 Open Access Article
Open Access ArticleSERS-assisted characterization of cell biomass from biofilm-forming Acinetobacter baumannii strains using chemometric tools†
Arslan Yousaf‡
a,
Muhammad Hafeez Ullah‡a,
Haq Nawaz *a,
Muhammad Irfan Majeed
*a,
Muhammad Irfan Majeed *a,
Nosheen Rashidb,
Abdulrahman Alshammaric,
Norah A. Albekairic,
Arslan Alia,
Munawar Hussaina,
Abu Bakar Salfia,
Muhammad Aamir Aslam*d,
Kinza Idreesd and
Allah Dittae
*a,
Nosheen Rashidb,
Abdulrahman Alshammaric,
Norah A. Albekairic,
Arslan Alia,
Munawar Hussaina,
Abu Bakar Salfia,
Muhammad Aamir Aslam*d,
Kinza Idreesd and
Allah Dittae
aDepartment of Chemistry, University of Agriculture Faisalabad, Faisalabad (38000), Pakistan. E-mail: haqchemist@yahoo.com; irfan.majeed@uaf.edu.pk
bDepartment of Chemistry, University of Education, Faisalabad Campus, Faisalabad (38000), Pakistan
cDepartment of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Post Box 2455, Riyadh, 11451, Saudi Arabia
dInstitute of Microbiology, Faculty of Veterinary Sciences, University of Agriculture Faisalabad, Faisalabad (38000), Pakistan. E-mail: aamir.aslam@uaf.edu.pk
eInstitute for Experimental Molecular Imaging, RWTH Aachen University Hospital, Aachen 52074, Germany
First published on 10th February 2025
Abstract
Acinetobacter baumannii (A. baumannii) is an emerging Gram-negative nosocomial pathogen responsible for infection on a global scale. It has the ability to develop biofilms on different surfaces, especially abiotic surfaces, which is considered a major contributor of its pathogenicity. Surface-enhanced Raman spectroscopy (SERS) holds great potential as an effective method for identifying and characterizing the biochemical composition of biofilm-forming species. In this study, cell mass samples from different strains of A. baumannii, categorized based on their biofilm-forming ability (strong, medium and non-biofilm forming) using a 96-well microtiter plate assay (MTP), were analyzed by SERS. The identified spectral features of the SERS spectra were used to characterize bacterial strains capable of producing biofilms. Silver nanoparticles (Ag-NPs) served as the SERS substrate to differentiate biofilm-forming strains of A. baumannii. Chemometric tools, such as principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA), were employed for the classification and differentiation of SERS spectra from bacterial strains with varying biofilm-producing capacities, achieving 100% sensitivity, 94.3% specificity, and an area under the curve (AUC) value of 0.81 through Monte Carlo cross-validation. Furthermore, K-fold (Leave-K-out cross-validation (LKOCV)) was applied to verify the robustness of the PLS-DA model, and the AUC value was found to be 0.90, with a sensitivity of 100% and specificity of 98%. These results demonstrate that the PLS-DA model is highly effective for the differentiation and classification of bacterial strains with varying capacities for biofilm production.
1 Introduction
A. baumannii is a globally emerging Gram-negative coccobacillus pathogen associated with antibiotic resistance. It is commonly found in soil, water, food products and medical devices and on human skin.1–3 An estimated 1 million cases of A. baumannii infections are recorded annually worldwide, with mortality rates ranging from 20% to 80%.4 Notably, A. baumannii has the ability to form biofilms on both biotic (such as fungal filaments and epithelial cells) and abiotic surfaces (polystyrene and glass), and this ability plays a crucial role in the onset of nosocomial infections. Common nosocomial infections caused by A. baumannii include skin and soft tissue infections, urinary tract infections (UTI), pneumonia, meningitis, wound infections, bacteremia, burns and endocarditis. Bloodstream infections and ventilator-associated pneumonia caused by A. baumannii have been linked to significant mortality, with rates reaching as high as 35%.5 Different strains of A. baumannii have developed resistance to numerous categories of drugs and demonstrate a high level of adaptability for genetic exchange. The phenomenon of biofilm development in A. baumannii is influenced by various factors, such as the presence of antibiotic resistant genes, the environmental conditions in which the bacteria grow and the bacterial cell density.6 These biofilms are composed of bacterial cells embedded in teichoic acid, polysaccharide intercellular adhesin (PIA), and a cell-surface-associated extracellular matrix composed of proteins.7Biofilms are complex microbial communities encased in an extracellular matrix (ECM) composed of extracellular polymeric substances (EPS), which include proteins, DNA and polysaccharides.8,9 These communities can comprise various microorganisms, including protozoa, fungi, algae and both Gram-positive and Gram-negative bacteria. The EPS matrix plays a key role in biofilm formation by facilitating cell-to-cell communication (quorum sensing), protecting bacteria from environmental stresses, and preventing desiccation, as water can constitute up to 98.5% of the biofilm mass.10 Recent research highlights a communication process known as auto-induction, where cells exchange signals within and between bacterial species. The Gram-negative bacteria typically use acylated homoserine lactones as auto-inducers, while the Gram-positive bacteria employ processed oligopeptides for communication.11
Biofilms are typically found on aortic and pulmonary valves, intrauterine devices, central venous catheters, contact lenses, urinary catheters, and water lines in dental units. Among the major causes of dangerous infections in patients, one is biofilms that grow on prosthetic heart valves or surrounding tissue. Microorganisms have been shown to smoothly attach and grow on the stitching cuff fibers of valves.12 The phenomenon of biofilm development in A. baumannii is influenced by various factors, such as the existence of antibiotic resistance genes, the environment in which bacteria grow, and the density of the bacterial cells.6 Quorum sensing is employed by both Gram-positive and Gram-negative bacteria to regulate several physiological functions, including biofilm formation, pathogenicity, antibiotic synthesis, motility and conjugation sporulation.11 Quorum sensing controls common steps in the development of biofilms, which include initial attachment to the biotic or abiotic surface, formation of a microcolony, maturation and formation of the architecture, and lastly, detachment of the biofilm.13 Due to its ability to develop resistance to all currently available antimicrobial agents, A. baumannii has been classified as a “red alert” human pathogen.14A. baumannii has been added to the drug-resistant and antimicrobial resistance bacteria research priority list of the World Health Organization (WHO), due to its rising multidrug-resistant strains, mortality and high morbidity.15
