Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Generation of topologically defined linear and cyclic DNA bottle brush polymers via a graft-to approach†
Nicholas G.
Pierini‡
a,
Wynter A.
Paiva‡
a,
Owen C.
Durant
a,
Aubrianna M.
Dobbins
a,
Ben B.
Wheeler
b,
Matthew E.
Currier
a,
James
Vesenka
b and
Nathan J.
Oldenhuis
 *a
*a
aDepartment of Chemistry, College of Engineering and Physical Science, University of New Hampshire, 23 Academic Way, Parsons Hall, Durham, NH 03824, USA. E-mail: nathan.oldenhuis@unh.edu
bSchool of Molecular and Physical Sciences, University of New England, 11 Hills Beach Road, Biddeford, ME 04005, USA
First published on 1st May 2025
Abstract
Herein, we report a graft-to approach for synthesizing linear and circular double-stranded DNA (dsDNA) bottlebrush polymers (BBPs). Using a bioreactor, plasmid DNA (pDNA) serves as an inexpensive and abundant source of circular, biodegradable, and unimolecular polymers. pDNA is easily converted to the linear isoform through enzymatic restriction, providing access to polymeric backbones with distinct topological states. DNA is grafted with polyethylene glycol monomethyl ether chloroethylamines (mPEGCEA) to yield DNA BBPs. Importantly this PEGylation occurs rapidly under ambient conditions in aqueous buffer. By varying the molecular weight of mPEGCEA (Mw = 750, 2000, 5000 Da) and the concentration relative to μmol of nucleotides, different brush arm densities and lengths were achieved with both linear and macrocyclic DNA backbones. Analysis of the DNA BBPs was achieved through agarose gel electrophoresis, which showed graft densities of up to ~68% and ~74% for linear and ring DNA respectively. The grafting process does not alter base pairing or circularity as determined using atomic force microscopy. Shear rheology was used to compare the mechanical response of 1% wt/wt solutions of the ring and linear DNA BBPs to their un-alkylated forms. Linear DNA BBPs exhibited a lower shear modulus versus linear DNA, which is expected due to the increased persistence length and decreased ability to interpenetrate associated with the attachment of polymer arms. However, the circular DNA BBPs exhibited a universally higher shear modulus versus the un-alkylated sample suggesting an increase in interchain interaction via addition of polymer arms. Finally, the increased steric encumbrance of the DNA BBPs slows enzymatic degradation, potentially providing a general method to increase stability of DNA constructs towards nuclease.
Introduction
Bottle brush polymers (BBPs) are complex macromolecules featuring grafted side chains attached to a polymeric backbone.1–3 By manipulating side chain length, density, and composition, BBPs can achieve remarkable rigidity, low entanglement and viscoelastic moduli, controllable self-assembly, and integrated structural functionality.1–3 Their unique architecture has enabled a wide variety of applications, including biomimetic materials, soft-elastomers, and drug-delivery. Recently, BBPs with cyclic backbones have become increasingly popular synthetic targets due to their altered molecular and mechanical properties such as a smaller hydrodynamic radius, lower intrinsic viscosities, lower inter-chain entanglements, increased chemical stability, and higher glass transition temperatures.4–11Typically, the synthesis of the backbone is constrained through the method used to append the sidechains, e.g. graft-to, graft-from, or graft-through.11–13 While each of the aforementioned methods has been employed to make cyclic BBPs, they commonly suffer from low yields, high dispersity, linear impurities, and poor biodegradability due to the inherent challenges associated with making any cyclic polymers.12,14–17 Low dispersity and lack of linear impurities are incredibly important to achieve when investigating cyclic systems as small contaminates can dominate the overall solution or material properties.18 This need is compounded when investigating cyclic BBPs as the side chains introduce another layer of complexity that impacts dynamics and rheological properties.6,19,20 Thus, as the complexity of the BBP topology increases, purification and production of pure rings becomes vital to their study.7,12,19–22
Considering the challenges associated with the synthesis and purity of cyclic polymers, an intriguing solution lies in the utilization of biologically sourced plasmid DNA (pDNA). pDNA natively exists as a double-stranded supercoiled (SC) cyclic polymer that can be relaxed or cut enzymatically to yield the open-circular (OC) or linear (L) form respectively.23 Interestingly, each isoform exhibits altered biological properties, such as circular DNA's increased resistance to thermal and enzymatic degradation versus L counterparts.23–26 Beyond topology, pDNA varies widely in length (2–20 kilobase pairs, kbp, 0.340 nm per bp) while remaining unimolecular (Đ = 1) which can greatly simplify determination of structure–property relationships.22,24–27 Accordingly, DNA is routinely used as a model polymer and was instrumental in developing models to describe the dynamics of entangled solutions of linear polymers.27–34 Leveraging the unique topological forms, unimolecular dispersity, and wealth of physical data available, DNA is an ideal polymeric backbone to generate linear and cyclic BBPs.35–38 However, significant challenges exist in the synthesis and functionalization of biologically derived DNA.
First, DNA must be obtained on a sufficient scale for study. Synthetic methods (e.g. solid phase oligonucleotide synthesis) and molecular biology methods (e.g. rolling circle amplification or polymerase chain reaction) remain prohibitively costly at scale and only produce linear DNA.39–41 Sheared genomic DNA has recently gained traction as a sustainable and inexpensive source of nucleic acids from biologic sources but only produces samples with high dispersity.28,42–44 Alternatively, pDNA can be obtained through simple and inexpensive fermentation with optimized protocols yielding up to 1–2 mg of pDNA per liter of culture.45 Unfortunately, this would require hundreds of liters to obtain a sufficient amount of pDNA for bulk mechanical study and excludes the significant purification steps required to isolate pDNA from the bacterium without shearing or introducing significant linear contaminants. To circumvent this issue, our group has recently reported a method to isolate and purify pDNA from bioreactors which can inexpensively and efficiently produce up to 1 g of unimolecular pDNA per batch.46–48 Access to gram scale quantities of linear and cyclic DNA provides topologically defined polymers which could be subsequently transformed into BBPs after grafting.
Another major challenge in DNA BBP synthesis is the small number of chemistries available to append functionality to native DNA. Typically, solid phase oligonucleotide synthesis is employed to add non-native functionality to DNA or RNA for subsequent functionalization with polymers or initiators.49 Wiel, Ng, and many others have reported unique and useful DNA and RNA containing block copolymers leveraging functionality installed through solid phase oligonucleotide synthesis (Fig. 1).50–52 Similarly, Matyjaszewski has reported examples of DNA and RNA backbone BBPs synthesized using a graft-from approach and ATRP (Fig. 1). Unfortunately, both cases require a pre-functionalized single stranded oligonucleotide backbone or employs alkylating agents that are selective only for RNA.53–56 The de Vries group demonstrated a dsDNA BBP using electrostatic attraction to graft the chains to the backbone, but the dynamic nature of this interaction is not amenable to many applications of BBPs (Fig. 1).36
As we are constrained to using biologically sourced DNA to achieve control over BBP topology, we needed to develop a graft-to or graft-from approach that will efficiently attach polymer chains without compromising the base pairing or topology. While generally considered to be relatively inert, DNA is weakly nucleophilic and strong electrophiles could serve as potential candidates to append polymers or initiators. Non-specific DNA alkylators, such as nitrosamines, epoxides, acrylates, and alkyl sulfonates have all been employed to attach dyes, pharmaceuticals, and fluorescent probes to random nucleotides.44,57–64 However, these reagents can degrade DNA, have low selectivity for DNA over other biomolecules, or alkylate in the base pairing face which compromises any instilled topology.44,57–64 A review of DNA therapeutics revealed that chloroethylamines (CEAs) are capable of more selectively reacting with dsDNA, work in biologically relevant conditions, and do not denature dsDNA. The reactivity of chloroethylamines with dsDNA has been shown to react preferentially with the external guanine N7 position and presented as an attractive option for attaching side chains to our DNA backbone (Fig. 2A).57,60
Herein, we report on the development of topologically defined linear and cyclic DNA BBPs using pUC19 (2.686 kb) and pEYFP (5.045 kb) as our backbones and mPEG functionalized with chloroethylamines (CEA) as side chains. High graft density (>60% of base pairs) is accessible through excess stoichiometric alkylator concentration. Subsequent investigation of the rheological properties of linear (L-BBP) and ring (R-BBP) BBPs compared to their unmodified counterparts revealed topology dependent mechanical properties. Finally, as the stability of DNA BBPs is important for many end use applications, we explored their enzymatic resistance to DNase I and found that PEGylation of DNA provides some protection at higher graft densities.
