Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chimeric fungal–firefly luciferins exhibit red shifted fungal bioluminescence
Jude I.
Ayogu
a,
Minyan
Lyu
a,
Aleksei D.
Barykin
bc,
Anastasia A.
Fadeeva
b,
Zinaida M.
Kaskova
 bd and
James C.
Anderson
bd and
James C.
Anderson
 *a
*a
aDepartment of Chemistry, University College London, 20 Gordon Street, London WC1H 0AJ, UK. E-mail: j.c.anderson@ucl.ac.uk
bShemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow 117997, Russia
cMoscow Center for Advanced Studies, Kulakova str. 20, Moscow 123592, Russian Federation
dPirogov Russian National Research Medical University, Moscow 117997, Russia
First published on 22nd October 2025
Abstract
A rational design of new luciferins based upon the combination of luciferins from different lineages is exemplified by the synthesis of chimeric luciferins from a combination of fungal and firefly luciferin. Two new chimeric fungal–firefly luciferins exhibited the furthest red shifted bioluminescence of a fungal luciferin analogue.
Introduction
Bioluminescence is the emission of light by living organisms and is one of nature's most beautiful spectacles on land and in the sea. The light is generated from chemical energy, in most cases, by the catalytic oxidation of a small molecule, luciferin, by a respective enzyme, luciferase. Each different bioluminescent organism has a specific structure of a luciferin and luciferase pair, and a mechanistically different chemical pathway to giving light. There are a few known structures of luciferins and their structures are diverse (Fig. 1).1 The structures were elucidated mostly in the 20th century, with fungal luciferin 1 being one of the most recent in 2015.2 A small part of each luciferin molecule, the ‘reactive’ core, is oxidised to give an excited state species which is conjugated to the remaining bulk, the chromophore, of the luciferin molecule which characterises the subsequent light emission. The light emission can also be affected to some extent by the microenvironment of the luciferase active site.Luciferases are widely used for imaging biological processes in vitro, in live cells and in animal models. The most popular bioluminescent imaging system uses firefly's D-luciferin (2) which bioluminesces with λmax = 558 nm (Fig. 1).3 The technique does not require an external light source, unlike fluorescence imaging, so there are no background photons, leading to a higher signal/noise ratio, which is particularly attractive for bioanalytical methods that require enhanced sensitivity. It has proven easy to use, cheap, non-invasive, highly sensitive and has a wide dynamic range.4 However, one of the main limitations of using bioluminescence in vivo is that only a fraction of the light typically reaches the detector, because the photons are absorbed and/or scattered by haemoglobin, melatonin and tissue. This has a detrimental effect on image resolution and signal penetration depth. It is well accepted that light beyond the visible range (infrared) is more tissue penetrant.5
The development of new imaging techniques has relied upon manipulating the natural capabilities of a bioluminescence system, most often to red shift the wavelength of the emitted light for improved tissue penetration. For the most popular bioluminescent imaging system D-luciferin this was approached first through mutations of the luciferase enzyme,6 but was found to be limited by the inherent structure and electronic properties of the D-luciferin itself. Derivatives of D-luciferin (Fig. 2) have led to enhanced properties in terms of colour modulation, most desirably towards the red end of the spectrum.7
The most successful analogues for imaging have been based upon substitution of the hydroxyl group of D-luciferin with an amino group (CycLuc8 and AkaLumine,9Fig. 2). Akalumine was also the first example of extended conjugation leading to red shifted bioluminescent emission.10 This concept was in turn used to develop some of the most red shifted luciferins to date, infraluciferin11 and napthylluciferins12 (λmax = 730 and 750 nm respectively),13 along with other examples that have shown the beneficial effects of extending π-conjugation.14–17 Akalumine was also an example of complete substitution of the benzothiazole nucleus of D-luciferin. Other examples have substituted for new heterocylces,18,19 benzothiophene,20 quinoline,21 coumarin derivatives22–24 and benzobisthiazole ‘V’-shaped motifs.25 They all suffer from much lower quantum yields than D-luciferin, but some are nevertheless efficacious due to more tissue penetrant red light being produced and specific luciferase matching.13,26 This has been shown to be the case for CycLuc,8 akalumine,9 napthylluciferin12 and coumarin luciferins.23
There is still a pressing need for bright, multicoloured, nr-IR emitting luciferins for improved in vivo tissue penetration and new multiparametric bioluminescence analytical/imaging techniques for a wide range of applications. The opportunities to substitute and adorn the skeleton of the natural core of D-luciferin are dwindling and could be considered exhausted (Fig. 2). The coupling of other chromophores such as dye molecules (BODIPY, squaraine, rhodamine dye etc.) to the ‘reactive’ thiazoline unit has given useful bioluminescent species,9,10,12,18–24 but future examples of this concept are not obvious. Computer aided design is possible, but only when the three dimensional structure of the luciferase is known, which is not the case for all luciferases. We postulate that to rationally diversify the structures of new luciferins it would be beneficial to look beyond the structure of D-luciferin and its structurally exhausted analogues. We propose the design of chimeric luciferins that combine the ‘reactive’ core of one luciferin with the chromophore of a luciferin from another lineage to identify new luciferin structures that may form the basis for further development of bioluminescent systems for imaging.