Different techniques, including confocal laser scanning microscopy (CLSM),16 scanning electron microscopy (SEM),17 X-ray crystallography,18 transmission electron microscopy (TEM),19 Fourier transform infrared spectroscopy (FTIR)20 and nuclear magnetic resonance (NMR) spectroscopy,21 are widely used for the characterization of bacterial biofilms. Notably, CLSM, SEM, TEM and AFM are the most important microscopic techniques for the visualization of biofilm morphology from the nanometer scale to the micrometer scale on sample surfaces. However, these techniques are limited due to their low axial resolution, time-consuming sample preparation and high cost.22–24 Moreover, TEM, FTIR, X-ray crystallography and NMR provide information associated with biochemical composition; however, they have some drawbacks. TEM is a pre-treatment technique involving procedures, such as freezing and fixations, which may modify the biofilm structure.22,23 FTIR, NMR and IR are not suitable for aqueous environments, time-consuming experiments, and need extensive sample preparation.25–27 X-ray crystallography requires a large quantity of powdered sample, and its high energy can destroy the sample.28,29 In order to overcome these problems, the development of a rapid identification method is essential for the differentiation and classification of bacterial strains having different capacities of producing biofilms.30
Raman spectroscopy is a practical technique that can be used to overcome the limitations of the abovementioned techniques.31 This is a non-destructive, suitable analytical technique used for the rapid identification and characterization of samples in aqueous environments. However, it faces challenges due to its weak signal, which can cause sample degradation at high laser intensities and long exposure time.32 SERS has effectively resolved the challenges with Raman scattering by enhancing the Raman signal by up to 1014 through adherence of sample molecules in close proximity to a roughened surface of metallic nanostructures.33 Silver nanoparticles (Ag-NPs) with suitable size and morphology can serve as a SERS platform to increase the detection sensitivity.34 Due to its great sensitivity, low cast, and ease of sample preparation, SERS has great potential for quickly and accurately identifying bacterial species and the molecular structure of bacteria, including carbohydrates, proteins, nucleic acids and lipids, and can be responsible for their characteristic SERS spectral features.35
In the present study, SERS has been employed using Ag NPs as a substrate for the rapid, sensitive and cost-effective discrimination among three different bacterial strains having different capacities for producing biofilms, which are categorized as strong, medium and non-biofilm forming on the basis of a 96-well microtiter plate assay (MTP). Moreover, SERS spectral data of different biofilm forming and non-biofilm forming strains are compared to identify specific SERS features that are directly or indirectly related to biochemical changes that occur due to the development of biofilm matrix and the unique features that correspond to individual strains. Moreover, different chemometric techniques such as PCA are used to identify the individual strains and PLS-DA to classify the biofilm forming and non-biofilm forming strains.
2 Materials and methods
2.1 Cell mass pellet isolation of bacteria from culture media
The A. baumannii strains are grown at 37 °C for 48 hours to produce biofilms in 96-well polystyrene microtiter plates. The optical density (OD) at 570 nm was measured by using the 96-well microtiter plate assay (MTP) prior to staining with crystal violet (0.1%) W/V.36 This assay helped to categorize the bacterial strains on the basis of their ability to produce biofilms. In this classification, the mean optical density (OD) of the negative control is three standard deviations lower than the optical density cutoff value (ODc), which is measured at 570 nm. Using their relative ODc, isolates of A. baumannii biofilm formers are classified into three categories based on their ability to form biofilms. A non-biofilm former revealed OD ≤ ODc, medium biofilm shown ODc < OD ≤ 2 × ODc and strong biofilm former shown 4 × ODc < OD. The bacterial isolates of fifteen different A. baumannii strains were collected from institutional stock cultures for this investigation. Five isolates were identified in each category using the above described criteria. OD values for the negative control were 0.091 ± 0.008. Based on these values, ODc was calculated to be 0.102. For non-biofilm producers: OD ≤ 0.102, for medium biofilm producers: 0.102 < OD ≤ 0.204, for strong biofilm former 0.408 < OD. Five isolates for each of the previously identified biofilm-forming categories were revived overnight in tryptic soy broth (TSB) at 37 °C. On nutrient agar plates, the bacterial growth was streaked to observe the colony morphology. For this purpose, 1 ml of sterile TSB was used to inoculate a separate colony, which was stirred at 180 rpm for 16 hours at 37 °C. The bacterial cell mass pellets were isolated after the growth centrifugation for consecutive 5 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. For further spectroscopic investigation, the cell mass pellets were stored separately at 4 °C.29
000 rpm. For further spectroscopic investigation, the cell mass pellets were stored separately at 4 °C.29
2.2 Silver nanoparticles (Ag-NPs) preparation
The materials used in the chemical reduction process to produce Ag-NPs include trisodium citrate and silver nitrate. Briefly 500 ml of deionized water and 0.085 g of silver nitrate were combined, followed by heating to 100 °C. After that, 0.125 g of trisodium citrate was added as a reducing agent and capping ligand. In order to get monodispersed grey-colored silver nanoparticles, the solution was heated on a hot plate with continuous magnetic stirring for an hour.37 For the characterization of these nanoparticles, SEM38 and TEM39 are utilized to determine the size (65 × 45 nm) and morphology (oval-shaped) of Ag NPs.2.3 SERS spectral acquisition from bacterial strains
For SERS spectral measurements, 40 μl of bacterial cell mass pellets suspended in an equal volume of the saline solution were incubated with 40 μl of Ag-NPs in a microcentrifuge tube, and left to interact for 40 min. The purpose of this incubation is to enhance the close binding of nanoparticles with the cell mass/pellet samples. The spectral measurements were performed by utilizing the (OPTOSKY ATR8300) Raman spectrometer instrument equipped with a 40× objective lens and a 785 nm laser source. For SERS analysis through the Raman spectrometer, approximately 80 μl of the incubated mixture was positioned on an aluminum substrate. A total of 15 spectra were collected for all bacterial pellet samples, covering the spectral range of 300–1800 cm−1, with an integration time of 8 seconds and laser power of 50 mW. Five samples of all three noticeable classes of clinical isolates of A. baumannii bacteria categorized into non, medium and strong biofilm-forming bacteria are used in this study. So, a total of 15 samples are used for the purpose of sample collection, resulting in a total of 225 (15 × 15 spectra) SERS spectra, as 15 SERS spectra are collected per sample.372.4 Pre-processing of the SERS spectral data
MATLAB 7.8a software was employed to preprocess the SERS spectral raw data that provide significant and useful information related to the sample, but also have undesired backgrounds such as baseline, noise and substrate contributions, which can affect the usefulness of the data related to the sample. The vector normalization, baseline correction, smoothing and background removal methods were used for pre-processing of the spectra. To remove the undesired components, various methods were implemented, such as Savitzky–Golay for smoothing purpose, vector normalization for normalizing data, polynomial and the Rubber band method for baseline correction, and subtraction to remove the substrate.292.5 Data analysis
In the present study, two multivariate chemometric methods, PCA and PLS-DA, were used to analyze the multivariate spectral datasets.40 PCA is an unsupervised chemometric tool that is used to identify the structural changes of biomolecules in high biofilm-forming bacterial strains in comparison to those bacterial strains that do not form biofilms. The distinguishing SERS features in the spectra acquired from pellets of both high biofilm (strong, medium) and non-biofilm forming of A. baumannii strains can be associated with these biochemical differences. The analysis of discrimination and variability in several spectral data sets of bacterial strains was conducted using the PCA scatter score. The reasons for this discrimination, as revealed by PCA loadings, were also examined.41 PCA is used for dimensionality reduction, and to assess the differentiation and variability within different data sets and convert the covariance variables into the variance. The first principal component (PC-1) can be used to calculate the maximum variability, and then the second maximum from the remaining variability is revealed by the second principal component (PC-2) and so on.42 PCA analysis is used to form the separate clusters of the different data sets of SERS spectra of different samples based on their variability from one another, but initial knowledge of the samples is not required for it.PLS-DA is a supervised chemometric method that is applied in the current study on SERS spectra for calibration and validation of the preprocessed spectral dataset in order to categorize the respective bacterial strains into non, medium, and strong based on their biofilm forming ability.38 Monte Carlo and K-fold cross validation through LOOC (Leave one sample out cross validation) were used to build the PLS-DA model. Monte Carlo and LKOCV were generally implemented for the purpose to ensure maximum statistical relevancy, and also to select the minimum number of latent variables, but avoiding the over fitting under the given spectral data. It can also be used to demonstrate that the analysis is not biased, as all spectra collected from a sample are either included in the calibration or the test sets, but cannot be represented in both. Particularly, calibration was performed on 60% of data sets by selecting it randomly, and the remaining 40% of data sets were utilized for prediction.43 In order to determine the specificity and sensitivity of the classification of SERS data sets, the ROC curve was used to display the results of the SERS spectral group classification as false positive, true positive, false negative, and true negative.37
3 Results and discussion
3.1 Mean of SERS spectra
In Fig. 1, the SERS mean plot of all three classes of biofilm-forming A. baumannii strains shows some characteristic SERS spectral features, which differentiated the biofilm-forming A. baumannii strains from the ones that are not capable of doing so on the basis of the difference in their biomolecular components. The differentiating SERS bands of the cell mass/pellet samples are associated with the biomolecular changes in the DNA/RNA, amino acids, proteins, carbohydrates and lipids. Major distinguishing SERS spectral features are labeled with solid lines, while intensity-based differences in peaks are labeled with dashed lines. The peak assignments, according to literature relevant to the SERS mean plot of the high biofilm and non-biofilm forming A. baumannii strains in Fig. 1, are described in Table 1, along with the relevant citations. | ||
| Fig. 1 Mean plot of the SERS spectra of non, medium and strong biofilm-forming strains of A. baumannii. | ||
| Raman shift (cm−1) | Assignments | Components | References |
|---|---|---|---|
| a ν = Stretching (vs = symmetric, vas = antisymmetric), δ = bending/scissoring, ρt = twisting. | |||
| 326 | β (C–C–O) ring vibration | Carbohydrates | 37 |
| 343 | C–C–O δ, C–OH ρt | Carbohydrates | 44 |
| 393 | S–S ν | Carbohydrates | 45 |
| 420 | (C–C–C deformation) | Carbohydrates | 38 |
| 467 | C–C–O δ, C–OH ρt, β(CCC), β(CCO), β(OCO) | Carbohydrates | 44 and 46 |
| 516 | S–S ν, disulfide | Proteins | 47 and 48 |
| 633 | C–C twisting of guanine ring region | DNA | 49 and 47 |
| 679 | Thymine | DNA | 50 |
| 699 | (Guanine (COO deformation) | DNA | 51 and 52 |
| 720 | Adenine, glycosidic ring mode of adenine | DNA | 53 and 54 |
| 729 | Adenine | DNA | 29 |
| 747 | (C–S) ν | Proteins | 55 |
| 762 | Ring breathing of tryptophan | Proteins | 56 |
| 861 | C–C ν, C–O–C 1,4-glycosidic linkage | Proteins | 57 and 53 |
| CCH deformation in ring breathing of tyrosine | |||
| 953 | υ(CO) and C–N of proteins | Proteins | 58 |
| 990 | C–C ring breathing in phenylalanine | Proteins | 59 and 60 |
| 1030 | C–H δ | Proteins | 61 |
| 1070 | C–N ν. (protein) | Proteins | 62 |
| 1097 | PO2− vs | DNA/RNA | 29 |
| 1130 | (δ (CC, CO, –COH) | Carbohydrates | 38 |
| 1327 | δ(CH) | Carbohydrates | 7 |
| 1395 | COO−1vs | Proteins | 63 |
| 1445 | CH2 δ vibration (lipid band) | Lipids | 46 |
| 1550 | Tryptophan | Proteins | 64 |
| 1693 | Amide I α-helix | Proteins | 49 |
The major differentiating SERS bands in the mean spectral plot of bacterial strains include 420 (C–C–C deformation), 467 (C–OH ρt, β(CCC), β(CCO), β(OCO)), 516 (S–S ν, disulfide), 633 (the guanine ring region and the tyrosine amino acid chain include aromatic rings with a C–C configuration), 699 (guanine (COO deformation), 720 (adenine), 729 (the glycosidic ring conformation of adenine), 762 (ring breathing of tryptophan), 953 (υ(CO) and C–N of proteins), 990 (C–C ring breathing of phenylalanine), 1030 (C–H δ), 1070 (C–N ν of proteins), 1130 (CC, CO, –COH), 1327 (CH), 1395 (COO−1vs), 1445 (CH2δ vibration (lipid band)) and 1693 cm−1 (amide I α-helix).
