-3), 119.24 (C-4′a), 119.33 (C-6), 124.13 (C-4), 124.19 (C-5′), 125.42 (C-6), 129.32 (C-2′′ and C-6′′), 129.47 (C-3′′ and C-5′′), 129.87 (C-4′′), 133.98 (C-5), 134.09 (C-1′′), 135.08 (C-8′), 135.85 (C-7′), 138.69 (C-8′a), 142.12 (C-1), 145.54 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.65 (C-2), 155.89 (C-2′), 160.89 (CO-quinazolinone), 168.56 (CONH); anal. calc. for C28H28N4O6S2 (580.67): C, 57.92; H, 4.86; N, 9.65. Found: C, 57.82; H, 4.99; N, 9.51.
N), 151.65 (C-2), 155.89 (C-2′), 160.89 (CO-quinazolinone), 168.56 (CONH); anal. calc. for C28H28N4O6S2 (580.67): C, 57.92; H, 4.86; N, 9.65. Found: C, 57.82; H, 4.99; N, 9.51.
3.1.1.10. 4-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)phenyl methanesulfonate (5j). Yield: 90%; mp 236–238 °C; IR (KBr) cm−1, ν: 3325 (OH, tautomer), 3170 (NH), 1671 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1620 (C
O), 1620 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1355 (SO2), 1214 (C–O, C–N); 1H NMR δ (ppm): 3.33 (s, 3H, SO2CH3), 3.98 (s, 2H, CH2, iminol tautomer), 4.46 (s, 2H, CH2, amide tautomer), 7.41 (brs, 1H, H-8′), 7.49 (brs, 2H, H-2′′ and H-6′′), 7.59 (brs, 2H, H-2 and H-6), 7.79–7.81 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.85–7.86 (m, 2H, H-3 and H-5), 7.98–8.00 (m, 2H, H-5′ and H-7′), 8.06 (s, 1H, CH
N), 1355 (SO2), 1214 (C–O, C–N); 1H NMR δ (ppm): 3.33 (s, 3H, SO2CH3), 3.98 (s, 2H, CH2, iminol tautomer), 4.46 (s, 2H, CH2, amide tautomer), 7.41 (brs, 1H, H-8′), 7.49 (brs, 2H, H-2′′ and H-6′′), 7.59 (brs, 2H, H-2 and H-6), 7.79–7.81 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.85–7.86 (m, 2H, H-3 and H-5), 7.98–8.00 (m, 2H, H-5′ and H-7′), 8.06 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.28 (s, 1H, CH
N, iminol tautomer), 8.28 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.71 (s, 1H, OH/NH, tautomer), 11.82 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 34.26 (CH2), 37.54 (SO2CH3), 120.77 (C-4′a), 122.70 (C-2 and C-6), 125.48 (C-4), 128.14 (C-5), 128.45 (C-2′′ and C-6′′), 128.72 (C-4′′), 129.27 (C-3′′ and C-5′′), 129.56 (C-3 and C-5), 130.07 (C-8′), 133.17 (C-6′), 134.92 (C-1′′), 135.55 (C-7′), 142.06 (C-8′a), 145.79 (HC
N, amide tautomer), 11.71 (s, 1H, OH/NH, tautomer), 11.82 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 34.26 (CH2), 37.54 (SO2CH3), 120.77 (C-4′a), 122.70 (C-2 and C-6), 125.48 (C-4), 128.14 (C-5), 128.45 (C-2′′ and C-6′′), 128.72 (C-4′′), 129.27 (C-3′′ and C-5′′), 129.56 (C-3 and C-5), 130.07 (C-8′), 133.17 (C-6′), 134.92 (C-1′′), 135.55 (C-7′), 142.06 (C-8′a), 145.79 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.85 (C-1), 157.84 (C-2′), 159.66 (CO-quinazolinone), 168.71 (CONH); C24H19ClN4O5S2 (543.01): C, 53.09; H, 3.53; N, 10.32. Found: C, 53.18; H, 3.41; N, 10.21
N), 149.85 (C-1), 157.84 (C-2′), 159.66 (CO-quinazolinone), 168.71 (CONH); C24H19ClN4O5S2 (543.01): C, 53.09; H, 3.53; N, 10.32. Found: C, 53.18; H, 3.41; N, 10.21
3.1.1.11. 4-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)phenyl ethanesulfonate (5k ). Yield: 81%; mp 170–172 °C; IR (KBr) cm−1, ν: 3321 (OH, tautomer), 3173 (NH), 1673 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1622 (C
O), 1622 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1360 (SO2), 1210 (C–O, C–N); 1H NMR δ (ppm): 1.38 (t, 3H, J = 7.0 Hz, CH2CH3), 3.55 (q, 2H, J = 7.5 Hz, CH2CH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.46 (s, 2H, CH2, amide tautomer), 7.38–7.39 (m, 1H, H-8′), 7.48–7.51 (m, 2H, H-2′′ and H-6′′), 7.58–7.61 (m, 2H, H-2 and H-6), 7.78–7.79 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82–7.87 (m, 3H, J = 8.5, 1.5 Hz, H-3, H-5 and H-7′), 8.00 (s, 1H, H-5′), 8.06 (s, 1H, CH
N), 1360 (SO2), 1210 (C–O, C–N); 1H NMR δ (ppm): 1.38 (t, 3H, J = 7.0 Hz, CH2CH3), 3.55 (q, 2H, J = 7.5 Hz, CH2CH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.46 (s, 2H, CH2, amide tautomer), 7.38–7.39 (m, 1H, H-8′), 7.48–7.51 (m, 2H, H-2′′ and H-6′′), 7.58–7.61 (m, 2H, H-2 and H-6), 7.78–7.79 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82–7.87 (m, 3H, J = 8.5, 1.5 Hz, H-3, H-5 and H-7′), 8.00 (s, 1H, H-5′), 8.06 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.28 (s, 1H, CH
N, iminol tautomer), 8.28 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.72 (s, 1H, OH/NH, tautomer), 11.83 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.00 (CH2CH3), 34.27 (CH2), 44.79 (CH2CH3), 120.74 (C-4′a), 122.54 (C-2 and C-6), 125.45 (C-4), 128.12 (C-5′), 128.41 (C-2′′ and C-6′′), 128.69 (C-4′′), 129.24 (C-3′′ and C-5′′), 129.53 (C-3 and C-5), 130.03 (C-8′), 133.04 (C-6′), 134.88 (C-1′′), 135.53 (C-7′), 142.05 (C-8′a), 145.75 (HC
N, amide tautomer), 11.72 (s, 1H, OH/NH, tautomer), 11.83 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.00 (CH2CH3), 34.27 (CH2), 44.79 (CH2CH3), 120.74 (C-4′a), 122.54 (C-2 and C-6), 125.45 (C-4), 128.12 (C-5′), 128.41 (C-2′′ and C-6′′), 128.69 (C-4′′), 129.24 (C-3′′ and C-5′′), 129.53 (C-3 and C-5), 130.03 (C-8′), 133.04 (C-6′), 134.88 (C-1′′), 135.53 (C-7′), 142.05 (C-8′a), 145.75 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.67 (C-1), 157.80 (C-2′), 159.66 (CO-quinazolinone), 168.66 (CONH); anal. calc. for C25H21ClN4O5S2 (557.04): C, 53.91; H, 3.80; N, 10.06. Found: C, 53.85; H, 3.65; N, 10.19.