Experimental
Materials and reagents
Chloroform and toluene were purchased from Fisher Chemical. Polyethylene glycol monomethyl ether (Mw = 750, 2000, and 5000 g mol−1), diethanolamine, anhydrous N,N-dimethylformamide (DMF), and Amicon Ultra 0.5 and 15 mL centrifugal filter units (10 kDa MWCO) were purchased from Sigma Aldrich. Triethylamine, sodium iodide, and thionyl chloride were purchased from Oakwood Chemical. 10× Tris-Acetate-EDTA (TAE) solution and methanesulfonyl chloride were purchased from Thermo Fisher Scientific. 10× TAE was diluted with ultrapure water from a Barnstead MicroPure water system to 1× before use. Unless stated otherwise, all reagents and materials were used as supplied and/or directed by the manufacturer.Instrumentation
Bacterial cultures were grown in a 7 L vessel using an Applikon ez-2 control bioreactor. Anion exchange chromatography was performed using a Bio-Rad NGC chromatography system. 1H NMR spectra were measured using a Bruker 700 MHz spectrometer. Chemical shifts are reported in parts per million using CDCl3. All spectra can be found in the ESI.† Electrophoresis images were obtained using a Syngene GeneFlash BioImaging system and processed using ImageJ. All rheological measurements were taken using a TA Instruments Discovery HR-20 rheometer with a 40 mm sandblasted parallel plate geometry and solvent trap.Production and purification of pDNA
pDNA samples were obtained using methods previously reported by our group which couples a fed-batch fermentation process with a modified alkaline lysis method and anion exchange chromatography to access gram-scale quantities of pure pDNA solutions.46 Briefly, E. coli transformed with the target plasmid were grown in a 7 L bioreactor using a semi-defined growth media. Upon reaching a target OD600 of 10, a chemically defined feed medium was added at a constant rate for the duration of the culture. Upon reaching stationary phase (OD600 ∼ 90), the culture was chilled to 25 °C for 30 min before being chilled to 15 °C. Cells were then harvested using centrifugation and lysed using a modified alkaline lysis method to create a crude pDNA solution in which the major contaminant is lower Mw RNA. Pure samples were then obtained using anion exchange chromatography and concentrated using isopropanol precipitation. Concentration and purity of samples are measured using a Nanodrop OneC spectrophotometer. Quality and isoform content of samples were characterized using AGE and gel band densitometry analysis in ImageJ.Synthesis of mPEGOMs
Polyethylene glycol monomethyl ether (mPEG-OH) was dissolved in chloroform and cooled to 0 °C using an ice bath. Triethylamine (5 eq.) was added to the solution and allowed to stir for 10 min. Methanesulfonyl chloride (5 eq.) was then added dropwise to the reaction mixture which was then allowed to warm to room temperature and stirred for 16 h. The reaction mixture was then diluted with chloroform (0.1 g mPEG mL−1) and washed with 1 M HCl (1× 1 vol) followed by brine (1× 1 vol). A majority of the solvent was removed under reduced pressure to yield the crude product as orange oil. Product was precipitated through the addition of ether (10× v/v). The mixture was cooled to −20 °C overnight before the solids were collected by vacuum filtration and washed with additional cold ether to yield the product as either a tacky beige solid (mPEG750OMs) or white powder (mPEG2kOMs and mPEG5kOMs). Detailed procedures and NMR spectra can be found in the ESI (Fig. S1–S3).†Synthesis of mPEGDEA
mPEGOMs was dried under reduced pressure with heating at 60 °C for 1 h. The dried compound was then dissolved in anhydrous DMF under nitrogen. Sodium iodide was then added to the solution and allowed to stir for an additional 10 min at 60 °C under nitrogen. Diethanolamine (5 eq.) was then added dropwise under nitrogen. The reaction mixture was then heated to 100 °C and allowed to stir for 72 h under nitrogen. The reaction mixture was cooled to room temperature and the solvent was removed under reduced pressure. The resulting dark orange oil was dissolved in chloroform (0.1 g mPEG mL−1) and washed with saturated bicarbonate solution (1× 1 vol). A majority of the solvent was removed under reduced pressure to yield the crude product as an orange oil. Product was precipitated through the addition of ether (10× v/v). The mixture was cooled to −20 °C overnight before the solids were collected by vacuum filtration and washed with additional cold ether to yield the product as either a tacky yellow solid (mPEG750DEA) or beige powder (mPEG2kDEA and mPEG5kDEA). Detailed procedures and NMR spectra can be found in the ESI (Fig. S4–S6).†Synthesis of mPEGCEA
mPEGDEA was dissolved in thionyl chloride (40 eq.) and refluxed at 80 °C for 16 h. The reaction was diluted with toluene (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and then the solvent was removed under reduced pressure. Excess thionyl chloride was then removed through co-evaporation with toluene (3 × 1 vol). The resulting dark orange oil was dissolved in chloroform (0.1 g mPEG mL−1) and washed with saturated bicarbonate solution (1× 1 vol). A majority of the solvent was removed under reduced pressure to yield the crude product as an orange oil. Product was precipitated through the addition of ether (10× v/v). The mixture was cooled to −20 °C overnight before the solids were collected by vacuum filtration and washed with additional cold ether to yield the product as either a tacky orange solid (mPEG750CEA) or beige powder (mPEG2kCEA and mPEG5kCEA). Detailed procedures and NMR spectra can be found in the ESI (Fig. S7–S9).†
10) and then the solvent was removed under reduced pressure. Excess thionyl chloride was then removed through co-evaporation with toluene (3 × 1 vol). The resulting dark orange oil was dissolved in chloroform (0.1 g mPEG mL−1) and washed with saturated bicarbonate solution (1× 1 vol). A majority of the solvent was removed under reduced pressure to yield the crude product as an orange oil. Product was precipitated through the addition of ether (10× v/v). The mixture was cooled to −20 °C overnight before the solids were collected by vacuum filtration and washed with additional cold ether to yield the product as either a tacky orange solid (mPEG750CEA) or beige powder (mPEG2kCEA and mPEG5kCEA). Detailed procedures and NMR spectra can be found in the ESI (Fig. S7–S9).†
Synthesis of DNA BBP conjugates