The dashed lines are used to represent the intensity-based differences of the SERS bands in the non-biofilm and high biofilm-forming strains, indicating that all sample strains belong to the same species of A. baumannii; hence, they have some similar biomolecular features. These SERS features include 326 (β (C–C–O) ring vibration), 343 (C–C–O δ, C–OH ρt), 393 (S–S ν), 679 (thymine), 747 ((C–S) ν), 861 (C–C ν, C–O–C 1,4-glycosidic linkage, CCH deformation in the ring breathing of tyrosine, namely in the CCH group; Gram-positive bacterium cell wall (teichuronic acid, which is associated with protein), 1097 (PO2-symmetrical ν) and 1550 cm−1 (tryptophan).
The SERS features observed that are related to carbohydrates include those at 326, 343, 393, 420, 467, 1130 and 1327 cm−1. The SERS bands at 420 (C–C–C deformation), 467 (C–OH ρt, β(CCC), β(CCO), β(OCO), corresponding to a specific sugar profile), 1130 ((δ (CC, CO, –COH) (specific SERS feature of glucose) and 1327 cm−1 (belonging to δ(CH)) appeared in biofilm-forming bacterial strains, and were absent in non-biofilm-forming strains.65,66 The SERS biomarkers at 326 (β (C–C–O) ring vibration), 343 (C–C–O δ, C–OH ρt), and 393 cm−1 (S–S ν) are observed in both strong and non-biofilm forming strains, but the increasing intensities of these SERS biomarkers indicate the high contents of polysaccharides in the cell mass/pellets of biofilm-forming strains in comparison to those that do not form biofilms. The peak observed at 393 cm−1 related to S–S ν has lower intensity in the biofilm-forming A. baumannii strains than that in those A. baumannii strains which do not form biofilms. In the process of biofilm formation and accumulation, the concentration of polysaccharide intercellular adhesion (PIA) is high, which mediates the attachment of A. baumannii bacteria to the fibrous net that builds the biofilm mass.67–69
The SERS bands related to proteins are 516 (S–S ν, disulfide), 747 ((C–S) ν), 762 (ring breathing of tryptophan), 861 (C–C ν, C–O–C 1,4-glycosidic linkage; cell wall of Gram-positive bacteria associated with protein), 953 (υ(CO) and C–N of proteins), 990 (C–C ring breathing in phenylalanine), 1070 (C–N ν of proteins), 1395 (COO−1 symmetric ν), 1550 (tryptophan) and 1693 cm−1 (alpha-helix of Amide I), of which the peaks at 762 (ring breathing of tryptophan), 990 (C–C ring breathing in phenylalanine), 1395 (COO−1 symmetric ν) and 1693 cm−1 (alpha-helix of Amide I) are present only in bacterial strains involved in biofilm formation. Moreover, the SERS band at 953 cm−1 (υ(CO) is present only in the non-biofilm-forming A. baumannii strains and absent in the biofilm-forming A. baumannii strains (medium, strong) due to alterations in the protein structure.70 The SERS features, including 516 (S–S ν, disulfide), 747 ((C–S) ν), 861 (C–C ν, C–O–C 1,4-glycosidic linkage, CCH deformation in tyrosine ring breathing; cell wall of Gram-positive bacteria) and 1550 cm−1 (tryptophan), are observed in all bacterial cell mass samples with intensity-based differences. There are thirteen cell wall anchor proteins (CWA) related to virulence in A. baumannii, which may be helpful in biofilm development and are covalently bound to peptidoglycan in the cell wall. Moreover, multimeric cellular proteins, structural proteins and surface proteins (such as pili, flagella, fimbriae and appendages proteins) are also present in the biofilm forming process. The function of these proteins in the biofilm-forming process is to connect the cells with one another through various microorganisms.71,72
SERS features observed at 633 (C–C twisting of the guanine ring region), 679 (thymine), 699 (guanine (COO deformation), 720 (adenine), 729 (the glycosidic ring conformation of adenine) and 1097 cm−1 (PO2− symmetrical ν) are linked to DNA. The SERS band at 633 (the guanine ring region and the tyrosine amino acid chain include aromatic rings with a C–C configuration), 699 (guanine (COO deformation), 720 (adenine) and 729 cm−1 (the glycosidic ring conformation of adenine) are found only in bacterial strains capable of producing biofilms, and not in ones that are unable to do so. This indicates the high contents of DNA in biofilm-forming strains due to the double strand chromosomal plasmid DNA utilized in New Delhi metallo beta-lactamase (NDM) synthesis, which transforms and replicates its genes in new colonies that serve as hosts and play a crucial role in bacterial cell adhesion to surfaces and biofilm formation. This plasmid genetic transformation of information changes the non-biofilm-forming bacterial strains into biofilm-forming bacterial strains.73–75 Moreover, the SERS band at 679 (thymine) and 1097 cm−1 (PO2− vs) are observed in all samples with intensity-based differences.
3.2 PCA (principal component analysis)
PCA is used for dimensionality reduction, and to assess the differentiation and variability within different data sets of preprocessed SERS spectra. The PCA scatter plot of strong, medium and non-biofilm forming strains of A. baumannii is illustrated in Fig. 2, showing the spectra of strong (green dots), medium (blue dots) and non-biofilm-forming strains (red dots). While the SERS spectra of non-biofilm forming A. baumannii strains cluster on the negative side of PC-1, the spectra of strong biofilm-forming strains cluster on the positive side of PC-1, with an explained variance of 75.70% and accounts for 13.53% variance for the medium biofilm-forming strains grouped along the positive axis of PC-2. Notably, the SERS spectral dots of the biofilms-forming (strong, medium) strains are far from the non-biofilm forming strain, indicating the clear spectral differences in the SERS spectral datasets of the A. baumannii biofilm forming strains Fig. 2.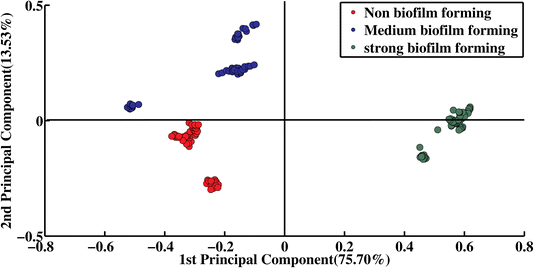 | ||
| Fig. 2 PCA scatter plot of the score for the SERS spectral groups of non, medium and strong biofilm-forming A. baumannii strains. | ||
Fig. 3a illustrates a pairwise comparison of the PCA scatter score of the SERS spectra of non- and medium biofilm-forming strains of A. baumannii bacteria, in which the SERS spectral datasets are differentiated along PC-2 with variability of 23.59%, while 60.68% variance was obtained, as explained by PC-1. The spectra of the medium biofilm A. baumannii strains concentrated along the positive axis of PC-2 are shown with blue dots, while the red dots indicate the spectra of the non-biofilm forming A. baumannii strain, which are grouped along the negative axis of PC-2. This indicates that the changes in the biomolecular contents of the two types of A. baumannii bacterial strains through their SERS spectral features. The basis for distinction in the SERS spectral datasets of the bacterial cell mass (pellets) is explained by the pairwise PCA loadings in Fig. 3b, where the negative loadings represent the SERS spectral bands of bacterial strains which do not form a biofilm as their SERS spectra are clustered on the negative side as red dots in the PCA scatter plot, including characteristic SERS bands at 326, 343, 467, 729, 990, 1327, 1395 and 1693 cm−1. The positive loadings represent the spectra of samples of medium biofilm strains, which appear on the positive side as blue dots in the PCA scatter plot and are observed at 393, 633, 679, 762, 861 and 1097 cm−1.
 | ||
| Fig. 3 Pairwise PCA: (a) scatter plot of scores and (b) loadings for SERS spectral groups of non-biofilm- versus medium biofilm-forming strains. | ||
Fig. 4a shows a PCA scatter score of the spectral data of cell mass samples of A. baumannii strains producing medium and strong biofilms, which clearly separates the spectral datasets of strains producing medium and strong biofilms, providing an explanation for 79.70% of the variation along PC-1. The spectra of the bacteria with a medium level of biofilm formation are clustered on the negative side of PC-1, which are represented by blue dots, whereas bacteria with a strong biofilm formation tendency are shown by green dots concentrated along the positive side of PC-1. The two clusters of spectral data are distant from each other, showing significant differences in the SERS spectral characteristics of these groups of samples at the biochemical level.
 | ||
| Fig. 4 Pairwise PCA: (a) scatter plot of scores and (b) loadings of the SERS spectra of strong biofilm- versus medium biofilm-forming A. baumannii strains. | ||
Fig. 4b presents data regarding distinct biochemical characteristics in the manner of pairwise comparison of the SERS spectra of strong versus medium biofilm-forming A. baumannii strains as PCA loadings. The loadings on the positive side of PC-1 are observed at 393 (S–S ν of carbohydrates), 679 (thymine related to DNA), 720 (adenine), 953 (υ(CO) and C–N of proteins) and 1395 cm−1 (the protein has COO−1 symmetry in its structure), as the SERS spectral features of the strong biofilm (which are green dots) are plotted on the positive side of the PC-1 scatter plot. Meanwhile, the negative loading indicates the SERS bands of medium biofilm-forming strains that include 467 (C–C–O δ, C–OH ρt, β(CCC), β(CCO), β(OCO) in carbohydrates), 861 (C–C ν, C–O–C 1,4-glycosidic linkage, CCH deformation in ring breathing of tyrosine, namely in the CCH group; Gram-positive bacterium cell wall (teichuronic acid, which is associated with protein), 1097 (PO2− vs in DNA), and 1550 cm−1 (tryptophan linked to protein), which are represented with blue dots scattered on the negative side of the PC-1.
The pairwise PCA of the SERS spectra of the non-biofilm forming A. baumannii strains (red dots), on the negative axis, and the strong biofilm-forming A. baumannii strains (green dots), along the positive axis of PC-1, is shown in Fig. 5a. PC-1 exhibits a variance of 79.70%, while 16.53% is shown along PC-2. The SERS spectra of both types of bacterial strains are differentiated from each other on the basis of biomolecular changes, which can be associated with their capacity of producing the biofilm.
 | ||
| Fig. 5 Pairwise PCA: (a) scatter plot of scores and (b) loadings of the SERS spectra of non-biofilm- versus strong biofilm-forming A. baumannii strains. | ||
Fig. 5b displays the PC-1 loadings for the PCA of the SERS spectra from the cell mass samples of the strong non-biofilm forming A. baumannii strains. The positive loadings correspond to spectra of strong biofilm-forming (green dots) strains grouped on the positive axis, and the negative loadings are associated with the spectra of the non-biofilm forming (red dots) strains clustered on the negative axis. The major SERS bands of the strong biofilm strains in the positive loadings include 393 (S–S ν in carbohydrates), 679 (related to guanosine, thymine in DNA), 720 (adenine), 953 (υ(CO) and C–N of proteins) and 1395 cm−1 (COO−1vs group that is commonly found in proteins), while the non-biofilm SERS features in negative loading are observed at 420, 467, 699, 861, 1097 and 1550 cm−1.
3.3 PLS-DA (partial least squares discriminant analysis)
PLS-DA is a statistical tool employed to discriminate and classify data sets of SERS spectra of A. baumannii strains based on their biofilm-forming capabilities, categorized as non-, medium and strong biofilm-forming. To prevent overfitting, the SERS spectral data were split into 60% for calibration and 40% for validation, ensuring unbiasedness. To construct the PLS-DA model, 14 latent variables were chosen as the optimum latent variables, which are shown in the ESI (Fig. S1).†Fig. 6 displays the scatter score of the PLS-DA model showing the SERS spectra of the strong (green dots), medium (blue dots), and non-biofilm forming bacterial strains (red dots) on the positive axis, negative axis, and the negative side of the PLS-DA scatter score plot, respectively. This clearly differentiates between the SERS spectra of the positive biofilm strains (blue clustered dots, green clustered dots) and the negative biofilm strains (red clustered dots) due to their characteristic SERS spectral features, which can be associated with the increasing concentration levels of DNA, protein, lipids and carbohydrates in the high biofilm-forming strains.
 | ||
| Fig. 6 PLS-DA scatter plots: (a) Monte Carlo and (b) K-fold cross-validation of the SERS spectra of non, medium and strong biofilm-forming strains of A. baumannii. | ||
The SERS features observed in the PLS-DA loading plot of the non-, medium and strong biofilm-forming strains of A. baumannii are shown in the ESI (Fig. S2),† corresponding to the SERS bands observed in the mean plot of Fig. 1. The SERS spectral features observed in the non-, medium and strong biofilm-forming strains include bands at 326, 343, 420, 516, 633, 679, 720, 861, 953, 990, 1097, 1130, 1327, 1445 and 1693 cm−1.
Fig. 7 displays the ROC curve of the PLS-DA model used to classify the SERS spectral data sets of the positive and negative biofilm strains. The ROC curve was drawn between 1-specificity (false positive rate) and sensitivity (true positive rate). The Monte Carlo validation method was employed, and the Area Under the Curve (AUC) value for the PLS-DA model is found to be 0.81 with a sensitivity of 100% and specificity of 94.3%. Moreover, LKOCV was applied to further verify the robustness of the classification model. The values of AUC (0.90), sensitivity (100%), and specificity (98%), respectively, are determined through LKOCV for the PLS-DA model. A value of AUC at close to 1 indicates that the model is a good fit with the high accuracy and validation, while a value at close to zero would suggest an inaccurate fit and bad performance of this model.
The biofilm-forming and non-biofilm-forming bacterial strains have common intensity-based different SERS spectral features that are labeled with dotted line, and some features that are present with the same intensity in the biofilm-forming and non-biofilm-forming strains are shown in Fig. 1. These features may complicate the chemometric model's ability to achieve 100% specificity. Although SERS provides high sensitivity, some biochemical features may be common across different strains, making it difficult for the chemometric models to differentiate between them with 100% accuracy. The chemometric model PLS-DA relies on spectral features to classify strains. This model is effective, but sometimes struggles with borderline cases where the spectra of the biofilm and non-biofilm strains have common features. This results in misclassification, which reduces the specificity of the model. Improvements can be achieved through optimized preprocessing, by minimizing the variation in the experimental conditions, and by addressing the class imbalance.76,77 These steps should help to enhance the model's performance and bring the specificity closer to 100%.
4 Conclusions
The SERS technique is found to be an effective method in recognizing the SERS spectral characteristic features related to the biofilm-forming ability of three types of A. baumannii strains, including biofilm-forming (medium and strong) and non-biofilm forming. Numerous SERS spectral features were identified to differentiate the SERS spectra of the high biofilm-forming bacterial strains from the non-biofilm one. The SERS spectra of biofilm and non-biofilm forming strains of A. baumannii bacterial cell mass samples are classified, and differentiated with chemometric stools like PCA and PLS-DA. The major SERS bands found to be solely present in biofilm-forming strains are observed at 420 (C–C–C deformation), 467 (C–OH ρt, β(CCC), β(CCO), β(OCO)), 516 (S–S ν, disulfide), 633 (C–C twisting of the guanine ring region), 699 (guanine (COO deformation), 720 (adenine), 729 (glycosidic ring conformation of the adenine), 762 (ring breathing of tryptophan), 953 (υ(CO) and C–N of proteins), 990 (C–C ring breathing exhibited in phenylalanine), 1030 (C–H δ), 1070 (C–N ν of proteins), 1130 ((δ (CC, CO, –COH), 1327 (δ(CH), 1395 (COO−1vs), 1445 (CH2δ vibration (lipid band)) and 1693 cm−1 (Amide I α-helix). These differentiating SERS features indicate high contents of proteins, carbohydrates, lipids and DNA biomolecules, which are considered to play a crucial role in the formation of biofilms in A. baumannii bacteria. The PLS-DA model also differentiates and discriminates the spectral features of SERS in the biofilm-forming and non-biofilm forming strains with 100% sensitivity, 94.3% specificity, and 81% AUC by Monte Carlo cross validation. To further enhance the robustness of the model, LKOCV validation was employed and the AUC, sensitivity and specificity of the PLS-DA model through LKOCV was found to be 90%, 100% and 98%. These results of the PLS-DA model indicate that the model is the best fit for the differentiation and classification of the SERS spectral groups of three different types of A. baumannii strains having different biofilm-forming capabilities and categorized as medium, strong and non-biofilm forming strains.Ethical statement
This study was performed in strict accordance with the NIH guidelines for the protection of human subjects (45 CFR 46), and was approved by the Institutional Ethical Review Board (IERB) of Nishtar Medical University Multan, Pakistan (Reference No. 2230, dated 02-02-24). Informed consent was obtained from all human subjects.Data availability
All data underlying the results are available within the article, and no additional source data are necessary.Conflicts of interest
The authors have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors are thankful to the Researchers Supporting Project number (RSP2025R491), King Saud University, Riyadh, Saudi Arabia.References
- M.-F. Lin and C.-Y. Lan, World J. Clin. Cases, 2014, 2, 787 CrossRef PubMed.
- A. Gedefie, W. Demsis, M. Ashagrie, Y. Kassa, M. Tesfaye, M. Tilahun, H. Bisetegn and Z. Sahle, Infect. Drug Resist., 2021, 3711–3719 Search PubMed.
- P. Espinal, S. Martí and J. Vila, J. Hosp. Infect., 2012, 80, 56–60 CAS.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Front. Microbiol., 2019, 10, 539 CrossRef PubMed.
- P. Nordmann, Pathol. Biol., 2004, 6, 301–303 CrossRef PubMed.
- J. A. Gaddy, A. P. Tomaras and L. A. Actis, Infect. Immun., 2009, 77, 3150–3160 CrossRef CAS PubMed.
- U. Neugebauer, U. Schmid, K. Baumann, W. Ziebuhr, S. Kozitskaya, V. Deckert, M. Schmitt and J. Popp, ChemPhysChem, 2007, 8, 124–137 CrossRef CAS PubMed.
- J. Wimpenny, W. Manz and U. Szewzyk, FEMS Microbiol. Rev., 2000, 24, 661–671 CrossRef CAS PubMed.
- H.-C. Flemming and J. Wingender, Nat. Rev. Microbiol., 2010, 8, 623–633 CrossRef CAS PubMed.
- N. P. Ivleva, P. Kubryk and R. Niessner, J. Phys. Chem. B, 2017, 409, 4353–4375 CAS.
- M. B. Miller and B. L. Bassler, Annu. Rev. Microbiol., 2001, 55, 165–199 CAS.
- E. Efeoglu and M. Culha, J. Appl. Spectrosc., 2013, 67, 498–505 CAS.
- M. Jamal, U. Tasneem, T. Hussain and S. Andleeb, Formation and Role in Human Infections Research & Reviews, Journal of Microbiology and Biotechnology, 2015, 4, 1–14 CAS.
- G. M. Cerqueira and A. Y. Peleg, IUBMB life, 2011, 63, 1055–1060 CrossRef CAS PubMed.