N), 149.67 (C-1), 157.80 (C-2′), 159.66 (CO-quinazolinone), 168.66 (CONH); anal. calc. for C25H21ClN4O5S2 (557.04): C, 53.91; H, 3.80; N, 10.06. Found: C, 53.85; H, 3.65; N, 10.19.
3.1.1.12. 4-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)phenyl propane-1-sulfonate (5l). Yield: 82.5%; mp 179–181 °C; IR (KBr) cm−1, ν: 3317 (OH, tautomer), 3168 (NH), 1679 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1630 (C
O), 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1360 (SO2), 1210 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 7.5 Hz, CH3CH2CH2), 1.86 (sextet, 2H, J = 7.5 Hz, CH3CH2CH2), 3.54 (t, 2H, J = 7.5 Hz, CH3CH2CH2), 3.98 (s, 2H, CH2, iminol tautomer), 4.46 (s, 2H, CH2, amide tautomer), 7.38 (d, 1H, J = 8.0 Hz, H-8′), 7.49–7.50 (m, 2H, H-2′′ and H-6′′), 7.59–7.61 (m, 2H, H-2 and H-6), 7.78–7.79 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82–7.87 (m, 3H, H-3, H-5 and H-7′), 8.00 (s, 1H, H-5′), 8.06 (s, 1H, CH
N), 1360 (SO2), 1210 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 7.5 Hz, CH3CH2CH2), 1.86 (sextet, 2H, J = 7.5 Hz, CH3CH2CH2), 3.54 (t, 2H, J = 7.5 Hz, CH3CH2CH2), 3.98 (s, 2H, CH2, iminol tautomer), 4.46 (s, 2H, CH2, amide tautomer), 7.38 (d, 1H, J = 8.0 Hz, H-8′), 7.49–7.50 (m, 2H, H-2′′ and H-6′′), 7.59–7.61 (m, 2H, H-2 and H-6), 7.78–7.79 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82–7.87 (m, 3H, H-3, H-5 and H-7′), 8.00 (s, 1H, H-5′), 8.06 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.27 (s, 1H, CH
N, iminol tautomer), 8.27 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.72 (s, 1H, OH/NH, tautomer), 11.83 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.32 (CH3CH2CH2), 16.94 (CH3CH2CH2), 34.27 (CH2), 51.49 (CH3CH2CH2), 120.73 (C-4′a), 122.56 (C-2 and C-6), 125.45 (C-4), 128.11 (C-5′), 128.40 (C-2′′ and C-6′′), 128.69 (C-4′′), 129.25 (C-3′′ and C-5′′), 129.53 (C-3 and C-5), 130.04 (C-8′), 133.02 (C-6′), 134.87 (C-1′′), 135.53 (C-7′), 142.05 (C-8′a), 145.74 (HC
N, amide tautomer), 11.72 (s, 1H, OH/NH, tautomer), 11.83 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.32 (CH3CH2CH2), 16.94 (CH3CH2CH2), 34.27 (CH2), 51.49 (CH3CH2CH2), 120.73 (C-4′a), 122.56 (C-2 and C-6), 125.45 (C-4), 128.11 (C-5′), 128.40 (C-2′′ and C-6′′), 128.69 (C-4′′), 129.25 (C-3′′ and C-5′′), 129.53 (C-3 and C-5), 130.04 (C-8′), 133.02 (C-6′), 134.87 (C-1′′), 135.53 (C-7′), 142.05 (C-8′a), 145.74 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.66 (C-1), 157.80 (C-2′), 159.66 (CO-quinazolinone), 168.66 (CONH); anal. calc. for C26H23ClN4O5S2 (571.06): C, 54.69; H, 4.06; N, 9.81. Found: C, 54.86; H, 4.17; N, 9.69.
N), 149.66 (C-1), 157.80 (C-2′), 159.66 (CO-quinazolinone), 168.66 (CONH); anal. calc. for C26H23ClN4O5S2 (571.06): C, 54.69; H, 4.06; N, 9.81. Found: C, 54.86; H, 4.17; N, 9.69.
3.1.1.13. 4-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)-2-methoxyphenyl methanesulfonate (5m). Yield: 85.6%; mp 232–234 °C; IR (KBr) cm−1, ν: 3327 (OH, tautomer), 3188 (NH), 1669 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1623 (C
O), 1623 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1359 (SO2), 1225 (C–O, C–N); 1H NMR δ (ppm): 3.38 (s, 3H, SO2CH3), 3.89 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.48 (s, 2H, CH2, amide tautomer), 7.31–7.36 (m, 2H, H-6 and H-8′), 7.48–7.49 (m, 3H, H-5′, H-2′′ and H-6′′), 7.59–7.60 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82 (d, 1H, J = 8.5 Hz, H-5), 7.85 (d, 1H, J = 8.5 Hz, H-7′), 8.00 (brs, 1H, H-3), 8.04 (s, 1H, CH
N), 1359 (SO2), 1225 (C–O, C–N); 1H NMR δ (ppm): 3.38 (s, 3H, SO2CH3), 3.89 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.48 (s, 2H, CH2, amide tautomer), 7.31–7.36 (m, 2H, H-6 and H-8′), 7.48–7.49 (m, 3H, H-5′, H-2′′ and H-6′′), 7.59–7.60 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82 (d, 1H, J = 8.5 Hz, H-5), 7.85 (d, 1H, J = 8.5 Hz, H-7′), 8.00 (brs, 1H, H-3), 8.04 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer). 8.26 (s, 1H, CH
N, iminol tautomer). 8.26 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.78 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 34.45 (CH2), 38.41 (SO2CH3), 55.99 (OCH3), 111.06 (C-3), 119.52 (C-4′a), 120.23 (C-6), 120.73 (C-5), 124.24 (C-5′), 125.49 (C-6′), 128.06 (C-4), 129.26 (C-2′′ and C-6′′), 129.55 (C-3′′ and C-5′′), 130.07 (C-4′′), 134.19 (C-1′′), 134.90 (C-8′), 135.54 (C-7′), 138.79 (C-1), 142.24 (C-8′a), 145.78 (HC
N, amide tautomer), 11.78 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 34.45 (CH2), 38.41 (SO2CH3), 55.99 (OCH3), 111.06 (C-3), 119.52 (C-4′a), 120.23 (C-6), 120.73 (C-5), 124.24 (C-5′), 125.49 (C-6′), 128.06 (C-4), 129.26 (C-2′′ and C-6′′), 129.55 (C-3′′ and C-5′′), 130.07 (C-4′′), 134.19 (C-1′′), 134.90 (C-8′), 135.54 (C-7′), 138.79 (C-1), 142.24 (C-8′a), 145.78 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.68 (C-2), 157.84 (C-2′), 159.69 (CO-quinazolinone), 168.71 (CONH); anal. calc. for C25H21ClN4O6S2 (573.04): C, 52.40; H, 3.69; N, 9.78. Found: C, 52.58; H, 3.59; N, 9.64.
