Open Access Article
Open Access ArticleCarbon dots with red emission as nanoprobe for sensing of heparin in biofluids and pharmaceutical samples†
Mohammad Safarpoora,
Rassoul Dinarvand *a,
Mehrorang Ghaedi
*a,
Mehrorang Ghaedi *b and
Arash Asfaram
*b and
Arash Asfaram c
c
aNanotechnology Research Center, Faculty of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran. E-mail: dinarvand@tums.ac.ir
bChemistry Department, Yasouj University, Yasouj 75918-74831, Iran. E-mail: m_ghaedi@mail.yu.ac.ir
cMedicinal Plants Research Center, Yasuj University of Medical Sciences, Yasuj, Iran
First published on 23rd January 2025
Abstract
Heparin (HEP) is one of the oldest anticoagulant drugs, widely used in clinical settings, particularly in surgery and dialysis machines. Despite its long history, it remains extensively employed in medical practice. This study introduces a selective and cost-effective method for the rapid detection of HEP using red-emission carbon dots (R-CDs). These R-CDs were synthesized through a one-pot hydrothermal method, utilizing neutral red and thiourea for photostability, biocompatibility, and low cytotoxicity. Key parameters affecting the sensing process, including nanoprobe volume, pH, buffer type, and incubation time, were optimized to achieve potential assay conditions. The fluorescence intensity of the nanoprobe at 625 nm gradually decreased as the concentration of HEP increased from 60 to 240 nM. These changes in fluorescence intensity showed a linear relationship with HEP concentration within this range, achieving a limit of detection (LOD) of 5 nM. The proposed nanoprobes facilitate both quantitative and qualitative non-invasive analysis of HEP in various human biofluids, suggesting their potential for broader bioanalytical applications.
1. Introduction
Heparin (HEP) is a linear polysaccharide with high sulfation, characterized by a primary disaccharide repeating unit composed of linked iduronic acid and glucosamine residues.1 With its abundance of sulfo and carboxyl groups, HEP possesses the highest negative charge density among known biomolecules.2,3 This biomolecule uniquely binds to antithrombin, accelerating the interaction between antithrombin and thrombin, leading to thrombin-inactivation blood coagulation.4 Consequently, HEP is extensively applied in clinical settings as an anticoagulant. However, the improper use of HEP can lead to adverse effects, such as heparin-induced thrombocytopenia (HIT).5 Moreover, the precise monitoring of HEP dosage is critical during treatment, as overdosing can result in severe complications, including hemorrhage and osteoporosis.6 For cardiovascular surgery, a dosage range of 2–8 U mL−1 is considered appropriate, while for postoperative and long-term treatment, a range of 0.2–1.2 U mL−1 is recommended.Currently, HEP dosage in clinical settings is typically monitored using activated clotting time (ACT), activated partial thromboplastin time (aPTT), or anti-factor Xa assays.7,8 Although these methods are well established, they have inherent limitations, such as indirect measurement and lack of precision, underscoring the need for novel analytical methods to measure HEP concentrations in blood samples.9,10
HEP injections prevent the formation of harmful blood clots in blood vessels, the heart, and the lungs, playing a vital role during surgery, such as open-heart and bypass procedures. They are also crucial for kidney dialysis and blood transfusions. Although commonly referred to as a “blood thinner”, HEP does not dissolve clots, but prevents their growth. HEP is also used in diagnosing and treating severe blood disorders.11 Recent studies suggest that HEP also exhibits anti-tumor properties and hypolipidemic effects. Given the varying dosages required for surgical cardiovascular treatment and long-term therapy, the accurate and sensitive measurement of HEP levels is essential.12
Several sensors for HEP detection have been reported (Table 1), including HPLC/resonance light scattering detection,13 fluorometry14–18 and colorimetry.19–21 While effective, these methods have limitations such as high cost, complexity, time consumption, environmental hazards, and the need for rare reagents, emphasizing the necessity for simpler analytical methods for HEP determination.22 HEP detection could benefit from newly developed sensing probes capable of measuring various non-fluorescent compounds.23–27
| Method | LOD | LDR | Time (min) | Sample | Ref. |
|---|---|---|---|---|---|
| HPLC | 0.2 mg L−1 | 0.5–10 mg L−1 | — | Serum | 13 |
| Spectrofluorimetry | 8.2 nM | 10–100 nM | — | Injection form | 18 |
| Spectrofluorimetry | 1.41 μg mL−1 | 6–20 μg mL−1 | 3.5 | Injection form | 17 |
| Spectrofluorimetry | 10 μg mL−1 | 14.4–72 μg mL−1 | 5 | Serum | 16 |
| Spectrofluorimetry | 0.01 mg mL−1 | 0.01–4 mg mL−1 | — | Urine | 15 |
| Spectrofluorimetry | 0.4 μM | 0–16 μM | — | Injection form | 14 |
| Colorimetry | 90 nM | 0–60 μM | — | Water | 21 |
| Colorimetry | 5 ng mL−1 | 0.02–0.28 μg mL−1 | — | Water | 20 |
| Colorimetry | 30 ng mL−1 | 0.09–3.12 μg mL−1 | 15 | Serum | 19 |
| Spectrofluorimetry | 5 nM | 60–240 nM | 4 | Urine, serum, injection form | This study |
Carbon dots (CDs), a novel class of quantum dots (QDs), are fluorescent, zero-dimensional carbon nanoparticles less than 10 nm in size. They show significant potential in various fields due to their excellent properties, such as low or non-toxicity,28 optical and chemical stability,29 biocompatibility, environmental friendliness,30 and high water solubility.31,32 CDs are especially valuable in sensing research, as their engineered surface functional groups provide specific molecular recognition capabilities. However, most reported CDs emit blue fluorescence, and their fluorescence is often quenched in the solid state due to aggregation. Red-emission carbon dots (R-CDs) offer advantages for various applications, but pure R-CDs with red emission in both solid state and aqueous solution have rarely been reported. Addressing these challenges, this study synthesizes R-CDs and applies them as nanoprobes for detecting and quantifying HEP in human biofluids such as serum and urine, as well as in injection samples.
2. Experimental
2.1. Materials and apparatus
Details of the materials and apparatus are provided in the ESI.†2.2. Synthesis of R-CDs
R-CDs were synthesized using a hydrothermal method. Briefly, a solution containing 0.1 g of thiourea and 0.1 g of neutral red (NR) dissolved in 20 mL of ultrapure water was prepared in a 50 mL Teflon-lined autoclave. The reaction mixture was heated at 180 °C for 3 hours. After the reaction, the crude product was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 minutes to remove impurities. The resulting clear supernatant was purified using dialysis bags (1000 Da) with ultrapure water for 48 hours. Finally, the R-CD solution was stored at 4 °C for further use.33
000 rpm for 20 minutes to remove impurities. The resulting clear supernatant was purified using dialysis bags (1000 Da) with ultrapure water for 48 hours. Finally, the R-CD solution was stored at 4 °C for further use.33
2.3. HEP detection procedure
Fluorescence analysis was conducted as follows: 800 μL of R-CDs was dissolved in phosphate buffer (PB) solution (10 mM, pH 6.0). Different aliquots (12–48 μL) of HEP stock solution (1000 nM) were added to the prepared solution, and the final volume was adjusted to 2 mL with Milli-Q grade DI water. Four minutes after the final addition of HEP, the solution was transferred to a cell, and the fluorescence spectrum was recorded under excitation at 530 nm.2.4. The measurement of interference
To evaluate the selectivity of the developed sensor for HEP, various substrates, including adenosine triphosphate (ATP), NaCl, Na2HPO4, Na2CO3, NaAc, glucose and sucrose, ascorbic acid, dopamine and cysteine (each at a concentration of 1200 nM), were mixed with 800 μL of R-CDs and 200 μL of phosphate buffer (PB) solution (pH 6.0, 10 mM). The final volume was adjusted to 2 mL with DI water. Under optimal conditions, the fluorescence spectrum was recorded with excitation at 530 nm.2.5. Preparation of real samples
Urine and serum samples were collected from healthy, fasting 30 year-old female participants in accordance with ethical guidelines. The samples were separately centrifuged at 4000 rpm for 15 minutes. Then, 100 μL of each centrifuged sample was diluted with deionized (DI) water in 10 mL volumetric flasks. HEP was added to these diluted samples to achieve final concentrations of 60, 160, and 240 nM. Additionally, a HEP sodium injection was diluted with deionized (DI) water to a final concentration of 1000 nM. All prepared samples were tested using the developed sensor, with each measurement performed in triplicate.3. Results and discussion
3.1. Characteristics of the R-CDs
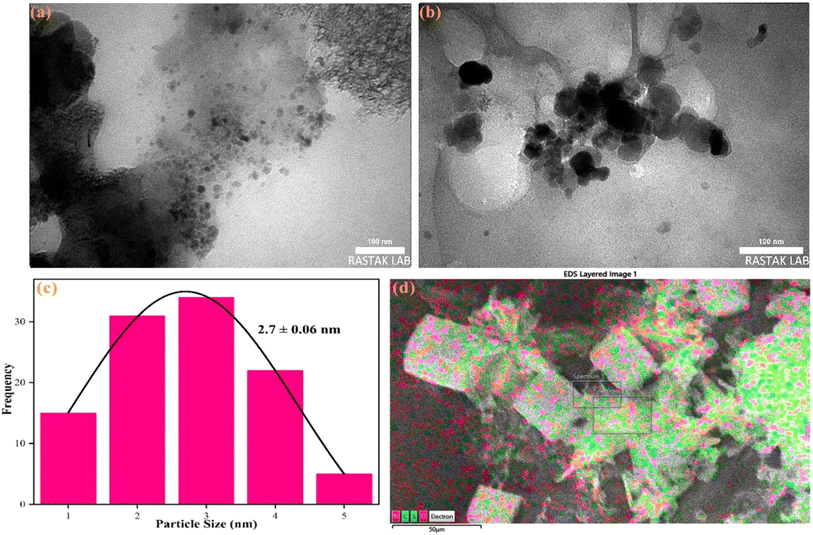
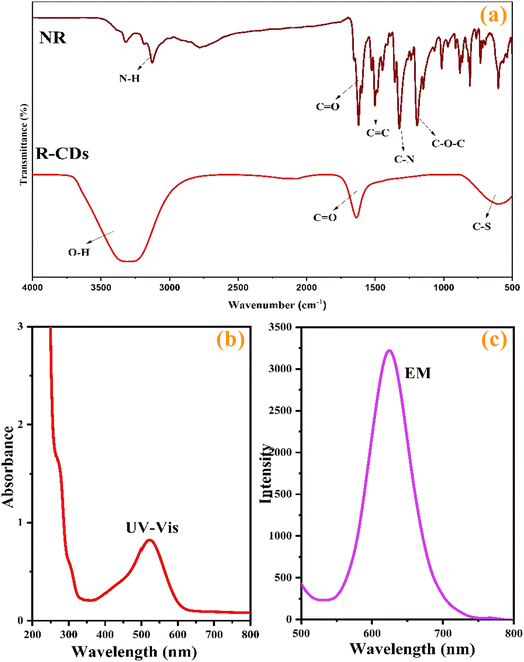
Various analyses were conducted to characterize the structural and morphological features of the synthesized nanoprobes. TEM images of R-CDs at different magnifications (Fig. 1a and b) show that the R-CDs are uniformly dispersed in spherical shapes without significant aggregation. This uniform distribution confirms the successful synthesis of the nanostructures. The average nanoparticle size was estimated to be 2.7 ± 0.06 nm, as depicted in the histogram of the R-CDs (Fig. 1c). EDX mapping (Fig. 1d) revealed the presence of carbon (C), oxygen (O), nitrogen (N), and sulfur (S) in the synthesized nanoprobes.To further investigate the functional groups in the synthesized product compared to the primary substances, FTIR spectra of natural red (NR) and the synthesized R-CDs were recorded and are shown in Fig. 2a. Peaks around 3496 cm−1, 1720 cm−1, and 1210 cm−1 correspond to the stretching vibrations of O–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and C–O–C, respectively.34 The peak at 3296 cm−1 represents the stretching vibrations of N–H. Additionally, peaks at 1551 cm−1 and 1412 cm−1 indicate the presence of C
O, and C–O–C, respectively.34 The peak at 3296 cm−1 represents the stretching vibrations of N–H. Additionally, peaks at 1551 cm−1 and 1412 cm−1 indicate the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C–N bonds.35 Absorption peaks around 618 cm−1, 1106 cm−1, and 2918 cm−1 correspond to the stretching vibrations of C–S, C–O, and C–H, respectively.33,36
C and C–N bonds.35 Absorption peaks around 618 cm−1, 1106 cm−1, and 2918 cm−1 correspond to the stretching vibrations of C–S, C–O, and C–H, respectively.33,36
The optical properties of the R-CDs were examined using UV-vis absorption (Fig. 2b) and emission spectra (Fig. 3b). As shown in Fig. 2b, the UV-vis absorption spectrum of the R-CDs displays a broad absorption band spanning from 400 to 600 nm, with a maximal absorption peak at 530 nm. This peak is attributed to the n–π* transitions of surface states containing C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–O/C–N, and C–S structures,37,38 The R-CDs display optimal emission at 625 nm when excited at 530 nm (Fig. 3b). Therefore, 530 nm was chosen as the excitation wavelength for all experiments to ensure high sensitivity.
O, C–O/C–N, and C–S structures,37,38 The R-CDs display optimal emission at 625 nm when excited at 530 nm (Fig. 3b). Therefore, 530 nm was chosen as the excitation wavelength for all experiments to ensure high sensitivity.
 | ||
| Fig. 3 PL spectra of the different solutions: R-CDs, HEP (100 nM), a mixture of R-CDs and PB (20 mM) with pH of 6.0 in the absence and the presence of HEP. | ||
3.2. HEP detection using R-CDs
The primary objective of this study is to develop a novel strategy for HEP measurement using R-CDs as the nanoprobe. To evaluate the role of R-CDs in this approach, photoluminescence (PL) spectra for each substrate and their mixtures were recorded within the range of 500 nm to 800 nm. As shown in Fig. 3, a solution containing 100 nM of HEP does not exhibit emission spectra under 530 nm excitation, whereas a solution of R-CDs shows a peak at 625 nm under the same excitation. A mixture of R-CDs and phosphate buffer (PB, 20 mM, pH 6.0) exhibits a peak at 620 nm. The addition of 180 nM of HEP to this mixture results in a decrease in the intensity of the emission spectra, likely due to the interaction between buffer ions and the surface states of R-CDs, indicating the aggregation of R-CDs upon the introduction of HEP. A schematic illustration suggesting the selective detection mechanism of HEP by R-CDs is shown in Scheme 1.3.3. Optimization of HEP detection using R-CDs
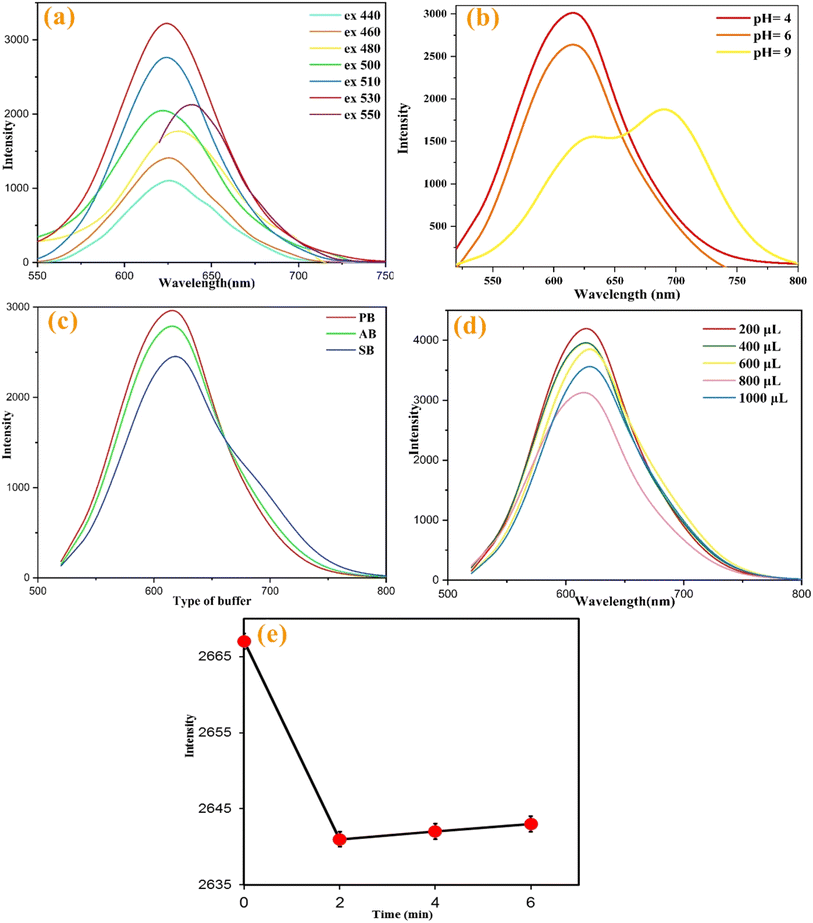
To achieve a stable maximum response, various analytical parameters, including excitation wavelength, pH, buffer type, nanoprobe volume, and incubation time for HEP detection, were optimized. Fig. 4a illustrates the fluorescence emission of the R-CD solution under different excitation wavelengths (λex). Increasing λexw from 440 to 530 nm enhances the fluorescence emission intensity (λem around 625 nm); thus 530 nm was selected for maximum sensitivity. According to a recent study,30 pH significantly influences the fluorescence intensity of R-CDs (Fig. 4b). Fluorescence intensity increases from pH 2.0, peaks at pH 6.0, and then decreases up to pH 9. Therefore, measurements for HEP were performed at pH 6.0. Different 20 mM buffers (phosphate (PB), citrate (CB), and acetate (AB) buffers), at pH 6.0 were tested in a mixture of R-CDs (800 μL) and HEP (12 μL, 60 nM). The fluorescence emission spectra indicated that PB provided the best medium for HEP detection (Fig. 4c). The effect of R-CD volume on sensor response in 20 mM PB (pH 6.0) containing HEP (60 nM) was examined, showing a decrease in intensity with the volume increasing from 200 μL to 800 μL, followed by an increase from 800 μL to 1000 μL. Since 800 μL of R-CDs provided a stable minimum signal and demonstrated good interaction with HEP, this volume was chosen for further experiments (Fig. 4d). The effect of reaction time on 20 mM PB (pH 6.0) containing HEP (140 nM) and 800 μL of R-CDs showed that the intensity decreased upon HEP addition up to 2 minutes, after which it remained unchanged. Thus, 4 minutes was deemed sufficient for HEP detection (Fig. 4e).3.4. HEP detection
One notable advantage of this study lies in its utilization of simple, cost-effective, and environmentally friendly materials for HEP assays. The detection process involved the straightforward addition of R-CDs as the nanoprobe under mildly acidic conditions. To assess the impact of increasing HEP concentration, varying concentrations of HEP were introduced to 800 μL of the buffered nanoprobe under optimized conditions. Fluorescence spectra were then recorded from 500 to 800 nm (Fig. 5a). As depicted in Fig. 5b, in the absence of HEP, the fluorescence intensity of the R-CDs remains robust in aqueous solution. However, the fluorescence intensity at 620 nm gradually diminishes with HEP concentration increasing from 60 nM to 240 nM, fitting the regression equation I (intensity in the presence of analyte)/I0 (intensity in the absence of analyte) = −0.0017 [HEP] + 1.1084, with a high correlation coefficient (R2) of 0.9927. The limit of detection (LOD) was determined to be 5 nM based on S/N = 3. These calculated linear dynamic range (LDR) and LOD values are comparable to or exceed those reported for several sensors used in HEP detection across various real samples. The intense fluorescence of R-CDs in aqueous solution could be attributed to surface groups, such as hydroxyl, carboxyl, sulfur, or nitrogen. These groups likely interact with the negative sulfate and carboxylate groups present in HEP via electrostatic interactions, resulting in R-CD aggregation and fluorescence quenching.18,333.5. Interference study
To evaluate selectivity, potential interfering substrates, including adenosine triphosphate (ATP), ATP, NaCl, Na2HPO4, Na2CO3, NaAc, glucose, sucrose, ascorbic acid, dopamine and cysteine, were prepared and tested using the proposed sensor. The results presented in Fig. 5c indicate that no species, at higher concentrations than HEP (60 nM), show any significant interference. Thus, the sensor is acceptably selective for HEP.3.6. Detection of HEP in actual samples
To assess the practicality of the sensor in authentic samples, urine, human serum, and tablet specimens were prepared as outlined. Subsequently, 200 μL of each prepared sample was introduced to the R-CDs under optimized conditions, and the corresponding fluorescence spectra were captured using the standard addition technique. As detailed in Table 2, the determined concentrations of HEP in all samples closely matched the actual concentrations. The calculated recovery rates ranged from 95.41% to 106.71%, with RSD values below 3.00%, confirming the satisfactory feasibility of the developed sensor for HEP detection in real-world samples.| Real sample | Spiked (nM) | Found (nM) | Recovery (%) |
|---|---|---|---|
| Urine | 0.00 | 0.00 | 0.00 |
| 100.0 | 99.78 | 99.78 | |
| 160.0 | 152.0 | 95.41 | |
| 240.0 | 244.0 | 102.0 | |
| Serum | 0.00 | 0.00 | 0.00 |
| 100.0 | 99.47 | 99.47 | |
| 160.0 | 159.0 | 99.39 | |
| 240.0 | 240.9 | 100.4 | |
| HEP injection form | 0.00 | 6.10 | — |
| 100.0 | 106.7 | 106.7 | |
| 160.0 | 166.4 | 104.0 | |
| 240.0 | 232.9 | 97.00 |
4. Conclusion
In summary, R-CDs were created as nanoscale probes, and their physical and chemical characteristics were examined using various methods. Multiple factors influencing the detection were optimized. These economical probes, which perform well in slightly acidic environments, were employed for the swift and easy identification of HEP over a wide concentration range. The straightforward and immediate monitoring of HEP in real human and drug samples showcases the promising analytical capabilities of the developed sensor.Data availability
Data will be made available if requested. We are ready to make data available at any time.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The authors appreciate the financial support from Tehran University of Medical Sciences, Tehran, Iran, to conduct this study.References
- J. Hirsh and R. Raschke, Chest, 2004, 126, 188–203 CrossRef PubMed.
- S. Faham, R. E. Hileman, J. R. Fromm, R. J. Linhardt and D. C. Rees, Science, 1996, 271, 1116–1120 CrossRef CAS PubMed.
- R. E. Hurst, M. C. Poon and M. J. Griffith, J. Clin. Invest., 1983, 72, 1042–1045 CrossRef.
- R. D. Rosenberg and P. S. Damus, J. Biol. Chem., 1973, 248, 6490–6505 CrossRef PubMed.
- T. E. Warkentin, M. N. Levine, J. Hirsh, P. Horsewood, R. S. Roberts, M. Gent and J. G. Kelton, N. Engl. J. Med., 1995, 332, 1330–1335 CrossRef.
- A. G. Turpie, J. G. Robinson, D. J. Doyle, A. S. Mulji, G. J. Mishkel, B. J. Sealey, J. A. Cairns, L. Skingley, J. Hirsh and M. Gent, N. Engl. J. Med., 1989, 320, 352–357 CrossRef.
- D. J. Guervil, A. F. Rosenberg, A. G. Winterstein, N. S. Harris, T. E. Johns and M. S. Zumberg, Ann. Pharmacother., 2011, 45, 861–868 CrossRef.
- A. Liveris, R. A. Bello, P. Friedmann, M. A. Duffy, D. Manwani, J. S. Killinger, D. Rodriquez and S. Weinstein, Pediatr. Crit. Care Med., 2014, 15, e72–e79 CrossRef PubMed.
- S. Amemiya, Y. Kim, R. Ishimatsu and B. Kabagambe, Anal. Bioanal. Chem., 2011, 399, 571–579 CrossRef PubMed.
- L. Fan, D. Jia, W. Zhang and Y. Ding, Analyst, 2020, 145, 7809–7824 RSC.
- D. W. Hommes, A. Bura, L. Mazzolai, H. R. Büller and J. W. ten Cate, Ann. Intern. Med., 1992, 116, 279–284 CrossRef PubMed.
- S.-Y. Hung and W.-L. Tseng, Biosens. Bioelectron., 2014, 57, 186–191 CrossRef PubMed.
- Q. G. Liao, W. H. Li and L. G. Luo, Chromatographia, 2013, 76, 1677–1682 CrossRef.
- L. Cai, R. Zhan, K.-Y. Pu, X. Qi, H. Zhang, W. Huang and B. Liu, Anal. Chem., 2011, 83, 7849–7855 CrossRef PubMed.
- D. Liu, X. Guo, H. Wu and X. Chen, Spectrochim. Acta, Part A, 2024, 304, 123255 CrossRef CAS PubMed.
- C.-Y. Kuo and W.-L. Tseng, Chem. Commun., 2013, 49, 4607–4609 RSC.
- A. A. Abdella, A. M. Zaki, S. Hammad and F. R. Mansour, Spectrochim. Acta, Part A, 2024, 306, 123609 CrossRef CAS PubMed.
- R. Wang, X. Wang and Y. Sun, Spectrochim. Acta, Part A, 2017, 184, 187–193 CAS.
- R. Cao and B. Li, Chem. Commun., 2011, 47, 2865–2867 RSC.
- X. Fu, L. Chen, J. Li, M. Lin, H. You and W. Wang, Biosens. Bioelectron., 2012, 34, 227–231 CrossRef CAS PubMed.
- R. Zhan, Z. Fang and B. Liu, Anal. Chem., 2010, 82, 1326–1333 CrossRef CAS PubMed.
- R. Tabaraki and N. Sadeghinejad, Ecotoxicol. Environ. Saf., 2018, 153, 101–106 CrossRef CAS PubMed.
- L.-C. Feng, X.-X. Li, R. Chen, L.-L. Man and W.-K. Dong, J. Mol. Struct., 2023, 1289, 135941 CrossRef CAS.
- Y. Zhang, L. Tong, X. Li, L. Dou, H. Cheng, W.-K. Dong and Y.-J. Ding, J. Mol. Struct., 2023, 1289, 135868 CrossRef CAS.
- Y. X. Sun, W.-Y. Han, Z.-P. Deng, Y.-G. Sun, Y.-H. Jia, Y. Sun and S.-Z. Zhang, Inorg. Chim. Acta, 2023, 556, 121643 CrossRef CAS.
- Y.-X. Sun, Y.-G. Sun, Z.-P. Deng, Y.-H. Jia, W.-Y. Han, J.-J. Wang and Y. Sun, J. Mol. Struct., 2023, 1291, 136069 CrossRef CAS.
- M. Mabrouk, S. F. Hammad, A. A. Abdella and F. R. Mansour, Colloids Surf., A, 2021, 614, 126182 CrossRef.
- M. J. Molaei, Anal. Methods, 2020, 12, 1266–1287 RSC.
- R. Kumari and S. K. Sahu, Langmuir, 2020, 36, 5287–5295 CrossRef.
- S. Wu, W. Li, Y. Sun, X. Pang, X. Zhang, J. Zhuang, H. Zhang, C. Hu, B. Lei and Y. Liu, J. Mater. Chem. C, 2020, 8, 8935–8941 RSC.
- X. Yuan, W. Jiang, J. Wang, H. Liu and B. Sun, ACS Appl. Mater. Interfaces, 2020, 12, 25150–25158 CrossRef.
- L. Li, X. Wang, Z. Ma, L. Liu, H. Wang, W. Zhang and X. Li, Appl. Surf. Sci., 2019, 475, 109–116 CrossRef.
- L. Li, L. Shi, J. Jia, O. Eltayeb, W. Lu, Y. Tang, C. Dong and S. Shuang, Sens. Actuators, B, 2021, 332, 129513 CrossRef.
- P. Arabkhani, N. Saeedi, H. Sadeghi, S. Nouripour-Sisakht, M. Gharaghani and A. Asfaram, J. Water Process Eng., 2023, 54, 104020 CrossRef.
- E. Kasiri, P. Arabkhani, H. Haddadi, A. Asfaram and R. S. Varma, New J. Chem., 2022, 46, 21704–21716 RSC.
- W. Gao, H. Song, X. Wang, X. Liu, X. Pang, Y. Zhou, B. Gao and X. Peng, ACS Appl. Mater. Interfaces, 2018, 10, 1147–1154 CrossRef.
- G. Zuo, A. Xie, J. Li, T. Su, X. Pan and W. Dong, J. Phys. Chem. C, 2017, 121, 26558–26565 CrossRef.
- H. Yang, Y. Liu, Z. Guo, B. Lei, J. Zhuang, X. Zhang, Z. Liu and C. Hu, Nat. Commun., 2019, 10, 1789 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06655c |
| This journal is © The Royal Society of Chemistry 2025 |