- B. Havenga, T. Ndlovu, T. Clements, B. Reyneke, M. Waso and W. Khan, BMC Microbiol., 2019, 19, 1–16 CrossRef PubMed.
- J. Lawrence, T. Neu and G. Swerhone, J. Microbiol. Methods, 1998, 32, 253–261 CrossRef CAS.
- M. Alhede, K. Qvortrup, R. Liebrechts, N. Høiby, M. Givskov and T. Bjarnsholt, FEMS Immunol. Med. Microbiol., 2012, 65, 335–342 CrossRef CAS PubMed.
- A. Diehl, Y. Roske, L. Ball, A. Chowdhury, M. Hiller, N. Molière, R. Kramer, D. Stöppler, C. L. Worth and B. Schlegel, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 3237–3242 CrossRef CAS PubMed.
- C. Li, S. Felz, M. Wagner, S. Lackner and H. Horn, Bioresour. Technol., 2016, 200, 128–136 CrossRef CAS PubMed.
- G. Sharma and A. Prakash, J. Microbiol., Biotechnol. Food Sci., 2014, 3, 310–314 Search PubMed.
- P. D. Majors, J. S. McLean, G. E. Pinchuk, J. K. Fredrickson, Y. A. Gorby, K. R. Minard and R. A. Wind, J. Microbiol. Methods, 2005, 62, 337–344 CrossRef CAS PubMed.
- P. Mhaske, S. Kasapis, A. Farahnaky and M. Dokouhaki, Food Biophys., 2022, 17, 165–170 CrossRef.
- L. Lu, Y. Zhao, M. Li, X. Wang, J. Zhu, L. Liao and J. Wang, J. Pharm. Anal., 2024, 14, 100906 CrossRef PubMed.
- Y. Huang, S. Chakraborty and H. Liang, Anal. MethodFood Biophys.s, 2020, 12, 416–432 CAS.
- R. P. Velamakanni, P. Vuppugalla and R. Merugu, Microbial Exopolysaccharides as Novel and Significant Biomaterials, 2021, pp. 19–43 Search PubMed.
- S. M. Wickramasinghe, Engineering Nanomaterials for Imaging and Antibiofilm Applications, Case Western Reserve University, 2020 Search PubMed.
- Y. T. Cheah and D. J. C. Chan, J. Appl. Microbiol., 2022, 132, 3490–3514 CrossRef PubMed.
- N. P. Ivleva, P. Kubryk and R. Niessner, Anal. Bioanal. Chem., 2017, 409, 4353–4375 CrossRef CAS PubMed.
- M. Shakeel, M. I. Majeed, H. Nawaz, N. Rashid, A. Ali, A. Haque, M. U. Akbar, M. Tahir, S. Munir and Z. Ali, Photodiagn. Photodyn. Ther., 2022, 40, 103145 CrossRef CAS PubMed.
- L. Fajir, A. E. Al-Niaame and N. Hussein, Eur. J. Biomed. Pharm. Sci., 2017, 4, 84–91 CAS.
- M. Tahira, M. Munir, H. Nawaz, M. I. Majeed, M. Wasim, M. Naz, M. Asghar, A. Shahzadi, N. Ghafoor and R. Umer, Anal. Lett., 2024, 1–14 Search PubMed.
- T. N. Zu, A. I. Athamneh, R. S. Wallace, E. Collakova and R. S. Senger, J. Bacteriol., 2014, 196, 3983–3991 CrossRef PubMed.
- E. Efeoglu and M. Culha, Spectroscopy, 2013, 28, 36–41 Search PubMed.
- S. Rafiq, M. I. Majeed, H. Nawaz, N. Rashid, U. Yaqoob, F. Batool, S. Bashir, S. Akbar, M. Abubakar, S. Ahmad, S. Ali, M. Kashif and I. Amin, Spectrochim. Acta, Part A, 2021, 259, 119908 CrossRef CAS PubMed.
- S. Bashir, H. Nawaz, M. I. Majeed, M. Mohsin, A. Nawaz, N. Rashid, F. Batool, S. Akbar, M. Abubakar and S. Ahmad, Spectrochim. Acta, Part A, 2021, 258, 119831 CAS.
- S. Stepanović, D. Vuković, I. Dakić, B. Savić and M. Švabić-Vlahović, J. Microbiol. Methods, 2000, 40, 175–179 Search PubMed.
- A. Naman, H. Tahseen, H. Nawaz, M. I. Majeed, A. Ali, A. Haque, M. U. Akbar, N. Mehmood, R. Nosheen and S. Nadeem, Spectrochim. Acta, Part A, 2024, 305, 123414 CrossRef CAS PubMed.
- N. Mehmood, M. W. Akram, M. I. Majeed, H. Nawaz, M. A. Aslam, A. Naman, M. Wasim, U. Ghaffar, A. Kamran and S. Nadeem, RSC Adv., 2024, 14, 5425–5434 RSC.
- R. Z. A. Bari, H. Nawaz, M. I. Majeed, N. Rashid, M. Iqbal, M. Akram, N. Yaqoob, S. Yousaf, A. Mushtaq and F. Almas, Photodiagn. Photodyn. Ther., 2022, 38, 102808 CrossRef CAS PubMed.
- M. Czaplicka, A. Kowalska, A. Nowicka, D. Kurzydłowski, Z. Gronkiewicz, A. Machulak, W. Kukwa and A. Kamińska, Anal. Chim. Acta, 2021, 1177, 338784 CrossRef CAS PubMed.
- J. Matera, A. Cruz, R. Raices, M. Silva, L. Nogueira, S. Quitério, R. Cavalcanti, M. Freiras and C. C. Júnior, Food Res. Int., 2014, 64, 380–386 CrossRef CAS PubMed.
- S. K. Jha, I. Uzunov and X. Zhang, Comput. Model. Eng. Sci., 2021, 126, 991–1009 Search PubMed.
- S. Rafiq, M. I. Majeed, H. Nawaz, N. Rashid, U. Yaqoob, F. Batool, S. Bashir, S. Akbar, M. Abubakar and S. Ahmad, Spectrochim. Acta, Part A, 2021, 259, 119908 CrossRef CAS PubMed.
- Y. Zhao, C.-Y. Ma, S.-N. Yuen and D. L. Phillips, J. Agric. Food Chem., 2004, 52, 1815–1823 CrossRef CAS PubMed.
- M. Kahraman, A. I. Zamaleeva, R. F. Fakhrullin and M. Culha, Anal. Bioanal. Chem., 2009, 395, 2559–2567 CrossRef CAS PubMed.
- A. Mushtaq, H. Nawaz, M. I. Majeed, N. Rashid, M. Tahir, M. Z. Nawaz, K. Shahzad, G. Dastgir, R. Z. A. Bari and A. ul Haq, Spectrochim. Acta, Part A, 2022, 278, 121315 CrossRef CAS PubMed.
- N. A. Mungroo, G. Oliveira and S. Neethirajan, Microchim. Acta, 2016, 183, 697–707 CrossRef CAS.
- A. Dong, P. Huang and W. S. Caughey, Biochemistry, 1990, 29, 3303–3308 CrossRef CAS PubMed.
- S. Bashir, H. Nawaz, M. I. Majeed, M. Mohsin, S. Abdullah, S. Ali, N. Rashid, M. Kashif, F. Batool and M. Abubakar, Photodiagn. Photodyn. Ther., 2021, 34, 102280 CrossRef CAS PubMed.
- C. C. Lin, C. H. Hung, C. W. Chang and L. R. Huang, Appl. Mech. Mater., 2013, 284, 523–527 Search PubMed.
- K. De Gussem, P. Vandenabeele, A. Verbeken and L. Moens, Spectrochim. Acta, Part A, 2005, 61, 2896–2908 CrossRef PubMed.
- K. Schenzel and S. Fischer, Cellulose, 2001, 8, 49–57 CrossRef CAS.
- K. Maquelin, C. Kirschner, L.-P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann and G. Puppels, J. Microbiol. Methods, 2002, 51, 255–271 CrossRef CAS PubMed.
- R. M. Jarvis and R. Goodacre, Anal. Chem., 2004, 76, 40–47 CrossRef CAS PubMed.
- M. Saleem, H. Nawaz, M. I. Majeed, N. Rashid, F. Anjum, M. Tahir, R. Shahzad, A. Sehar, A. Sabir and N. Rafiq, Photodiagn. Photodyn. Ther., 2023, 41, 103278 CrossRef PubMed.
- I. Notingher, S. Verrier, S. Haque, J. Polak and L. Hench, Biopolymers: Original Research on Biomolecules, 2003, 72, 230–240 CrossRef CAS PubMed.
- M. Harz, P. Rösch, K.-D. Peschke, O. Ronneberger, H. Burkhardt and J. Popp, Analyst, 2005, 130, 1543–1550 RSC.
- J. C. Ramirez-Perez and D. Durigo, J. Saudi Chem. Soc., 2022, 26, 101531 CrossRef CAS.
- A. Walter, A. März, W. Schumacher, P. Rösch and J. Popp, Lab Chip, 2011, 11, 1013–1021 RSC.
- Y. Liu, Y.-R. Chen, X. Nou and K. Chao, Appl. Spectrosc., 2007, 61, 824–831 CrossRef CAS PubMed.
- Y. Liu, H. Yu, Y. Cheng, Y. Guo, W. Yao and Y. Xie, Food Anal. Methods, 2020, 13, 1710–1716 CrossRef.
- A. ul Haq, M. I. Majeed, H. Nawaz, N. Rashid, M. R. Javed, M. A. Iqbal, A. Raza, S. T. Zahra, L. Meraj and A. Perveen, Photodiagn. Photodyn. Ther., 2023, 42, 103533 CrossRef PubMed.
- M. L. Laucks, A. Sengupta, K. Junge, E. J. Davis and B. D. Swanson, Appl. Spectrosc., 2005, 59, 1222–1228 CrossRef CAS PubMed.
- J. T. Pelton and L. R. McLean, Anal. Biochem., 2000, 277, 167–176 CrossRef CAS PubMed.
- M. Wagner, N. P. Ivleva, C. Haisch, R. Niessner and H. Horn, Water Res., 2009, 43, 63–76 CrossRef CAS PubMed.
- B. Gieroba, M. Krysa, K. Wojtowicz, A. Wiater, M. Pleszczyńska, M. Tomczyk and A. Sroka-Bartnicka, Int. J. Mol. Sci., 2020, 21, 3811 CrossRef CAS PubMed.
- E. Balducci, F. Papi, D. E. Capialbi and L. Del Bino, Int. J. Mol. Sci., 2023, 24, 4030 CrossRef CAS PubMed.
- N. N. Schommer, M. Christner, M. Hentschke, K. Ruckdeschel, M. Aepfelbacher and H. Rohde, Infect. Immun., 2011, 79, 2267–2276 CrossRef CAS PubMed.
- A. Flannery, M. Le Berre, G. B. Pier, J. P. O'Gara and M. Kilcoyne, Int. J. Mol. Sci., 2020, 21, 2465 CrossRef CAS PubMed.
- M. Wasim, U. Ghaffar, M. R. Javed, H. Nawaz, M. I. Majeed, A. Ijaz, S. Ishtiaq, N. Rehman, R. Razaq and S. Younas, ACS Omega, 2024, 9, 15202–15209 CAS.
- H. L. Tytgat, N. H. van Teijlingen, R. M. Sullan, F. P. Douillard, P. Rasinkangas, M. Messing, J. Reunanen, R. Satokari, J. Vanderleyden, Y. F. Dufrêne, T. B. Geijtenbeek, W. M. de Vos and S. Lebeer, PLoS One, 2016, 11, e0151824 Search PubMed.
- A. S. Pugazhendhi, F. Wei, M. Hughes and M. Coathup, in Musculoskeletal Infection, Springer, 2022, pp. 19–64 Search PubMed.
- G. M. Qader, K. K. Jarjees and R. K. Jarjees, J. Med. Life, 2022, 15, 1105–1109 CrossRef PubMed.
- A. Silva, V. Silva, M. López, B. Rojo-Bezares, J. A. Carvalho, A. P. Castro, Y. Sáenz, G. Igrejas and P. Poeta, Antibiotics, 2023, 12, 1248 CrossRef CAS PubMed.
- F. F. Tuon, P. H. Suss, J. P. Telles, L. R. Dantas, N. H. Borges and V. S. T. Ribeiro, Antibiotics, 2023, 12, 87 CrossRef CAS PubMed.
- S. Efrima and L. Zeiri, J. Raman Spectrosc., 2009, 40, 277–288 CrossRef CAS.
- A. Sharma and P. K. Mishra, Int. J. Inf. Technol., 2022, 14, 1949–1960 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06267a |
| ‡ The first two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |

