Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Pyreno-1,2,4-triazines as multifunctional luminogenic click reagents
Hind
Alshaikh
ab,
Jonathan
Parsons
b,
Gary
Askwith
b,
Michael E.
Deary
 b,
Catherine E.
Nicholson
b,
Catherine E.
Nicholson
 b,
Mark
Sims
b,
Mark
Sims
 *b and
Valery N.
Kozhevnikov
*b and
Valery N.
Kozhevnikov
 *b
*b
aDepartment of Chemistry, College of Science and Arts, King Abdulaziz University, Rabigh 21911, Saudi Arabia
bSchool of Geography and Natural Sciences, Northumbria University, Newcastle upon Tyne, NE1 8ST, UK. E-mail: mark.sims@northumbria.ac.uk; valery.kozhevnikov@northumbria.ac.uk
First published on 9th October 2025
Abstract
Several examples of 1,2,4-triazines that contain a pyrene fragment attached to the C5 position and a carbonyl functional group at the C3 position of the triazine ring have been prepared. While pyrene is a widely used luminophore, the carboxylic acid group at C3 is a convenient handle for further functionalization and significantly facilitates the Inverse Electron Demand Diels–Alder (IEDDA) reaction with strained alkenes and alkynes. The pyreno-1,2,4-triazine derivatives were investigated in reactions with the strained dienophiles BCN, TCO, and s-TCO. The rates were compared with reference to parent 1,2,4-triazine 10 with an unsubstituted C3 position which was synthesised through thermal decarboxylation of acid 6. The structure reactivity relationship was investigated theoretically showing that the predictive power of computational analysis benefits from extensive transition state conformer searches.
Introduction
Pyrene derivatives are useful in a variety of applications. Their bright luminescence and unique proximity-dependent emission have been widely exploited in materials science and bioimaging.1–3 In addition, the pyrene fragment has a high affinity for extended aromatic surfaces, which opens avenues for easy and efficient non-covalent functionalisation of carbon nanotubes and other 2D carbon materials.2,4–10 Combining more than one possible way of functionalisation is an even more convenient approach to fine-tune their properties for a particular application. In this regard, several pyrene derivatives functionalised with click groups have been previously developed.11–14 1,2,4-Triazines act as dienes in inverse electron demand Diels Alder (IEDDA) reactions with electron-rich or strained dienophiles.15 This well-known reaction was recently evaluated in the context of bioorthogonal chemistry16–26 where 1,2,4-triazines are complementary to commonly used 1,2,4,5-tetrazines and provide better stability under physiological conditions, although at the expense of slower kinetics. Luminescent bioorthogonal materials are in great demand because luminescence is arguably the most widely used methodology in biological investigations. However, while there are many examples of luminogenic 1,2,4,5-tetrazines, some of which are commercially available, there are only a few examples of luminogenic 1,2,4-triazines reported to date. Vrabel and co-workers19 described cationic 1,2,4-triazines that form a push–pull system after reaction with TCO with 50 fold increase in emission intensity. A large Stokes shift of 245 nm is certainly an advantage that will help to reduce background luminescence and separate excitation and emission channels. However, the 2.4% quantum yield of emission is low, which is a limitation in achieving high brightness. More recently, the same research group reported luminogenic 3-trifluoromethyl-1,2,4-triazines27 as well as 1,2,4-triazinium systems as much brighter luminogenic probes.28,29 Our group previously reported luminogenic behaviour of iridium(III) complexes of 1,2,4-triazines in reaction with BCN.23,30 Clearly there is a need to prepare more examples of luminescent and ideally luminogenic 1,2,4-triazines as click reagents to enrich the toolkit of chemical biology and other disciplines.In this paper, we report the synthesis, kinetics, and luminogenic behaviour of pyrene derivatives that have two distinct pathways for functionalisation: the carboxylic acid as well as the 1,2,4-triazine click group. Although a thorough evaluation of bioorthogonality is beyond the scope of the paper, we investigated in detail, both experimentally and computationally, the structure–reactivity relationship in IEDDA reactions with strained dienophiles TCO, s-TCO, and BCN providing important data for future adaptation of these molecules for bioorthogonal ligations.
Results and discussion
Synthesis
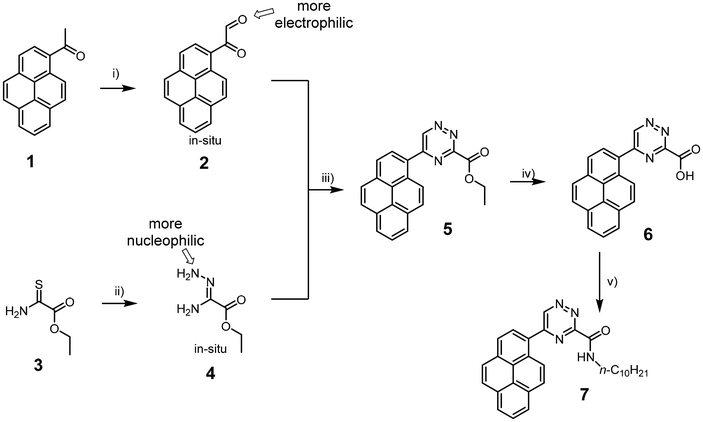
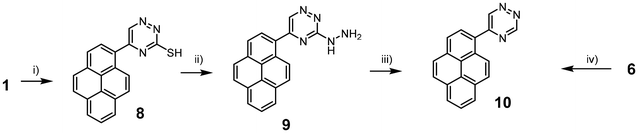
The synthesis of the acid derivative 3-carboxy-5-(pyren-1-yl)-1,2,4-triazine 6 is depicted in Scheme 1. The 1,2,4-triazine ring was constructed by the condensation of the pyrene glyoxal 2 and the amidrazone 4. Both building blocks were prepared in situ and were not isolated. The ester derivative 5 was isolated in 62% yield. The method provides an access to the key intermediate 5 on a 5 g scale over a period of just one day starting from inexpensive 1 and 3. Importantly, no other isomers form during the synthesis. This is the result of more electrophilic nature of the aldehyde than ketone groups in glyoxal 2 and more nucleophilic amine group next to the imine nitrogen in the intermediate 4. Hydrolysis of the ester 5 gave the target acid 6 in 69% yield. The carboxylic acid give opportunity for functionalisation. For example, the use of the acid 6 in an amidation reaction gives potential access to a great number of possible structures. We exemplified the methodology by the reaction with decylamine to obtain the lipophilic amide 7, but many more amines or other nucleophiles can potentially be used to tune the systems for a particular application.For the synthesis of the 5-(pyren-1-yl)-1,2,4-triazine 10 we initially used a known methodology based on the synthesis of mercapto-derivative 8, then hydrazino derivative 9 and finally dehydrazination in strongly basic conditions.23 Unfortunately, the last dehydrazination step proved to be low-yielding. We therefore explored an alternative route and found that heating the acid 6 to 200 °C is accompanied by decarboxylation and gives access to the desired triazine 10 in 56% yield (Scheme 2). Therefore, the decarboxylation is a convenient method for the synthesis of 1,2,4-triazines with unsubstituted position C3. The compound 10 is an important intermediate for the synthesis of functional metal complexes with a pyrene fragment.31 But maybe more importantly for this study, it is also a good model compound to investigate the influence of the substituents in the C3 position of the 1,2,4-triazine on the rate of the reaction, which is described in the next section.
High quality copies of NMR spectra for all compounds are available in the SI (Fig. S1–S26).
Kinetics
The use of 1,2,4-triazines as biorthogonal dienes in reactions with strained dienophiles was reviewed recently.32,33 The classical way to facilitate the IEDDA reaction is to introduce electron-withdrawing substituents into the 1,2,4-triazine.27,34 By comparing with unsubstituted triazine 10, the influence of the amide, ester and carboxy substituents on the rate of the reaction can be clearly identified.The structures of the dienophiles used in our studies are depicted in Fig. 1. The experiments were performed either in DMSO or DMSO-D6. Second order rate constants k2 (M−1 s−1) for the reaction between the triazine substrates (5, 6, 7 and 10) and the dienophiles were determined either spectrophotometrically under pseudo-first order conditions for TCO and s-TCO derivatives 11 and 12 or by NMR using second order kinetics for the BCN derivative 13 (Table 1).
 | ||
| Fig. 1 Structures of the dienophiles used in kinetics experiments as well as truncated structures used in computational analysis. | ||
| Second order rate constant k2/M−1 s−1 | |||
|---|---|---|---|
| 11 | 12 | 13 | |
| Kinetic plots are provided in the SI (Fig. S27–S37). | |||
| 5 (ester) | 0.133 ± 0.003 | 25.43 ± 0.29 | 0.143 ± 0.006 |
| 6 (acid) | 0.064 ± 0.002 | 18.99 ± 0.36 | 0.041 ± 0.001 |
| 7 (amide) | 0.051 ± 0.001 | 11.33 ± 0.04 | nd |
| 10 (parent) | 0.019 ± 0.001 | 2.28 ± 0.02 | 0.036 ± 0.000 |
The ester derivative 5 has the highest rate in the series, with the rates increasing approximately ten times, compared to 10, in reactions with all tested dienophiles. The acid 6 and the amide 7 gave a noticeable but less profound increase in the rate of the reaction than the ester derivative. It should be noted that the acid and the amide are somewhat more useful for biological applications as the acid provides better water solubility while the amide would be the bioconjugation product to biomolecules such as proteins. Although the overall increase in the reaction rate is in line with classical “tuning” of the reactivity of IEDDA reaction, computational analysis described below reveals a more intricate picture.
Computational analysis
In order to reduce computational cost, ethyl, decane and 4-nitrophenolate groups were replaced with methyl groups (Fig. 1). Conformer searches of all reactants were carried out using CREST 3.0 with an RMSD threshold of 1 Å and an energy window of 20 kJ mol−1.35 DFT optimisations and frequency calculations were performed for all identified conformers at the M06-2X-D3(0)/6-31G(d) level36 in the gas phase. This functional has been widely used in previous computational studies of Diels Alder reactions.37–39 All DFT calculations were carried out with Orca 6.0.40 The absence of negative frequencies was used to confirm the identification of minimum energy structures. Corrections to obtain Gibbs energies were determined at 298.15 K using the Quasi-RRHO approach for the entropy term. Subsequent single point energies were calculated using 6-311G(d,p) basis set as well as a DMSO solvent field described using the CPCM SMD solvation model,41 and the previously calculated Gibbs energy correction was added to the single point energy to obtain the final molecular energy.37Transition state conformer searches were performed in the same way as for the reactants, but with distances between reacting atoms constrained to 2.2 Å and with an RMSD threshold of 2 Å. DFT optimisations were carried out at the same level of theory as the reactants, with transition states identified by a single negative frequency corresponding to movement of the reacting atoms. When multiple transition states were identified from conformer searches, effective activation energies were determined.42
In the absence of experimental activation energies and pre-exponential factors and wanting to avoid normalising activation energies against a single arbitrarily chosen reaction, assessment of the quality of the calculated energies was performed by analysing all possible reaction pairs and comparing the difference in calculated energy difference with the equivalent experimental difference for each pair. As well as quantitatively predicting relative activation energies, it is also of significant value for a computational method to successfully predict trends between different reactions, primarily when changing one of the two reactants. To assess this, calculated trends in activation energies for all reaction pairs sharing a common diene or dienophile were compared with the experimental trends, giving a fraction of reactions for which the experimental trend (higher or lower) was successfully predicted.
Experimentally, these reactions are relatively straightforward, i.e. the more strained s-TCO reactions proceed the fastest, TCO and BCN exhibit relatively similar reactivities, and largely the reactivity of the dienes (specifically 5 > 6 > 7 > 10) is as may be expected from the electronic nature of the substituent groups. However, the relatively small differences in relative activation energies of these reactions make them an interesting test-case for calculations.
Initially, transition states were obtained by taking optimised geometries of the reactants, optimising transition state geometries for each of the four possible dienophile orientations, and using the calculated lowest energy transition state. This approach gave mean error, RMSE, and MUE values of 2.1 kJ mol−1, 7.2 kJ mol−1 and 5.9 kJ mol−1, respectively, vs. experimental relative activation energies. The calculations also successfully predicted trends in activation energies for 20 of 25 pairs of reactions sharing a common reactant (full details of these approaches in the methods).
Although the transition state structures described above were systematically obtained, the method does not consider alternative conformations such as those obtained via rotation about the bond between the triazine and pyrene moieties for which there are four possible energy minima per transition state. To gain an understanding of the importance or otherwise of such conformations, conformer searches were performed based on the initial four transition states for each reaction described above, the results of which were then optimised using the same approach. This methodology gave between 55 and 141 transition states per reaction, from which revised activation energies were determined, giving mean error, RMSE, and MUE values of 0.7 kJ mol−1, 7.9 kJ mol−1, and 6.2 kJ mol−1, respectively. In terms of predicting trends, 23 of the 25 pairs of reactions defined above were predicted correctly.
For these reactions there are evidently modest improvements to the mean error and trend predictions when using a thorough conformational search, but it is notable that even for these quite simple molecules the lowest energy transition states after the conformer searches were on average 6.0 kJ mol−1 lower in energy than the original transition states. Notably the largest differences were for reactions involving BCN, which reduced by between 9.9 kJ mol−1 and 12.1 kJ mol−1 after the conformer search, attributable to a favourable interaction of the BCN amide group with the pyrene ring, as shown in Fig. 2 for transition states of 10 with BCN derivative 13*. These differences are of a magnitude comparable to previously reported differences between calculated and experimental values, indicating the importance of considering possible reactant conformations in the calculation of transition state energies.38
A further advantage of considering possible conformations in a systematic and thorough manner is to minimise the impact of conformational variation on the results, and for this set of reactions, the lowest energy transition state conformations were found to be very consistent for each dienophile. All the calculated lowest-energy transition state geometries had the same orientation of the pyrene relative to the triazine ring, with the dienophile always approaching from the same face, as shown in Fig. 2.
Fig. 3 shows the calculated activation energies plotted against the experimentally derived energies, each relative to the average of the respective datasets, from which it is apparent that a significant source of error in this set of reactions appears to be an overestimation of the activation energies of reactions involving 10.
Photophysical properties
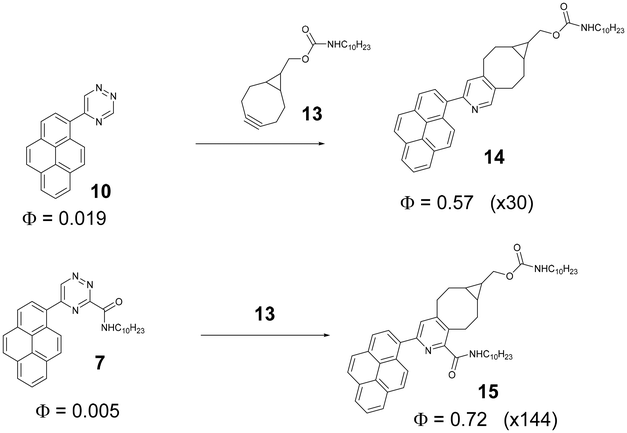
To demonstrate luminogenic behaviour of our pyrenotriazines we have chosen compounds 7 and 10 as dienes and BCN derivative 13 as a dienophile. We compared the luminescence of ethanolic solutions of 7 and 10 before and after the click reaction. Approximately 20 molar equivalents of BCN derivative 13 were used to drive the reaction to completion. During the IEDDA reaction the triazine is converted into aromatic pyridine (Scheme 3).The luminescence of 2-(2-pyridyl)pyrene is known to be sensitive to the environment with often monomer and the excimer emission simultaneously present in the sample.43Scheme 3 shows that before the click reaction, triazines 7 and 10 are very weekly emissive with quantum yields of 0.5% and 1.9% respectfully. After the click reaction with 13, however, there is a drastic increase in luminescence with quantum yields reaching values of 72% and 57% respectively. The emission spectra have a structured profile with emission λmax centred at around 390 nm and 405 nm attributed to monomeric emission of the pyrene unit. Interestingly, no excimer emission was registered for either compound after the click reaction. However, considering the hydrophobic nature of 14 and 15, different aggregation behaviour is expected in aqueous solutions. In case of future use of these systems in bioorthogonal applications, additional photophysical evaluation in aqueous solutions is therefore advisable.
Conclusion
In this paper we promoted the synthetic availability and flexibility of novel pyrene derivatives that contain a 1,2,4-triazine ring as a click group for IEDDA reaction. The presence of a carboxylic acid group is an important additional handle for further functionalisation as well as for increasing the reaction rates of IEDDA reactions with strained alkenes and alkynes. The luminogenic character of the click reaction benefits the use of these versatile pyrene reagents in biological applications and beyond.Experimental
3-Ethoxycarbonyl-5-(pyren-1-yl)-1,2,4-triazine 5
1-Acetylpyrene (5 g, 20.5 mmol) was dissolved in 1,4-dioxane (100 mL) at heating. To this solution, selenium dioxide (2.27 g, 20.5 mmol) was added. The mixture was heated under reflux for 14 hours and then filtered to remove Se. The filtrate was evaporated to dryness and dissolved in ethanol (40 mL) to give solution A. Separately, ethyl thiooxamate (2.73 g, 20.5 mmol) was dissolved in ethanol (30 mL). Hydrazine hydrate (1.03 g, 20.5 mmol) was dissolved in ethanol (10 mL) and added slowly to the ethyl thiooxamate solution. The mixture was left stirring at room temperature overnight (Caution evolution of toxic H2S, leave the flask open) and then diluted to a volume of 50 mL with ethanol to give solution B. Solution B was then added to the stirred solution A at room temperature. The mixture was heated under reflux for 15 minutes and then vacuum filtered while boiling hot. The filtrate was allowed to cool to room temperature. The precipitated solid was filtered, washed with ethanol to give the desired product. Yield 4.5 g. (62%). 1H NMR (400 MHz, CDCl3): δ 9.88 (s, 1H), 8.63 (d, J = 8.4 Hz, 1H), 8.38–8.25 (m, 5H), 8.14 (d, J = 8.4 Hz, 1H), 8.11 (t, J = 7.9 Hz, 1H), 4.68 (q, J = 7.4 Hz, 2H), 1.56 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, CDCl3): δ 162.8, 159.5, 156.9, 151.4, 133.95, 131.1, 130.6, 130.3, 130.0, 129.6, 128.3, 127.2, 127.1, 126.8, 126.7, 126.4, 125.2, 125.0, 124.3, 122.8, 63.4, 14.3.3-Carboxy-5-(pyren-1-yl)-1,2,4-triazine 6
To a boiling mixture of the ester 5 (2.24 g, 6.34 mmol) and ethanol (50 mL), a solution of LiOH (532 mg, 12.68 mmol, 2 eq.) in water (3 mL) was added. The mixture was heated under reflux for 60 minutes, then allowed to cool to room temperature and filtered. The solid on filter (Li salt) was washed with ethanol and transferred back to the reaction flask while still a little wet with ethanol. Acetic acid (20 mL) was added, and the mixture was briefly heated to reflux to dissolve the solid. The mixture was allowed to cool to room temperature. Water (20 mL) was added. Precipitated solid was filtered off by vacuum filtration and washed with water. The product was dried in oven (100 °C) for 15 minutes to give the acid 6 as a yellow solid. Yield 1.42 g (4.37 mmol, 69%). 1H NMR (400 MHz, CDCl3): δ 10.14 (s, 1H), 8.77 (d, J = 8.6 Hz, 1H), 8.56–8.29 (m, 7H), 8.19 (t, J = 7.8 Hz, 1H). 13C NMR (100 MHz, CDCl3): δ 164.7, 159.1, 157.3, 152.1, 133.6, 131.2, 130.7, 130.1, 130.0, 129.5, 129.2, 128.3, 127.8, 127.5, 127.1, 126.7, 125.7, 124.6, 124.4, 124.0. HRMS (ESI): m/z calcd for [M + H] 326.09252, found 326.09244.Amide derivative 7
A mixture of the acid 6 (325 mg, 1 mmol) and thionylchloride (6 mL) was heated under reflux until starting material dissolves (approximately 15 min). The mixture was then evaporated to dryness on rotary evaporator to give a dark glassy residue. The residue was dissolved in DCM (15 mL). To this mixture, a solution of the amine (157 mg, 1 mmol) and the Hunigs’ base (0.4 mL, 2.3 mmol, 2.3 eq.) in DCM (3 mL) was added in portions. After 30 min of stirring at room temperature, the mixture was evaporated to dryness. The product was purified by column chromatography using silica gel as stationary phase and a mixture of DCM/EA, 1/1 v/v as an eluent (Rf 0.7). The fractions containing product were combined and evaporated to a volume of approximately 5 mL. Methanol (15 mL) was added. The mixture was then evaporated to a volume of approximately 5 mL. First, oily precipitate formed, which then solidified after approximately 10 minutes at RT. The solid was filtered off, and washed with methanol to give 7 as a yellow solid. Yield 285 mg (61%) 1H NMR (400 MHz, CDCl3): δ 9.84 (s, 1H), 8.59 (d, J = 8.4 Hz, 1H), 8.38–8.10 (m, 8H), 3.64 (q, J = 7.8 Hz, 2H), 1.72 (quintet, J = 7.8 Hz, 2H), 1.5–1.2 (m, 12H), 0.87 (t, J = 7.8 Hz, 3H). 13C NMR (100 MHz, CDCl3): δ 161.0, 159.7, 156.9, 151.2, 133.9, 131.2, 130.6, 130.3, 130.0, 129.6, 128.3, 127.4, 127.2, 126.8, 126.7, 126.4, 125.2, 125.0, 124.4, 122.8, 40.1, 31.9, 29.6, 29.5, 29.3, 27.0, 22.7, 14.1. HRMS (ESI): m/z calcd for [M + H] 465.26543, found 465.26489.3-Mercapto-5-(pyren-1-yl)-1,2,4-triazine 8
1-Acetylpyrene 1 (2.51 g 10.3 mmol) was placed in a 100 mL round-bottom flask and dissolved in 1,4-dioxane (22 mL) at 50 °C. Selenium dioxide (1.14 g, 10.3 mmol) was added to the flask and the mixture was stirred under reflux for 14 hours. The mixture was filtered. The filtrate was evaporated to dryness to give crude glyoxal derivative. Thiosemicarbazide (0.91 g, 10 mmol) was added followed by 1 M aqueous potassium carbonate. The reaction mixture was heated with stirring at ∼50 °C for two hours. The mixture was filtered. To the filtrate, glacial acetic acid (2 mL) was added causing formation of bright red solid. The solid was filtered off via Büchner filtration and washed with water, to give after drying triazine 8. 2.62 g (84%). 1H NMR (400 Hz, DMSO-D6): 8.89 (s, 1H), 8.74 (d, J = 9.2 Hz, 1H), 8.45–8.39 (m, 4H), 8.33 (d, J = 9.6 Hz, 1H), 8.25 (d, J = 8.8 Hz, 1H), 8.13 (t, J = 7.6 Hz, 1H). 13C NMR (100 MHz, CDCl3): δ 181.3, 161.6, 139.6, 133.9, 131.2, 130.6, 130.4, 130.0, 129.6, 128.8, 128.1, 127.8, 127.5, 127.3, 126.9, 125.5, 124.6, 124.5, 123.9.3-Hydrazino-5-(pyren-1-yl)-1,2,4-triazine 9
In a 50 mL round-bottom flask the starting mercaptotriazine 8 (2.0 g, 6.39 mmol) was mixed with hydrazine monohydrate (4 mL, 63.9 mmol, 10 eq.). The flask was placed in an oil bath that had been preheated to 150 °C. The mixture was stirred for 15 minutes. Ethanol (25 mL) was carefully added through the top of the condenser. The mixture was heated under reflux for 25 minutes. The mixture was cooled to room temperature and filtered. The solid on filter was washed with ethanol to give 9. Yield 1.97 g (98%). 1H NMR (400 Hz, DMSO-D6): 9.11 (s, 1H), 8.93 (br.s, 1H), 8.67 (br.d, J = 9.2 Hz, 1H) 8.41–8.24 (m, 7H), 8.16 (t, J = 7.6 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ 158.5, 142.0 br.s, 132.8, 131.3, 130.7, 129.9, 129.5, 129.4, 129.1, 128.2, 127.8, 127.3, 126.7, 126.4, 125.5, 124.7, 124.6, 124.1.5-(Pyren-1-yl)-1,2,4-triazine 10
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ethyl Acetate (1
Ethyl Acetate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as an eluent. Yield 74 mg (13%).
1) as an eluent. Yield 74 mg (13%).
Author contributions
HA, JP, GA, VNK: synthesis; MED: kinetics experiments; CEN: photophysical characterisation; MS: computational approach development and implementation; MED, CEN, MS, VNK: prepared manuscript for publication; VNK: conceptualisation, supervision.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ob01512j.Acknowledgements
The authors thank King Abdulaziz University for the financial support during the scientific collaboration of Dr Hind Alshaikh with Northumbria University.References
- M. E. Østergaard and P. J. Hrdlicka, Chem. Soc. Rev., 2011, 40, 5771–5788 RSC.
- O. A. Krasheninina, D. S. Novopashina, E. K. Apartsin and A. G. Venyaminova, Molecules, 2017, 22, 2108 CrossRef PubMed.
- K. Murayama, A. Ikeda, F. Sato and H. Asanuma, Org. Biomol. Chem., 2025 10.1039/D5OB00924C.
- N. Nakashima, Y. Tomonari and H. Murakami, Chem. Lett., 2002, 31, 638–639 CrossRef.
- K. Maehashi, T. Katsura, K. Kerman, Y. Takamura, K. Matsumoto and E. Tamiya, Anal. Chem., 2007, 79, 782–787 CrossRef CAS PubMed.
- V. Georgakilas, J. N. Tiwari, K. C. Kemp, J. A. Perman, A. B. Bourlinos, K. S. Kim and R. Zboril, Chem. Rev., 2016, 116, 5464–5519 CrossRef CAS PubMed.
- D.-W. Lee, T. Kim and M. Lee, Chem. Commun., 2011, 47, 8259–8261 RSC.
- Y. Zhang, C. Liu, W. Shi, Z. Wang, L. Dai and X. Zhang, Langmuir, 2007, 23, 7911–7915 CrossRef CAS PubMed.
- X. V. Zhen, E. G. Swanson, J. T. Nelson, Y. Zhang, Q. Su, S. J. Koester and P. Bühlmann, ACS Appl. Nano Mater., 2018, 1, 2718–2726 CrossRef CAS.
- D. Kwong Hong Tsang, T. J. Lieberthal, C. Watts, I. E. Dunlop, S. Ramadan, A. E. del Rio Hernandez and N. Klein, Sci. Rep., 2019, 9, 13946 CrossRef PubMed.
- H.-C. Hung, C.-W. Cheng, I. T. Ho and W.-S. Chung, Tetrahedron Lett., 2009, 50, 302–305 CrossRef CAS.
- X. Liu, X. Yang, Y. Fu, C. Zhu and Y. Cheng, Tetrahedron, 2011, 67, 3181–3186 CrossRef CAS.
- J. Nakazawa and T. D. P. Stack, J. Am. Chem. Soc., 2008, 130, 14360–14361 CrossRef CAS PubMed.
- J. Nicks and J. A. Foster, Nanoscale, 2022, 14, 6220–6227 RSC.
- W. Dittmar, J. Sauer and A. Steigel, Tetrahedron Lett., 1969, 10, 5171–5174 CrossRef.
- D. N. Kamber, Y. Liang, R. J. Blizzard, F. Liu, R. A. Mehl, K. N. Houk and J. A. Prescher, J. Am. Chem. Soc., 2015, 137, 8388–8391 CrossRef CAS PubMed.
- K. A. Horner, N. M. Valette and M. E. Webb, Chem. – Eur. J., 2015, 21, 14376–14381 CrossRef CAS PubMed.
- K. Peewasan and H.-A. Wagenknecht, ChemBioChem, 2017, 18, 1473–1476 CrossRef CAS PubMed.
- S. J. Siegl, R. Dzijak, A. Vazquez, R. Pohl and M. Vrabel, Chem. Sci., 2017, 8, 3593–3598 RSC.
- M. Baalmann, M. J. Ziegler, P. Werther, J. Wilhelm and R. Wombacher, Bioconjugate Chem., 2019, 30, 1405–1414 CrossRef CAS PubMed.
- Z. Chen, N. Ren, X. Ma, J. Nie, F.-G. Zhang and J.-A. Ma, ACS Catal., 2019, 4600–4608, DOI:10.1021/acscatal.9b00846.
- D. N. Kamber, S. S. Nguyen, F. Liu, J. S. Briggs, H.-W. Shih, R. D. Row, Z. G. Long, K. N. Houk, Y. Liang and J. A. Prescher, Chem. Sci., 2019, 10, 9109–9114 RSC.
- V. N. Kozhevnikov, M. E. Deary, T. Mantso, M. I. Panayiotidis and M. T. Sims, Chem. Commun., 2019, 55, 14283–14286 RSC.
- U. Reisacher, B. Groitl, R. Strasser, G. B. Cserép, P. Kele and H.-A. Wagenknecht, Bioconjugate Chem., 2019, 30, 1773–1780 CrossRef CAS PubMed.
- U. Reisacher, D. Ploschik, F. Rönicke, G. B. Cserép, P. Kele and H.-A. Wagenknecht, Chem. Sci., 2019, 10, 4032–4037 RSC.
- J. Li, Q.-L. Hu, J.-S. Liu and X.-F. Xiong, Org. Lett., 2024, 26, 10928–10933 CrossRef CAS PubMed.
- V. Šlachtová, V. Motornov, P. Beier and M. Vrabel, Chem. – Eur. J., 2024, 30, e202400839 CrossRef PubMed.
- V. Šlachtová, S. Bellová and M. Vrabel, J. Org. Chem., 2024, 89, 14634–14640 CrossRef PubMed.
- V. Šlachtová, S. Bellová, A. La-Venia, J. Galeta, M. Dračínský, K. Chalupský, A. Dvořáková, H. Mertlíková-Kaiserová, P. Rukovanský, R. Dzijak and M. Vrabel, Angew. Chem., Int. Ed., 2023, 62, e202306828 CrossRef PubMed.
- L. Cooke, K. Gristwood, K. Adamson, M. T. Sims, M. E. Deary, D. Bruce, A. N. Antoniou, H. R. Walden, J. C. Knight, T. Antoine-Brunet, M. Joly, D. Goyard, P.-H. Lanoë, N. Berthet and V. N. Kozhevnikov, Dalton Trans., 2024, 53, 15501–15508 RSC.
- A. J. Howarth, M. B. Majewski and M. O. Wolf, Coord. Chem. Rev., 2015, 282–283, 139–149 CrossRef CAS.
- F.-G. Zhang, Z. Chen, X. Tang and J.-A. Ma, Chem. Rev., 2021, 121, 14555–14593 CrossRef CAS PubMed.
- Y. Fang, A. S. Hillman and J. M. Fox, Top. Curr. Chem., 2024, 382, 15 CrossRef CAS PubMed.
- Z. Chen, N. Ren, X. Ma, J. Nie, F.-G. Zhang and J.-A. Ma, ACS Catal., 2019, 9, 4600–4608 CrossRef CAS.
- P. Pracht, S. Grimme, C. Bannwarth, F. Bohle, S. Ehlert, G. Feldmann, J. Gorges, M. Müller, T. Neudecker, C. Plett, S. Spicher, P. Steinbach, P. A. Wesołowski and F. Zeller, J. Chem. Phys., 2024, 160, 114110 CrossRef CAS PubMed.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 Search PubMed.
- Y. Liang, J. L. Mackey, S. A. Lopez, F. Liu and K. N. Houk, J. Am. Chem. Soc., 2012, 134, 17904–17907 CrossRef CAS PubMed.
- F. Liu, Y. Liang and K. N. Houk, Acc. Chem. Res., 2017, 50, 2297–2308 CrossRef CAS PubMed.
- P. Ma, D. Svatunek, Z. Zhu, D. L. Boger, X.-H. Duan and K. N. Houk, J. Am. Chem. Soc., 2024, 146, 18706–18713 CrossRef CAS PubMed.
- F. Neese, Wiley Interdiscip. Rev.:Comput. Mol. Sci., 2012, 2, 73–78 CAS.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS PubMed.
- F. Giralt and R. W. Missen, Can. J. Chem. Eng., 1974, 52, 781–783 CrossRef.
- V. Kachwal, P. Alam, H. R. Yadav, S. S. Pasha, A. R. Choudhury and I. R. Laskar, New J. Chem., 2018, 42, 1133–1140 RSC.
| This journal is © The Royal Society of Chemistry 2025 |