N), 151.68 (C-2), 157.84 (C-2′), 159.69 (CO-quinazolinone), 168.71 (CONH); anal. calc. for C25H21ClN4O6S2 (573.04): C, 52.40; H, 3.69; N, 9.78. Found: C, 52.58; H, 3.59; N, 9.64.
3.1.1.14. 4-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)-2-methoxyphenyl ethanesulfonate (5n). Yield: 78.5%; mp 210–212 °C; IR (KBr) cm−1, ν: 3316 (OH, tautomer), 3185 (NH), 1670 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1630 (C
O), 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1349 (SO2), 1207 (C–O, C–N); 1H NMR δ (ppm): 1.39 (t, 3H, J = 5.5 Hz, CH2CH3), 3.50 (q, 2H, J = 6.5 Hz, CH2CH3), 3.88 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.48 (s, 2H, CH2, amide tautomer), 7.31–7.34 (m, 2H, H-6 and H-8′), 7.48 (brs, 3H, H-6, H-2′′ and H-6′′), 7.59 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.82 (d, 1H, J = 8.5 Hz, H-5), 7.86 (d, 1H, J = 8.5 Hz, H-7′), 8.00 (s, 1H, H-3), 8.03 (s, 1H, CH
N), 1349 (SO2), 1207 (C–O, C–N); 1H NMR δ (ppm): 1.39 (t, 3H, J = 5.5 Hz, CH2CH3), 3.50 (q, 2H, J = 6.5 Hz, CH2CH3), 3.88 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.48 (s, 2H, CH2, amide tautomer), 7.31–7.34 (m, 2H, H-6 and H-8′), 7.48 (brs, 3H, H-6, H-2′′ and H-6′′), 7.59 (brs, 3H, H-3′′, H-4′′ and H-5′′), 7.82 (d, 1H, J = 8.5 Hz, H-5), 7.86 (d, 1H, J = 8.5 Hz, H-7′), 8.00 (s, 1H, H-3), 8.03 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer). 8.26 (s, 1H, CH
N, iminol tautomer). 8.26 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.77 (s, 1H, OH/NH, tautomer), 11.83 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.02 (CH2CH3), 34.43 (CH2), 45.70 (CH2CH3), 55.96 (OCH3), 111.00 (C-3), 119.47 (C-4′a), 120.18 (C-6), 120.72 (C-5), 124.12 (C-5′), 125.46 (C-6′), 128.05 (C-4), 129.24 (C-2′′ and C-6′′), 129.93 (C-3′′ and C-5′′), 130.03 (C-4′′), 134.03 (C-1′′), 134.88 (C-8′), 135.53 (C-7′), 138.70 (C-1), 142.19 (C-8′a), 145.74 (HC
N, amide tautomer), 11.77 (s, 1H, OH/NH, tautomer), 11.83 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.02 (CH2CH3), 34.43 (CH2), 45.70 (CH2CH3), 55.96 (OCH3), 111.00 (C-3), 119.47 (C-4′a), 120.18 (C-6), 120.72 (C-5), 124.12 (C-5′), 125.46 (C-6′), 128.05 (C-4), 129.24 (C-2′′ and C-6′′), 129.93 (C-3′′ and C-5′′), 130.03 (C-4′′), 134.03 (C-1′′), 134.88 (C-8′), 135.53 (C-7′), 138.70 (C-1), 142.19 (C-8′a), 145.74 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.61 (C-2), 157.82 (C-2′), 159.65 (CO-quinazolinone), 168.67 (CONH); anal. calc. for C26H23ClN4O6S2 (587.06): C, 53.19; H, 3.95; N, 9.54. Found: C, 53.30; H, 3.80; N, 9.39.
N), 151.61 (C-2), 157.82 (C-2′), 159.65 (CO-quinazolinone), 168.67 (CONH); anal. calc. for C26H23ClN4O6S2 (587.06): C, 53.19; H, 3.95; N, 9.54. Found: C, 53.30; H, 3.80; N, 9.39.
3.1.1.15. 4-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)- hydrazono)methyl)-2-methoxyphenyl propane-1-sulfonate (5o). Yield: 82%; mp 206–208 °C; IR (KBr) cm−1, ν: 3319 (OH, tautomer), 3189 (NH), 1682 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1622 (C
O), 1622 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1360 (SO2), 1219 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 7.0 Hz, CH3CH2CH2), 1.87 (sextet, 2H, J = 7.0 Hz, CH3CH2CH2), 3.48 (t, 2H, J = 7.5 Hz, CH3CH2CH2), 3.88 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.47 (s, 2H, CH2, amide tautomer), 7.32–7.34 (m, 2H, H-6 and H-8′), 7.48–7.49 (m, 3H, H-5′, H-2′′ and H-6′′), 7.58–7.60 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82 (dd, 1H, J = 8.5, 2.5 Hz, H-5), 7.86 (dd, 1H, J = 8.5, 2.5 Hz, H-7′), 8.00 (s, 1H, H-3), 8.03 (s, 1H, CH
N), 1360 (SO2), 1219 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 7.0 Hz, CH3CH2CH2), 1.87 (sextet, 2H, J = 7.0 Hz, CH3CH2CH2), 3.48 (t, 2H, J = 7.5 Hz, CH3CH2CH2), 3.88 (s, 3H, OCH3), 3.99 (s, 2H, CH2, iminol tautomer), 4.47 (s, 2H, CH2, amide tautomer), 7.32–7.34 (m, 2H, H-6 and H-8′), 7.48–7.49 (m, 3H, H-5′, H-2′′ and H-6′′), 7.58–7.60 (m, 3H, H-3′′, H-4′′ and H-5′′), 7.82 (dd, 1H, J = 8.5, 2.5 Hz, H-5), 7.86 (dd, 1H, J = 8.5, 2.5 Hz, H-7′), 8.00 (s, 1H, H-3), 8.03 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.25 (s, 1H, CH
N, iminol tautomer), 8.25 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.77 (s, 1H, OH/NH, tautomer), 11.82 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.41 (CH3CH2CH2), 17.05 (CH3CH2CH2), 34.46 (CH2), 52.46 (CH3CH2CH2), 56.02 (OCH3), 111.03 (C-3), 119.55 (C-4′a), 120.28 (C-6), 120.76 (C-5), 124.27 (C-5′), 125.50 (C-6′), 128.10 (C-4), 129.28 (C-2′′ and C-6′′), 129.58 (C-3′′ and C-5′′), 130.09 (C-4′′), 134.08 (C-1′′), 134.94 (C-8′), 135.56 (C-7′), 138.74 (C-1), 142.28 (C-8′a), 145.80 (HC
N, amide tautomer), 11.77 (s, 1H, OH/NH, tautomer), 11.82 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.41 (CH3CH2CH2), 17.05 (CH3CH2CH2), 34.46 (CH2), 52.46 (CH3CH2CH2), 56.02 (OCH3), 111.03 (C-3), 119.55 (C-4′a), 120.28 (C-6), 120.76 (C-5), 124.27 (C-5′), 125.50 (C-6′), 128.10 (C-4), 129.28 (C-2′′ and C-6′′), 129.58 (C-3′′ and C-5′′), 130.09 (C-4′′), 134.08 (C-1′′), 134.94 (C-8′), 135.56 (C-7′), 138.74 (C-1), 142.28 (C-8′a), 145.80 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.66 (C-2), 157.86 (C-2′), 159.71 (CO-quinazolinone), 168.74 (CONH); anal. calc. for C27H25ClN4O6S2 (601.09): C, 53.95; H, 4.19; N, 9.32. Found: C, 53.81; H, 4.10; N, 9.19.
N), 151.66 (C-2), 157.86 (C-2′), 159.71 (CO-quinazolinone), 168.74 (CONH); anal. calc. for C27H25ClN4O6S2 (601.09): C, 53.95; H, 4.19; N, 9.32. Found: C, 53.81; H, 4.10; N, 9.19.
3.1.1.16. 5-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)- hydrazono)methyl)-2-methoxyphenyl methanesulfonate (5p). Yield: 82%; mp 240–242 °C; IR (KBr) cm−1, ν: 3310 (OH, tautomer), 3185 (NH), 1672 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1636 (C
O), 1636 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1345 (SO2), 1218 (C–O, C–N); 1H NMR δ (ppm): 3.40 (s, 3H, SO2CH3), 3.90 (s, 3H, OCH3), 3.98 (s, 2H, CH2, iminol tautomer), 4.48 (s, 2H, CH2, amide tautomer), 7.27 (brs, 1H, H-3), 7.49–7.53 (m, 3H, H-8′, H-2′′ and H-6′′), 7.59–7.61 (m, 4H, H-6, H-3′′, H-4′′ and H-5′′), 7.66 (brs, 1H, H-5′), 7.80 (d, 1H, J = 7.5 Hz, H-4), 7.86 (d, 1H, J = 7.5 Hz, H-7′), 8.00 (s, 1H, CH
N), 1345 (SO2), 1218 (C–O, C–N); 1H NMR δ (ppm): 3.40 (s, 3H, SO2CH3), 3.90 (s, 3H, OCH3), 3.98 (s, 2H, CH2, iminol tautomer), 4.48 (s, 2H, CH2, amide tautomer), 7.27 (brs, 1H, H-3), 7.49–7.53 (m, 3H, H-8′, H-2′′ and H-6′′), 7.59–7.61 (m, 4H, H-6, H-3′′, H-4′′ and H-5′′), 7.66 (brs, 1H, H-5′), 7.80 (d, 1H, J = 7.5 Hz, H-4), 7.86 (d, 1H, J = 7.5 Hz, H-7′), 8.00 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.20 (s, 1H, CH
N, iminol tautomer), 8.20 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.66 (s, 1H, OH/NH, tautomer), 11.74 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 34.60 (CH2), 38.40 (SO2CH3), 56.21 (OCH3), 113.67 (C-3), 120.75 (C-4′a), 121.36 (C-6), 125.43 (C-5), 127.16 (C-5′), 127.39 (C-6′), 127.50 (C-4), 128.15 (C-1′′), 129.24 (C-2′′ and C-6′′), 129.51 (C-3′′ and C-5′′), 130.02 (C-4′′), 134.81 (C-8′), 135.54 (C-7′), 138.09 (C-1), 142.05 (C-8′a), 145.77 (HC
N, amide tautomer), 11.66 (s, 1H, OH/NH, tautomer), 11.74 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 34.60 (CH2), 38.40 (SO2CH3), 56.21 (OCH3), 113.67 (C-3), 120.75 (C-4′a), 121.36 (C-6), 125.43 (C-5), 127.16 (C-5′), 127.39 (C-6′), 127.50 (C-4), 128.15 (C-1′′), 129.24 (C-2′′ and C-6′′), 129.51 (C-3′′ and C-5′′), 130.02 (C-4′′), 134.81 (C-8′), 135.54 (C-7′), 138.09 (C-1), 142.05 (C-8′a), 145.77 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 152.64 (C-2), 157.84 (C-2′), 159.67 (CO-quinazolinone), 168.48 (CONH); anal. calc. for C25H21ClN4O6S2 (573.04): C, 52.40; H, 3.69; N, 9.78. Found: C, 52.58; H, 3.51; N, 9.69.
N), 152.64 (C-2), 157.84 (C-2′), 159.67 (CO-quinazolinone), 168.48 (CONH); anal. calc. for C25H21ClN4O6S2 (573.04): C, 52.40; H, 3.69; N, 9.78. Found: C, 52.58; H, 3.51; N, 9.69.
3.1.1.17. 5-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)-2-methoxyphenyl ethanesulfonate (5q). Yield: 81%; mp 208–210 °C; IR (KBr) cm−1, ν: 3314 (OH, tautomer), 3187 (NH), 1683 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1624 (C
O), 1624 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1366 (SO2), 1220 (C–O, C–N); 1H NMR δ (ppm): 1.40 (t, 3H, J = 7.0 Hz, CH3CH2), 3.53 (q, 2H, J = 7.0 Hz, CH3CH2), 3.89 (s, 3H, OCH3), 3.97 (s, 2H, CH2, iminol tautomer), 4.47 (s, 2H, CH2, amide tautomer), 7.24–7.27 (m, 1H, H-3), 7.49 (d, 2H, J = 7.5 Hz, H-2′′ and H-6′′), 7.53 (d, 1H, J = 8.5 Hz, H-8′), 7.59–7.62 (m, 4H, H6, H-3′′, H-4′′ and H-5′′), 7.65 (d, 1H, J = 1.5 Hz, H-5′), 7.80 (dd, 1H, J = 9.0, 2.0 Hz, H-4), 7.86 (dd, 1H, J = 8.5, 2.0 Hz, H-7′), 8.00 (s, 1H, CH
N), 1366 (SO2), 1220 (C–O, C–N); 1H NMR δ (ppm): 1.40 (t, 3H, J = 7.0 Hz, CH3CH2), 3.53 (q, 2H, J = 7.0 Hz, CH3CH2), 3.89 (s, 3H, OCH3), 3.97 (s, 2H, CH2, iminol tautomer), 4.47 (s, 2H, CH2, amide tautomer), 7.24–7.27 (m, 1H, H-3), 7.49 (d, 2H, J = 7.5 Hz, H-2′′ and H-6′′), 7.53 (d, 1H, J = 8.5 Hz, H-8′), 7.59–7.62 (m, 4H, H6, H-3′′, H-4′′ and H-5′′), 7.65 (d, 1H, J = 1.5 Hz, H-5′), 7.80 (dd, 1H, J = 9.0, 2.0 Hz, H-4), 7.86 (dd, 1H, J = 8.5, 2.0 Hz, H-7′), 8.00 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.20 (s, 1H, CH
N, iminol tautomer), 8.20 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.66 (s, 1H, OH/NH, tautomer), 11.73 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.17 (CH3), 34.82 (CH2), 45.91 (CH2CH3), 56.38 (OCH3), 113.76 (C-3), 120.83 (C-4′a), 121.41 (C-6), 121.76 (C-4), 125.60 (C-5′), 127.27 (C-6′), 127.58 (C-5), 127.79 (C-1′′), 128.36 (C-8′), 129.37 (C-2′′ and C-6′′), 129.76 (C-3′′ and C-5′′), 130.29 (C-4′′), 135.66 (C-7′), 138.14 (C-1), 142.43 (C-8′a), 145.93 (HC
N, amide tautomer), 11.66 (s, 1H, OH/NH, tautomer), 11.73 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 8.17 (CH3), 34.82 (CH2), 45.91 (CH2CH3), 56.38 (OCH3), 113.76 (C-3), 120.83 (C-4′a), 121.41 (C-6), 121.76 (C-4), 125.60 (C-5′), 127.27 (C-6′), 127.58 (C-5), 127.79 (C-1′′), 128.36 (C-8′), 129.37 (C-2′′ and C-6′′), 129.76 (C-3′′ and C-5′′), 130.29 (C-4′′), 135.66 (C-7′), 138.14 (C-1), 142.43 (C-8′a), 145.93 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 152.78 (C-2), 157.95 (C-2′), 159.92 (CO-quinazolinone), 168.75 (CONH); anal. calc. for C26H23ClN4O6S2 (587.06): C, 53.19; H, 3.95; N, 9.54. Found: C, 53.25; H, 3.79; N, 9.69.
N), 152.78 (C-2), 157.95 (C-2′), 159.92 (CO-quinazolinone), 168.75 (CONH); anal. calc. for C26H23ClN4O6S2 (587.06): C, 53.19; H, 3.95; N, 9.54. Found: C, 53.25; H, 3.79; N, 9.69.
3.1.1.18. 5-((2-(2-((6-Chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetyl)-hydrazono)methyl)-2-methoxyphenyl propane-1-sulfonate (5r). Yield: 85%; mp 216–218 °C; IR (KBr) cm−1, ν: 3311 (OH, tautomer), 3185 (NH), 1683 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1637 (C
O), 1637 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1358 (SO2), 1215 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 6.5 Hz, CH3CH2CH2), 1.87 (sextet, 2H, J = 6.0 Hz, CH3CH2CH2), 3.50 (t, 2H, J = 6.0 Hz, CH3CH2CH2), 3.89 (s, 3H, OCH3), 3.98 (s, 2H, CH2, iminol tautomer), 4.47 (s, 2H, CH2, amide tautomer), 7.25–7.28 (m, 1H, H-3), 7.49 (brs, 2H, H-2′′ and H-6′′), 7.53 (d, 1H, J = 8.5 Hz, H-8′), 7.56–7.64 (m, 4H, H-6, H-3′′, H-4′′ and H-5′′), 7.65 (brs, 1H, H-5), 7.80 (d, 1H, J = 8.0, Hz, H-4), 7.86 (d, 1H, J = 8.5 Hz, H-7′), 8.00 (s, 1H, CH
N), 1358 (SO2), 1215 (C–O, C–N); 1H NMR δ (ppm): 1.04 (t, 3H, J = 6.5 Hz, CH3CH2CH2), 1.87 (sextet, 2H, J = 6.0 Hz, CH3CH2CH2), 3.50 (t, 2H, J = 6.0 Hz, CH3CH2CH2), 3.89 (s, 3H, OCH3), 3.98 (s, 2H, CH2, iminol tautomer), 4.47 (s, 2H, CH2, amide tautomer), 7.25–7.28 (m, 1H, H-3), 7.49 (brs, 2H, H-2′′ and H-6′′), 7.53 (d, 1H, J = 8.5 Hz, H-8′), 7.56–7.64 (m, 4H, H-6, H-3′′, H-4′′ and H-5′′), 7.65 (brs, 1H, H-5), 7.80 (d, 1H, J = 8.0, Hz, H-4), 7.86 (d, 1H, J = 8.5 Hz, H-7′), 8.00 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, iminol tautomer), 8.20 (s, 1H, CH
N, iminol tautomer), 8.20 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, amide tautomer), 11.67 (s, 1H, OH/NH, tautomer), 11.71 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.36 (CH3CH2CH2), 17.02 (CH3CH2CH2), 34.67 (CH2), 52.39 (CH3CH2CH2), 56.22 (OCH3), 113.57 (C-3), 120.72 (C-4′a), 121.29 (C-6), 121.72 (C-4), 125.43 (C-5′), 127.13 (C-6′), 127.33 (C-5), 128.15 (C-1′′), 129.24 (C-2′′ and C-6′′), 129.53 (C-3′′ and C-5′′), 130.03 (C-4′′), 134.78 (C-8′), 135.60 (C-7′), 137.86 (C-1), 142.10 (C-8′a), 145.78 (HC
N, amide tautomer), 11.67 (s, 1H, OH/NH, tautomer), 11.71 (s, 1H, OH/NH, tautomer); 13C NMR δ (ppm): 12.36 (CH3CH2CH2), 17.02 (CH3CH2CH2), 34.67 (CH2), 52.39 (CH3CH2CH2), 56.22 (OCH3), 113.57 (C-3), 120.72 (C-4′a), 121.29 (C-6), 121.72 (C-4), 125.43 (C-5′), 127.13 (C-6′), 127.33 (C-5), 128.15 (C-1′′), 129.24 (C-2′′ and C-6′′), 129.53 (C-3′′ and C-5′′), 130.03 (C-4′′), 134.78 (C-8′), 135.60 (C-7′), 137.86 (C-1), 142.10 (C-8′a), 145.78 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 152.60 (C-2), 157.86 (C-2′), 159.68 (CO-quinazolinone), 168.48 (CONH); anal. calc. for C27H25ClN4O6S2 (601.09): C, 53.95; H, 4.19; N, 9.32. Found: C, 53.88; H, 4.31; N, 9.49.
N), 152.60 (C-2), 157.86 (C-2′), 159.68 (CO-quinazolinone), 168.48 (CONH); anal. calc. for C27H25ClN4O6S2 (601.09): C, 53.95; H, 4.19; N, 9.32. Found: C, 53.88; H, 4.31; N, 9.49.
3.2. Investigation of the antimicrobial susceptibility of the synthesized compounds
The efficiency of the prepared molecules to serve as bactericidal and fungicidal was evaluated following the standard procedure.44–48 More details about the experiment used were provided in the ESI file†.The study was approved by the Medical Research Ethical Committee of the National Research Centre (04461123).
3.3. Effect of the targeted synthesized compounds on the bacterial lipid peroxidation (LPO)
Oxidative stress on the bacterial cells was usually indicated by the oxidation of the fatty acid contents in the bacterial cell membrane.49 Additional information is provided in the supplemental data.3.4. Biofilm inhibition activity of the synthesized molecules
The compounds being inhibit the biofilm formation were also determined by the crystal violet method.50 More details about the procedure are provided in the supplemental data.3.5. Confocal laser scanning microscope
The effect of the most potent compounds against microbial pathogens was analyzed by confocal laser scanning microscopy (Leica Microsystems DMi8, GmbH, Wetzlar, Germany). Following the standard process.51,52 More details about the procedure are provided in the supplemental data.3.6. Docking studies on DNA gyrase enzyme
We conducted a molecular docking investigation to assess how a series of newly synthesized quinazolinone derivatives (5a–r) interact with the DNA gyrase enzyme (PDB code: 6F86). These synthesized compounds' binding interactions and affinities towards the DNA gyrase enzyme were analyzed. The study commenced by retrieving the X-ray crystal structure of the DNA gyrase enzyme from the Protein Data Bank (https://www.rcsb.org) using the specified PDB code (6F86). To prepare the receptor for docking simulations, Biovia Discovery Studio was employed to streamline the enzyme structure by removing redundant chains and water molecules. This process included the addition of polar hydrogens and the adjustment of partial charges. Subsequently, docking studies were performed using Autodock 4.2 software, with a detailed protocol provided in the supplemental data. The grid box used for docking was set to dimensions of 60 × 60 × 60 points with a grid spacing of 0.375 Å. The box was centered on the coordinates of the native ligand (CWW) in the crystal structure to ensure that the entire binding pocket was encompassed. The center of the grid was positioned at X: 61.8154, Y: 28.8269, and Z: 63.8559 Å. This setup ensures the grid adequately covers the active site for accurate docking predictions. For each docking run, 100 poses were generated using the Lamarckian Genetic Algorithm in AutoDock 4.2. These poses were ranked based on their binding free energy, and the lowest-energy pose was selected as the most likely binding conformation.4. Conclusion
In conclusion, the present study describes the development of a novel series of quinazolinone–alkanesulfonate derivatives (5a–r) via the attachment of aryl alkanesulfonates to the quinazolinone core through thioacetohydrazide azomethine linkers. The antimicrobial evaluation of these newly synthesized compounds revealed reasonable bactericidal activities compared to standard drugs. Notably, derivatives 5g and 5k exhibited the greatest minimum inhibitory concentration (MIC) values against Candida albicans, while 5g was the most effective against Staphylococcus aureus with MIC = 12.5 μg mL−1, which is half the value recorded for the standard drug Sulfadiazine. Furthermore, 5k significantly prevented biofilm formation for all tested bacterial pathogens, surpassing the performance of the standard drug Ciprofloxacin. Additionally, 5k caused elevated lipid peroxidation (LPO) when added to the tested microbial pathogens, and confocal laser scanning microscopy (CLSM) visualization revealed fewer live cells after treatment. The molecular docking studies against the DNA gyrase enzyme revealed strong binding interactions, particularly with the acid hydrazide core, which effectively interacted with key residues GLU50, ASN46, GLY77, and ASP136. These interactions highlight the core's critical role in binding affinity and align well with the observed antimicrobial efficacy of the compounds, suggesting their potential as effective antimicrobials.5 Limitation of the study
It is important to interpret the findings of this study with caution. Specifically, assessments for genotoxicity, mutagenicity, developmental toxicity, reproductive toxicity, neurotoxicity, and immunotoxicity must be conducted before considering the results for further studies, including in vivo, preclinical, or clinical applications. These additional assessments will ensure the reliability and applicability of the findings in real-world scenarios, thereby enhancing the validity of subsequent research and potential therapeutic use.Data availability
The authors declare that the data supporting the findings of this study are available within the paper. Should any raw data files be needed in another format they are available from the corresponding author upon a reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors extend their appreciation to Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2024R89) for funding this work.References
- E. Toner, A. Adalja, G. K. Gronvall, A. Cicero and T. V. Inglesby, Antimicrobial resistance is a global health emergency, Health Secur., 2015, 13, 153–155, DOI:10.1089/hs.2014.0088.
- G. Mancuso, A. Midiri, E. Gerace and C. Biondo, Bacterial Antibiotic Resistance: The Most Critical Pathogens, Pathogens, 2021, 10, 1310, DOI:10.3390/pathogens10101310.
- J. Scheres and K. Kuszewski, Ten Threats to Global Health in 2018 and 2019. A welcome and informative communication of WHO to everybody, Zeszyty Naukowe Ochrony Zdrowia, Zdrowie Publiczne i Zarzadzanie, 2019, 17, 2–8 CrossRef.
- S. K. Ahmed, S. Hussein, K. Qurbani, R. H. Ibrahim, A. Fareeq, K. A. Mahmood and M. G. Mohamed, Antimicrobial resistance: Impacts, challenges, and future prospects, J. Med. Surg. Public Health, 2024, 2, 100081–100089, DOI:10.1016/j.glmedi.2024.100081.
- M. A. Salam, M. Y. Al-Amin, M. T. Salam, J. S. Pawar, N. Akhter, A. A. Rabaan and M. A. A. Alqumber, Antimicrobial Resistance: A Growing Serious Threat for Global Public Health, Healthcare, 2023, 11(13), 1946–1966, DOI:10.3390/healthcare11131946.
- A. M. Almansour, M. A. Alhadlaq, K. O. Alzahrani, L. E. Mukhtar, A. L. Alharbi and S. M. Alajel, The Silent Threat: Antimicrobial-Resistant Pathogens in Food-Producing Animals and Their Impact on Public Health, Microorganisms, 2023, 11(9), 2127, DOI:10.3390/microorganisms11092127.
- C. Miranda, V. Silva, R. Capita, C. Alonso-Calleja, G. Igrejas and P. Poeta, Implications of antibiotics use during the COVID-19 pandemic: present and future, J. Antimicrob. Chemother., 2020, 75, 3413–3416, DOI:10.1093/jac/dkaa350.
- R. Nieuwlaat, L. Mbuagbaw, D. Mertz, L. L. Burrows, D. M. Bowdish, L. Moja, G. D. Wright and H. J. Schünemann, Coronavirus disease 2019 and antimicrobial resistance: parallel and interacting health emergencies, Clin. Infect. Dis., 2021, 72, 1657–1659, DOI:10.1093/cid/ciaa773.
- M. Bassetti and D. R. Giacobbe, A look at the clinical, economic, and societal impact of antimicrobial resistance in 2020, Expert Opin. Pharmacother., 2020, 21, 2067–2071, DOI:10.1080/14656566.2020.1802427.
- L. Toro-Alzate, K. Hofstraat and D. H. de Vries, The pandemic beyond the pandemic: a scoping review on the social relationships between COVID-19 and antimicrobial resistance, Int. J. Environ. Res. Public Health, 2021, 18, 8766, DOI:10.3390/ijerph18168766.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review, Front. Microbiol., 2019, 10, 539, DOI:10.3389/fmicb.2019.00539.
- G. M. Rossolini, F. Arena, P. Pecile and S. Pollini, Update on the antibiotic resistance crisis, Curr. Opin. Pharmacol., 2014, 18, 56, DOI:10.1016/j.coph.2014.09.006.
- R. J. Fair and Y. Tor, Antibiotics and bacterial resistance in the 21st century, Perspect. Medicin. Chem., 2014, 6 DOI:10.4137/PMC.S14459 PMC. S14459, doi:.
- A. M. Borcea, I. Ionuţ, O. Crişan and O. Oniga, An overview of the synthesis and antimicrobial, antiprotozoal, and antitumor activity of thiazole and bisthiazole derivatives, Molecules, 2021, 26, 624, DOI:10.3390/molecules26030624.
- M. Miethke, M. Pieroni, T. Weber, M. Brönstrup, P. Hammann, L. Halby, P. B. Arimondo, P. Glaser, B. Aigle and H. B. Bode, Towards the sustainable discovery and development of new antibiotics, Nat. Rev. Chem., 2021, 5, 726–749, DOI:10.1038/s41570021003131.
- B. Liu, D. Jiang and G. Hu, The antibacterial activity of isatin hybrids, Curr. Top. Med. Chem., 2022, 22, 25–40, DOI:10.2174/1568026621666211116090456.
- S. Kumar, S. M. Lim, K. Ramasamy, V. Mani, S. A. A. Shah and B. Narasimhan, Design, synthesis, antimicrobial and cytotoxicity study on human colorectal carcinoma cell line of new 4,4-(1,4-phenylene) bis (pyrimidin-2-amine) derivatives, Chem. Cent. J., 2018, 12, 1–13, DOI:10.1186/s1306501804403.
- A. M. Alsibaee, H. M. Al-Yousef and H. S. Al-Salem, Quinazolinones, the Winning Horse in Drug Discovery, Molecules, 2023, 28(3), 978, DOI:10.3390/molecules28030978.
- A. M. Srour, D. H. Dawood and D. O. Saleh, Synthesis, 3D-pharmacophore modelling and 2D-QSAR study of new pyridine-3-carbonitriles as vasorelaxant active agents, New J. Chem., 2021, 45, 7731–7740, 10.1039/D0NJ06319C.
- A. A. Radwan, F. K. Alanazi, Biological Activity of Quinazolinones. Quinazolinone and Quinazoline Derivatives, IntechOpen, 2019, DOI:10.5772/intechopen.90621.
- I. Khan, S. Zaib, S. Batool, N. Abbas, Z. Ashraf, J. Iqbal and A. Saeed, Quinazolines and quinazolinones as ubiquitous structural fragments in medicinal chemistry: An update on the development of synthetic methods and pharmacological diversification, Bioorg. Med. Chem., 2016, 24(11), 2361–2381, DOI:10.1016/j.bmc.2016.03.031.
- A. Urbina, R. Lira, G. Visbal and J. Bartrolí, In vitro antiproliferative effects and mechanism of action of the new triazole derivative UR-9825 against the protozoan parasite trypanosoma (Schizotrypanum) cruzi, Antimicrob. Agents Chemother., 2000, 44(9), 2498–2502, DOI:10.1128/aac.44.9.2498-2502.2000.
- S. J. Marcroft and T. D. Potter, The fungicide fluquinconazole applied as a seed dressing to canola reduces Leptosphaeria maculans (blackleg) severity in south-eastern Australia. Australas, Plant Pathol., 2008, 37, 396–401, DOI:10.1071/AP08016.
- S. Gatadi, T. V. Lakshmi and S. Nanduri, 4(3H)-Quinazolinone derivatives: Promising antibacterial drug leads, Eur. J. Med. Chem., 2019, 15, 157–172, DOI:10.1016/j.ejmech.2019.03.018.
- S. Rollas and S. G. Küçükgüzel, Biological Activities of Hydrazone Derivatives, Molecules, 2007, 12, 1910–1939, DOI:10.3390/12081910.
- M. A. Omar, R. A. El-Shiekh, D. H. Dawood, A. Temirak and A. M. Srour, HydrazoneSulfonate Hybrids as Potential Cholinesterase Inhibitors: Design, Synthesis and Molecular Modelling Simulation, Fut, Med. Chem., 2023, 15(24), 2269–2287, DOI:10.4155/fmc-2023-0238.
- S. G. Komurc, S. Rollas, M. Ulgen, J. W. Gorrod and A. Çevikbas, Evaluation of some arylhydrazones of p-aminobenzoic acid hydrazide as antimicrobial agents and their in vitro hepatic microsomal metabolism, Boll. Chim. Farm., 1995, 134, 375–379 Search PubMed PMID: 7546542..
- M. Ulgen, B. B. Durgun, S. Rollas and J. W. Gorrod, The in vitro hepatic microsomal metabolism of benzoic acic benzylidenehydrazide, Drug Metab. Drug Interact., 1997, 13, 285–294, DOI:10.1515/DMDI.1997.13.4.285.
- L. Popiołek, Hydrazide–hydrazones as potential antimicrobial agents: overview of the literature since 2010, Med. Chem. Res., 2017, 26, 287–301, DOI:10.1007/s00044-016-1756-y.
- S. Chen, Y. Zhang, Y. Liu and Q. Wang, Design, synthesis, acaricidal activities, and structure-activity relationship studies of novel oxazolines containing sulfonate moieties, J. Agric. Food Chem., 2019, 11 67(49), 13544–13549, DOI:10.1021/acs.jafc.9b05547.
- E. H. Glass, Current Status of Pesticide Resistance in Insects and Mites Attacking Deciduous Orchard Crops, Research Progress on Insect Resistance, Chapter 5, 1974, DOI:10.4182/ECEM7264.II-1.17.
- S. Ramalingam, S. Periandy, S. Sugunakala, T. Prabhu and M. Bououdina, Insilico molecular modeling, docking and spectroscopic [FT-IR/FT-Raman/UV/NMR] analysis of Chlorfenson using computational calculations, Spectrochim Acta A Mol. Biomol. Spectrosc., 2013, 115, 118–135, DOI:10.1016/j.saa.2013.06.034.
- R. Patel and T. P. Busulfan, [Updated 2022 Oct 17], n. StatPearls [Internet], Treasure Island (FL), StatPearls Publishing, 2024 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK555986/ Search PubMed.
- I. V. Nechepurenko, E. D. Shirokova, M. V. Khvostov, T. S. Frolova, O. I. Sinitsyna and A. M. Maksimov, Synthesis, hypolipidemic and antifungal activity of tetrahydroberberrubine sulfonates, Russ. Chem. Bull., 2019, 68, 1052–1060, DOI:10.1007/s11172-019-2519-y.
- S. S. Ragab, A. M. Sweed and A. Srour, Synthesis and antiproliferative properties of schiff base hydrazones based on a spirocyclic pyrimidine, Chemistryselect, 2024, e202400161, DOI:10.1002/slct.202400161.
- E. A. Kazancioğlu, M. Güney, M. Şentürk and C. T. Supuran, Simple methanesulfonates are hydrolyzed by the sulfatase carbonic anhydrase activity, J. Enzyme Inhib. Med. Chem., 2012, 27(6), 880–885, DOI:10.3109/14756366.2011.637202.
- S. Gatadi, T. V. Lakshmi and S. Nanduri, 4(3H)-Quinazolinone derivatives: Promising antibacterial drug leads, Eur. J. Med. Chem., 2019, 170, 157–172, DOI:10.1016/j.ejmech.2019.03.018.
- D. Murugesan, P. C. Ray and T. Bayliss, et al., 2-Mercapto-Quinazolinones as Inhibitors of Type II NADH Dehydrogenase and Mycobacterium tuberculosis: Structure-Activity Relationships, Mechanism of Action and Absorption, Distribution, Metabolism, and Excretion Characterization, ACS Infect Dis, 2018, 4(6), 954–969, DOI:10.1021/acsinfecdis.7b00275.
- F. M. Sroor, A. F. El-Sayed and M. Abdelraof, Design, synthesis, structure elucidation, antimicrobial, molecular docking, and SAR studies of novel urea derivatives bearing tricyclic aromatic hydrocarbon rings, Arch. Pharm., 2024, e2300738, DOI:10.1002/ardp.202300738.
- H. T. Abdel-Mohsen, M. A. Omar, O. Kutkat, A. M. ElKerdawy, A. A. Osman, M. GabAllah, A. Mostafab, M. A. Ali and H. I. ElDiwani, Discovery of novel thioquinazoline-N-aryl-acetamide/N-arylacetohydrazide hybrids as anti-SARS-CoV-2 agents: Synthesis, in vitro biological evaluation, and molecular docking studies, J. Mol. Struct., 2023, 1276, 134690–134706, DOI:10.1016/j.molstruc.2022.134690.
- S. Narramore, C. E. M. Stevenson, A. Maxwell, D. M. Lawson and C. W. G. Fishwick, New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase, Bioorg. Med. Chem., 2019, 27(16), 3546–3550, DOI:10.1016/j.bmc.2019.06.015.
- BIOVIA, Dassault Systèmes, [BIOVIA Discovery Studio Visualizer], [2021], San Diego: Dassault Systèmes, [ 2023] Search PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. BelewK, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem., 2009, 30(16), 2785–2791, DOI:10.1002/jcc.21256.
- S. S. Ragab, A. M. Sweed, Z. K. Hamza, E. Shaban and A. A. El-Sayed, Design, synthesis, and antibacterial activity of spiropyrimidinone derivatives incorporated azo sulfonamide chromophore for polyester printing application, Fibers Polym., 2022, 23, 2114–2122, DOI:10.1007/s12221-022-4032-4.
- S. S. Ragab, M. Abdelraof, A. A. Elrashedy and A. M. Sweed, Design, synthesis, molecular dynamic simulation studies, and antibacterial evaluation of new spirocyclic aminopyrimidines, J. Mol. Struct., 2023, 1278, 134912, DOI:10.1016/j.molstruc.2023.134912.
- S. S. Ragab, N. E. Ibrahim, M. S. Abdel-Aziz, A. A. Elrashedy and A. K. Allayeh, Synthesis, biological activity, and molecular dynamic studies of new triazolopyrimidine derivatives, Results Chem., 2023, 6, 101163, DOI:10.1016/j.rechem.2023.101163.
- M. Mahran, N. A. Hassan, D. A. A. Osman, S. S. Ragab and A. A. Hassan, Synthesis and biological evaluation of novel pyrimidines derived from 6-aryl-5-cyano-2-thiouracil, Z. Naturforsch. C. J. Biosci., 2016, 71(5–6), 133–140, DOI:10.1515/znc-2015-0265.
- A. Sabt, M. T. Abdelrahman, M. Abdelraof and H. R. M. Rashdan, Investigation of novel mucorales fungal inhibitors: synthesis, In-silico study and anti-fungal potency of novel class of coumarin-6-sulfonamides-thiazole and thiadiazole hybrids, ChemistrySelect, 2022, 7(17), e202200691, DOI:10.1002/slct.202200691.
- S. X. T. Lianga, L. S. Wongb, Y. M. Limc, P. F. Leed and S. Djearamanea, Effects of Zinc Oxide nanoparticles on Streptococcus pyogenes, S. Afr. J. Chem. Eng., 2020, 34, 63–71, DOI:10.1016/j.sajce.2020.05.009.
- M. Abdelraof, M. S. Hasanin, M. M. Farag and H. Y. Ahmed, Green synthesis of bacterial cellulose/bioactive glass nanocomposites: Effect of glass nanoparticles on cellulose yield, biocompatibility and antimicrobial activity, Int. J. Biol. Macromol., 2019, 138, 975–985, DOI:10.1016/j.ijbiomac.2019.07.144.
- M. A. El-Bendary, M. Abdelraof, M. E. Moharam, E. M. Elmahdy and M. A. Allam, Potential of silver nanoparticles synthesized using low active mosquitocidal Lysinibacillus sphaericus as novel antimicrobial agents, Prep. Biochem. Biotech., 2021, 51(9), 925–935, DOI:10.1080/10826068.2021.1875236.
- M. M. Qader, A. A. Hamed, S. Soldatou, M. Abdelraof, M. E. Elawady, A. S. I. Hassane, L. Belbahri, R. Ebel and M. E. Rateb, Antimicrobial and antibiofilm activities of the fungal metabolites isolated from the marine Endophytes Epicoccum nigrum M13 and Alternaria alternata 13A, Mar. Drugs., 2021, 19(4), 323, DOI:10.3390/md19040232.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra07563c |
| This journal is © The Royal Society of Chemistry 2025 |