A stock solution of mPEGCEA (50 mM) was made using 1× TAE and diluted as needed. Alkylator solutions were made fresh each time and used within 10 minutes of complete dissolution. pDNA solution, 1× TAE, and alkylator solutions were combined in either a 1.5 mL Eppendorf tube or a 15 mL conical tube. Samples were homogenized by gently inverting and vortexing for 3 s and then centrifuged in either a microcentrifuge (1.5 mL) or a hand-crank centrifuge (15 mL) for 15 s. Reactions were then incubated at 37 °C for 1–2 h. Success of alkylation was assessed using agarose gel electrophoresis. Final pDNA concentration was 0.1 mg mL−1 for small-scale reactions and 1 mg mL−1 for large-scale reactions. Excess unreacted mPEGCEA was removed from large-scale reactions using Amicon Ultra-15 centrifugal filter units with a molecular weight cutoff of 10 kDa. Detailed procedure can be found in the ESI.†DNase I activity tests
DNase I (1 U μL−1) was freshly diluted with 1× reaction buffer + MgCl2 to a final concentration of 0.001 U μL−1. 0.1 μg of DNA or DNA BBP was diluted with nuclease-free water (to 20 μL total volume) and 10× reaction buffer with MgCl2 (2 μL). Diluted DNase I (2 μL, 0.0001 U μL−1) was added to initiate the reaction and samples were incubated at 22 °C for 5, 10, 20, 40, or 60 minutes on a dry heat block. Reactions were stopped by adding EDTA (25 mM) to inactivate enzymes. Extent of degradation in each sample was analyzed using agarose gel electrophoresis. Percentage of higher molecular weight DNA (>1 kb or 617 kDa) was determined using gel band densitometry analysis in ImageJ.Agarose gel electrophoresis
Unless stated otherwise, DNA BBP conjugates were characterized using 12 cm, 0.5% agarose gels that contained 0.35 g mL−1 of ethidium bromide. Gels were loaded with 0.25 μg of the GeneRuler 1 kb ladder and 0.05 μg of standards and samples. The running buffer was a 40 mM tris, 20 mM sodium acetate, 2 mM EDTA, pH 8.3 solution that was purchased as a 10× stock. Gels were run at 75 V for 1–2 h and visualized using a UV transilluminator to determine completion before imaging on Syngene GeneFlash BioImaging.Results and discussion
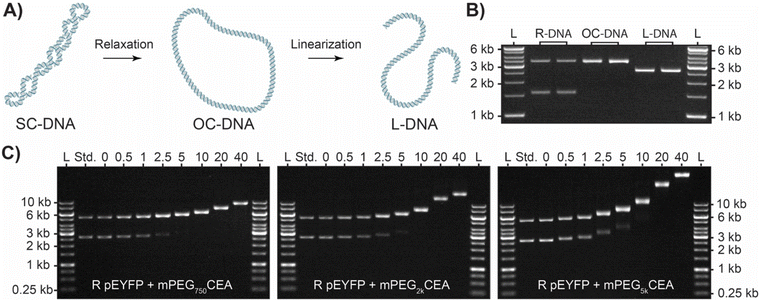
To develop a graft-to agent, bis-CEA was appended to the end of polyethylene glycol monomethyl ether to yield mPEGCEA (Fig. 2A, see ESI† for details). As the pDNA solutions from the bioreactor contain a variable mixture of SC and OC DNA, a standard protocol for DNA BBP production and characterization was first established using linearized pDNA (L-DNA). A freshly made solution of mPEG2kCEA was combined with L pEYFP at 37 °C for 1.5 h in 1× TAE buffer, pH 8.3. mPEG2kCEA was added relative to μmole of nucleotides in DNA (0.5–40 eq.) and alkylation was analyzed by agarose gel electrophoresis (Fig. 2B). As increasing equivalence of mPEG2kCEA were added, a single band of increasing apparent molecular weight was observed, suggesting successful conjugation and production of linear DNA BBPs (L-BBP) (Fig. 2B). UV-Vis absorbance at 260 nm before and after grafting confirmed no loss in double stranded nature (Table S1†). This was then repeated with mPEG750CEA and mPEG5kCEA which yielded similar results. As expected, L-BBP made with the shortest mPEGCEA produced the smallest shifts while L-BBPs made with the longest mPEGCEA produced the greatest shift in apparent molecular weight (Fig. 2B). Graft density was estimated using the retention factor (Rf) of the L-BBP relative to the Mw of the ladder and the standard run with each sample (Table 1). Large decreases in Rf indicate relatively high levels of grafting, with over 60% estimated grafting density observed for each sample at 40 eq. A larger molecular weight ladder was used to confirm that the Rf estimations for the largest BBPs could be extrapolated from the smaller ladder (Fig. S1†). We note that a large excess of reagent is necessary to achieve reasonable graft densities likely due to both the inefficiency of the graft-to approach, and the hydrolysis of CEAs in aqueous media (t1/2 ∼ 1 h).1,60,65 Additionally, the grafting of PEG to DNA likely shields charge therefore, migration of DNA BBP conjugates is not due to increase of size alone. However, until better methods are established, this approach provides a quantitative method for comparing DNA BBPs.| Equivalence | Apparent Mw![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (kDa) a (kDa) |
Graft densityb (%) |
|---|---|---|
| a Apparent Mw calculated by extrapolation based on the 1 kb DNA ladder reference. b Grafting density calculated based on the difference in extrapolated Mw values for each DNA BBP conjugate. | ||
| L pEYFP + mPEG750CEA | ||
| 0 | 3158 | 0.0 |
| 0.5 | 3272 | 2.6 |
| 1 | 3310 | 3.5 |
| 2.5 | 3511 | 8.1 |
| 5 | 3680 | 12.0 |
| 10 | 4090 | 21.5 |
| 20 | 4994 | 42.3 |
| 40 | 6098 | 67.8 |
| L pEYFP + mPEG2kCEA | ||
| 0 | 3187 | 0.0 |
| 0.5 | 3350 | 1.5 |
| 1 | 3407 | 2.0 |
| 2.5 | 3778 | 5.5 |
| 5 | 4232 | 9.7 |
| 10 | 5310 | 19.7 |
| 20 | 7846 | 43.2 |
| 40 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 594 594 |
68.7 |
| L pEYFP + mPEG5kCEA | ||
| 0 | 3216 | 0.0 |
| 0.5 | 3441 | 0.9 |
| 1 | 3619 | 1.6 |
| 2.5 | 4164 | 3.7 |
| 5 | 4994 | 6.9 |
| 10 | 6775 | 13.7 |
| 20 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 614 614 |
36.3 |
| 40 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 511 511 |
62.9 |
Having established a graft-to method for production and characterization of L-BBPs using mPEGCEAs we next sought to produce their cyclic analogs (R-BBP). pDNA obtained from our bioreactor process contains a variable mixture of SC/OC DNA (Ring DNA or R-DNA) resulting from chemical and mechanical nicking during alkaline lysis and purification process (Fig. 3A). It should be noted that SC, OC, and L isoforms of DNA are easily distinguishable from each other via their relative Rf as they have unique electromotive forces based on their topology (Fig. 3B). When the R-DNA solution was mixed with each of the mPEGCEAs, a similar decrease in Rf was observed and used to estimate graft density (Fig. 3C and Table 2). Interestingly, as graft density increases, chemical nicking and subsequent relaxation of SC to OC DNA increases until only OC-BBP is observed. We hypothesize that this is due to the increased strain energy SC DNA experiences during alkylation and chemical induced nicking of the phosphate backbone.57–60,66,67 As only the disappearance of the SC-BBP is observed, we can deduce no linearization and contamination from L-BBPs is occurring.