The luciferases and luciferin extracted from different species within the same phylogenetic lineage can be cross-reacted, resulting in light emission, because they are essentially identical bioluminescent systems within each lineage.27,28 Even though bioluminescence from each system requires the oxidation of a small molecule luciferin to its excited state, due to the subtly different mechanisms of bioluminescent light production, it is not possible for a luciferin from one lineage to give light with the luciferase from another lineage. However, luciferases can be promiscuous. For example and as already discussed, firefly luciferase can generate light with a multitude of unnatural D-luciferins (Fig. 2).7 This indicates that if the ‘reactive’ core of the molecule is present in an unnatural luciferin then the enzyme normally used to give light with that particular ‘reactive’ molecular feature, will most probably still give light with the new luciferin. The remaining bulk of the molecule can have a modified structure, which has been proven by the substitution of the benzothiazole of D-luciferin with other aromatic groups. Luciferin metabolites in nature have presumably evolved to gain selective advantages in their environment, in this specific case to give chemical light. By extension, each part of a luciferin, the ‘reactive’ core and the chromophore, have been selected for efficient bioluminescence. We wanted to demonstrate that for a rational synthesis of diverse new luciferins, a ‘reactive’ core of a luciferin from one lineage could be combined with a chromophore from a luciferin from another lineage. The wild type luciferase matched to the ‘reactive’ partner of the chimeric luciferin should be able to generate bioluminescence. This cross-lineage, modular approach allows extended π-conjugation and rational tuning of emission wavelengths while retaining enzymatic compatibility. We report our preliminary investigation of this chimeric approach by the synthesis and bioluminescent evaluation of two chimeric fungal–firefly luciferins, that demonstrate their potential to expand the spectral and structural diversity of luciferins.
Results and discussion
Design, calculations and synthesis
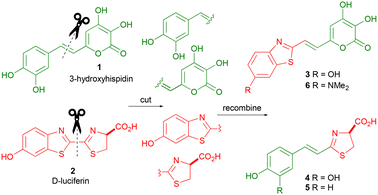
As a rational design concept, the chimeric luciferin should possess the character of both parent luciferins, and the two conjoined parts should be in conjugation with each other. By analysing the structures and mechanisms of bioluminescence from natural luciferins (Fig. 1) potential partners can be identified. In this preliminary study we focussed on the ‘reactive’ part of fungi's luciferin, 3-hydroxyhispidin (1) (Scheme 1).29 In fungal luciferin 1, and like D-luciferin (2), conjugation between the 3-hydroxyl group of catechol and the ‘reactive’ functional group is essential for bioluminescence. Hypothetically conjoining the two separate halves of fungal and D-luciferin could lead to two chimeric luciferins 3 and 4, both of which retain conjugation of a hydroxyl group with the ‘reactive’ partner (Scheme 1). Chimeric structure 3 retains the active core of fungal luciferin 1 conjugated with the benzothiazole chromophore from D-luciferin (2). While chimera 4 retains the active core from D-luciferin (2) with the catechol chromophore from fungal luciferin 1.We thought that chimera 3 would be more red shifted than fungal luciferin 1 as D-luciferin (2) (λmax = 558 nm) bioluminesces with a more red shifted emission than fungal luciferin 1 (λmax ∼ 530 nm). We also thought that chimera 4 would most likely be a poor bioluminescence emitter and unlikely to have significantly red shifted bioluminescence compared to that of fungal luciferin. This was based upon the seminal work on Akalumine (Fig. 2) which showed that the 4-substiuted phenol analogue of chimera 4, analogue 5, was a weak bioluminescence emitter (λmax = 530 nm).10 Amino luciferins are often red shifted compared to their hydroxyl parents and increased lipohilicity can yield advantages in terms of lowering Km.7–9,12 Therefore this study focussed on the synthesis of chimera 3 and due to the beneficial tuning effect of substituting the hydroxyl group in D-luciferin with a dimethylamine,7–9,12 we also chose to additionally synthesise chimera 6.
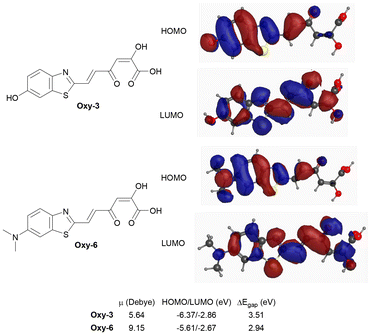
DFT calculations at the B3LYP/6-311+G(2d,p) level30,31 were performed to interrogate the proposed red shifting effect of dimethylamine substitution in chimera 6 over the hydroxyl derivative chimera 3. The calculations supported our prediction that of the light giving oxidised forms of the luciferins, that would be produced by fungal luciferase, Oxy-6 had a smaller HOMO–LUMO gap (2.94 eV) and a higher dipole moment (μ = 9.15 D) than Oxy-3 (3.51 eV; μ = 5.64 D) which should manifest itself in red shifted bioluminescence of chimera analogue 6 compared to chimera 3. The calculated values (Fig. 3) are consistent with increased charge delocalisation and enhanced intramolecular charge transfer (ICT).32,33 Orbital density mapping showed HOMO localisation on the aryl donor and LUMO concentration on the reactive backbone (Fig. 3), supporting directional ICT upon excitation.
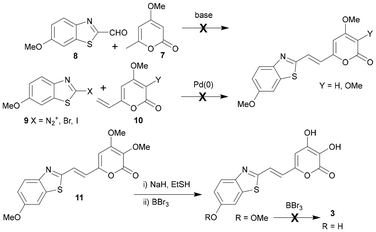
We investigated a synthetic strategy for synthesising the chimeric fungal–firefly luciferins that focused on the central alkene function as a convergent joining point. Although a series of condensation reactions between 4-methoxy-6-methyl-2H-pyran-2-one (7) and a range of aryl aldehydes has been reported,34 analogous attempts with 6-methoxybenzo[d]thiazole-2-carbaldehyde (8), with a view to using the fully oxygenated 3,4-dimethoxy-pyranone under similar conditions if successful, led to degradation (Scheme 2). The exploration of a range of deprotonating conditions using LDA, t-BuOK, NaOAc, piperidine and DBU were also unfruitful. Inspired by the reported palladium catalysed coupling of arene diazonium tetrafluoroborates with a vinyl-2-pyrone,35 investigation of a Heck type approach with suitable coupling partners 9 and 10 was also unsuccessful under a range of different conditions (Scheme 2). Resorting to Wittig based technology that had already been shown by Kaskova et al. to be effective for the synthesis of fungal luciferin analogues,29 a fully tris-methyl protected analogue 11 was prepared. Unfortunately global deprotection of the methyl groups to give target chimera 3 was unsuccessful due to the 6-MeO group being recalcitrant to unmasking under a range of more forcing conditions that ultimately led to degradation. It was decided to repeat this latter approach with a more compliant 6-phenolic protecting group.
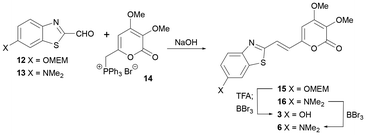
The approach was repeated with a 6-OMEM group as it had been used successfully in luciferin synthesis.36,37 Wittig reaction of known aldehydes 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 and 13
37 and 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38 with phosphonium salt 14
38 with phosphonium salt 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 gave good yields of alkene products 15 and 16 (Scheme 3). Global deprotection of 15 was achieved in two steps, first by removal of the MEM protecting group with TFA and then treatment with an excess of BBr3 to give target 3 in a 60% yield after reverse phase HPLC purification. After optimisation global deprotection of 16 was achieved by stirring with an excess of BBr3 for 4 days to give 6 in 69% yield after reverse phase HPLC purification.
29 gave good yields of alkene products 15 and 16 (Scheme 3). Global deprotection of 15 was achieved in two steps, first by removal of the MEM protecting group with TFA and then treatment with an excess of BBr3 to give target 3 in a 60% yield after reverse phase HPLC purification. After optimisation global deprotection of 16 was achieved by stirring with an excess of BBr3 for 4 days to give 6 in 69% yield after reverse phase HPLC purification.
Spectroscopic and bioluminescent properties
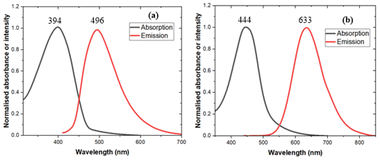
Fluorescence spectra (DMSO) showed maximum absorption for 3λ = 394 nm and 6λ = 444 nm with corresponding emissions for 3λ = 496 nm and 6λ = 633 nm (Fig. 4). The greater red shifted emission of 6 was in line with our DFT calculations.The bioluminescence of the chimeric fungal–firefly luciferins, 3 and 6 were assayed with recombinant Neonothopanus nambi luciferase. Both compounds produced red shifted emission profiles compared to wild type fungal luciferin 1, with λmax values for 3 at 630 nm more red shifted than 6 at 600 nm (Fig. 5).
 | ||
| Fig. 5 Bioluminescence emission spectra of chimeric fungal–firefly luciferins 3 and 6 with recombinant fungal luciferase, showing red shifted λmax values relative to fungal luciferin 1. | ||
While the red shifted bioluminescence emission of 3 compared to 6 contradicted our DFT calculations, fluorescence measurements and the results from amino firefly luciferins in the literature,7–9,12 the results did fit the trend seen for fungal luciferin analogues,29 and those of coumarin luciferins23 with respect to substitution of hydroxyl groups for dimethylamino substituents. Hydroxyl groups can induce bathochromic shifts through microenvironmental interactions within the active site of the luciferase that are not captured by in vitro DFT or optical measurements.
Kinetic analyses (Fig. 6 and Table 1) revealed low Michaelis constants (Km = 0.40 μM for 3 and 0.25 μM for 6) compared to fungal luciferin (Km = 1.09 μM), indicative of high luciferase affinity. The lower Km value of dimethylamino chimera analogue 6 over that of chimera 3 is in line with previous observations for amino substituted luciferins.7–9,12 However, the Vmax values were markedly reduced compared to fungal luciferin with relative bioluminescence intensities of ∼1.5% for 3 and ∼2.8% for 6 (Fig. 7). This suggested tight binding, but poor catalytic turnover possibly due to steric or electronic misalignment in the active site. These magnitudes of relative bioluminescence are expected for luciferin analogues.7
| Substrate | Relative activity (%) | K m (μM) | Emission wavelength maximum (nm) |
|---|---|---|---|
| 1 | 100 | 1.09 ± 0.06 | 548 ± 10 |
| 6 | 2.8 | 0.25 ± 0.05 | 600 ± 10 |
| 3 | 1.5 | 0.40 ± 0.05 | 630 ± 10 |
Conclusions
This work has shown proof of concept that the combination of luciferins from different lineages can lead to bioluminescent chimeric luciferins. In this preliminary work, careful selection of luciferin partners from fungal and firefly bioluminescence systems resulted in the design and synthesis of chimeric fungal–firefly luciferins that exhibited the farthest red shifted fungal luciferin bioluminescence recorded. The initial structural designs were validated by DFT calculations. The synthesis of the new chimeric fungal–firefly luciferins used the existing route used by Kaskova for the synthesis of fungal luciferin analogues and proved to be efficient in terms of yield and purity. The chimeric fungal–firefly luciferin 3 exhibited bioluminescence of λmax = 630 nm compared to wild type fungal luciferin of λmax = 548 nm. Substitution of the hydroxyl group in chimera 3 for a dimethylamino group gave chimera analogue 6 with λmax = 600 nm. DFT calculations and optical measurements had suggested that chimera 6 possessed physical properties that could make it more red shifted than its parent chimera 3. However, subtle interactions with the oxidised light giving forms of their luciferins in the microenvironment of the luciferase active site, particularly with hydroxyl groups,7 may alter the character of the frontier molecular orbitals of the luciferins that are not captured by in vitro DFT or optical measurements. This has previously been noted for fungal luciferin analogues,29 and those of coumarin luciferins23 with respect to substitution of hydroxyl groups for dimethylamino substituents. The relative activities of chimera 3 and 6 are 2 orders of magnitude lower than that of fungal luciferin 1. In bioluminescence imaging brightness is not the absolute requirement for better penetration and resolution because of a reduced signal to noise effect caused by light scattering. Scatter of light is inversely proportional to the fourth power of wavelength and so is less for red shifted light. The two new luciferin structures 3 and 6 may thus offer a starting point for the further development of fungal luciferin bioluminescence imaging39 through the engineering of fungal luciferase mutants and simple chemical structure modifications as has been achieved in firefly bioluminescence imaging. In addition the design of chimeric luciferins from other lineages offers a rational design concept for new luciferins that may be tuned for other bioluminescence applications.Experimental
General experimental details – please see SI
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 20 (1.99 g, 78%, lit.3 34%) as a yellow solid; Rf 0.29 (pet. ether/EtOAc 1
1) to give 20 (1.99 g, 78%, lit.3 34%) as a yellow solid; Rf 0.29 (pet. ether/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); 1H NMR (400 MHz, CDCl3) δ 6.29 (1H, s, CH), 4.14 (2H, s, CH2), 3.97 (3H, s, CH3), 3.83 (3H, s, CH3). 13C NMR (100 MHz, CDCl3) δ 160.9 (C
1); 1H NMR (400 MHz, CDCl3) δ 6.29 (1H, s, CH), 4.14 (2H, s, CH2), 3.97 (3H, s, CH3), 3.83 (3H, s, CH3). 13C NMR (100 MHz, CDCl3) δ 160.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 157.8 (CCH2), 154.2 (COCH3), 128.5 (C(OCH3)C
O), 157.8 (CCH2), 154.2 (COCH3), 128.5 (C(OCH3)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 99.6 (CH), 60.3 (OCH3), 57.8 (OCH3), 26.9 (CH2). The data was in agreement with the literature.29
O), 99.6 (CH), 60.3 (OCH3), 57.8 (OCH3), 26.9 (CH2). The data was in agreement with the literature.29
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 (24.0 mg, 0.0471 mmol) in DCM (1.0 mL) was added aldehyde 12
29 (24.0 mg, 0.0471 mmol) in DCM (1.0 mL) was added aldehyde 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 (18.8 mg, 0.0704 mmol) and the reaction mixture was allowed to stir at room temperature. After 4 h, the reaction mixture was diluted with DCM (2 mL) and washed with water (2 × 2 mL). The aqueous layer was re-extracted with DCM (2 mL), and the combined organic layers were dried (Na2SO4), filtered and concentrated in vacuo. The residue was purified by flash column chromatography (pet. ether/EtOAc 1
37 (18.8 mg, 0.0704 mmol) and the reaction mixture was allowed to stir at room temperature. After 4 h, the reaction mixture was diluted with DCM (2 mL) and washed with water (2 × 2 mL). The aqueous layer was re-extracted with DCM (2 mL), and the combined organic layers were dried (Na2SO4), filtered and concentrated in vacuo. The residue was purified by flash column chromatography (pet. ether/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 → 1
1 → 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) to afford the product 10 (46.0 mg, quant.) as a fluorescent green syrup; Rf 0.26 (pet. ethe/EtOAc 1
4) to afford the product 10 (46.0 mg, quant.) as a fluorescent green syrup; Rf 0.26 (pet. ethe/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); λmax (acetonitrile) = 389 nm (c = 0.27 mM, ε = 14
1); λmax (acetonitrile) = 389 nm (c = 0.27 mM, ε = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 967 L mol−1 cm−1); IR (neat) 1692 (C
967 L mol−1 cm−1); IR (neat) 1692 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1247 (C–O), 1029 cm−1; 1H NMR (500 MHz, CD3CN) δ 7.81 (1H, d, J = 8.9, ArCH), 7.62 (1H, d, J = 2.4, ArCH), 7.33 (1H, d, J = 15.8, HC
C), 1247 (C–O), 1029 cm−1; 1H NMR (500 MHz, CD3CN) δ 7.81 (1H, d, J = 8.9, ArCH), 7.62 (1H, d, J = 2.4, ArCH), 7.33 (1H, d, J = 15.8, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.16–7.12 (2H, m, HC
CH), 7.16–7.12 (2H, m, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH, ArCH), 6.78 (1H, s, CHCOCH3), 5.27 (2H, s, OCH2O), 3.95 (3H, s, OCH3), 3.74–3.72 (2H, m, OCH2), 3.70 (3H, s, OCH3), 3.45–3.43 (2H, m, OCH2), 3.18 (3H, s, OCH3); 13C NMR (125 MHz, CD3CN) δ 162.3 (COC
CH, ArCH), 6.78 (1H, s, CHCOCH3), 5.27 (2H, s, OCH2O), 3.95 (3H, s, OCH3), 3.74–3.72 (2H, m, OCH2), 3.70 (3H, s, OCH3), 3.45–3.43 (2H, m, OCH2), 3.18 (3H, s, OCH3); 13C NMR (125 MHz, CD3CN) δ 162.3 (COC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 159.2 (C
O), 159.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 158.1 (C
N), 158.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.0 (ArC), 152.4 (ArC), 149.3 (ArC), 136.4 (COCH3), 129.2 (HC
O), 156.0 (ArC), 152.4 (ArC), 149.3 (ArC), 136.4 (COCH3), 129.2 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 125.8 (HC
CH), 125.8 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 125.5 (ArCH), 123.7 (ArCH), 117.5 (C(OCH3)C
CH), 125.5 (ArCH), 123.7 (ArCH), 117.5 (C(OCH3)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 107.6 (ArCH), 101.6 (HCCOCH3), 93.8 (OCH2O), 71.4 (OCH2), 67.7 (OCH2), 59.1 (OCH3), 57.8 (OCH3), 57.0 (OCH3); HRMS C20H22NO732S [M + H]+ calcd 420.1102, found 420.1112.
O), 107.6 (ArCH), 101.6 (HCCOCH3), 93.8 (OCH2O), 71.4 (OCH2), 67.7 (OCH2), 59.1 (OCH3), 57.8 (OCH3), 57.0 (OCH3); HRMS C20H22NO732S [M + H]+ calcd 420.1102, found 420.1112.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford the mono deprotected 6-OH (231 mg, 95%) as a pale green-fluorescent solid; Rf 0.14 (pet. ether/EtOAc 1
1) to afford the mono deprotected 6-OH (231 mg, 95%) as a pale green-fluorescent solid; Rf 0.14 (pet. ether/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); mp 85–86 °C; IR (neat) cm−1 3099, 2995, 2930 (C–H), 1684 (C
1); mp 85–86 °C; IR (neat) cm−1 3099, 2995, 2930 (C–H), 1684 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1606 (C
O), 1606 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1410, 1347 (O–H bend), 1265 (C–O) cm−1; 1H NMR (500 MHz, DMSO) δ 10.14 (1H, s, OH), 7.81 (1H, d, J = 8.9, ArCH), 7.64–7.53 (1H, m, ArCH), 7.35 (1H, d, J = 15.8, HC
C), 1410, 1347 (O–H bend), 1265 (C–O) cm−1; 1H NMR (500 MHz, DMSO) δ 10.14 (1H, s, OH), 7.81 (1H, d, J = 8.9, ArCH), 7.64–7.53 (1H, m, ArCH), 7.35 (1H, d, J = 15.8, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.23 (1H, J = 15.9, HC
CH), 7.23 (1H, J = 15.9, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.09 (1H, s, HCCOCH3), 7.01 (1H, dd, J = 8.8, 2.4, ArCH), 3.94 (3H, s, OCH3), 3.71 (3H, s, OCH3); 13C NMR (125 MHz, DMSO) δ 162.3 (COC
CH), 7.09 (1H, s, HCCOCH3), 7.01 (1H, dd, J = 8.8, 2.4, ArCH), 3.94 (3H, s, OCH3), 3.71 (3H, s, OCH3); 13C NMR (125 MHz, DMSO) δ 162.3 (COC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 159.2 (C
O), 159.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 158.1 (C
N), 158.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.0 (ArC), 152.4 (ArC), 149.3 (ArC), 131.5 (COCH3), 128.7 (HC
O), 156.0 (ArC), 152.4 (ArC), 149.3 (ArC), 131.5 (COCH3), 128.7 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 125.5 (HC
CH), 125.5 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 125.4 (ArCH), 123.8 (ArCH), 116.7 (COCH3), 106.8 (ArCH), 101.6 (CHCOCH3), 59.5 (OCH3), 57.4 (OCH3); HRMS C16H14O5N32S [M + H]+ calcd 332.0587, found 332.0580.
CH), 125.4 (ArCH), 123.8 (ArCH), 116.7 (COCH3), 106.8 (ArCH), 101.6 (CHCOCH3), 59.5 (OCH3), 57.4 (OCH3); HRMS C16H14O5N32S [M + H]+ calcd 332.0587, found 332.0580.
To a solution of the mono deprotected 6-OH compound prepared above (51.0 mg 0.142 mmol) in anhydrous DCM (40 mL) was added a solution of BBr3 (7.10 mL of 1.0 M in DCM, 30 eq.) and the reaction mixture was allowed to stir for 3 days under N2. The solvent and the excess BBr3 were removed in vacuo, and the residue was partitioned between phosphate buffer (100 mL, pH 7.4) and EtOAc (100 mL). The layers were separated, the aqueous layer was further extracted with EtOAc (3 × 100 mL), and the combined organic layers dried (Na2SO4), filtered and concentrated in vacuo. The residue was dissolved in DMSO (6.00 mL) and purified by reverse-phase HPLC to give the product 3 (162 mg, 58%) as a green solid; Rt 13.4 min (5–95% acetonitrile in water); λmax (DMSO) = 394 nm (c = 0.33 μM, ε = 2666 L mol−1 cm−1); IR (neat) 2925, 2856 (C–H), 1732 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1457, 1376 (O–H bend), 1265 (C–O) cm−1; 1H-NMR (700 MHz, DMSO-d6) δ 10.80 (1H, br s, OH), 9.96 (1H, s, OH), 9.23 (1H, br s, OH), 7.78 (1H, d, J = 8.8, ArCH), 7.37 (1H, d, J = 2.5, ArCH), 7.19 (1H, d, J = 15.9, HC
O), 1457, 1376 (O–H bend), 1265 (C–O) cm−1; 1H-NMR (700 MHz, DMSO-d6) δ 10.80 (1H, br s, OH), 9.96 (1H, s, OH), 9.23 (1H, br s, OH), 7.78 (1H, d, J = 8.8, ArCH), 7.37 (1H, d, J = 2.5, ArCH), 7.19 (1H, d, J = 15.9, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.18 (1H, d, J = 15.9, HC
CH), 7.18 (1H, d, J = 15.9, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 6.97 (1H, dd, J = 8.8, 2.5, ArCH), 6.59 (1H, s, CHC(OH)); 13C NMR (150 MHz, acetone-d6) δ 162.3 (COH), 161.0 (COC
CH), 6.97 (1H, dd, J = 8.8, 2.5, ArCH), 6.59 (1H, s, CHC(OH)); 13C NMR (150 MHz, acetone-d6) δ 162.3 (COH), 161.0 (COC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 157.1 (C
O), 157.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.8 (C
N), 149.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 149.0 (ArC), 137.5 (ArC), 132.7 (C(OH)C
O), 149.0 (ArC), 137.5 (ArC), 132.7 (C(OH)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 129.5 (ArC), 129.4 (HC
O), 129.5 (ArC), 129.4 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 126.4 (HC
CH), 126.4 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 124.7 (ArCH), 117.2 (ArCH), 107.4 (ArCH), 107.0 (CHC(OH)); HRMS C14H10O5N32S [M + H]+ calcd 304.0274, found 304.0278.
CH), 124.7 (ArCH), 117.2 (ArCH), 107.4 (ArCH), 107.0 (CHC(OH)); HRMS C14H10O5N32S [M + H]+ calcd 304.0274, found 304.0278.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 (204 mg, 0.400 mmol) in DCM (14 mL) was added the aldehyde 1338 (124 mg, 0.602 mmol). After 3 h, the reaction mixture was diluted with DCM (2 × 50 mL), washed with H2O (2 × 50 mL), dried (Na2SO4), filtered and concentrated in vacuo. The residue was purified by flash column chromatography (pet. ether/EtOAc 1
29 (204 mg, 0.400 mmol) in DCM (14 mL) was added the aldehyde 1338 (124 mg, 0.602 mmol). After 3 h, the reaction mixture was diluted with DCM (2 × 50 mL), washed with H2O (2 × 50 mL), dried (Na2SO4), filtered and concentrated in vacuo. The residue was purified by flash column chromatography (pet. ether/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 16 (123 mg, 86%.) as a fluorescent deep orange solid; Rf 0.41 (pet. ether/EtOAc 1
1) to afford 16 (123 mg, 86%.) as a fluorescent deep orange solid; Rf 0.41 (pet. ether/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); mp >200 °C (dec.); λmax (acetonitrile) = 415 (c = 0.22 M, ε = 17
1); mp >200 °C (dec.); λmax (acetonitrile) = 415 (c = 0.22 M, ε = 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312 L mol−1 cm−1); IR (neat) 3087, 2918 (C–H), 1684 (C
312 L mol−1 cm−1); IR (neat) 3087, 2918 (C–H), 1684 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1611 (C
O), 1611 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1348 (C–N) 1278 (C–O), 1156 (C–O) cm−1; 1H NMR (500 MHz, CDCl3) δ 7.84 (1H, d, J = 9.0, ArCH), 7.50 (1H, d, J = 15.6, HC
C), 1348 (C–N) 1278 (C–O), 1156 (C–O) cm−1; 1H NMR (500 MHz, CDCl3) δ 7.84 (1H, d, J = 9.0, ArCH), 7.50 (1H, d, J = 15.6, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.03 (1H, d, J = 2.6, ArCH), 6.97–6.92 (2H, m, HC
CH), 7.03 (1H, d, J = 2.6, ArCH), 6.97–6.92 (2H, m, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH, ArCH), 6.21 (1H, s, CHC(OCH3)), 3.99 (3H, s, OCH3), 3.89 (3H, s, OCH3), 3.05 (6H, s, N(CH3)2); 13C NMR (125 MHz, CDCl3) δ 160.5 (COC
CH, ArCH), 6.21 (1H, s, CHC(OCH3)), 3.99 (3H, s, OCH3), 3.89 (3H, s, OCH3), 3.05 (6H, s, N(CH3)2); 13C NMR (125 MHz, CDCl3) δ 160.5 (COC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 159.0 (C
O), 159.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 158.2 (C
N), 158.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 153.4 (ArC), 149.4 (ArC-NMe2), 146.2 (ArC), 138.0 (COCH3), 129.3 (HC
O), 153.4 (ArC), 149.4 (ArC-NMe2), 146.2 (ArC), 138.0 (COCH3), 129.3 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 127.3 (HC
CH), 127.3 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 123.8 (ArCH), 123.1 (ArCH), 114.0 (ArCH), 102.4 (ArCH), 100.2 (CHC(OCH3)), 60.5 (OCH3), 57.6 (OCH3), 41.0 (N(CH3)2); HRMS C18H18N2O432S [M + H]+ calcd 359.1060, found 359.1057.
CH), 123.8 (ArCH), 123.1 (ArCH), 114.0 (ArCH), 102.4 (ArCH), 100.2 (CHC(OCH3)), 60.5 (OCH3), 57.6 (OCH3), 41.0 (N(CH3)2); HRMS C18H18N2O432S [M + H]+ calcd 359.1060, found 359.1057.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1561 (C
O), 1561 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1354 (C–N), 1153 (C–O) cm−1; 1H NMR (700 MHz, acetone-d6) δ 7.77 (1H, d, J = 9.1, ArCH), 7.30 (1H, d, J = 15.8, HC
C), 1354 (C–N), 1153 (C–O) cm−1; 1H NMR (700 MHz, acetone-d6) δ 7.77 (1H, d, J = 9.1, ArCH), 7.30 (1H, d, J = 15.8, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.24 (1H, d, J = 2.6, ArCH), 7.05 (1H, d, J = 15.8, HC
CH), 7.24 (1H, d, J = 2.6, ArCH), 7.05 (1H, d, J = 15.8, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.03 (1H, dd, J = 9.1, 2.6, ArCH), 6.52 (1H, s, CHC(OH)), 3.06 (6H, s, (N(CH3)2); 13C NMR (150 MHz, acetone-d6) δ 162.4 (COH), 160.1 (COC
CH), 7.03 (1H, dd, J = 9.1, 2.6, ArCH), 6.52 (1H, s, CHC(OH)), 3.06 (6H, s, (N(CH3)2); 13C NMR (150 MHz, acetone-d6) δ 162.4 (COH), 160.1 (COC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 150.4 (C
O), 150.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 150.1 (C
N), 150.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 146.8 (ArC), 138.2 (ArC), 128.0 (C(OH)C
O), 146.8 (ArC), 138.2 (ArC), 128.0 (C(OH)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 125.4 (ArC), 125.2 (HC
O), 125.4 (ArC), 125.2 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 124.1 (HC
CH), 124.1 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 122.4 (ArCH)), 114.4 (ArCH), 103.3 (ArCH), 103.0 (CHC(OH)), 40.8 (N(CH3)2); HRMS C16H15N2O432S [M + H]+ calcd 331.0742, found 331.0747.
CH), 122.4 (ArCH)), 114.4 (ArCH), 103.3 (ArCH), 103.0 (CHC(OH)), 40.8 (N(CH3)2); HRMS C16H15N2O432S [M + H]+ calcd 331.0742, found 331.0747.
Author contributions
JIA and ML developed the syntheses, ADB and AAF determined the bioluminescence data, ZMK interpreted the results, JCA conceived the project, directed the research and wrote the paper with input from all authors.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: general experimental, kinetic data collection, computational data files, 1H and 13C NMR spectra for all new compounds and HPLC traces with fraction collection. See DOI: https://doi.org/10.1039/d5ob01589h.Acknowledgements
We thank the Leverhulme Trust RPG-2019-360, UCL and the Russian Science Foundation project number 25-76-30006 for funding.References
- Z. M. Kaskova, A. S. Tsarkova and I. V. Yampolsky, Chem. Soc. Rev., 2016, 45, 6048 RSC.
- K. V. Purtov, V. N. Petushkov, M. S. Baranov, K. S. Mineev, N. S. Rodionova, Z. M. Kaskova, A. S. Tsarkova, A. I. Petunin, V. S. Bondar, E. K. Rodicheva, S. E. Medvedeva, Y. Oba, Y. Oba, A. S. Arseniev, S. Lukyanov, J. I. Gitelson and I. V. Yampolsky, Angew. Chem., Int. Ed., 2015, 54, 8124 CrossRef CAS.
- M. Deluca, Adv. Enzymol., 1976, 44, 37 CAS.
- L. Mezzanotte, M. van't Root, H. Karatas, E. A. Goun and C. W. G. M. Löwick, Trends Biotechnol., 2017, 35, 640 CrossRef CAS PubMed.
- W. B. Rice and C. H. Contag, Nat. Biotechnol., 2009, 27, 624 CrossRef.
- See for example; B. R. Branchini, D. M. Ablamsky, A. L. Davis, T. L. Southworth, B. Butler, F. Fan, A. P. Jathoul and M. A. Pule, Biochem, 2010, 396, 290 CAS.
- R. Podsiadly, A. Grzelakowska, J. Modrzejewska, P. Siarkiwicz, D. Slowinski and M. Szala, Dyes Pigm., 2019, 170, 107627 CrossRef CAS.
- M. S. Evans, J. P. Chaurette, S. T. Adams, G. R. Reddy, M. A. Pauley, N. Aronin, J. A. Prescher and S. C. Miller, Nat. Methods, 2014, 11, 393 CrossRef CAS PubMed.
- T. Kuchimaru, S. Iwano, M. Kiyama, S. Mitsumata, T. Kadonosono, H. Niwa, S. Maki and S. Kizaka-Kondon, Nat. Commun., 2016, 7, 11856 CrossRef CAS.
- S. Iwano, R. Obata, C. Miura, M. Kiyama, K. Hama, M. Nakurama, Y. Amano, S. Kojima, T. Hirano, S. Maki and H. Niva, Tetrahedron, 2013, 69, 3847 CrossRef CAS.
- A. P. Jathoul, H. Grounds, J. C. Anderson and M. A. Pule, Angew. Chem., Int. Ed., 2014, 53, 13059 CrossRef CAS PubMed.
- M. P. Hall, C. C. Woodroofe, M. G. Wood, I. Que, M. V. Root, Y. Ridwan, C. Shi, T. A. Kirkland, L. P. Encell, K. V. Wood, C. Löwik and L. Mezzanotte, Nat. Commun., 2018, 9, 132 CrossRef PubMed.
- B. R. Branchini, D. M. Fontaine, D. Kohrt, B. P. Huta, A. R. Racela, B. R. Fort, T. L. Southworth and A. Roda, Int. J. Mol. Sci., 2022, 23, 2451 CrossRef CAS.
- G. R. Reddy, W. C. Thomson and S. C. Miller, J. Am. Chem. Soc., 2010, 132, 13586 CrossRef CAS PubMed.
- C. Miura, M. Kiyama, S. Iwano, K. Ito, R. Obata, T. Hirano, S. Maki and H. Niwa, Tetrahedron, 2013, 69, 9726 CrossRef.
- Y. Ikeda, T. Nomoto, Y. Hiruta, N. Nishiyama and D. Citterio, Anal. Chem., 2020, 92, 4235 CrossRef PubMed.
- Z. Yao, B. S. Zhang, R. C. Steinhardt, J. H. Mills and J. A. Prescher, J. Am. Chem. Soc., 2020, 142, 14080 CrossRef PubMed.
- D. C. McCutcheon, M. A. Paley and J. A. Prescher, J. Am. Chem. Soc., 2012, 134, 7604 CrossRef PubMed.
- B. S. Zhang, K. A. Jones, D. C. McCutcheon and J. A. Prescher, ChemBioChem, 2018, 19, 470 CrossRef.
- C. C. Woodroofe, P. L. Meisenheimer, D. H. Klaubert, Y. Kovic, J. C. Rosenberg, C. E. Behney, T. L. Southworth and B. R. Branchini, Biochemistry, 2012, 51, 9807 CrossRef.
- B. R. Branchini, Methods Enzymol., 2000, 305, 188 Search PubMed.
- H. Takakura, K. Sasakura, T. Ueno, Y. Urano, T. Terai, K. Hanaoka, T. Tsuboi and T. Nagano, Chem. – Asian J., 2010, 5, 2053 CrossRef PubMed.
- Z. Yao, D. R. Cadwell, A. C. Love, B. Kolbaba-Kartchner, J. H. Mills, M. J. Schnermann and J. A. Prescher, Chem. Sci., 2021, 12, 11684 RSC.
- A. C. Love, D. R. Cadwell, B. Kolbaba-Katchner, K. M. Townsend, L. P. Halbers, Z. Yao, C. K. Brennan, J. Ivanic, T. Hadjian, J. H. Mills, M. J. Schnermann and J. A. Prescher, J. Am. Chem. Soc., 2023, 145, 335 CrossRef PubMed.
- C.-H. Chang, D. M. Fontaine, B. R. Branchini, J. C. Anderson and S. Gómez, Chem. – Eur. J., 2023, 29, e202302204 CrossRef PubMed.
- A. C. Love and J. A. Prescher, Cell Chem. Biol., 2020, 27, 904 CrossRef PubMed.
- C. Szent-Gyorgyi, B. T. Ballou, E. Dagnal and B. Bryan, in Proceedings of SPPIE – The International Society for Optical Engineering (Biomedical Imaging: Reporters, Dyes and Instrumentation), 1999, p. 4.
- A. G. Oliveira, D. E. Desjardin, B. A. Perry and C. V. Stevani, Photochem. Photobiol. Sci., 2012, 11, 848 CrossRef PubMed.
- Z. M. Kaskova, F. A. Dörr, V. N. Petushkov, K. V. Purtov, A. S. Tsarkova, N. S. Rodionova, K. S. Mineev, E. B. Guglya, A. Kotlobay, N. S. Baleeva, M. S. Baranov, A. S. Arseniev, J. I. Gitelson, S. Lukyanov, Y. Suzuki, S. Kanie, E. Pinto, P. D. Mascio, H. E. Waldenmaier, T. A. Pereira, R. P. Carvalho, A. G. Oliveira, Y. Oba, E. L. Bastos, C. V. Stevani and I. L. Yampolsky, Sci. Adv., 2017, 3, e1602847 CrossRef PubMed.
- A. Becke, J. Chem. Phys., 1993, 98, 5648 CrossRef.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B:Condens. Matter Mater. Phys., 1988, 37, 785 CrossRef CAS PubMed.
- M. Zemmouche, C. García-Iriepa and I. Navizet, Phys. Chem. Chem. Phys., 2020, 22, 82 RSC.
- R. Winkler and S. Pantelides, J. Chem. Phys., 1997, 106, 7714 CrossRef CAS.
- C. Li, B. Cheng, S. Fang, H. Zhou, Q. Gu and J. Xu, Eur. J. Med. Chem., 2018, 143, 114 CrossRef CAS PubMed.
- C. Soldi, A. V. Moro, M. G. Pizzolatti and C. R. D. Correia, Eur. J. Org. Chem., 2012, 3607 CrossRef CAS.
- B. R. Branchinni, M. H. Murtiashaw, J. N. Carmody, E. Mygatt and T. L. Southworth, Bioorg. Med. Chem. Lett., 2005, 15, 3860 CrossRef.
- J. C. Anderson, H. Grounds, A. P. Jathoul, J. A. H. Murray, S. J. Pacman and L. Tisi, RSC Adv., 2017, 7, 3975 RSC.
- P. Hrobárik, I. Sigmundová and P. Zahradník, Synthesis, 2005, 600 Search PubMed.
- E. S. Shakova, T. A. Karataeva, N. M. Markena, T. Mitiouchkina, K. A. Palkina, M. M. Perfilov, M. Wood, T. T. Hoang, M. P. Hall, L. I. Fakhranurova, A. E. Alekberova, A. K. Malyshevskaia, D. A. Gorbachev, E. N. Bugaeva, L. K. Pletneva, V. V. Babenko, D. I. Boldreva, A. Y. Gorokhovatsky, A. V. Balakireva, F. Gao, V. V. Choob, L. P. Encell, K. V. Wood, I. L. Yampolsky, K. S. Sarkisyan and A. S. Mishin, Nat. Methods, 2024, 21, 406 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |