Open Access Article
Open Access ArticleDesign, synthesis and biological activity of peptidyl β-nitrostyrenes as cysteine protease inhibitors against Leishmania donovani†
Sweta Sharmaa,
Mirza A. Begb,
Insha Latiefa,
Jyoti Abotia,
Samra Jamalc,
Pallavi Junejad,
Supriya Tanward,
Kalicharan Sharma e,
Sayeed ur Rehman*d,
Angamuthu Selvapandiyan*b and
Syed Shafi
e,
Sayeed ur Rehman*d,
Angamuthu Selvapandiyan*b and
Syed Shafi *a
*a
aDepartment of Chemistry, School of Chemical and Life Sciences, Jamia Hamdard, New Delhi, India. E-mail: syedshafi@jamiahamdard.ac.in
bDepartment of Molecular Medicine, School of Interdisciplinary Sciences and Technology, Jamia Hamdard, New Delhi 110062, India. E-mail: selvapandiyan@jamiahamdard.ac.in
cDepartment of Biotechnology, School of Chemical and Life Sciences, Jamia Hamdard, New Delhi, India
dDepartment of Biochemistry, School of Chemical and Life Sciences, Jamia Hamdard, New Delhi, India. E-mail: Sayeed.rehman@jamiahamdard.ac.in
eDepartment of Pharmaceutical Chemistry, ISF College of Pharmacy, Moga, Punjab, India
First published on 19th February 2025
Abstract
Cysteine proteases are essential for the survival of Leishmania parasites that cause several clinical forms of leishmaniases. Inhibiting cysteine protease can be a promising strategy against parasitic diseases because of their essential functions in the life cycles of these pathogens. The aim of the present study was to synthesize and evaluate peptidyl -nitrostyrenes as antipromastigote inhibitors against Leishmania donovani promastigotes. A library of 12 peptidyl β-nitrostyrenes was synthesized and evaluated for anti-promastigote activity. Most of the compounds exhibited comparable activity to the standard, with IC50 values ranging from 1.468 to 16.81 μM. Notably, compounds 14a, 14e, 14f, and 14g showed significant activity against both L. donovani promastigotes and intracellular amastigotes. Compounds 14e and 14f displayed superior anti-promastigote activity with IC50 values of 1.468 μM and 1.551 μM, respectively, compared to the standard (IC50 = 3.073 μM). Moreover, compounds 14e and 14f demonstrated better inhibitory potential against intracellular amastigotes, with IC50 values of 1.28 μM and 0.64 μM, respectively, outperforming AmphoB (IC50 = 3.07 μM). Additionally, compounds 14a and 14g showed negligible cytotoxicity to mammalian macrophages even at a concentration of 28 μM. Given their high activity, favorable safety profiles, and cost-effective synthesis, this class of compounds holds promise for the development of anti-leishmanial drugs.
1. Introduction
Leishmaniasis is a neglected tropical disease caused by parasites from the genus Leishmania. The WHO has identified it as one of 17 such diseases that pose a significant threat to mankind in tropical regions where over a billion people are directly exposed to tropical parasites.1 Major pharmaceutical companies have largely overlooked such diseases in their drug discovery programs due to the lack of financial return.1 The burden of Leishmaniasis is concentrated in a few countries, and environmental factors like massive migrations, urbanization, deforestation, and new irrigation schemes, as well as individual risk factors like HIV, malnutrition, and genetics, contribute to its continued prevalence.2,3 Leishmaniasis can manifest in various ways, with the most common forms being cutaneous and visceral, ranging from asymptomatic to chronic disease. The type of Leishmaniasis a person gets depends on various factors relating to the patient and the parasite causing the disease.4Cysteine proteases are important enzymes in parasitic organisms and are critical to their replication, virulence, and survival. They are particularly essential to Trypanosoma and Leishmania species, making them attractive targets for developing therapeutic agents against parasitic infections.5 Inhibiting cysteine proteases can be a promising strategy against parasitic diseases because of their essential functions in the life cycles of these pathogens. Researchers have been investigating this approach for years and have made significant strides in developing cysteine protease inhibitors.6,7
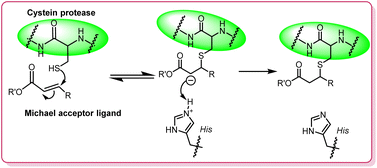
Animal trials have shown the potential of these inhibitors in treating diseases caused by parasites like malaria, Leishmania, and Trypanosoma cruzi. Leishmania secretes several cysteine proteases, including cathepsin L-like proteases, cathepsin B-like proteases, and papain-like proteases, which are involved in nutrient acquisition, host cell invasion, and immune evasion.8–10 The mechanism of action of cysteine proteases involves the deprotonation of a nucleophilic cysteine thiol in the enzyme's active site. Michael acceptors, which interact with these thiol groups through a Michael-type addition (Fig. 1), can inhibit the protease's activity, making them potent covalent inhibitors. This property has been exploited in designing inhibitors for leishmaniasis.11,12
The versatility and therapeutic potential of β-nitrostyrenes have been demonstrated across a range of applications, from antifungal agents to inhibitors of vital proteases like the SARS-CoV-2 3CL protease.7,13–15 Their effectiveness in disrupting the cellular processes of pathogens positions β-nitrostyrenes as valuable leads in the development of new antimicrobial agents.16,17 Syed Shafi and colleagues created a library of thirty β-nitrostyrenes and evaluated their efficacy against L. donovani, uncovering potent anti-leishmanial activity in both promastigote and amastigote forms. Notably, compound 1a, featuring a methoxy group at the para-position and a methyl substituent at the β-position, demonstrated remarkable anti-leishmanial effects, with IC50 values of 40.50 ± 1.47 nM (promastigotes) and 26.43 ± 2.71 nM (amastigotes). Similarly, compound 1b, distinguished by a hydroxy group at the para-position and an ethyl group at the β-position (Fig. 2), displayed significant activity, with IC50 values of 55.66 ± 2.84 nM (promastigotes) and 61.63 ± 8.02 nM (amastigotes).18 Ze-jun Jia et al. have reported β-nitrostyrenes as inhibitors of SARS-CoV-2 3CL protease, which is a key enzyme in the replication of the COVID-19 virus. The study identified seven β-nitrostyrene derivatives and found that 4-nitro-β-nitrostyrene, 2 showed the lowest IC50 values of 0.7297 μM.19 These findings position these β-nitrostyrenes as promising candidates for further development into anti-leishmanial therapeutics.
Recent studies have further established the significance of peptidyl hybrids as promising cysteine protease inhibitors (Fig. 3). In the realm of anti-leishmanial research, Ahmed H. E. Hassan and his team have made significant contributions with the synthesis of chromone-peptidyl hybrids as potential antileishmanial hits against visceral leishmaniasis. Three hybrids demonstrated notable IC50 values against L. donovani, with values of 9.8, 10, and 12 μM, respectively, rivalling that of erufosine (IC50 = 9.8 μM). Importantly, preliminary cytotoxicity assessments revealed that hybrids 3a and 3b were non-cytotoxic up to 100 μM, unlike erufosine, which exhibited a CC50 value of 19.4 μM. Furthermore, Hassan's group also identified rosmarinic acid-β-amino-α-ketoamide hybrids, with two notable compounds, 3c and 3d, showing promising antileishmanial activity and negligible cytotoxicity, marking them as potential candidates for further development. Additionally, Mayara Castro de Morais and colleagues evaluated the antileishmanial potential of cinnamic acid derivatives against L. infantum. Compounds 3e and 3f stood out, showing strong antileishmanial activity with IC50 values of 33.71 μM and 42.80 μM, respectively, and high selectivity indices, indicating their potential as safer, more targeted treatments compared to traditional drugs like amphotericin B. Compound S9 (3g) displayed excellent leishmanicidal activity20 against L. major promastigotes (IC50 = 37.4 μM) and amastigotes (IC50 = 2.3 μM). Compounds CPB2.8DCTE (3h) exhibited potential anti-leishmanial activity against Leishmania Mexicana cysteine protease (IC50 = 3.7 μM).21 The exploration of peptidyl β-nitrostyrenes and their biological evaluation as cysteine protease inhibitors represents a cutting-edge approach in the ongoing quest for effective treatments against parasitic diseases.
The rational approach for the designing the new molecules is shown in Fig. 4. The design of the final molecules was inspired by previous studies demonstrating the anti-leishmanial potential of β-nitrostyrenes. Syed Shafi and colleagues synthesized a library of β-nitrostyrenes and identified 4-hydroxy β-nitrostyrene with significant activity against Leishmania donovani.7,18 Building on these findings, we aimed to enhance their therapeutic potential by incorporating the peptidyl motif, which is known to increase selectivity for cysteine proteases. Previous reports have shown that Michael acceptor-type inhibitors, such as peptidyl nitroalkenes, effectively inhibit cysteine proteases like cruzipain and rhodesain with nanomolar potency.22 Additionally, compounds like K11777 and other α,β-unsaturated esters have been successfully used to target viral and parasitic proteases, further validating the use of Michael acceptors for enzyme inhibition.23–27
In the present study, we applied the hybrid conjugation of β-nitro styrene with different amino acids with diverse chemical structures allowing detailed characterization of the residue preference of the binding pocket within the substrate-binding site of the protease (Fig. 4).
2. Results and discussion
2.1 Chemistry
The designed peptidyl β-nitrostyrenes were prepared by employing a multistep synthetic strategy. The retrosynthetic analysis of the target molecule results in two important building blocks viz. fragment A, non-peptidyl β-nitrostyrene attached to a carboxylic acid linker and fragment B, a dipeptidyl moiety (Scheme 1).Fragment A (10) was prepared by reacting (E)-4-(2-nitrobut-1-en-1-yl)phenol (9) with bromoacetic acid in the presence of K2CO3. (E)-4-(2-Nitrobut-1-en-1-yl)phenol (9) was obtained by Henry reaction by reacting p-hydroxybenzaldehyde (8) with nitropropane using ammonium acetate as a base (Scheme 2).
Boc-protected fragment B was synthesised by coupling various Boc-protected amino acids (12a–k) with morpholine/thiomorpholine/amino acid methyl esters under EDC coupling conditions (Scheme 3). Finally, deprotection of the Boc group resulted in fragment B (13a–k) with the free –NH2 group which was directly used for further coupling reaction, without any purification.
Both the fragments were finally combined by employing EDC coupling to obtain the peptidyl β-nitrostyrenes (14a–k) as shown below in Scheme 4.
By employing the above multistep synthetic strategy, a library of eleven peptidyl β-nitrostyrenes were prepared, as depicted in Fig. 5. The formation of all the synthesized compounds was confirmed by various analytical techniques, including 1H NMR, 13C NMR, Mass and IR.
The formation of compound 9 was confirmed by the appearance of a characteristic vinyl proton signal at δ 8.0 ppm, ethyl signals at δ 2.88 and 1.28 ppm along with the aromatic signals in 1H NMR. The 1H NMR values for compound 10 are in correlation with the literature values.18 The appearance of two singlets, one corresponding to –COOH proton and the other for -CH2 at δ 13.2 and 4.7 ppm, respectively along with aromatic/aliphatic protons in the 1H NMR confirmed the formation of compound, 10. The formation of peptide fragments 12a–g was confirmed by the appearance of two multiplets between δ 3.73–3.56 and 2.87–2.70 ppm. Further, the formation of dipeptide fragments 12h–k was confirmed by the peaks in the aliphatic region and the –NH peak in 1H NMR spectrum. Finally, the formation of peptidyl β-nitrostyrenes (14a–k) through EDC coupling of 10 and 13(a–k) was confirmed by the missing peak for –COOH proton and the appearance of a new amide peak in the range of δ 3.00–4.00 ppm along with signals belonging to both fragments. The presence of signals at δ 169.51–115.45 ppm and 67.27–12.52 ppm in the 13C NMR further confirms the formation of a new amide bond between both fragments. Finally, the formation of all the peptidyl β-nitrostyrenes, 14a–k was confirmed by mass spectroscopy.
2.2 Pharmacology
| S no. | Test samples | % inhibition (48 h) |
|---|---|---|
| 1 | 14a | 84.8 |
| 2 | 14b | 83.7 |
| 3 | 14c | 82.4 |
| 4 | 14d | 80.8 |
| 5 | 14e | 80.4 |
| 6 | 14f | 77.9 |
| 7 | 14g | 78.1 |
| 8 | 14h | 82.2 |
| 9 | 14i | 84.5 |
| 10 | 14j | 65.5 |
| 11 | 14k | 54.8 |
| 12 | AmphoB | 78.5 |
| Molecules | IC50 (μM) |
|---|---|
| ● 14a | 11.87 |
| ■ 14b | 16.81 |
| ▲ 14c | 7.236 |
| ▼ 14d | 7.563 |
| ♦ 14e | 1.468 |
| ● 14f | 1.551 |
| ■ 14g | 14.42 |
| ▲ 14h | 10.01 |
| ▼ 14i | 5.298 |
| ♦ AmphoB | 3.073 |
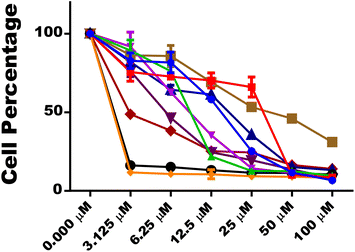
Compounds 14e and 14f demonstrated better anti-promastigote activity with IC50 values of 1.468 μM and 1.551 μM, respectively, compared to standard (IC50 = 3.073 μM).
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CC50 CC50 |
CC50 | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CC50 CC50 |
CC50 | ||
|---|---|---|---|---|---|
| 14a | 1.458 | 28.68 | 14f | 0.4741 | 2.979 |
| 14b | 0.7734 | 5.934 | 14g | 1.670 | 46.79 |
| 14c | 0.5684 | 3.701 | 14h | 1.109 | 12.86 |
| 14d | 0.5025 | 3.180 | 14i | 0.7120 | 5.153 |
| 14e | 0.3743 | 2.367 | STD | 1.029 | 10.69 |
Based on determined IC50 values against L. donovani promastigotes and cytotoxicity studies (Fig. 8), it might be stated that derivatives 14a, 14e, 14f and, 14g could act as non-toxic and potent antileishmanial hit compounds for further research.
| SAMPLE | Percentage Reductions (%) | |||
|---|---|---|---|---|
| 3.125 μM | 6.25 μM | 12.5 μM | IC50 (μM) | |
| 14a | 37.2 | 44.1 | 57.5 | 8.07 |
| 14e | 65.4 | 75.1 | 88.5 | 1.28 |
| 14f | 63.2 | 73.4 | 90.3 | 0.64 |
| 14g | 36.7 | 46.8 | 63.2 | 6.37 |
| AmphoB | 52.3 | 63.2 | 85.5 | 3.07 |
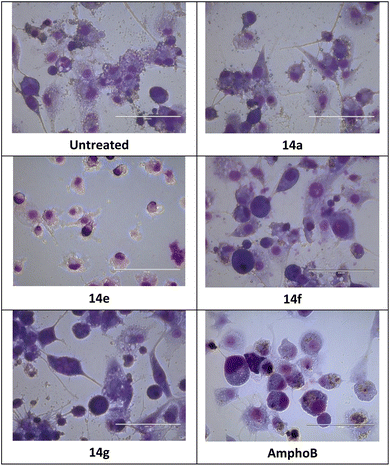
The intracellular amastigotes of L. donovani were inhibited by all four compounds, 14a, 14e, 14f and 14g. The inhibitory potential of compounds 14e and 14f with IC50 values of 1.28 μM and 0.64 μM was better than AmphoB with IC50 values of 3.07 μM.
The selectivity data in Table 5 demonstrates that 14a (SI 3.55), 14f (SI 4.65), and 14g (SI 7.34), were equally or more selective for L. donovani amastigotes than the host macrophages as compared to amphoterecin B (SI 3.48).
| Molecules | IC50 (μM) | CC50 (μM) | SI |
|---|---|---|---|
| 14a | 8.07 | 28.68 | 3.55 |
| 14e | 1.28 | 2.367 | 1.85 |
| 14f | 0.64 | 2.979 | 4.65 |
| 14g | 6.37 | 46.79 | 7.34 |
| AmphoB | 3.07 | 10.69 | 3.48 |
| Molecules | 14a | 14e | 14f | 14g |
|---|---|---|---|---|
| IC50 (μM) | 9.63 ± 0.28 | 4.44 ± 0.59 | 7.30 ± 0.39 | 11.74 ± 0.48 |
| Code | Docking score [kcal mol−1] | Code | Docking score [kcal mol−1] |
|---|---|---|---|
| 14a | −4.195 | 14g | −4.620 |
| 14b | −4.463 | 14h | −6.140 |
| 14c | −5.300 | 14i | −5.108 |
| 14d | −4.452 | 14j | −5.979 |
| 14e | −5.480 | 14k | −6.105 |
| 14f | −5.515 | Standard | −6.007 |
Among all the docked compounds 14e, 14f, 14h, 14j and 14k exhibited good docking scores against the targets (cysteine protease). All docked compounds possess in similar catalytic domain of cysteine protease (Fig. 12 and 13). In the Leishmania mexicana CPB structure (PDB ID 6P4E), both non-covalent hydrophilic and hydrophobic interactions were observed between these compounds and the protein. The compounds demonstrate pi-interactions with TRP 186 and form hydrogen bonds with LEU 162 and GLY 67. Furthermore, they exhibit hydrophobic interactions with LEU 68, MET 69, TRP 27, ALA 140, LEU 162, and TYR 210 amino acid residues. Additionally, they display some polar interactions with SER 65 in chain A, as shown in the 2D & 3D poses in Fig. 12.
 | ||
| Fig. 12 (A) and (B) The docking pose of molecule 14e is shown, in PDB ID 6P4E in 2D, and 3D, where it binds with LEU 162 and GLY 67 amino acid residues via hydrogen-bonding as represented by pink line; protein is displayed in ribbon representation (C) and (D) docking pose of 14f is shown, in PDB ID 6P4E in a 2D and 3D structure, where it binds with GLY 67 amino acid residue via hydrogen bonding as represented by the pink line. (E) and (F) docking pose of 14h is shown, in PDB ID 6P4E in a 2D and 3D structure, where it binds with GLY 67 amino acid residue via hydrogen bonding as represented by a pink line (G) The superimposable images of ligand 14h and standard with the catalytic site (H) distance measurement from CYS26 catalytic site (I) superimposition of 14e with E64. | ||
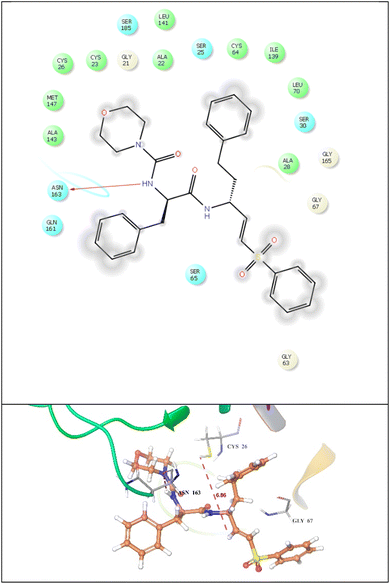 | ||
| Fig. 13 2d and 3D Binding interaction of standard ligand (dipeptidyl aza-nitrile) against cysteine protease (PDBID: 6P4E), where it binds with ASN 163 amino acid residue via hydrogen bonding as represented by a pink line. Protein is displayed in ribbon representation. | ||
Similarly, the standard exhibited hydrogen bonding with ASN163 amino acid residues in catalytic domain of cysteine protease and all our potent compounds possess similar binding pattern as per standard ligand. It also demonstrated hydrophobic interaction with TRP 27, MET 60, LEU 68, MET 69, ALA 140, LEU 162, TRP 186, and TYR 210 amino acids. Additionally, it displayed some polar interactions with GLN 20, SER 25, SER 65, GLN 71, ASN 163, and HIE 164 in chain A, and the 2D &3D poses are given in Fig. 13. Further, we have superimposed all our potent compounds with standard ligand and found similar binding pattern against catalytic domain of cysteine protease. We have done the distance measurement from CYS26 catalytic site (Fig. 12H) and found that all residues were present within range. After that also compare the known inhibitor E64 with compound 14e (Fig. 12I) and found similar catalytic domain.
These varying peptide linkers significantly affect biological activity based on factors like steric hindrance and chain flexibility. For instance, the presence of bulky or flexible linkers, as observed in 14e and 14h, alters the binding geometry, indicating that a more rigid structure could enhance interactions with the target protease's active site.
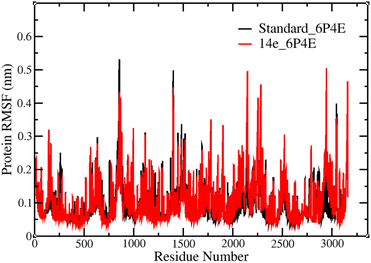
These modifications collectively emphasize the importance of specific peptide groups in optimizing the biological activity of these peptidyl β-nitrostyrenes. The MD simulations demonstrated that 14e, featuring a combination of tryptophan and morpholine, exhibited remarkable stability, as evidenced by low root mean square fluctuations and consistent hydrogen bond interactions with the protein. This confirms its structural integrity and compatibility within the binding pocket under simulated physiological conditions.
| Code | 14a | 14e | 14f | 14g | STD |
|---|---|---|---|---|---|
a van der Waals surface area of polar nitrogen and oxygen atoms and carbonyl carbon atoms (range 7–200).b Predictedoctanol/water partition coefficient (<5).c Predicted aqueous solubility, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S. S in mol dm3 is the concentration of the solute in a saturated solution that is in equilibrium with the crystalline solid (range −6.5 to 0.5).d Lipinski's violations (≤1).e % human oral absorption >80% is high, <25% is low.f Donor HB (0–6).g Acceptor HB (2–20). S. S in mol dm3 is the concentration of the solute in a saturated solution that is in equilibrium with the crystalline solid (range −6.5 to 0.5).d Lipinski's violations (≤1).e % human oral absorption >80% is high, <25% is low.f Donor HB (0–6).g Acceptor HB (2–20). |
|||||
| PSAa | 117.272 | 133.044 | 110.681 | 114.779 | 107.84 |
QP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P o/wb P o/wb |
2.27 | 2.759 | 3.38 | 2.083 | 4.81 |
QP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sc Sc |
−2.386 | −4.657 | −5.365 | −1.766 | −6.57 |
| Rule of fived | 0 | 0 | 0 | 0 | 1 |
| Donor HBf | 0.25 | 1.25 | 0.25 | 0.25 | 1.25 |
| Acceptor HBg | 9.2 | 9.2 | 8 | 9.2 | 9.45 |
| % HOAe | 85.924 | 78.384 | 89.464 | 74.957 | 89.169 |
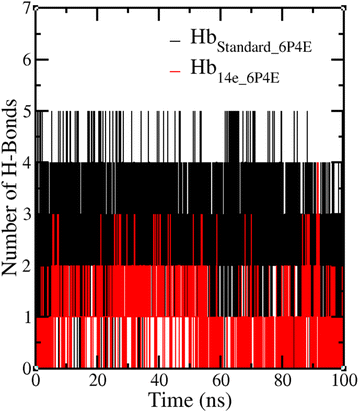
The radius of gyration for the protein–ligand complexes with 14e and standard were analyzed. The average radius of gyration values was found to be 1.66 ± 0.03 nm for all protein–ligand complexes and can be represented as the arrangement of atoms around the axis and found to be quite stable throughout the 100 ns MD simulation run, depicted in Fig. 17.
3 Conclusion
A library of 12 peptidyl β-nitrostyrenes was synthesized and tested for their efficacy against L. donovani promastigotes and amastigotes. Most of the compounds demonstrated comparable anti-promastigote activity to the standard with IC50 values ranging between 1.468–16.81 μM. Compounds 14a, 14e, 14f, and 14g exhibited significant activity against both L. donovani promastigotes and intracellular amastigotes. Compounds 14e and 14f demonstrated better anti-promastigote activity with IC50 values of 1.468 μM and 1.551 μM respectively as compared to standard (IC50 = 3.073 μM). The inhibitory potential of 14e and 14f against intracellular amastigotes with IC50 values of 1.28 μM and 0.64 μM was better than AmphoB with IC50 value of 3.07 μM. Additionally, insignificant cytotoxicity was seen for compounds 14a and 14g against mammalian macrophages even at a concentration of 28 μM. Given their high activity, safety profiles, and the fact that they can be synthesized cost-effectively, this class of compounds could be potentially valuable for the development of anti-leishmanial drugs.The use of peptidyl β-nitrostyrenes as inhibitors of cysteine proteases to treat Leishmania donovani represents a new approach compared to current anti-leishmanial therapies like miltefosine and amphotericin B, which target different biological processes. Miltefosine, the first oral drug for leishmaniasis, disrupts cell membrane integrity by affecting lipid metabolism, leading to a type of cell death in the parasite like apoptosis.28,29 While effective, its use is becoming limited due to emerging resistance.29 On the other hand, amphotericin B, which binds to ergosterol in the parasite's membrane, causes cell lysis but has significant nephrotoxicity, making it less practical for widespread use, especially in resource-limited settings.29,30 In contrast, peptidyl β-nitrostyrenes act selectively by inhibiting cysteine proteases that are crucial for the parasite's survival, particularly in intracellular amastigotes. Compounds such as 14e and 14f in this study showed promising efficacy, with IC50 values of 1.28 μM and 0.64 μM against intracellular amastigotes. Additionally, these compounds displayed low toxicity to mammalian macrophages, suggesting a favourable safety profile compared to miltefosine and amphotericin B. Although further optimization is necessary, the selective mechanism and initial efficacy of peptidyl β-nitrostyrenes make them promising candidates for future anti-leishmanial drug development.
4 Materials and methods
4.1 General
All reagents and chemicals used in this research were obtained from GLR, Sigma Aldrich, and Merck (India), and were of AR (analytical reagent) grade quality. The 0.25 mm silica gel 60–120 TLC plates were used to monitor the reactions. UV cabinet was used to visualize spots on the TLC to monitor the reactions. Bruker NMR spectrometer was used for 1H and 13C NMR spectral analysis at 400 MHz and 100 MHz. The spectra were analyzed using Mestrenova software. The HRMS and ESI-MS were obtained on ESI-MS using an Agilent mass spectrometer. The IR spectra were obtained on a Bruker ALPHA FT-IR spectrometer. The Buchi Labortechnik AG 9230 automated melting point apparatus was used for determining the melting points of the derivatives. The identities of the compounds prepared in this study were confirmed through spectral analysis. The compounds were purified through column chromatography using 60–120 and 230–400 mesh sizes of silica gel and recrystallized in ethanol. Additional chemicals used in the in vitro assay including 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were from BPS Biosciences.4.2 Chemistry
5 Characterization data
5.1 (E)-N-(1-Morpholino-1-oxo-3-phenylpropan-2-yl)-2-(4-(2-nitrobut-1-en-1-yl) phenoxy)acetamide (14a)
Light yellowish white solid 91% yield; melting point 119.7–120.8 °C; 1H NMR (400 MHz, CDCl3) δ 8.00 (s, 1H), 7.42 (d, J = 8.0 Hz, 3H), 7.31 (d, J = 8.0 Hz, 3H), 7.20 (d, J = 8.0 Hz, 2H), 7.01 (d, J = 8.0 Hz, 2H), 5.22 (q, J = 8.0 Hz, 1H), 4.54 (s, 2H), 3.63–3.56 (m, 2H), 3.52–3.40 (m, 3H), 3.32–3.27 (m, 1H), 3.10 (d, J = 8.0 Hz, 1H), 3.06 (d, J = 8.0 Hz, 1H), 3.06–2.97 (m, 1H), 2.96–2.85 (m, 3H), 1.28 (t, J = 8.0 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 169.51, 166.99, 158.55, 152.17, 135.79, 132.74, 131.96, 129.67, 128.87, 127.57, 12.28, 115.45, 67.27, 66.52, 66.11, 49.22, 46.14, 42.43, 40.19, 20.91, 12.52; IR (KBr) ν 3392, 3158, 2933, 2744, 1647, 1609, 1563, 1530, 1511, 1462, 1346, 1285, 1247, 1238, 1131, 1054, 976, 875, 769, 623 cm−1; HRMS (Q-TOF, ESI) calcd for C25H29N3O6 [M + H]+ 468.2056, found 468.2062. Anal. calcd for C25H29N3O6: C, 64.23; H, 6.25; N, 8.99. Found: C, 64.29; H, 6.18; N, 9.04.5.2 (E)-1-(2-(Morpholine-4-carbonyl)pyrrolidin-1-yl)-2-(4-(2-nitrobut-1-en-1-yl)phenoxy) ethan-1-one (14b)
Light yellowish white solid 81% yield; melting point 108.4–109.1 °C; 1H NMR (400 MHz, CDCl3) δ 7.99 (s, 1H), 7.40 (d, J = 8.0 Hz, 2H), 7.01 (d, J = 8.0 Hz, 2H), 4.91 (dd, J = 4.0, 4.0 Hz, 1H), 4.74 (d, J = 4.0 Hz, 2H), 3.82–3.76 (m, 2H), 3.74–3.60 (m, 6H), 3.57–3.49 (m, 3H), 2.87 (q, J = 8.0 Hz, 2H), 1.27 (t, J = 8.0 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 170.15, 166.34, 159.66, 151.66, 133.16, 131.98, 129.85, 125.55, 115.41, 115.26, 67.05, 66.92, 56.67, 46.52, 46.28, 42.68, 28.70. 25.70, 20.86, 20.73, 12.47; IR (KBr) ν 3372, 3218, 2927, 2741, 1651, 1609, 1563, 1530, 1511, 1462, 1346, 1285, 1247, 1238, 1131, 1054, 946, 874, 779, 621 cm−1; HRMS (Q-TOF, ESI) calcd for C21H27N3O6 [M + H]+ 484.1861, found 484.1905. Anal. calcd for C21H27N3O6: C, 60.42; H, 6.52; N, 10.07. Found: C, 60.38; H, 6.57; N, 10.11.5.3 (E)-2-(4-(2-Nitrobut-1-en-1-yl)phenoxy)-N-(1-oxo-3-phenyl-1-thiomorpholinopropan-2-yl)acetamide (14c)
Light yellowish white solid 74% yield; Melting point 86.8–87.9 °C; 1H NMR (400 MHz, CDCl3) δ 8.00 (s, 1H), 7.42 (d, J = 8.0 Hz, 3H), 7.31 (d, J = 8.0 Hz, 3H), 7.18 (d, J = 8.0 Hz, 2H), 7.00 (d, J = 8.0 Hz, 2H), 5.24 (q, J = 8.0 Hz, 1H), 4.52 (s, 2H), 3.83–3.75 (m, 2H), 3.56–3.49 (m, 1H), 3.42–3.37 (m, 1H), 3.04 (d, J = 8.0 Hz, 2H), 2.88 (q, J = 8.0 Hz, 2H), 2.58–2.54 (m, 1H), 2.47–2.38 (m, 2H), 1.90–1.86 (m, 1H), 1.29 (t, J = 8.0 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 169.56, 166.99, 158.53, 152.18, 135.75, 132.73, 131.96, 129.65, 128.56, 127.55, 126.28, 115.45, 67.24, 49.36, 48.46, 44.97, 40.06, 27.47, 27.34, 20.90, 12.52; IR (KBr) ν 3282, 3148, 2911, 2744, 1647, 1637, 1563, 1530, 1515, 1447, 1346, 1285, 1247, 1238, 1131, 1052, 913, 874, 769, 540 cm−1; HRMS (Q-TOF, ESI) calcd for C25H29N3O5S [M + H]+ 484.1828, found 484.1837. Anal. calcd for C25H29N3O5S: C, 62.09; H, 6.04; N, 8.69; S, 6.63. Found: C, 62.12; H, 5.99; N, 8.73; S, 6.68.5.4 (E)-2-(4-(2-Nitrobut-1-en-1-yl)phenoxy)-1-(2-(thiomorpholine-4-carbonyl)pyrrolidin-1-yl)ethan-1-one (14d)
Light yellow solid 81% yield; melting point 119.1–120.3 °C; 1H NMR (500 MHz, DMSO) δ 8.06 (s, 1H), 7.56 (d, J = 5.0 Hz, 2H), 7.06 (d, J = 5.0 Hz, 2H), 5.77 (s, 1H), 4.94 (d, J = 15 Hz, 1H), 4.84 (m, 2H), 3.88–3.72 (m, 3H), 3.64–3.57 (m, 3H), 3.47 (d, J = 5.0 Hz, 1H), 2.85 (q, J = 10.0 Hz, 2H), 2.77–2.66 (m, 1H), 2.17–2.13 (m, 1H), 1.98–1.91 (m, 2H), 1.75–1.71 (m, 1H), 1.21 (t, J = 10 Hz, 3H); 13C NMR (100 MHz, DMSO): δ 170.19, 165.52, 160.43, 160.37, 151.12, 133.74, 132.51, 124.66, 115.88, 56.87, 55.40, 45.99, 44.88, 31.52, 28.83, 24.88, 22.04, 20.88, 12.49; IR (KBr) ν 3417, 3391, 3157, 3062, 2931, 2742, 2541, 2211. 1929, 1647, 1609, 1563, 1514, 1462, 1346, 1285, 1238, 1131, 1052, 976, 875, 769 cm−1; HRMS (Q-TOF, ESI) calcd for C21H27N3O5S [M + H]+ 434.1671, found 434.1677. Anal. Calcd for C21H27N3O5S: C, 58.18; H, 6.28; N, 9.69; S, 7.40. Found: C, 58.22; H, 6.26; N, 9.73; S, 7.37.5.5 (E)-N-(1-(1H-indol-3-yl)-2-morpholino-2-oxoethyl)-2-(4-(2-nitrobut-1-en-1-yl) phenoxy)acetamide (14e)
Light yellow solid 85% yield; melting point 83.1–84.9 °C; 1H NMR (400 MHz, CDCl3) δ 8.14 (s, 1H), 7.99 (s, 1H), 7.66 (d, J = 8.0 Hz, 1H), 7.50 (d, J = 8.0 Hz, 1H), 7.41–7.36 (m, 3H), 7.21 (t, J = 8.0 Hz, 1H), 7.13 (t, J = 8.0 Hz, 1H), 7.07 (s, 1H), 7.00 (d, J = 8.0 Hz, 2H), 5.35 (q, J = 8.0 Hz, 1H), 4.54 (s, 2H), 3.83–3.75 (m, 2H), 3.56–3.49 (m, 1H), 3.42–3.37 (m, 1H), 3.04 (d, J = 8.0 Hz, 2H), 2.88 (q, J = 8.0 Hz, 2H), 2.58–2.54 (m, 1H), 2.47–2.38 (m, 2H), 1.90–1.86 (m, 1H), 1.29 (t, J = 8.0 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 170.27, 167.05, 158.56, 152.11, 136.18, 132.79, 131.94, 127.54, 126.20, 122.98, 122.67, 120.12, 118.75, 115.46, 111.50, 110.32, 67.29, 48.86, 48.44, 44.86, 30.20, 27.28, 27.21, 20.90, 12.51; IR (KBr) ν 3362, 3178, 2943, 2754, 1637, 1609, 1563, 1516, 1462, 1346, 1284, 1237, 1132, 1054, 975, 871, 769, 591 cm−1; HRMS (Q-TOF, ESI) calcd for C26H28N4O6 [M + H]+ 493.2009, found 493.2012. Anal. calcd for C26H28N4O6: C, 63.40; H, 5.73; N, 11.38. Found: C, 63.45; H, 5.72; N, 11.34.5.6 (E)-2-(4-(2-nitrobut-1-en-1-yl)phenoxy)-N-(2-oxo-1-phenyl-2-thiomorpholinoethyl) acetamide (14f)
Light yellow solid 82% yield; melting point 107.0–108.2 °C; 1H NMR (400 MHz, CDCl3) δ 8.10 (d, J = 8.0 Hz, 1H), 7.98 (s, 1H), 7.41–7.29 (m, 7H), 6.98 (d, J = 8.0 Hz, 2H), 5.89 (d, J = 8.0 Hz, 1H), 4.51 (q, J = 8.0 Hz, 2H), 4.19–4.15 (m, 1H), 3.73–3.3.60 (m, 3H), 2.91–2.84 (m, 2H), 2.58 (t, J = 8.0 Hz, 2H), 2.32 (dd, J = 4.0, J = 12.0, Hz, 1H), 1.90–1.86 (m, 1H), 1.42–1.35 (m, 1H). 13C NMR (100 MHz, CDCl3) δ 167.69, 166.67, 158.58, 152.02, 136.92, 132.79, 131.88, 129.45, 128.87, 128.01, 126.09, 115.45, 67.29, 53.86, 48.17, 45.26, 29.79, 27.23, 26.92, 20.86, 12.47; IR (KBr) ν 3312, 3154, 2923, 2735, 1800, 1644, 1603, 1583, 1530, 1511, 1440, 1346, 1285, 1247, 1238, 1179, 1054, 976, 873, 779, 613 cm−1; HRMS (Q-TOF, ESI) calcd for C24H27N3O5S [M + H]+ 470.1671, found 470.1678. Anal. Calcd for C24H27N3O5S: C, 61.39; H, 5.80; N, 8.95; S, 6.83. found: C, 61.37; H, 5.83; N, 9.01; S, 6.78.5.7 (E)-N-(2-morpholino-2-oxo-1-phenylethyl)-2-(4-(2-nitrobut-1-en-1-yl)phenoxy) acetamide (14g)
Light yellow solid 79% yield; melting point 128.5–129.4 °C; 1H NMR (400 MHz, CDCl3) δ 8.08 (d, J = 8.0 Hz, 1H), 7.98 (s, 1H), 7.41–7.31 (m, 7H), 6.98 (d, J = 8.0 Hz, 2H), 5.90 (d, J = 8.0 Hz, 1H), 4.51 (q, J = 8.0 Hz, 2H), 3.78–3.62 (m, 2H), 3.60–3.51 (m, 3H), 3.50–3.42 (m, 1H), 3.29–3.26 (m, 1H), 3.14–3.10 (m, 1H), 2.86 (q, J = 8.0 Hz, 2H), 1.27 (t, J = 4.0 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 167.81, 166.70, 158.61, 152.06, 136.94, 132.80, 131.90, 129.40, 128.86, 127.95, 126.14, 115.48, 115.16, 67.34, 66.64, 66.07, 53.64, 45.95, 42.86, 20.88, 12.43; IR (KBr) ν 3484.27, 3460.18, 3424.85, 3385.04, 3249.24, 3322.91, 3209.25, 3173.12, 3124.24, 3039.54, 3007.66, 2965.80, 2932.01, 2867.52, 1777.73, 1750.78, 1677.18, 1652.31, 1602.42, 1501.41, 1471.97, 1437.08, 1365.68, 1321.19, 1248.71, 1180.87, 1108.57, 1063.88, 1032.59, 990.68, 945.59, 912.54, 836.85, 811.03, 764.45, 708.25, 662.07, 586.35, 529.62, 484.69, 464.20, 422.40 cm-1; HRMS (Q-TOF, ESI) calcd for C24H27N3O6 [M + H]+ 454.1900, found 454.1906. Anal. Calcd for C24H27N3O6: C, 63.56; H, 6.00; N, 9.27. found: C, 63.53; H, 5.98; N, 9.32.5.8 Methyl (E)-(2-(1H-indol-3-yl)-2-(2-(4-(2-nitrobut-1-en-1-yl)phenoxy)acetamido)acetyl)valinate (14h)
Off white solid 89% yield; melting point 128.5–129.4 °C; 1H NMR (400 MHz, CDCl3) δ 8.11 (s, 1H), 7.98 (s, 1H), 7.76 (d, J = 8.0 Hz, 1H), 7.37 (d, J = 8.0 Hz, 4H), 7.21 (t, J = 8.0 Hz, 1H), 7.13 (t, J = 8.0 Hz, 2H), 6.90 (d, J = 8.0 Hz, 2H), 6.11 (d, J = 8.0 Hz, 1H), 4.82 (q, J = 8.0 Hz, 1H), 4.56 (s, 2H), 4.39–4.36 (m, 1H), 3.68 (s, 3H), 3.41–3.36 (m, 1H), 3.22–3.16 (m, 1H), 2.87 (q, J = 8.0 Hz, 2H), 2.05–2.00 (m, 1H), 1.29 (t, J = 4.0 Hz, 3H), 0.79 (d, J = 8.0 Hz, 3H), 0.74 (d, J = 8.0 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 167.81, 166.70, 158.61, 152.06, 136.94, 132.80, 131.90, 129.40, 128.86, 127.95, 126.14, 115.48, 115.16, 67.34, 66.64, 66.07, 53.64, 45.95, 42.86, 20.88, 12.43; IR (KBr) ν 3322, 3148, 2922, 2742, 1645, 1603, 1563, 1516, 1440, 1346, 1285, 1247, 1238, 1180, 1154, 1034, 946, 809, 701, 530 cm−1; HRMS (Q-TOF, ESI) calcd for C29H34N4O7 [M + H]+ 551.2427, found 551.2434. Anal. Calcd for C29H34N4O7: C, 63.26; H, 6.22; N, 10.18. Found: C, 63.23; H, 6.26; N, 10.15.5.9 Methyl (E)-(2-(4-(2-nitrobut-1-en-1-yl)phenoxy)acetyl)phenylalanylglycinate (14i)
Light yellow white solid 85% yield; melting point 122.5–124.7 °C; 1H NMR (400 MHz, CDCl3) δ 7.99 (s, 1H), 7.40 (d, J = 8.0 Hz, 2H), 7.34 (d, J = 8.0 Hz, 2H), 7.31–7.23 (m, 3H), 7.19 (t, J = 8.0 Hz, 2H), 7.13 (d, J = 8.0 Hz, 1H), 6.94 (d, J = 8.0 Hz, 2H), 4.79 (d, J = 8.0 Hz, 1H), 4.58–4.37 (m, 2H), 4.04–3.89 (m, 2H), 3.73 (s, 2H), 3.16–3.05 (m, 2H), 2.87 (q, J = 8.0 Hz, 2H), 1.28 (t, J = 8 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 170.95, 169.95, 168.04, 158.43, 152.04, 135.99, 132.63, 131.82, 129.26, 128.63, 127.10, 115.29, 115.25, 67.08, 53.75, 52.40, 41.16, 38.21, 20.74, 12.32. IR (KBr) ν 3475, 3440, 3330, 3170, 3134, 3081, 2924, 2853, 1777, 1750, 1646, 1460, 1402, 1337, 1285, 944, 838, 811, 476 cm−1; HRMS (Q-TOF, ESI) calcd for C24H27N3O7 [M + H]+ 470.1849, found 470.1889. Anal. Calcd for C24H27N3O7: C, 61.40; H, 5.80; N, 8.95. Found: C, 61.41; H, 5.77; N, 8.98.5.10 Methyl (E)-(2-(4-(2-nitrobut-1-en-1-yl)phenoxy)acetyl)tryptophylalaninate (14j)
Light yellow white solid 85% yield; melting point 108.5–109.4 °C; 1H NMR (400 MHz, CDCl3) δ 8.01 (s, 2H), 7.47 (q, J = 4.0 Hz, 2H), 7.42 (d, J = 8.0 Hz, 2H), 7.37 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 4.0 Hz, 2H), 7.23 (d, J = 8.0 Hz, 2H), 6.97 (d, J = 8.0 Hz, 2H), 4.98 (s, 1H), 4.79 (q, J = 8.0 Hz, 1H), 4.54 (s, 1H), 4.45 (q, J = 4.0 Hz, 1H), 3.73 (s, 3H), 3.20–3.08 (m, 2H), 2.12 (q, J = 8.0 Hz, 2H), 1.30 (m, 3H), 0.87 (d, J = 8.0 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 171.64, 170.28, 167.72, 158.36, 152.13, 135.95, 132.53, 131.84, 131.01, 129.29, 128.78, 127.21, 126.23, 121.94, 116.22, 115.27, 67.17, 57.51, 54.17, 38.18, 31.11, 20.77, 12.39. IR (KBr) ν 3397, 3293, 3220, 3199, 3174, 2975, 2924, 2853, 1779, 1742, 1649, 1602, 1401, 1364, 1177, 1059, 1025, 944, 837, 811, 742, 699, 475, 423 cm−1; HRMS (Q-TOF, ESI) calcd for C27H30N4O7 [M + H]+ 523.2114, found 523.2148. Anal. Calcd for C27H30N4O7: C, 62.06; H, 5.79; N, 10.72. found: C, 62.10; H, 5.82; N, 10.68.5.11 Methyl (E)-(2-(4-(2-nitrobut-1-en-1-yl)phenoxy)acetyl)phenylalanylalaninate (14k)
Light yellow white solid 85% yield; melting point 118.2–120.4 °C; 1H NMR (400 MHz, CDCl3) δ 8.35 (s, 1H), 7.99 (s, 2H), 7.69 (d, J = 8 Hz, 1H), 7.55 (d, J = 4 Hz, 1H), 7.20 (t, J = 8 Hz, 1H), 7.12 (d, J = 12 Hz, 2H), 7.00 (d, J = 8.0 Hz, 2H), 6.89 (d, J = 8.0 Hz, 2H), 4.85 (q, J = 8.0 Hz, 1H), 4.70 (s, 2H), 4.46 (t, J = 8 Hz, 1H), 3.69 (s, 2H), 3.38 (m, 1H), 3.24 (m, 1H), 2.88 (q, J = 8 Hz, 2H), 1.34–1.24 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 172.77, 170.69, 168.15, 159.12, 151.82, 136.28, 133.49, 132.80, 131.79, 123.59, 122.32, 119.88, 118.63, 116.25, 115.18, 111.37, 77.39, 77.07, 76.75, 67.04, 52.55, 48.42, 28.34, 20.77, 18.05, 12.31. IR (KBr) ν 3328, 3277, 3249, 3198, 2979, 2441, 1776, 1744, 1651, 1602, 1510, 1457, 1402, 1365, 1181, 1063, 1026, 945, 837, 810, 739, 505, 470, 423 cm−1; HRMS (Q-TOF, ESI) calcd for C25H29N3O7 [M + H]+ 483.2006, found 483.2039. Anal. Calcd for C25H29N3O7: C, 62.10; H, 6.05; N, 8.69. found: C, 62.08; H, 6.01; N, 8.63.5.12 Pharmacology
5.13 Cell line culture
THP-1 human macrophages were cultured in RPMI based medium at 37 °C in presence of 5% CO2 for 48 to 72 hours in RPMI-1640 medium (pH 7.4) with 10% heat-inactivated FBS in a 5% CO2 humidified atmosphere (Avishek et al. 2024). Afterwards, an average density of 2 × 105 cells per mL of the cells were transferred to new RPMI-1640 medium.5.14 MTT assay for anti-promastigote activity
The anti-leishmanial activity of peptidyl β-nitrostyrenes (14a–k) was performed using MTT assay (Sharma et al. 2023). This assay is based on the mitochondrial enzymes NAD and NADH-dehydrogenases forming formazan crystals from the tetrazolium soluble salt. 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 THP-1 human monocytic cells were seeded at per well in a 96-well plate. The cells were exposed to the synthesized molecules at varying concentrations starting from 100, 50, 25, 12.5, and 6.25 μM, for 24 hours after they reached normal morphology. The 96-well plate was then kept in an incubator with humidified CO2 at 37 °C. Following this, the supernatant was discarded, followed by addition of 0.5 mg mL−1 of MTT solution. It was then incubated for 4 hours at 37 °C in a humidified environment. The media was taken out, followed by the addition of DMSO to dissolve the formazan crystals, and an i3x spectramax molecular device was used to record the absorbance at 570 nm. Compounds showing greater than 50% inhibition were selected for cytotoxicity assay (IC50).31
000 THP-1 human monocytic cells were seeded at per well in a 96-well plate. The cells were exposed to the synthesized molecules at varying concentrations starting from 100, 50, 25, 12.5, and 6.25 μM, for 24 hours after they reached normal morphology. The 96-well plate was then kept in an incubator with humidified CO2 at 37 °C. Following this, the supernatant was discarded, followed by addition of 0.5 mg mL−1 of MTT solution. It was then incubated for 4 hours at 37 °C in a humidified environment. The media was taken out, followed by the addition of DMSO to dissolve the formazan crystals, and an i3x spectramax molecular device was used to record the absorbance at 570 nm. Compounds showing greater than 50% inhibition were selected for cytotoxicity assay (IC50).31
5.15 Anti-leishmanial activity against L. donovani promastigotes
The synthesized molecules were tested for their in vitro activity against L. donovani promastigotes. A concentration of 2 × 106 cells per mL of L. donovani promastigotes were cultured in RPMI medium for 72 hours at 26 °C with 100 μM concentration of each compound. Amphoterecin B (AmphoB) was used as a reference, and DMSO (0.2%) was the solvent. Parasites in media alone were the control experiment. The viability of parasites was measured using the MTT assay after 72 hours.325.16 Determination of IC50 from dose-dependent anti-promastigote activity
The promastigotes of L. donovani were incubated using 2 × 106 cells per mL in the presence and absence of the most active compounds (14a–i) of the series at serial dilutions starting at 25 μM for 72 hours at 26 °C, using AmphoB was as a standard anti-leishmanial drug control. Cell viability was measured using by MTT assay, and the following formula was used to calculate the mean percentage viability: (Mean cell number of treated promastigotes/Mean cell number of untreated promastigotes) × 100. The IC50 values were determined by extrapolation of graph in the graph of % viability vs. concentration of the drug.335.17 Cytotoxicity assay
THP-1 human monocytic cells were cultured with 10% FBS in RPMI media, and 1 × 106 cells per mL concentration of cells were seeded. The plate was kept in a humidified CO2 incubator for 24 h at 37 °C. After 24 h, media was removed, and varying concentrations of standard and compounds were added like 100, 50, 25, 12.5, and 6.25 μM for 72 h, before performing the MTT assay.345.18 Intracellular amastigotes assay
Macrophages of L.donovani were cultivated on eight-chamber tissue culture slides with 5 × 104 cells in each well and were allowed to adhere in a CO2 incubator containing 5% CO2 for 2 h at 37 °C. RPMI 1640 medium without serum was used to wash the wells twice to eliminate the nonadherent macrophages. The adherent macrophages were subsequently exposed to the metacyclic stage of Leishmania donovani, with a Leishmania to macrophage ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which was sustained overnight in a 200 μL final solution of complete RPMI 1640 medium. Following a 24 hours incubation period, RPMI 1640 medium with no serum was used to wash free promastigotes. Infected macrophages were then incubated with compounds in duplicate for 72 hours at 37 °C with 5% CO2, except the control well. The number of amastigotes in 100 macrophages were counted at each concentration to evaluate IC50 values of test compounds. Infected macrophage percentages were determined by counting the infected macrophages per 100 macrophages.35
1, which was sustained overnight in a 200 μL final solution of complete RPMI 1640 medium. Following a 24 hours incubation period, RPMI 1640 medium with no serum was used to wash free promastigotes. Infected macrophages were then incubated with compounds in duplicate for 72 hours at 37 °C with 5% CO2, except the control well. The number of amastigotes in 100 macrophages were counted at each concentration to evaluate IC50 values of test compounds. Infected macrophage percentages were determined by counting the infected macrophages per 100 macrophages.35
5.19 Calculation of percentage of inhibition and ED50
Using a Neubauer chamber, the extracellular parasites present in treated wells as well as control wells were counted after 24 hours of treatment. For every duplicate well, a microscopic enumeration was conducted at various concentrations. Dead parasites with deteriorated body surfaces were not counted. All counts at varying concentrations were converted to percentages by using the control well mean, which was taken to represent 100% survival. Using Zeiss, AXIO, and Imager A1 light microscope, the slides of the infected macrophage cell line which were stained with Giemsa, were examined, and the infected macrophages/amastigotes per 100 cells were counted.365.20 Statistical analysis
IC50 values against L. donovani promastigotes and amastigotes were determined from the triplicate's experiments for the mean and standard error of the mean (SEM). To analyze the differences among groups GraphPad Prism 8.0 software was used. Using the two-way ANOVA, the correlation between drug and their effect was determined. P < 0.05 was regarded as being statistically important for difference and correlation.375.21 In Vitro cysteine protease inhibition assay
Inhibition of papain (Roche cat #10108014001) was studied using an increasing concentration of the derivatives. Nα-Benzoyl-L-arginine-7-amido-4-methyl coumarin hydrochloride (Bz-Arg-AMC) from Sigma (cat #B7260), a fluorogenic peptide substrate was used for the papain activity assay. The inhibitors were dissolved in DMSO and an activation buffer containing sodium acetate buffer (pH 5.5), 8 mM DTT, and 4 mM EDTA was used. The reaction mixture (100 μl) also included papain (4 mM), increasing concentration of inhibitor (0–32 μM) and 10 μM substrate. The enzymatic hydrolysis of substrate by papain was monitored using SpectraMax Multi-Mode Microplate Readers with an emission wavelength of 440 nm and an excitation wavelength of 345 nm. The IC50 value for the inhibitor was calculated from the plot of the % residual activity against the log[inhibitor], as described previously.385.22 Molecular docking
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We would like to show our gratitude to SERB (ECR/2017/001067/CS) for financial support and Jamia Hamdard for providing the laboratory facilities.References
- E. Torres-Guerrero, M. Romano Quintanilla-Cedillo, J. Ruiz-Esmenjaud, R. Arenas, F. Bravo-Puccio, P. Cayetano Heredia and R. J. Hay, F1000Research, 2017, 6, 750 Search PubMed.
- P. Desjeux, Comp. Immunol., Microbiol. Infect. Dis., 2004, 27, 305–318 CrossRef CAS PubMed.
- A. Selvapandiyan, S. L. Croft, S. Rijal, H. L. Nakhasi and N. K. Ganguly, PLoS Neglected Trop. Dis., 2019, 13, e0007616 CrossRef PubMed.
- P. Bhattacharya, R. Dey, P. K. Dagur, A. B. Joshi, N. Ismail, S. Gannavaram, A. Debrabant, A. D. Akue, M. A. KuKuruga, A. Selvapandiyan, J. P. McCoy and H. L. Nakhasi, PLoS Neglected Trop. Dis., 2016, 10, e0004963 CrossRef PubMed.
- M. M. S. Andrade, L. C. Martins, G. V. L. Marques, C. A. Silva, G. Faria, S. Caldas, J. S. C. Dos Santos, V. G. Maltarollo, R. S. Ferreira and R. B. Oliveira, Future Med. Chem., 2020, 12, 571–581 CrossRef CAS PubMed.
- A. Latorre, T. Schirmeister, J. Kesselring, S. Jung, P. Johé, U. A. Hellmich, A. Heilos, B. Engels, R. L. Krauth-Siegel, N. Dirdjaja, L. Bou-Iserte, S. Rodríguez and F. V. González, ACS Med. Chem. Lett., 2016, 7, 1073–1076 CrossRef CAS PubMed.
- S. Sharma, P. A. Yakkala, J. Aboti, I. Latief, M. A. Ansari, W. H. Khan and S. Shafi, ChemistrySelect, 2023, 8, e202300912 CrossRef CAS.
- P. J. Rosenthal, G. K. Lee and R. E. Smith, J. Clin. Invest., 1993, 91, 1052–1056 CrossRef CAS PubMed.
- J. H. McKerrow, Int. J. Parasitol., 1999, 29, 833–837 CrossRef CAS PubMed.
- H. Mahmoudzadeh-Niknam and J. H. McKerrow, Exp. Parasitol., 2004, 106, 158–163 CrossRef CAS PubMed.
- S. T. Liang, C. Chen, R. X. Chen, R. Li, W. L. Chen, G. H. Jiang and L. L. Du, Front. Pharmacol, 2022, 13, 1033003 CrossRef CAS PubMed.
- S. Sharma, P. Anjaneyulu Yakkala, M. A. Beg, S. Tanwar, I. Latief, A. Khan, K. Sharma, S. ur Rehman, A. Selvapandiyan and S. Shafi, ChemistrySelect, 2023, 8, e202302415 CrossRef CAS.
- Evaluation of nitrostyrenes as antifungal agents. I. In vitro studies - PubMed, https://pubmed.ncbi.nlm.nih.gov/24544016/, (accessed 24 March 2024).
- Antifungal activity of beta-nitrostyrenes and some cyclohexane derivatives - PubMed, https://pubmed.ncbi.nlm.nih.gov/24543964/, (accessed 24 March 2024).
- A. Ramzan, S. A. Padder, K. Z. Masoodi, S. Shafi, I. Tahir, R. U. Rehman, R. Prasad and A. H. Shah, Eur. J. Med. Chem., 2022, 240, 114609 CrossRef CAS PubMed.
- Y. Mikami, K. Yazawa, A. Maeda, J. Uno, A. Kubo, N. Saito and N. Kawakami, J. Antibiot., 1991, 44, 1454–1456 CrossRef CAS PubMed.
- N. Milhazes, R. Calheiros, M. P. M. Marques, J. Garrido, M. N. D. S. Cordeiro, C. Rodrigues, S. Quinteira, C. Novais, L. Peixe and F. Borges, Bioorg. Med. Chem., 2006, 14, 4078–4088 CrossRef CAS PubMed.
- S. Shafi, F. Afrin, M. Islamuddin, G. Chouhan, I. Ali, F. Naaz, K. Sharma and M. S. Zaman, Front. Microbiol., 2016, 7, 195169 Search PubMed.
- Z. jun Jia, X. wei Lan, K. Lu, X. Meng, W. jie Jing, S. ru Jia, K. Zhao and Y. jie Dai, J. Mol. Struct., 2023, 1284, 135409 CrossRef PubMed.
- C. Schad, U. Baum, B. Frank, U. Dietzel, F. Mattern, C. Gomes, A. Ponte-Sucre, H. Moll, U. Schurigt and T. Schirmeister, Antimicrob. Agents Chemother., 2015, 60, 797–805 CrossRef PubMed.
- A. Scala, N. Micale, A. Piperno, A. Rescifina, T. Schirmeister, J. Kesselring and G. Grassi, RSC Adv., 2016, 6, 30628–30635 RSC.
- A. Latorre, T. Schirmeister, J. Kesselring, S. Jung, P. Johé, U. A. Hellmich, A. Heilos, B. Engels, R. L. Krauth-Siegel, N. Dirdjaja, L. Bou-Iserte, S. Rodríguez and F. V. González, ACS Med. Chem. Lett., 2016, 7, 1073–1076 CrossRef CAS PubMed.
- Y. Zhou, P. Vedantham, K. Lu, J. Agudelo, R. Carrion, J. W. Nunneley, D. Barnard, S. Pöhlmann, J. H. McKerrow, A. R. Renslo and G. Simmons, Antiviral Res., 2015, 116, 76–84 CrossRef CAS PubMed.
- J. J. Shie, J. M. Fang, T. H. Kuo, C. J. Kuo, P. H. Liang, H. J. Huang, Y. T. Wu, J. T. Jan, Y. S. E. Cheng and C. H. Wong, Bioorg. Med. Chem., 2005, 13, 5240–5252 CrossRef CAS PubMed.
- A. Citarella, A. Scala, A. Piperno and N. Micale, Biomolecules, 2021, 11, 607 CrossRef CAS PubMed.
- K. Akaji, H. Konno, H. Mitsui, K. Teruya, Y. Shimamoto, Y. Hattori, T. Ozaki, M. Kusunoki and A. Sanjoh, J. Med. Chem., 2011, 54, 7962–7973 CrossRef CAS PubMed.
- S. Previti, R. Ettari, E. Calcaterra, S. Di Maro, S. J. Hammerschmidt, C. Müller, J. Ziebuhr, T. Schirmeister, S. Cosconati and M. Zappalà, Eur. J. Med. Chem., 2023, 247, 115021 CrossRef CAS PubMed.
- T. P. C. Dorlo, M. Balasegaram, J. H. Beijnen and P. J. de vries, J. Antimicrob. Chemother., 2012, 67, 2576–2597 CrossRef CAS PubMed.
- The treatment of visceral leishmaniasis: safety and efficacy - PubMed, https://pubmed.ncbi.nlm.nih.gov/25327244/, (accessed 17 October 2024).
- J. D. Herman, Rev. Infect. Dis., 1988, 10, 560–586 CrossRef PubMed.
- P. A. Yakkala, S. R. Panda, S. Shafi, V. G. M. Naidu, M. S. Yar, P. N. Ubanako, S. A. Adeyemi, P. Kumar, Y. E. Choonara, E. v. Radchenko, V. A. Palyulin and A. Kamal, Molecules, 2022, 27, 7642 CrossRef CAS PubMed.
- O. Gupta, T. Pradhan, R. Bhatia and V. Monga, Eur. J. Med. Chem., 2021, 223, 113606 CrossRef CAS PubMed.
- G. Chouhan, M. Islamuddin, M. Y. Want, M. Z. Abdin, H. A. Ozbak, H. A. Hemeg, D. Sahal and F. Afrin, Parasites Vectors, 2015, 8, 183 CrossRef PubMed.
- R. N. Duffin, V. L. Blair, L. Kedzierski and P. C. Andrews, J. Inorg. Biochem., 2020, 203, 110932 CrossRef CAS PubMed.
- K. Vats, R. Tandon, Roshanara, M. A. Beg, R. M. Corrales, A. Yagoubat, E. Reyaz, T. H. Wani, M. S. Baig, A. Chaudhury, A. Krishnan, N. Puri, P. Salotra, Y. Sterkers and A. Selvapandiyan, Biochim. Biophys. Acta, Mol. Cell Res., 2023, 1870, 119416 CrossRef CAS PubMed.
- K. Das Manandhar, T. P. Yadav, V. K. Prajapati, S. Kumar, M. Rai, A. Dube, O. N. Srivastava and S. Sundar, J. Antimicrob. Chemother., 2008, 62, 376–380 CrossRef CAS PubMed.
- V. V. Andrade-Neto, K. M. Rebello, T. M. Pereira and E. C. Torres-Santos, Antimicrob. Agents Chemother., 2021, 65, 1–11 CrossRef PubMed.
- E. Pitsillou, J. Liang, K. Ververis, A. Hung and T. C. Karagiannis, J. Mol. Graphics Modell., 2021, 104, 107851 CrossRef CAS PubMed.
- H. Krishna, P. Anchi, K. Lakshmi, J. Prakash, C. Godugu, N. Shankaraiah and A. Kamal, Bioorg. Chem., 2021, 117, 105461 CrossRef PubMed.
- P. A. Yakkala, S. R. Panda, S. Shafi, V. G. M. Naidu, M. S. Yar, P. N. Ubanako, S. A. Adeyemi, P. Kumar, Y. E. Choonara, E. V. Radchenko, V. A. Palyulin and A. Kamal, Molecules, 2022, 27, 7642 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedure and characterization of new compounds (1H and 13C NMR spectra).See DOI: https://doi.org/10.1039/d4ra06510g |
| This journal is © The Royal Society of Chemistry 2025 |