| OC | SC | |||
|---|---|---|---|---|
| Equivalence | Apparent Mw![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (kDa) a (kDa) |
Graft densityb (%) | Apparent Mw![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (kDa) a (kDa) |
Graft densityb (%) |
| a Apparent Mw calculated by extrapolation based on the 1 kb DNA ladder reference. b Grafting density calculated based on the difference in extrapolated Mw values for each DNA BBP conjugate. | ||||
| R pEYFP + mPEG750CEA | ||||
| 0 | 3627 | 0.0 | 1667 | 0.0 |
| 0.5 | 3663 | 0.8 | 1717 | 1.2 |
| 1 | 3735 | 2.5 | 1751 | 1.9 |
| 2.5 | 3847 | 5.1 | 1876 | 4.8 |
| 5 | 4081 | 10.5 | ||
| 10 | 4503 | 20.2 | ||
| 20 | 5323 | 39.1 | ||
| 40 | 6418 | 64.3 | ||
| R pEYFP + mPEG2kCEA | ||||
| 0 | 3589 | 0.0 | 1639 | 0.0 |
| 0.5 | 3743 | 1.4 | 1696 | 0.3 |
| 1 | 3828 | 2.2 | 1754 | 0.9 |
| 2.5 | 4026 | 4.1 | 1881 | 2.1 |
| 5 | 4430 | 7.8 | ||
| 10 | 5332 | 16.2 | ||
| 20 | 8691 | 47.3 | ||
| 40 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 519 519 |
64.3 | ||
| R pEYFP + mPEG5kCEA | ||||
| 0 | 3796 | 0.0 | 1710 | 0.0 |
| 0.5 | 4102 | 1.2 | 1852 | 0.5 |
| 1 | 4295 | 1.9 | 2139 | 1.7 |
| 2.5 | 5036 | 4.8 | 2395 | 2.6 |
| 5 | 5979 | 8.4 | 2947 | 4.8 |
| 10 | 8271 | 17.3 | ||
| 20 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 093 093 |
47.5 | ||
| 40 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 165 165 |
74.8 | ||
To further confirm the topology and successful formation of the different DNA BBPs, select samples were dry imaged using atomic force microscopy (AFM). First, R and L pUC19 were imaged to serve as a point of comparison (Fig. 4A and D). We next performed dry imaging of L pUC19 grafted with mPEG2kCEA (∼10% grafting) (Fig. 4B). This sample revealed a pearl necklace appearance where the areas of high graft density are observed as “pearls” and unreacted polymer can be observed in the background. When trying to dry image sample with a higher grafting density, no BBP could be observed. Typically, when imaging DNA, freshly cleaved mica is pretreated with a NiCl2 solution to ensure electrostatic adherence of the sample to the substrate before subsequent washing, drying, and imaging. We hypothesize that the steric shielding of PEG brushes was blocking the backbone from interacting with the mica surface.68–73 To image a sample with higher graft density, direct liquid imaging was used. R pUC19 with a higher graft density (mPEG750CEA, ∼30% grafting) was successfully imaged to confirm that no linearization was observed (Fig. 4D, E and F). Based on our combined imaging and electrophoresis results, we can confirm the production and purity of both linear and cyclic DNA BBPs. Imaging the DNA BBPs grafted with larger Mw PEG at higher densities has proven challenging and is the focus of future studies.
To determine the mechanical properties of DNA BBP solutions it was necessary to scale up the synthesis to obtain enough sample for measurement via shear rheology. This was accomplished using pEYFP (5.045 kb) and mPEG2kCEA. R pEYFP (10.6 mg) and L pEYFP (9.1 mg) were each alkylated with 10 eq. of mPEG2kCEA (Fig. 5A). On a larger scale the functionalization performed similarly to the small-scale preparation however, the concentration of DNA in these reactions was increased from 0.1 mg mL−1 to 1 mg mL−1 to reduce the required volume. Under these conditions, a higher graft density is achieved at a given stoichiometric equivalence (Fig. S2†). To prevent interference in subsequent rheological and enzymatic studies it was necessary to purify the large excess of grafting agent from the samples. Typically, precipitation of DNA in ethanol or isopropanol can remove soluble chemical contaminants, but the DNA BBPs remained in solution thus requiring an alternative method of purification. Interestingly, the increased solubility of DNA BBPs in organic solvents could greatly expand their potential applications. The DNA BBPs were ultimately purified and concentrated via centrifugal spin filters to yield solutions with a concentration of ∼10 mg mL−1 which is above the overlap concentration of ∼3 mg mL−1 for pDNA of this size.46 The graft density of the resulting L-BBP and R-BBP was 40.1% and 42.2%, respectively. For the larger pEYFP plasmid and the resulting BBPs, the observed Rf of the linear and OC isoforms are closer, but still distinguishable confirming they are indeed the OC and L isoforms (Fig. 5A).
Due to the ability to make large amounts of sample, we investigated the mechanical properties of the DNA BBP samples using shear rheology at a concentration of 10 mg mL−1. The integrity of samples before and after rheological analysis was characterized using AGE to ensure that sample aging or shear induced linearization did not influence the observed viscoelastic properties (Fig. S3†). Due to the extreme length of DNA, a physical gel is formed at relatively low sample concentration (<1 mg mL−1).29,74 We first sought to explore the dynamics of our linear systems which exhibit predicted behavior of entangled polymers and BBPs (Fig. 5B).1,3,29,74–76 In our L-DNA sample, the system is predominantly in the transition zone where G′ < G′′ and scaling of G′(ω) ∼ ω0.9 and G′′(ω) ∼ ω0.6 is observed. Conversely, in the L-BBP sample we see a decrease in the viscoelastic moduli at low frequencies (ω < 100 rad s−1) compared to the unmodified L-DNA. The steric repulsion of the grafted side chains hinders chain packing and decreases interaction between polymer chains, acting as a diluting agent, which manifests as a decrease in the observed viscoelastic moduli.1,3,65,75 This is suggested by the steeper scaling of G′(ω) ∼ ω1.2 and G′′(ω) ∼ ω0.8 observed in the L-BBP.76 Interestingly, at high frequencies G′(ω) coalesces for both samples as both samples crossover into an elastic dominant regime. Typically, the bottlebrush backbone becomes more rigid due to the steric repulsion of the side chains, while the dilution effect decreases entanglements, lowering G′(ω) across all frequencies.1,3,65,75 However, as the DNA backbone is already relatively stiff (persistence length = 50 nm), grafting of relatively small PEG2k will likely have little effect on entanglement density at higher frequencies. When observing the R-DNA and R-BBP, an elastic plateau modulus is observed in the R-DNA sample at lower frequencies where G′ > G′′ and G′(ω) ∼ ω0.1 and G′′(ω) ∼ ω0.7 (Fig. 5C). At higher frequencies, we see a crossover (ωc = 1.6 rad s−1) into a viscous dominated regime where G′ < G′′ and scaling of G′(ω) ∼ G′′(ω) ∼ ω0.7 is observed. The plateau zone is not observed in the R-BBP sample which is predominantly in the transition zone where G′(ω) ∼ ω0.7 and G′′(ω) ∼ ω0.6 before crossing over (ωe = 6.3 rad s−1) to an elastic dominant regime. We note that this may not be a “true” change in the plateau modulus, but intrinsic to the bottle brushes polymers having a longer relaxation time. We hypothesize that there are two contributing causes to this phenomenon: (1) transition from a SC/OC mixture to exclusively OC, and (2) increased interaction of R-BBPs via their sidechains. Of the limited data reported for supercoiled and open circle solutions, blends exhibit dynamics like those of entangled linear polymers which is supported by the elastic plateau modulus observed in our ring pEYFP frequency sweeps.77 The exact fraction of supercoiled species is not quantified and also may introduce a higher entanglement density. While the impact of the SC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OC ratio has not been explored in detail, data out of our lab has shown that an increase in the OC content of the sample enhances the observed elastic plateau modulus (Fig. S4†). While the dynamics of ring polymers are still not fully understood, their ability to entangle and undergo “modified” reptation is an area of extreme debate and active investigation within the polymer's community.18,24,46,74,77–80 At this point, it is unclear whether change in OC
OC ratio has not been explored in detail, data out of our lab has shown that an increase in the OC content of the sample enhances the observed elastic plateau modulus (Fig. S4†). While the dynamics of ring polymers are still not fully understood, their ability to entangle and undergo “modified” reptation is an area of extreme debate and active investigation within the polymer's community.18,24,46,74,77–80 At this point, it is unclear whether change in OC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SC ratio or addition of brushes is driving the observed rheological differences. To truly investigate the properties of cyclic BBPs we will need to study solutions of pure OC and SC BPPs with much greater graft densities and brush arm lengths and thus is the subject of future studies.
SC ratio or addition of brushes is driving the observed rheological differences. To truly investigate the properties of cyclic BBPs we will need to study solutions of pure OC and SC BPPs with much greater graft densities and brush arm lengths and thus is the subject of future studies.
Next, due to the increased steric encumbrance of the DNA BBP, we investigated their degradation in the presence of DNAase I. Synthetic BBPs typically have all carbon backbones which suffer from limited ability to be degraded which complicates biological applications.15,81,82 On the contrary, DNA constructs, DNA origami, and DNA nanomachines are all rapidly degraded by endogenous DNase hindering their use. We hoped that PEGylation of DNA would increase the lifetime a of the DNA BBP, while retaining biodegradability on a longer timescale. To test this, we prepared L-BBP and R-BBP samples with varying graft densities and exposed them to a non-specific endonuclease: DNAase I. 50 μg of L and R pEYFP was alkylated with mPEG2kCEA at 5, 10, 20, and 40 eq. to achieve DNA BBPs with target graft densities of ∼10, 20, 40 and 70% (Fig. 6). Purified DNA and DNA BBPs were then incubated at 22 °C in the presence of DNase I (0.0001 U μL−1) for either 5, 10, 20, 40, or 60 minutes (Fig. 7A and B). Extent of degradation was assessed by quantifying the percent of higher molecular weight DNA observed at the end of the incubation period (Fig. 7C and D). We defined our higher molecular weight cut off as 1 kb or 617 kDa, similar to previous studies.83 Unsurprisingly DNA BBPs with low graft densities (<20%) exhibited little to no resistance towards enzymatic activity and were degraded almost as quickly as the unmodified DNA samples. In both linear and ring systems, almost no higher molecular weight DNA was observed beyond 10 minutes. At higher graft densities, our BBPs exhibited an increased resistance towards degradation which is supported by the observation of higher molecular weight DNA in these samples even after 300 minutes (Fig. S4†). Complete degradation of DNA BBPs with higher graft densities was not observed under the time scales assessed. These results highlight a major advancement for the use of oligonucleotides in the context of drug delivery and DNA origami as pre-mature degradation is a major obstacle in the field. This strategy is similar to PEGylation of proteins and can potentially be used to increase the half-life of DNA constructs like DNA Origami in blood.38,83,84
Conclusions
In summary, we have developed a methodology for synthesizing and characterizing both linear and cyclic DNA BBPs. We estimated the graft density of chains attached to these conjugates using Rf. Atomic force microscopy (AFM) provided direct observational evidence of successful conjugation of mPEG to DNA backbones and preservation of topology. By employing spin filtration, we efficiently purified the DNA–PEG conjugates and explored their unique rheological properties. Finally, we showed that at high graft densities, DNA BBPs can partially resist the action of DNase. Expanding the synthetic toolbox and refining the purification methods for DNA adducts under mild conditions will enable the exploration of their properties and applications in the fields of biomedical research and drug delivery. We anticipate that this orthogonal chemistry approach can be extended to other DNA conjugates for generating various nanostructures and investigating the bulk properties of these materials.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
N. J. O. would like to acknowledge the support from CIBBR (NIH, P20GM113131), NH Biomade (NSF, #IIA 1757371), the NSF DMR (NSF CAREER 2340569), and the NIH (NIGMS, 1R35GM154998).References
- R. Verduzco, X. Li, S. L. Pesek and G. E. Stein, Structure, Function, Self-Assembly, and Applications of Bottlebrush Copolymers, Chem. Soc. Rev., 2015, 44(8), 2405–2420, 10.1039/C4CS00329B.
- G. Xie, M. R. Martinez, M. Olszewski, S. S. Sheiko and K. Matyjaszewski, Molecular Bottlebrushes as Novel Materials, Biomacromolecules, 2019, 20(1), 27–54, DOI:10.1021/acs.biomac.8b01171.
- M. Abbasi, L. Faust and M. Wilhelm, Comb and Bottlebrush Polymers with Superior Rheological and Mechanical Properties, Adv. Mater., 2019, 31(26), 1806484, DOI:10.1002/adma.201806484.
- Z. Miao, D. Pal, W. Niu, T. Kubo, B. S. Sumerlin and A. S. Veige, Cyclic Poly(4-Methyl-1-Pentene): Efficient Catalytic Synthesis of a Transparent Cyclic Polymer, Macromolecules, 2020, 53(18), 7774–7782, DOI:10.1021/acs.macromol.0c01366.
- G. Chen and E. E. Dormidontova, Cyclic vs Linear Bottlebrush Polymers in Solution: Side-Chain Length Effect, Macromolecules, 2023, 56(9), 3286–3295, DOI:10.1021/acs.macromol.3c00362.
- D. Pal, Z. Miao, J. B. Garrison, A. S. Veige and B. S. Sumerlin, Ultra-High-Molecular-Weight Macrocyclic Bottlebrushes via Post-Polymerization Modification of a Cyclic Polymer, Macromolecules, 2020, 53(22), 9717–9724, DOI:10.1021/acs.macromol.0c01797.
- M. A. Cortez, W. T. Godbey, Y. Fang, M. E. Payne, B. J. Cafferty, K. A. Kosakowska and S. M. Grayson, The Synthesis of Cyclic Poly(Ethylene Imine) and Exact Linear Analogues: An Evaluation of Gene Delivery Comparing Polymer Architectures, J. Am. Chem. Soc., 2015, 137(20), 6541–6549, DOI:10.1021/jacs.5b00980.
- B.-H. Duan, J.-X. Yu, R.-T. Gao, S.-Y. Li, N. Liu and Z.-Q. Wu, Controlled Synthesis of Cyclic Helical Polyisocyanides and Bottlebrush Polymers Using a Cyclic Alkyne–Pd(ii) Catalyst, Chem. Commun., 2023, 59(87), 13002–13005, 10.1039/D3CC04095J.
- M. Kruteva, J. Allgaier and D. Richter, Topology Matters: Conformation and Microscopic Dynamics of Ring Polymers, Macromolecules, 2023, 56(18), 7203–7229, DOI:10.1021/acs.macromol.3c00560.
- L. Xu, B. Gao, X. Xu, L. Zhou, N. Liu and Z. Wu, Controlled Synthesis of Cyclic–Helical Polymers with Circularly Polarized Luminescence, Angew. Chem., Int. Ed., 2022, 61(28), e202204966, DOI:10.1002/anie.202204966.
- Z. Miao, S. A. Gonsales, C. Ehm, F. Mentink-Vigier, C. R. Bowers, B. S. Sumerlin and A. S. Veige, Cyclic Polyacetylene, Nat. Chem., 2021, 13(8), 792–799, DOI:10.1038/s41557-021-00713-2.
- C. W. Bielawski, D. Benitez and R. H. Grubbs, An “Endless” Route to Cyclic Polymers, Science, 2002, 297(5589), 2041–2044, DOI:10.1126/science.1075401.
- F. M. Haque and S. M. Grayson, The Synthesis, Properties and Potential Applications of Cyclic Polymers, Nat. Chem., 2020, 12(5), 433–444, DOI:10.1038/s41557-020-0440-5.
- M. Schappacher and A. Deffieux, Synthesis of Macrocyclic Copolymer Brushes and Their Self-Assembly into Supramolecular Tubes, Science, 2008, 319(5869), 1512–1515, DOI:10.1126/science.1153848.
- J. Hu and S. Liu, Topological Effects of Macrocyclic Polymers: From Precise Synthesis to Biomedical Applications, Sci. China: Chem., 2017, 60(9), 1153–1161, DOI:10.1007/s11426-017-9083-1.
- Z. Miao, D. Konar, B. S. Sumerlin and A. S. Veige, Soluble Polymer Precursors via Ring-Expansion Metathesis Polymerization for the Synthesis of Cyclic Polyacetylene, Macromolecules, 2021, 54(17), 7840–7848, DOI:10.1021/acs.macromol.1c00938.
- Y. Xia, A. J. Boydston and R. H. Grubbs, Synthesis and Direct Imaging of Ultrahigh Molecular Weight Cyclic Brush Polymers, Angew. Chem., Int. Ed., 2011, 50(26), 5882–5885, DOI:10.1002/anie.201101860.
- F. M. Haque and S. M. Grayson, The Synthesis, Properties and Potential Applications of Cyclic Polymers, Nat. Chem., 2020, 12(5), 433–444, DOI:10.1038/s41557-020-0440-5.
- D. Pal, J. B. Garrison, Z. Miao, L. E. Diodati, A. S. Veige and B. S. Sumerlin, Nanobowls from Amphiphilic Core–Shell Cyclic Bottlebrush Polymers, Macromolecules, 2022, 55(17), 7446–7453, DOI:10.1021/acs.macromol.2c01232.
- M. A. Cortez, W. T. Godbey, Y. Fang, M. E. Payne, B. J. Cafferty, K. A. Kosakowska and S. M. Grayson, The Synthesis of Cyclic Poly(ethylene imine) and Exact Linear Analogues: An Evaluation of Gene Delivery Comparing Polymer Architectures, J. Am. Chem. Soc., 2015, 137(20), 6541–6549 CrossRef CAS.
- F. M. Haque and S. M. Gayson, The synthesis, properties and potential applications of cyclic polymers, Nat. Chem., 2020, 12, 433–444 CrossRef CAS.
- D. Kong, S. Banik, M. J. San Francisco, M. Lee, R. M. Robertson-Anderson, C. M. Schroeder and G. B. McKenna, Rheology of Entangled Solutions of Ring–Linear DNA Blends, Macromolecules, 2022, 55(4), 1205–1217, DOI:10.1021/acs.macromol.1c01672.
- P. Balagurumoorthy, S. J. Adelstein and A. I. Kassis, Method to Eliminate Linear DNA from Mixture Containing Nicked-Circular, Supercoiled, and Linear Plasmid DNA, Anal. Biochem., 2008, 381(1), 172–174, DOI:10.1016/j.ab.2008.06.037.
- J. Smrek, J. Garamella, R. Robertson-Anderson and D. Michieletto, Topological Tuning of DNA Mobility in Entangled Solutions of Supercoiled Plasmids, Sci. Adv., 2021, 7(20), eabf9260, DOI:10.1126/sciadv.abf9260.
- N. P. Higgins and A. V. Vologodskii, Topological Behavior of Plasmid DNA, Microbiol. Spectrum, 2015, 3(2), 3.2.17, DOI:10.1128/microbiolspec.PLAS-0036-2014.
- D. Michieletto, P. Neill, S. Weir, D. Evans, N. Crist, V. A. Martinez and R. M. Robertson-Anderson, Topological Digestion Drives Time-Varying Rheology of Entangled DNA Fluids, Nat. Commun., 2022, 13(1), 4389, DOI:10.1038/s41467-022-31828-w.
- Y. Li, R. Chen, B. Zhou, Y. Dong and D. Liu, Rational Design of DNA Hydrogels Based on Molecular Dynamics of Polymers, Adv. Mater., 2023, 2307129, DOI:10.1002/adma.202307129.
- R. Bandyopadhyay and A. Sood, Rheology of Semi-Dilute Solutions of Calf-Thymus DNA, Pramana, 2002, 58(4), 685–694, DOI:10.1007/s12043-002-0025-4.
- S. Banik, D. Kong, M. J. San Francisco and G. B. McKenna, Monodisperse Lambda DNA as a Model to Conventional Polymers: A Concentration-Dependent Scaling of the Rheological Properties, Macromolecules, 2021, 54(18), 8632–8654, DOI:10.1021/acs.macromol.0c02537.
- R. E. Teixeira, A. K. Dambal, D. H. Richter, E. S. G. Shaqfeh and S. Chu, The Individualistic Dynamics of Entangled DNA in Solution, Macromolecules, 2007, 40(7), 2461–2476, DOI:10.1021/ma062932e.
- R. G. Larson, The Structure and Rheology of Complex Fluids; Topics in chemical engineering, Oxford University Press, New York, 1999 Search PubMed.
- P.-G. de Gennes, Scaling Concepts in Polymer Physics, Cornell university press, Ithaca (N.Y.) London, 1991 Search PubMed.
- M. Rubinstein and R. H. Colby, Polymer Physics, Oxford university press, Oxford New York, 2003 Search PubMed.
- C. M. Schroeder, Single Polymer Dynamics for Molecular Rheology, J. Rheol., 2018, 62(1), 371–403, DOI:10.1122/1.5013246.
- F. Jia, H. Li, R. Chen and K. Zhang, Self-Assembly of DNA-Containing Copolymers, Bioconjugate Chem., 2019, 30(7), 1880–1888, DOI:10.1021/acs.bioconjchem.9b00067.
- R. J. Dalal, R. Kumar, M. Ohnsorg, M. Brown and T. M. Reineke, Cationic Bottlebrush Polymers Outperform Linear Polycation Analogues for pDNA Delivery and Gene Expression, ACS Macro Lett., 2021, 10(7), 886–893, DOI:10.1021/acsmacrolett.1c00335.
- Y. Wang, D. Wang, F. Jia, A. Miller, X. Tan, P. Chen, L. Zhang, H. Lu, Y. Fang, X. Kang, J. Cai, M. Ren and K. Zhang, Self-Assembled DNA–PEG Bottlebrushes Enhance Antisense Activity and Pharmacokinetics of Oligonucleotides, ACS Appl. Mater. Interfaces, 2020, 12(41), 45830–45837, DOI:10.1021/acsami.0c13995.
- F. Jia, X. Lu, X. Tan, D. Wang, X. Cao and K. Zhang, Effect of PEG Architecture on the Hybridization Thermodynamics and Protein Accessibility of PEGylated Oligonucleotides, Angew. Chem., Int. Ed., 2017, 56(5), 1239–1243, DOI:10.1002/anie.201610753.
- C. B. Reese, Oligo- and Poly-Nucleotides: 50 Years of Chemical Synthesis, Org. Biomol. Chem., 2005, 3(21), 3851, 10.1039/b510458k.
- B. I. Andrews, F. D. Antia, S. B. Brueggemeier, L. J. Diorazio, S. G. Koenig, M. E. Kopach, H. Lee, M. Olbrich and A. L. Watson, Sustainability Challenges and Opportunities in Oligonucleotide Manufacturing, J. Org. Chem., 2021, 86(1), 49–61, DOI:10.1021/acs.joc.0c02291.
- S. Kosuri and G. M. Church, Large-Scale de Novo DNA Synthesis: Technologies and Applications, Nat. Methods, 2014, 11(5), 499–507, DOI:10.1038/nmeth.2918.
- M. Yamada, M. Kawamura and T. Yamada, Preparation of Bioplastic Consisting of Salmon Milt DNA, Sci. Rep., 2022, 12(1), 7423, DOI:10.1038/s41598-022-11482-4.
- L. Bravo-Anaya, M. Rinaudo and F. Martínez, Conformation and Rheological Properties of Calf-Thymus DNA in Solution, Polymers, 2016, 8(2), 51, DOI:10.3390/polym8020051.
- D. Wang, J. Cui, M. Gan, Z. Xue, J. Wang, P. Liu, Y. Hu, Y. Pardo, S. Hamada, D. Yang and D. Luo, Transformation of Biomass DNA into Biodegradable Materials from Gels to Plastics for Reducing Petrochemical Consumption, J. Am. Chem. Soc., 2020, 142(22), 10114–10124, DOI:10.1021/jacs.0c02438.
- S. Laib, R. M. Robertson and D. E. Smith, Preparation and Characterization of a Set of Linear DNA Molecules for Polymer Physics and Rheology Studies, Macromolecules, 2006, 39(12), 4115–4119, DOI:10.1021/ma0601464.
- W. A. Paiva, S. D. Alakwe, J. Marfai, M. V. Jennison-Henderson, R. A. Achong, T. Duche, A. A. Weeks, R. M. Robertson-Anderson and N. J. Oldenhuis, From Bioreactor to Bulk Rheology: Achieving Scalable Production of Highly Concentrated Circular DNA, Adv. Mater., 2024, 36(35), 2405490, DOI:10.1002/adma.202405490.
- K. J. Prather, S. Sagar, J. Murphy and M. Chartrain, Industrial Scale Production of Plasmid DNA for Vaccine and Gene Therapy: Plasmid Design, Production, and Purification, Enzyme Microb. Technol., 2003, 33(7), 865–883, DOI:10.1016/S0141-0229(03)00205-9.
- G. M. Borja, E. Meza Mora, B. Barrón, G. Gosset, O. T. Ramírez and A. R. Lara, Engineering Escherichia Coli to Increase Plasmid DNA Production in High Cell-Density Cultivations in Batch Mode, Microb. Cell Fact., 2012, 11(1), 132, DOI:10.1186/1475-2859-11-132.
- N. Z. Fantoni, A. H. El-Sagheer and T. Brown, A Hitchhiker's Guide to Click-Chemistry with Nucleic Acids, Chem. Rev., 2021, 121(12), 7122–7154, DOI:10.1021/acs.chemrev.0c00928.
- N. Alleva, K. Eigen, D. Y. W. Ng and T. Weil, A Versatile and Efficient Method to Isolate DNA–Polymer Conjugates, ACS Macro Lett., 2023, 1257–1263, DOI:10.1021/acsmacrolett.3c00371.
- T. Lueckerath, T. Strauch, K. Koynov, C. Barner-Kowollik, D. Y. W. Ng and T. Weil, DNA–Polymer Conjugates by Photoinduced RAFT Polymerization, Biomacromolecules, 2019, 20(1), 212–221, DOI:10.1021/acs.biomac.8b01328.
- T. Lückerath, K. Koynov, S. Loescher, C. J. Whitfield, L. Nuhn, A. Walther, C. Barner-Kowollik, D. Y. W. Ng and T. Weil, DNA–Polymer Nanostructures by RAFT Polymerization and Polymerization–Induced Self–Assembly, Angew. Chem., Int. Ed., 2020, 59(36), 15474–15479, DOI:10.1002/anie.201916177.
- J. Jeong, X. Hu, H. Murata, G. Szczepaniak, M. Rachwalak, A. Kietrys, S. R. Das and K. Matyjaszewski, RNA-Polymer Hybrids via Direct and Site-Selective Acylation with the ATRP Initiator and Photoinduced Polymerization, J. Am. Chem. Soc., 2023, 145(26), 14435–14445, DOI:10.1021/jacs.3c03757.
- M. F. Fouz, S. K. Dey, K. Mukumoto, K. Matyjaszewski, B. A. Armitage and S. R. Das, Accessibility of Densely Localized DNA on Soft Polymer Nanoparticles, Langmuir, 2018, 34(49), 14731–14737, DOI:10.1021/acs.langmuir.8b02038.
- J. Jeong, G. Szczepaniak, S. R. Das and K. Matyjaszewski, Expanding the Architectural Horizon of Nucleic-Acid-Polymer Biohybrids by Site-Controlled Incorporation of ATRP Initiators in DNA and RNA, Chem, 2023, 9(11), 3319–3334, DOI:10.1016/j.chempr.2023.07.013.
- M. F. Fouz, K. Mukumoto, S. Averick, O. Molinar, B. M. McCartney, K. Matyjaszewski, B. A. Armitage and S. R. Das, Bright Fluorescent Nanotags from Bottlebrush Polymers with DNA-Tipped Bristles, ACS Cent. Sci., 2015, 1(8), 431–438, DOI:10.1021/acscentsci.5b00259.
- D. Gillingham, S. Geigle and O. Anatole von Lilienfeld, Properties and Reactivity of Nucleic Acids Relevant to Epigenomics, Transcriptomics, and Therapeutics, Chem. Soc. Rev., 2016, 45(9), 2637–2655, 10.1039/C5CS00271K.
- D. T. Beranek, Distribution of Methyl and Ethyl Adducts Following Alkylation with Monofunctional Alkylating Agents, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1990, 231(1), 11–30, DOI:10.1016/0027-5107(90)90173-2.
- B. Singer, The Chemical Effects ofNucleic Acid Alkylation and Their Relation to Mutagenesis and Carcinogen Esis, in Progress in Nucleic Acid Research and Molecular Biology, Elsevier, 1975, vol. 15, pp. 219–284. DOI:10.1016/S0079-6603(08)60121-X.
- B. Sauter and D. Gillingham, DNA Damaging Agents in Chemical Biology and Cancer, Chimia, 2020, 74(9), 693, DOI:10.2533/chimia.2020.693.
- M. Wenqi, S. Mingxue, X. Kai, P. Zhipeng, Z. Xinyue, C. Jiasheng, J. Xiang, X. Qingqiang, M. Guanchao and C. Jinfeng, Nitrogen mustard active probe and application thereof, CN116375586A, 2023.
- O. Okay, DNA Hydrogels: New Functional Soft Materials, J. Polym. Sci., Part B:Polym. Phys., 2011, 49(8), 551–556, DOI:10.1002/polb.22213.
- N. Orakdogen, B. Erman and O. Okay, Evidence of Strain Hardening in DNA Gels, Macromolecules, 2010, 43(3), 1530–1538, DOI:10.1021/ma902558f.
- F. Topuz and O. Okay, Rheological Behavior of Responsive DNA Hydrogels, Macromolecules, 2008, 41(22), 8847–8854, DOI:10.1021/ma801414p.
- Z. Li, M. Tang, S. Liang, M. Zhang, G. M. Biesold, Y. He, S.-M. Hao, W. Choi, Y. Liu, J. Peng and Z. Lin, Bottlebrush Polymers: From Controlled Synthesis, Self-Assembly, Properties to Applications, Prog. Polym. Sci., 2021, 116, 101387, DOI:10.1016/j.progpolymsci.2021.101387.
- J. M. Soll, R. W. Sobol and N. Mosammaparast, Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities, Trends Biochem. Sci., 2017, 42(3), 206–218, DOI:10.1016/j.tibs.2016.10.001.
- A. D. Bates and A. Maxwell, DNA Topology; In focus, IRL Press at Oxford University Press, Oxford; New York, 1993 Search PubMed.
- J. Vesenka, M. Guthold, C. L. Tang, D. Keller, E. Delaine and C. Bustamante, Substrate Preparation for Reliable Imaging of DNA Molecules with the Scanning Force Microscope, Ultramicroscopy, 1992, 42–44, 1243–1249, DOI:10.1016/0304-3991(92)90430-R.
- T. Thundat, D. P. Allison and R. J. Warmack, Stretched DNA Structures Observed with Atomic Force Microscopy, Nucleic Acids Res., 1994, 22(20), 4224–4228, DOI:10.1093/nar/22.20.4224.
- K. Rippe, N. Mucke and J. Langowski, Superhelix Dimensions of a 1868 Base Pair Plasmid Determined by Scanning Force Microscopy in Air and in Aqueous Solution, Nucleic Acids Res., 1997, 25(9), 1736–1744, DOI:10.1093/nar/25.9.1736.
- C. Bustamante, J. Vesenka, C. L. Tang, W. Rees, M. Guthold and R. Keller, Circular DNA Molecules Imaged in Air by Scanning Force Microscopy, Biochemistry, 1992, 31(1), 22–26, DOI:10.1021/bi00116a005.
- Y. L. Lyubchenko and L. S. Shlyakhtenko, Visualization of Supercoiled DNA with Atomic Force Microscopy in Situ, Proc. Natl. Acad. Sci. U. S. A., 1997, 94(2), 496–501, DOI:10.1073/pnas.94.2.496.
- J. Reed, C. Hsueh, B. Mishra and J. K. Gimzewski, Atomic Force Microscope Observation of Branching in Single Transcript Molecules Derived from Human Cardiac Muscle, Nanotechnology, 2008, 19(38), 384021, DOI:10.1088/0957-4484/19/38/384021.
- K. Regan, S. Ricketts and R. Robertson-Anderson, DNA as a Model for Probing Polymer Entanglements: Circular Polymers and Non-Classical Dynamics, Polymers, 2016, 8(9), 336, DOI:10.3390/polym8090336.
- S. Nian and L.-H. Cai, Dynamic Mechanical Properties of Self-Assembled Bottlebrush Polymer Networks, Macromolecules, 2022, 55(18), 8058–8066, DOI:10.1021/acs.macromol.2c01204.
- R. M. Robertson-Anderson, Biopolymer Networks: Design, Dynamics and Discovery, IOP Publishing, Bristol [England] (Temple Circus, Temple Way, Bristol BS1 6HG, UK), 2024 Search PubMed.
- K. R. Peddireddy, M. Lee, Y. Zhou, S. Adalbert, S. Anderson, C. M. Schroeder and R. M. Robertson-Anderson, Unexpected Entanglement Dynamics in Semidilute Blends of Supercoiled and Ring DNA, Soft Matter, 2020, 16(1), 152–161, 10.1039/C9SM01767D.
- M. Q. Tu, O. Davydovich, B. Mei, P. K. Singh, G. S. Grest, K. S. Schweizer, T. C. O'Connor and C. M. Schroeder, Unexpected Slow Relaxation Dynamics in Pure Ring Polymers Arise from Intermolecular Interactions, ACS Polym. Au, 2023, 3(4), 307–317, DOI:10.1021/acspolymersau.2c00069.
- B. W. Soh, A. R. Klotz, R. M. Robertson-Anderson and P. S. Doyle, Long-Lived Self-Entanglements in Ring Polymers, Phys. Rev. Lett., 2019, 123(4), 048002, DOI:10.1103/PhysRevLett.123.048002.
- Q. Huang, J. Ahn, D. Parisi, T. Chang, O. Hassager, S. Panyukov, M. Rubinstein and D. Vlassopoulos, Unexpected Stretching of Entangled Ring Macromolecules, Phys. Rev. Lett., 2019, 122(20), 208001, DOI:10.1103/PhysRevLett.122.208001.
- Y. Yamauchi, N. N. Horimoto, K. Yamada, Y. Matsushita, M. Takeuchi and Y. Ishida, Two–Step Divergent Synthesis of Monodisperse and Ultra–Long Bottlebrush Polymers from an Easily Purifiable ROMP Monomer, Angew. Chem., 2021, 133(3), 1552–1558, DOI:10.1002/ange.202009759.
- M. Müllner, Molecular Polymer Bottlebrushes in Nanomedicine: Therapeutic and Diagnostic Applications, Chem. Commun., 2022, 58(38), 5683–5716, 10.1039/D2CC01601J.
- F. Jia, D. Wang, X. Lu, X. Tan, Y. Wang, H. Lu and K. Zhang, Improving the Enzymatic Stability and the Pharmacokinetics of Oligonucleotides via DNA-Backboned Bottlebrush Polymers, Nano Lett., 2018, 18(11), 7378–7382, DOI:10.1021/acs.nanolett.8b03662.
- A. Hernandez-Garcia, N. A. Estrich, M. W. T. Werten, J. R. C. Van Der Maarel, T. H. LaBean, F. A. de Wolf, M. A. Cohen Stuart and R. de Vries, Precise Coating of a Wide Range of DNA Templates by a Protein Polymer with a DNA Binding Domain, ACS Nano, 2017, 11(1), 144–152, DOI:10.1021/acsnano.6b05938.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5py00082c |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |