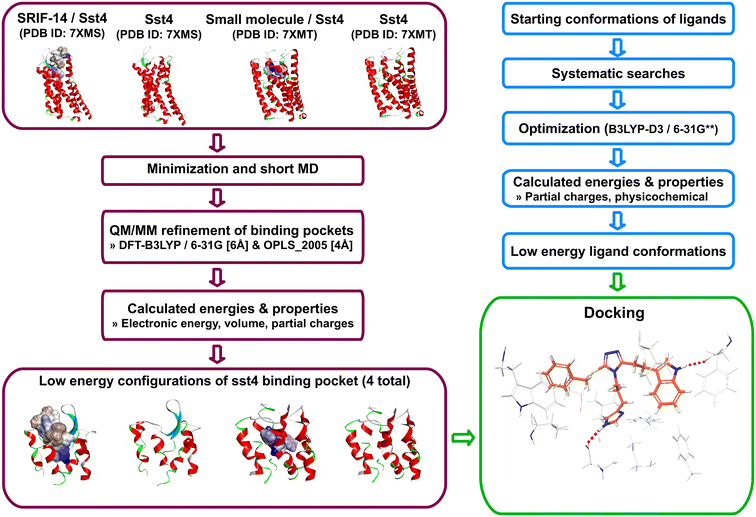
Open Access Article
Open Access ArticleElucidating structural and molecular requirements of somatostatin subtype-4 agonist bound complexes using quantum mechanics approaches
Olivia
Slater
and
Maria
Kontoyianni
*
Department of Pharmaceutical Sciences, School of Pharmacy, Southern Illinois University, Edwardsville, IL 62026, USA. E-mail: mkontoy@siue.edu
First published on 8th August 2025
Abstract
Somatotropin-release Inhibiting Factor (SRIF) binds somatostatin subtype-4 receptor (sst4) within the neocortex to increase amyloid beta catabolism, and possibly effects neuronal plaque formation and disease progression of Alzheimer's disease. Recently, sst4 in complex with SRIF or a small molecule agonist were resolved using cryo-EM, and mutagenesis identified amino acid residues that contributed to activity. In the present study, we used mixed Quantum Mechanics/Molecular Mechanics (QM/MM) to refine the experimental sst4 complexes and decipher agonist-sst4 interactions. SRIF rendered the complex more stable, while interactions with asparagines 199 and 293 seen in the experimental complexes were lost and replaced with Gln279. We also addressed long-standing questions related to sst4 agonist binding. Toward that end, we used quantum mechanics, molecular docking, and QM/MM refinement methods and employed pair-wise comparisons or matched molecular pairs to explore the effects of chemical substitutions on ligand properties and energetics. We identified amino acid interactions that were consistent among all refined complexes, and those that differentiated high affinity binders. Finally, we considered several parameters to discern which correlated best with affinity, and highlighted aspects often overlooked or yet to be explored in order to enhance lead optimization outcomes.
1. Introduction
Alzheimer's disease (AD) is the most common form of age-related dementia that affects over 55 million people globally. Disease management costs an estimated 1.3 trillion US dollars annually, according to the World Health Organization. Main neuropathological hallmarks of AD include loss of neurons and synapses, neurofibrillary tangles, and neuronal plaques deposited between neurons due to accumulated amyloid-beta (Aβ) peptide starting in the neocortex. Brain neurons express amyloid-precursor protein (APP) which is cleaved by β- and γ-secretases to release Aβ. Under normal physiological conditions, Aβ is rapidly catabolized by neprilysin and cleared from the brain. Neprilysin levels decrease with age to promote Aβ dyshomeostasis and plaque formation.1–3Somatostatin, or Somatotropin-Release Inhibiting Factor (SRIF), is a cyclic peptide of either 14 or 28 amino acid residues that affects the Aβ degradation pathway.4 SRIF-14 is the main isoform expressed in the central nervous system (CNS) where it modulates neuroplasticity and neurotransmission by interacting with inhibitory G-protein coupled receptors of the somatostatin receptor (sst) family. Within the neocortex, SRIF binds sst4 to stimulate neprilysin and Aβ catabolism.5–8
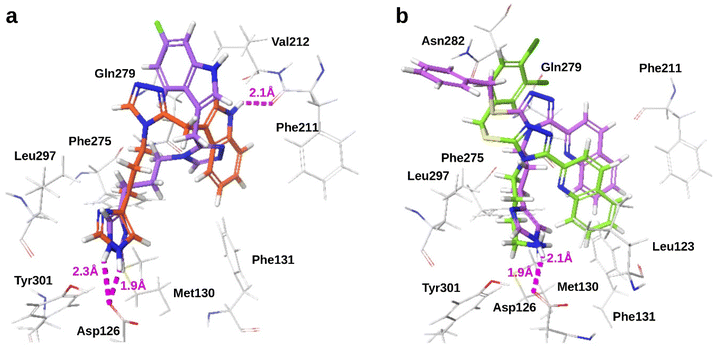
Poor bioavailability due to high molecular weight and short plasma half-life limit the potential of SRIF-14 as a CNS drug. Researchers have developed peptidomimetics and non-peptide agonists with chemical features mimicking SRIF residues important for potency (Phe7), and target binding (Trp8 and Lys9).9,10 Recently, sst4 complexes bound by SRIF-14 and a small molecule agonist were resolved using cryo-Electron Microscopy (cryo-EM).11 Both ligands formed electrostatic interactions with Asp126 through either the Lys9 of SRIF-14 or the moiety mimicking Lys9 of the small molecule, and Asn 199 with the main chain atoms of SRIF or the terminal amide of the small molecule (Fig. 1).
 | ||
| Fig. 1 Dipole interactions observed in experimental complexes of sst4 bound by (a) SRIF-14 and (b) small molecule agonists (PDB IDs: 7XMS and 7XMT). | ||
Internal ligands were developed through a structure-based design strategy. The earlier non-peptide agonists incorporated aromatic, heteroaromatic, and imidazole to mimic Phe7, Trp8, and Lys9 attached to the 1,2,4-triazole scaffold, whereas our most recent work incorporated 3-thio-1,2,4-triazole.12–14 Initial efforts modelled agonist binding to sst4 using model-built structures with the β2-adrenergic and μ-opioid X-ray structures as templates.15–19 Most recently we employed the cryo-EM sst4 structures to dock the thio-triazole series.20 High and moderate affinity ligands formed bidentate interactions between Asp126 and the imidazolyl and indolyl hydrogens. Alternatively, the Lys9 mimetic imidazole hydrogen-bonded with Asp126, whereas the indole or sulfur interacted with Gln279. Hydrophobic and van der Waals (vdW) interactions involved Leu123, Leu200, Trp202, Phe211, Phe275, and Leu297. In certain cases, the 3-thio-1,2,4-triazole was positioned near Asn199. For the most part, low affinity binders formed fewer interactions and adopted more strained conformations.20
In this work, we capitalized on experimental information and computational resources to address long-standing questions about the chemical features of agonists that render high affinity for sst4. We employed quantum mechanics (QM) and Quantum Mechanics/Molecular Mechanics (QM/MM) calculations to investigate quantum effects on agonist and receptor static structures. The questions we aimed to address were: (1) Can we decipher which amino acids are essential for binding with sst4? (2) What is the significance of the sulfur atom? (3) Does affinity correlate with certain parameters? (4) What are the best practices moving forward? Toward 1, we used experimental bound complexes (PDB IDs: 7XMS and 7XMT),11 or with the ligands removed. We performed QM/MM calculations of sst4's binding pocket in the bound and unbound configurations to observe changes energetically, and to identify which interactions were maintained, lost, or gained due to refinement. Toward 2, we compared QM-optimized ligand geometries of matched molecular pairs. Toward 3, optimized ligand conformations were docked into the refined sst4 binding pockets and docked solutions were optimized with QM/MM. Refined complexes were compared in a pair-wise manner to identify consistent binding patterns of high affinity ligands. We looked at energies of refined complexes and properties of optimized ligands to explore correlations with affinity. Collectively, these results gave insights toward 4 and how to best approach future lead optimization endeavors for this particular system.
2. Methods
2.1. Structure preparation
Sst4 in complex with SRIF or small molecule were extracted from the Protein Data Bank (PDB IDs: 7XMS and 7XMT). Hydrogens were added with ProPka21 and bond orders were reassigned using the Cambridge Crystallographic Database.22 Missing side-chains were added using Prime and tautomeric states were assigned with Epik at pH 7.4 ± 2.0. Ligands were either maintained to represent bound configurations or extracted to obtain unbound configurations of sst4. To relax the structures and to alleviate crystal packing, while maintaining observed conformations, agonist-bound and unbound structures were minimized with 1000 steps of steepest descent, followed by 2000 conjugate gradient with rms of 0.1 to resolve poor contacts. Subsequently, short molecular dynamics (MD) simulations were carried out using the CHARMM force field within Discovery Studio (BIOVIA, Dassault Systèmes, Discovery Studio Modeling Environment 2023, San Diego). The systems were heated to target temperature 300 K for 10 ps, followed by 100 ps equilibration. The final production stage was 1 ns and used the constant-volume thermodynamic ensemble with Leapfrog Verlet integration and SHAKE23 constraints applied.2.2. Optimizing conformations of sst4 binding pockets
Schrodinger's Qsite was used to perform QM/MM calculations (QSite, Schrödinger, LLC, New York, NY, 2024).24,25 We defined the binding pockets as amino acids within 10 Å of SRIF or the small molecule agonist which totaled 107 or 129 residues, respectively. We considered three definitions of the QM region that included (1) ligand atoms only, (2) ligand atoms plus side-chain atoms within 2 Å of the ligand, and (3) ligand atoms plus side-chain atoms within 6 Å of the ligand. We have reported values for the largest definition (ligand plus 6 Å). The DFT-B3LYP method with 6-31G basis set was used to treat the QM region, which employed the minimization method with a maximum of 100 iterations. The net charge of the QM region was equal to zero with a spin multiplicity of one. The OPLS_2005 force field was used to define the potential energy function for the MM region, with dielectric constant set to 1.0. We employed a maximum of 1000 minimization cycles using conjugate gradient algorithm with initial step size of 0.05 or 0.10 and a maximum step size of 1.0. An energy change criterion of 0.1 and a gradient criterion of 0.01 defined convergence for the MM region.2.3. Calculating properties of sst4 binding pockets
MM, electronic, and the total QM/MM energies of the binding pockets were taken directly from QM/MM calculations. We performed single point energy calculations of the optimized conformations using MOPAC (Molecular Orbital PACkage).26,27 Briefly, MOPAC performs semiempirical calculations using NDDO (neglect of diatomic differential overlap) theory. We employed the PM7 Hamiltonian and RHF wavefunction to calculate atomic electrostatic potential (ESP) charges fit to atom centers and total self-consistent field (SCF) energies. Calculations included all amino acid residues that defined the binding cavities (107 for SRIF-sst4 and 129 for small molecule-sst4 complexes). We compared the bound and unbound conformations, each before and after optimization, to visually inspect changes in conformation, and calculated RMSD's of side-chain heavy atoms using per-residue alignments of side-chain heavy atoms.2.4. QM optimization of small molecule conformations
Ligand conformations were optimized within Biovia's Discovery Studio 2023. Atom types were assigned using MMFF, and the CHARMM forcefield was applied. Initial structures were energy minimized using steepest descent followed by conjugate gradient, with a maximum of 200 steps each and rms gradients set to 0.1 and 0.01, respectively. Minimized conformations were used as initial structures for systematic searches to obtain low energy conformations. A maximum of 1000 conformations per ligand were generated with an energy threshold of 20 kcal mol−1, RMSD cutoff of zero and torsion increments of 15 degrees. Each conformation was energy minimized using conjugate gradient with rms 0.1. Conformations with the highest solvent accessible surface area were analyzed. Selected conformations underwent geometry optimization using Schrodinger's Jaguar.28 We employed density functional theory (DFT) calculations using B3LYP-D3/6-31G** basis set. The usual nonrelativistic Hamiltonian was used and the SCF spin was treated automatically. Accurate SCF level was used and a maximum 48 iterations and energy change of 5 × 10−5 Hartrees defined convergence. The QM optimized ligands were subsequently docked into the four configurations of sst4's binding pockets obtained as described above. We allowed a maximum of 30 poses per ligand. In addition, we extracted SRIF and the small molecule ligands from refined complexes and performed single point energy (SPE) calculations in vacuum to study electrostatic properties of bound conformations. We calculated ESP partial charges, net molecular charges, and mapped ESPs onto the isodensity surfaces. We included SRIF-14 resolved from solution NMR in these experiments because Thr10 was not resolved in the cryo-EM complex (PDB ID: 2MI1).29 Both methods give similar conformations of the peptide, and we found properties of either structure led to the same conclusions.2.5. Calculating ligand properties
We tested other methods and basis sets to compare energies with results of B3LYP-D3/6-31G**, and the same conclusions were drawn. Therefore, conformational energies were taken from the final geometry optimization step. Gibbs free energy was calculated using the equation ΔG = RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kd, where Kd = 1/10−pKa. Temperature was set to 310 Kelvin, and pKa's were calculated using quantum chemical approaches within Jaguar, as described above, with the acidity constant corresponding to the most ionizable group in each molecule. Differences in binding free energy (ΔΔG) between ligands within a pair (L1 and L2) were calculated as ΔΔG = ΔGL2 − ΔGL1, where ΔΔG > 0 kcal mol−1 indicated L1 had higher affinity. Atomic ESPs fit to atom centers were calculated using a spherical grid, and ESP surface maps were generated with grid density set to 20 pts per Å. ESP-properties that employed statistical measures of variance, maximum, minimum, and mean values were calculated and included local polarity (Πpol) and charge balance (υbal) parameters. To calculate solvation energies, optimized ligands were submitted for single-point energy (SPE) calculations that employed the Poisson Boltzmann Finite element method solvent model.30 Acidity constants (pKa's) were calculated using Jaguar's Micro-pKa method31 within Schrodinger's pKa prediction module. Briefly, Micro-pKa calculations include solvation free energies of protonated and deprotonated species, fitting charged species to experimental data with surface tension corrections trained on common functional groups applied for neutral species. Gas-phase deprotonation energy represents the free energy difference (ΔGg) between the ligand's protonated and deprotonated states. The summation of ΔGg and solvation free energies of neutral and ionic species equals the free energy of deprotonation (ΔGa) which is used to calculate the molecule's pKa as pKa = ΔGa/2.303RT. An empirical correction is applied after ab initio calculations of pKa's to better agree with experimental. We employed the automatic search to consider all pKa atoms, and compared accurate and quick SCF levels which gave approximately equivalent pKa's for optimized ligand geometries.32 A schematic representation of the workflow is shown in Fig. 2.
Kd, where Kd = 1/10−pKa. Temperature was set to 310 Kelvin, and pKa's were calculated using quantum chemical approaches within Jaguar, as described above, with the acidity constant corresponding to the most ionizable group in each molecule. Differences in binding free energy (ΔΔG) between ligands within a pair (L1 and L2) were calculated as ΔΔG = ΔGL2 − ΔGL1, where ΔΔG > 0 kcal mol−1 indicated L1 had higher affinity. Atomic ESPs fit to atom centers were calculated using a spherical grid, and ESP surface maps were generated with grid density set to 20 pts per Å. ESP-properties that employed statistical measures of variance, maximum, minimum, and mean values were calculated and included local polarity (Πpol) and charge balance (υbal) parameters. To calculate solvation energies, optimized ligands were submitted for single-point energy (SPE) calculations that employed the Poisson Boltzmann Finite element method solvent model.30 Acidity constants (pKa's) were calculated using Jaguar's Micro-pKa method31 within Schrodinger's pKa prediction module. Briefly, Micro-pKa calculations include solvation free energies of protonated and deprotonated species, fitting charged species to experimental data with surface tension corrections trained on common functional groups applied for neutral species. Gas-phase deprotonation energy represents the free energy difference (ΔGg) between the ligand's protonated and deprotonated states. The summation of ΔGg and solvation free energies of neutral and ionic species equals the free energy of deprotonation (ΔGa) which is used to calculate the molecule's pKa as pKa = ΔGa/2.303RT. An empirical correction is applied after ab initio calculations of pKa's to better agree with experimental. We employed the automatic search to consider all pKa atoms, and compared accurate and quick SCF levels which gave approximately equivalent pKa's for optimized ligand geometries.32 A schematic representation of the workflow is shown in Fig. 2.
3. Results and discussion
3.1. Changes in experimental structures after refinement
| Small molecule/sst4 (PDB ID: 7XMT) | SRIF-14/sst4 (PDB ID: 7XMS) | |||
|---|---|---|---|---|
| Bound | Unbound | Bound | Unbound | |
| a MM energy included QM and MM regions. b Electronic energy included atoms within the QM region only. c Semiempirical SCF total energy included all atoms within the binding pocket. | ||||
| MM energya (kcal mol−1) | −2.6 × 103 | −2.6 × 103 | −3.7 × 103 | −2.4 × 103 |
| Electronic energyb (kcal mol−1) | −4.6 × 107 | −3.2 × 107 | −7.2 × 107 | −2.9 × 107 |
| SCF total energyc (kcal mol−1) | −5.7 × 106 | −4.6 × 106 | −7.9 × 106 | −4.5 × 106 |
| Cavity volume (Å3) | 1.8 × 104 | 1.9 × 104 | 1.5 × 104 | 1.8 × 104 |
To determine residues most essential for agonist binding, we assessed which interactions were maintained or gained upon refinement and identified the amino acids that interacted with SRIF and the small molecule. Interactions with Asp126 were maintained, interactions with Gln279 were gained, and both agonists formed hydrophobic interactions with Leu123 and Leu297 in both refined complexes. SRIF's Lys9 maintained the interaction with Asp126 and the bond distance decreased from 2.7 Å to 1.8 Å. Leu297 formed a hydrophobic interaction with the hydrocarbon side-chain of Lys9. Tryptophan 8 hydrogen-bonded with the backbone carbonyl of Leu123. There was also a polar-π interaction formed between Gln279 and Trp8 (Fig. 3a). The small molecule maintained the salt-bridge with Asp126 and the bond distance decreased from 2.5 Å to 1.8 Å. The sulfonamide and Gln279 formed a polar vdW side-chain interaction also. Naphthyl and Phe211's side-chain became oriented edge-to-face (Fig. 3b). Backbone atoms of residues within ECL2 underwent small translations, and asparagine residues had different rotamers in refined complexes, hence interactions with Asn199 were not maintained. The side-chain RMSD's for the SRIF- and small molecule-bound complexes before and after refinement are in Table S1. Amino acid positioning changes are larger in the SRIF-complex when compared to the small molecule, suggesting tighter binding.
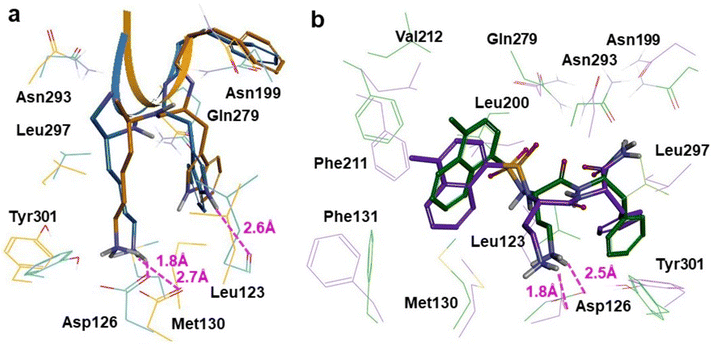 | ||
| Fig. 3 Superimpositions of (a) experimental (orange) and QM/MM treated (blue) complexes of sst4 bound by SRIF-14 (PDB ID: 7XMS) and (b) experimental (green) and QM/MM treated (purple) complexes of sst4 bound by a small molecule agonist (PDB ID: 7XMT). | ||
We extracted SRIF and the small molecule ligand from refined complexes and performed single point energy calculations in vacuum to study electrostatic properties of bound conformations. We calculated ESP partial charges, net molecular charges, and mapped ESPs onto the isodensity surfaces. SRIF had net molecular charge of +3 whereas the small molecule had +1. The Trp8 groups carried similar positive partial charges equal to 0.152 (indole) and 0.188 (naphthyl) in SRIF and small molecule, respectively. The distribution of charge was affected by the indole-NH which had a more localized positive charge and the rest of the atoms were neutral. In contrast, the naphthyl had slight positive charge across both rings because of the high electron withdrawing effect of the sulfonamide. The basic functionalities (NH3+) had equal positive partial charges of 0.560 (SRIF) and 0.581 (small molecule). Similar observations were made for the bound SRIF and small molecule. Specifically, charges for the Trp8 and Lys9 groups in SRIF were 0.026 and 0.967, respectively, whereas in the small molecule charges were 0.031 and 0.851 for the Trp8 and Lys9 mimetic groups. These results show net atomic charges were similar for functional groups that interacted with sst4.
In summary, SRIF-sst4 was the more stable complex. QM/MM refinement alleviated packing at the bottom of the cavity, and improved lengths and angles of non-bonded interactions between ligands and receptor residues. The binding cavities were highly similar except for ECL2. Residues Leu123, Asp126, Gln279, and Leu297 formed interactions with Trp8 and Lys9 moieties of bound agonists. Interactions with Asn199 and Asn293, seen in the experimental structures, were not observed in the refined complexes. Localized positive charge density about Lys9 was deemed essential, and positive partial charges of Trp8 contributed to affinity.
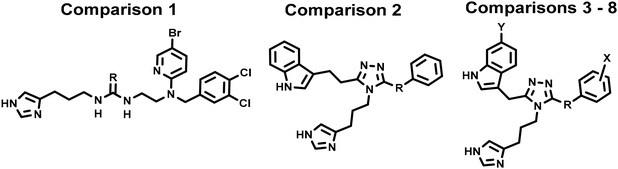
3.2. Sulfur atom effects on physicochemical properties
We used matched molecular pairs to explore the effects of sulfur. Chemical structures and properties are given in comparisons 1–8 of Table 2. We compared low energy ligand conformations using ESP partial charges, solvation free energy (ΔGsolv), acidity constant (pKa), and A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P. Affinity is affected by solvation/desolvation and we addressed it using ΔGsolv, which measured the free energy difference between the gas and solution phases, where more negative ΔGsolv indicated higher solubility. Given that ionization affects ligand partitioning and permeability, we considered pKa as well. pKa Represented the propensity of Lys9 mimetic (imidazole) to become deprotonated in water and was calculated by subtracting the gas-phase deprotonation energy from the summed free energy changes of solvation of the neutral and charged species
P. Affinity is affected by solvation/desolvation and we addressed it using ΔGsolv, which measured the free energy difference between the gas and solution phases, where more negative ΔGsolv indicated higher solubility. Given that ionization affects ligand partitioning and permeability, we considered pKa as well. pKa Represented the propensity of Lys9 mimetic (imidazole) to become deprotonated in water and was calculated by subtracting the gas-phase deprotonation energy from the summed free energy changes of solvation of the neutral and charged species 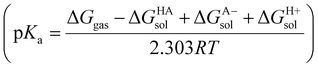 . We anticipated sulfur would affect electronic character because of the difference in electronegativity versus carbon or oxygen, reflected by differences in ESPs, ΔGsolv, or pKa. We also anticipated conformational effects due to increased size of the sulfur atom. Optimized ligands were docked into the refined cryo-EM structures of sst4 to observe whether sulfur affected how ligands interacted with the macromolecule. Sulfur increases solubility in thiourea 7versus urea 2 and results in a greater tendency of imidazole to become deprotonated when compared to urea (comparison 1). 3-Oxy-triazole 230 displays same solubility as 3-thio-triazole 42; however, oxygen's electronegativity increases the propensity of imidazole to deprotonate, contrary to lower electron-withdrawing sulfur (comparison 2). Notably, sulfur-containing ligands in Table 2 display higher solubility unless the benzene carries strong electron-withdrawing substituents such as trifluoromethyl or fluorine (comparisons 6 and 8). On the other hand, sulfur being more electronegative than carbon lowers the pKa, with the exception of highly electronegative substituents on the benzene such as mesylate and trifluoromethyl. The average summed partial charges of functional groups presumed to interact with Asp126 were affected by the sulfur, but the atoms acting as hydrogen bond donors (Nε2 hydrogen or urea hydrogens) maintained similar positive partial charges between 0.310 and 0.401. Sulfur compounds had uncharged imidazoles at brain pH (pKa < 7.0), versus urea 2 of comparison 1 with pKa equal to 7.9, and triazole 207 of comparison 4 with pKa of 7.5.
. We anticipated sulfur would affect electronic character because of the difference in electronegativity versus carbon or oxygen, reflected by differences in ESPs, ΔGsolv, or pKa. We also anticipated conformational effects due to increased size of the sulfur atom. Optimized ligands were docked into the refined cryo-EM structures of sst4 to observe whether sulfur affected how ligands interacted with the macromolecule. Sulfur increases solubility in thiourea 7versus urea 2 and results in a greater tendency of imidazole to become deprotonated when compared to urea (comparison 1). 3-Oxy-triazole 230 displays same solubility as 3-thio-triazole 42; however, oxygen's electronegativity increases the propensity of imidazole to deprotonate, contrary to lower electron-withdrawing sulfur (comparison 2). Notably, sulfur-containing ligands in Table 2 display higher solubility unless the benzene carries strong electron-withdrawing substituents such as trifluoromethyl or fluorine (comparisons 6 and 8). On the other hand, sulfur being more electronegative than carbon lowers the pKa, with the exception of highly electronegative substituents on the benzene such as mesylate and trifluoromethyl. The average summed partial charges of functional groups presumed to interact with Asp126 were affected by the sulfur, but the atoms acting as hydrogen bond donors (Nε2 hydrogen or urea hydrogens) maintained similar positive partial charges between 0.310 and 0.401. Sulfur compounds had uncharged imidazoles at brain pH (pKa < 7.0), versus urea 2 of comparison 1 with pKa equal to 7.9, and triazole 207 of comparison 4 with pKa of 7.5.
| Comparison | Compound | R | X | Y | Sst4 Ki (nM) | ΔGsolv (kcal mol−1) | Imidazole, pKa |
A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
|---|---|---|---|---|---|---|---|---|
| Comparison 1 | 2 | O | — | — | 14 | −21 | 7.9 | 4.5 |
| 7 | S | — | — | 70 | −26 | 5.9 | 4.8 | |
| Comparison 2 | 230 | OCH2 | — | — | 387 | −24 | 5.0 | 3.5 |
| 42 | SCH2 | — | — | 31 | −24 | 6.2 | 4.0 | |
| Comparison 3 | 180 | C | H | H | 1.7 | −21 | 5.8 | 3.9 |
| 93 | S | H | H | 0.9 | −23 | 5.7 | 4.6 | |
| Comparison 4 | 207 | C | 3-SO2CH3 | H | 1.0 | −21 | 7.5 | 3.4 |
| 95 | S | 3-SO2CH3 | H | 0.7 | −27 | 6.5 | 4.1 | |
| Comparison 5 | 208 | C | 3-SO2CH3 | 6-F | 0.7 | −27 | 5.8 | 3.6 |
| 96 | S | 3-SO2CH3 | 6-F | 1.0 | −30 | 6.7 | 4.3 | |
| Comparison 6 | 196 | C | 3-CF3 | 6-F | 0.7 | −25 | 4.1 | 5.0 |
| 98 | S | 3-CF3 | 6-F | 1.3 | −21 | 6.1 | 5.7 | |
| Comparison 7 | 201 | C | 3-OCH3 | 6-F | 0.6 | −26 | 5.4 | 5.1 |
| 97 | S | 3-OCH3 | 6-F | 0.6 | −27 | 4.7 | 4.6 | |
| Comparison 8 | 184 | C | 4-F | 6-F | 0.9 | −27 | 6.5 | 4.3 |
| 99 | S | 4-F | 6-F | 0.6 | −20 | 5.0 | 5.0 | |
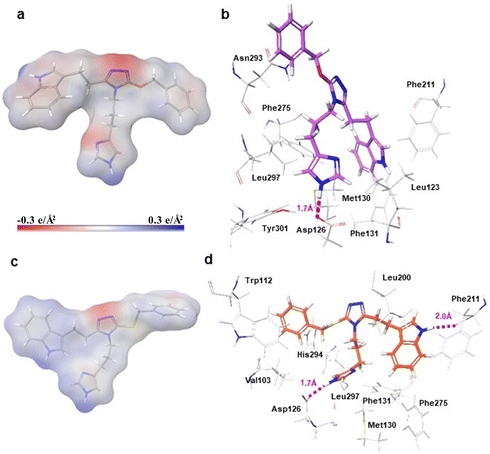
Docking results showed ligand pairs had similar conformations and orientations within the binding pocket, with the exception of comparison 2 ligands (Fig. 4). In vacuum, oxy-triazole 230's conformation was extended (Fig. 4a). Within the docked complex, the imidazole hydrogen-bonded with Asp126 with bond distance equal to 1.7 Å and the indole was positioned near Leu123. The benzyl side-chain was positioned higher and did not form vdW interactions. It should be noted that the binding mode of 230 (Fig. 4b) was similar to refined SRIF-sst4 with Trp8 moiety hydrogen bonding to Leu123 backbone carbonyl and Lys9 moiety donating a hydrogen to the carboxylate of Asp126. The conformations of thio-triazole 42 were similar in vacuum and within the receptor cavity, but imidazole side-chain was rotated 120° (Fig. 4c and d). Compound 42's imidazole hydrogen-bonded with the side-chain of Asp126 with bond distance equal to 1.7 Å, the indole NH hydrogen-bonded with the backbone carbonyl of Phe211 at approximately 2.0 Å distance. The benzyl group was positioned lower in the pocket and was involved in vdW interactions with Trp112 and His294.
In summary, sulfur compounds maintained similar net atomic charges to SRIF and small molecule agonists, with positive partial charges of functional groups meant to interact with Asp126. The sulfur atom affected solubility for the most part, acid–base character of the Lys9 mimetic, and binding modes of oxy- versus thio-triazole ligands. In contrast, sulfur did not affect how ligands bound sst4 when comparing sulfur versus carbon.
3.3. Determinants of affinity for sst4 agonists
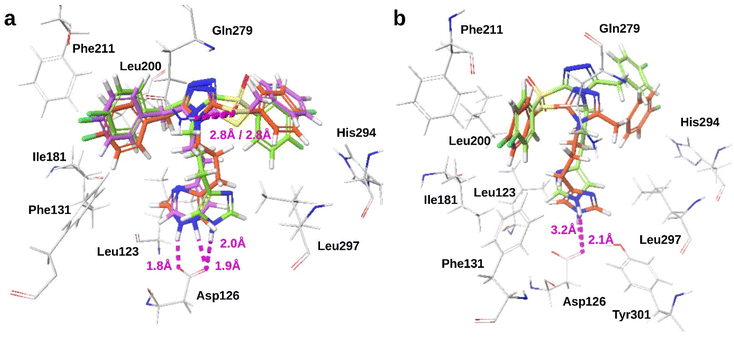
It is possible for ligands to have more than one binding mode, which we observed for ligands with phenyl or benzyl at positions 3 and 5 of the thiotriazole. In the first binding pattern, compounds 90 (Ki = 2.6 nM) and 233 (Ki = 6 nM) had the thiol or carbonyl oxygen of the sulfone forming a hydrogen bond with Gln279 with bond distances of approximately 2.8 Å, and the Lys9 mimetic hydrogen-bonded with Asp126 with bond distances approximately 1.8 Å and 1.9 Å, respectively. The 4-fluoro-benzyl groups formed edge-to-face π−π interactions with Phe211, and additional vdW and hydrophobic interactions involved Leu123, Phe131, Ile181, Leu200, and Leu297. The sulfur atom of compound 122 (Ki = 243 nM) was positioned slightly lower and pointed down, thus it did not interact with Gln279 but other interactions were maintained (Fig. 5a). In the second binding pattern, the positions of the benzyl or phenyl groups were flipped. The phenylthiol of compound 90 and the phenylsulfonyl of 233 oriented away from Gln279 and formed hydrophobic and vdW interactions with Ile181, Leu200, and Phe211, and the imidazoles maintained hydrogen bonds with Asp126 (Fig. 5b). The second binding pattern was not observed for compound 122 which had moderate affinity.
We were able to explain observed affinities for ligand pairs that we could not explain previously using the identified amino acids. Disubstituted triazoles 213 (Ki = 99 nM) and 214 (Ki = 717 nM) differed by a 6-fluoro substitution to the indole which lowered affinity sevenfold. Ligand conformations were similar and both ligands had imidazole forming a hydrogen bond with Asp126, but triazole and indole groups were positioned differently within the binding pocket (Fig. 6a). Unsubstituted indole hydrogen-bonded with Phe211 backbone carbonyl with bond distance of 2.1 Å, and formed vdW interactions with Phe131 and Phe211 side-chains. The triazole of 214 was positioned lower in the pocket, and 6F-indole was extended up toward the pocket entrance and thus it did not hydrogen bond with amino acid residues. Thiotriazoles 105 (Ki = 945 nM) and 107 (Ki = 959 nM) had quinoline at position 5 of the triazole as the Trp8 mimetic, and either an amine or imidazole as the basic functional group. Within the binding pocket, ligand conformations were similar, but 105 was positioned slightly lower compared with 107 and the benzyl-thiol moieties were positioned perpendicular to one another (Fig. 6b). These differences did not affect amino acid interactions. Both ligands had the basic functionality hydrogen bonding with Asp126, and forming hydrophobic or vdW interactions with Phe131, Phe275, and Leu297. Quinoline and triazole aromatic rings were within the same plane (flat) and parallel, and the quinoline nitrogen pointed down and away from Gln279 or Phe211.
| K i < 2 nM (n = 12) | 2 nM ≤ Ki < 20 nM (n = 12) | 20 nM ≤ Ki < 100 nM (n = 8) | ||
|---|---|---|---|---|
| Charge separation (Πpol) | Range | 15–20 | 15–21 | 15–22 |
| Average | 18 | 17 | 16 | |
| Charge balance (νbal) | Range | 0.21–0.25 | 0.20–0.25 | 0.21–0.25 |
| Average | 0.23 | 0.24 | 0.24 | |
| pKa (Lys9 mimetic) | Range | 4.1–7.5 | 4.0–8.4 | 4.2–9.1 |
| Average | 6.1 | 7.5 | 6.3 | |
In summary, we identified the same binding modes reported in our previous studies,14,20 but additional refinement of docked complexes with QM/MM helped clarify which interactions were most important or showed an additional interaction that helped differentiate high from medium affinity binders. The bidentate interaction we had reported previously was slightly altered by QM/MM refinement, which revealed that the indole interaction with Asp126 was replaced by a hydrogen bond with the backbone carbonyl of Leu123, similar to refined SRIF-sst4 complex. Amino acids Gln279 or Phe211, Leu123, and Leu297 differentiated high affinity agonists, whereas electrostatics with Asp126 and hydrophobic interactions with Leu200 were consistent across affinity ranges. The ligand property Πpol correlated with affinity, while calculated energies did not.
3.4. Best practices moving forward
When we initiated this project we expected: (1) more energetically favorable configurations for the SRIF- and small molecule-bound complexes when compared to unbound ones, and decreased cavity volumes for the bound complexes; (2) optimized bonding interactions with the main electrostatics residues, namely Asp126 and Asn199, reported in the refined experimental complexes; (3) sulfur would impact ligand conformations and to an extent binding modes, lipophilicity, and possibly electronic character; (4) higher affinity triazole and thiotriazole derivatives should form tighter complexes versus lower affinity ones. Even though we indeed saw favorable energetics for the refined experimental complexes, Asn199 did not participate in binding interactions after refinement. Sulfur and carbon triazoles displayed similar binding modes, which differed from oxy-triazoles. Sulfur increased solubility and lowered the pKa, not consistently but contingent upon substitutions on the nearby benzene. Finally, the highest affinity ligands showed additional electrostatics and vdW interactions when compared to those with lower affinity.Delving deeper beyond our expectations, and contrary to popular belief that post-docking refinement is necessary to achieve accurate docked solutions, our results indicate that extensive conformational sampling does not enhance accuracy. What matters instead are physically feasible starting conformations. Understanding molecular recognition is complex and multi-dimensional. Experimental structures are accurate to a degree, but not absolute representations of reality. Besides packing and unresolved parts of the structure, density mapping is approximated at times. On the other hand, high level theory such as the calculations presented herein promise accuracy for the particular area of the receptor under consideration, however they are not reflective of the actual binding events necessarily. Thus, factoring in the stage of the project, i.e. hit versus lead versus late candidate, against the computational cost of high level methods would be advisable. Besides, a global minimum is not representative of the bioactive ligand conformation and attaining the lowest energy complexes with QM/MM may give the investigator confidence in their approach, but changes occurring beyond the MM region are not considered. Drug discovery enterprises very seldom focus on protonation state changes of receptors and ligands and/or the pH-dependence of ligand–receptor binding. However, these changes impact ionizable groups in macromolecules and small molecules and could affect the nature of the final docked complex. For this system in particular, refinement helped identify critical amino acids but did not offer a correlation to affinity. It is our belief there is a lot to be uncovered at the physiological level for sst4. Given that electrostatics and even vdW required to render affinity are rather limited, conformational rearrangements upon ligand binding likely result in revised interactions of a residue with the ligand and within the receptor partial charges. These changes may extend beyond the immediate vicinity of the cavity. The fact we observed a trend between charge separation and affinity is indicative of the relevance of ionizable groups and local or distant electrostatic scenarios. Lead optimization efforts include optimizing ligand physicochemical properties related to pharmacokinetics, while ensuring potency is not impacted greatly. What we have seen here is that additional ligand characteristics such as Πpol would be invaluable in ascertaining which lead scaffolds should be advanced in the pipeline during lead optimization campaigns.
4. Conclusions
QM/MM refinement of the cryo-EM structures led to more stable bound complexes when compared to unbound ones, while the pocket volume decreased upon binding. Between SRIF and the small molecule, SRIF rendered a more stable complex. Binding interactions with Asn199 and Asn293 seen in the experimental structures were lost and replaced with Gln279. In an effort to identify critical amino acids for agonist binding, we compared bound configurations of several known sst4 ligands and identified Asp126, Gln279, and Leu297 interactions were consistent among refined complexes. To shed light on the effect of sulfur in the compound series under investigation, we considered several physicochemical properties of matched molecular pairs. Results showed sulfur affected solubility, ligand pKa's and binding modes. We also aimed at correlating affinity with certain parameters for sst4 agonists. Gibbs free energies, properties of ligand ESPs and acidity constants were considered to discern which correlated best with affinity. There was no correlation between energetic properties of ligands or protein–ligand complexes and affinity, but we identified a strong relationship between Πpol and affinity for sst4 agonists of different chemical classes. Further, Asp126, Gln279 or Phe211, Leu123, and Leu297 were important for high affinity of the triazole and thio-triazole derivatives considered herein. Moving forward, we advise: (1) less focus be given in post-docking refinement and more on physically viable starting conformations for better docking accuracy; (2) balancing the computational cost of high level theory and the extent to which models reflect reality with the stage of the drug discovery project; (3) deeper understanding of the system, protonation changes, and effects of altered interactions upon binding and beyond the cavity; and (4) consideration of ligand properties in lead optimization beyond the classical physicochemical ones.Conflicts of interest
There are no conflicts to declare.Data availability
All relevant data are within the paper and SI. The datasets supporting this article have been uploaded as part of the SI and are presented in Fig. S1. Table S1 provides the side-chain RMSD’s for the SRIF-sst4 and small molecule-sst4 complexes before and after refinement. Table S2 shows the differences in binding free energies for representative ligand pairs, with compounds in Fig. S2 used to validate the correlations between ligand properties and affinity.Supplementary information is available. See DOI: https://doi.org/10.1039/d5ob00775e.
Acknowledgements
This research was supported in part by the National Institute on Aging of the National Institutes of Health under Award Number R01AG047858 and SIUE's URCA program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
- U. Kumar, Expression of somatostatin receptor subtypes (SSTR1-5) in Alzheimer's disease brain: an immunohistochemical analysis, Neuroscience, 2005, 134(2), 525–538 CrossRef CAS PubMed
.
- C. Viollet, G. Lepousez, C. Loudes, C. Videau, A. Simon and J. Epelbaum, Somatostatinergic systems in brain: networks and functions, Mol. Cell. Endocrinol., 2008, 286(1–2), 75–87 CrossRef CAS PubMed
.
- T. Saito, N. Iwata, S. Tsubuki, Y. Takaki, J. Takano, S. M. Huang, T. Suemoto, M. Higuchi and T. C. Saido, Somatostatin regulates brain amyloid beta peptide Abeta42 through modulation of proteolytic degradation, Nat. Med., 2005, 11(4), 434–439 CrossRef CAS PubMed
.
- P. Brazeau, W. Vale, R. Burgus, N. Ling, M. Butcher, J. Rivier and R. Guillemin, Hypothalamic polypeptide that inhibits the secretion of immunoreactive pituitary growth hormone, Science, 1973, 179(68), 77–79 CrossRef CAS PubMed
.
- U. Kumar and M. Grant, Somatostatin and somatostatin receptors, Results Probl. Cell Differ., 2010, 50, 137–184 CAS
.
- L. N. Moller, C. E. Stidsen, B. Hartmann and J. J. Holst, Somatostatin receptors, Biochim. Biophys. Acta, 2003, 1616(1), 1–84 CrossRef CAS PubMed
.
- J. F. Bruno, Y. Xu, J. Song and M. Berelowitz, Molecular cloning and functional expression of a brain-specific somatostatin receptor, Proc. Natl. Acad. Sci. U. S. A., 1992, 89(23), 11151–11155 CrossRef CAS PubMed
.
- E. Hama and T. C. Saido, Etiology of sporadic Alzheimer's disease: somatostatin, neprilysin, and amyloid beta peptide, Med. Hypotheses, 2005, 65(3), 498–500 CrossRef CAS PubMed
.
- I. Lewis, W. Bauer, R. Albert, N. Chandramouli, J. Pless, G. Weckbecker and C. Bruns, A novel somatostatin mimic with broad somatotropin release inhibitory factor receptor binding and superior therapeutic potential, J. Med. Chem., 2003, 46(12), 2334–2344 CrossRef CAS PubMed
.
- A. M. Crider and K. A. Witt, Somatostatin sst4 ligands: chemistry and pharmacology, Mini-Rev. Med. Chem., 2007, 7(3), 213–220 CrossRef CAS PubMed
.
- W. Zhao, S. Han and N. Qiu,
et al., Structural insights into ligand recognition and selectivity of somatostatin receptors, Cell Res., 2022, 32, 761–772 CrossRef CAS PubMed
.
- M. Ankersen, A. M. Crider, S. Liu, B. Ho, H. S. Andersen and C. E. Stidsen, Discovery of a novel non-peptide somatostatin agonist with SST4 selectivity, J. Am. Chem. Soc., 1998, 120(7), 1368–1373 CrossRef CAS
.
- I. Daryaei, K. Sandoval, K. Witt, M. Kontoyianni and A. Michael Crider, Discovery of a 3,4,5-trisubstituted-1,2,4-triazole agonist with high affinity and selectivity at the somatostatin subtype-4 (sst(4)) receptor, MedChemComm, 2018, 9(12), 2083–2090 RSC
.
- W. L. Neumann, K. E. Sandoval, S. Mobayen, M. Minaeian, S. G. Kukielski, K. N. Srabony, R. Frare, O. Slater, S. A. Farr, M. L. Niehoff, A. Hospital, M. Kontoyianni, A. M. Crider and K. A. Witt, Synthesis and structure-activity relationships of 3,4,5-trisubstituted-1,2,4-triazoles: high affinity and selective somatostatin receptor-4 agonists for Alzheimer's disease treatment, RSC Med. Chem., 2021, 12(8), 1352–1365 RSC
.
- Z. Liu, A. M. Crider, D. Ansbro, C. Hayes and M. Kontoyianni, A structure-based approach to understanding somatostatin receptor-4 agonism (sst4), J. Chem. Inf. Model., 2012, 52(1), 171–186 CrossRef CAS PubMed
.
- O. Slater and M. Kontoyianni, A computational study of somatostatin subtype-4 agonist binding, SN Appl. Sci., 2022, 4, 140 CrossRef
.
- S. G. Rasmussen, H. J. Choi, J. J. Fung, E. Pardon, P. Casarosa, P. S. Chae, B. T. Devree, D. M. Rosenbaum, F. S. Thian, T. S. Kobilka, A. Schnapp, I. Konetzki, R. K. Sunahara, S. H. Gellman, A. Pautsch, J. Steyaert, W. I. Weis and B. K. Kobilka, Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor, Nature, 2011, 469(7329), 175–180 CrossRef CAS PubMed
.
- W. Huang, A. Manglik, A. J. Venkatakrishnan, T. Laeremans, E. N. Feinberg, A. L. Sanborn, H. E. Kato, K. E. Livingston, T. S. Thorsen, R. C. Kling, S. Granier, P. Gmeiner, S. M. Husbands, J. R. Traynor, W. I. Weis, J. Steyaert, R. O. Dror and B. K. Kobilka, Structural insights into micro-opioid receptor activation, Nature, 2015, 524(7565), 315–321 CrossRef CAS PubMed
.
- A. Negi, J. Zhou, S. Sweeney and P. V. Murphy, Ligand design for somatostatin receptor isoforms 4 and 5, Eur. J. Med. Chem., 2019, 163, 148–159 CrossRef CAS PubMed
.
- A. M. Crider, A. Hospital, K. E. Sandoval, W. L. Neumann, S. Kukielski, L. Garic, K. Ingold, M. Dunahoo, K. N. Srabony, R. Frare, O. Slater, N. Peel, M. Kontoyianni and K. A. Witt, 3-Thio-3,4,5-trisubstituted-1,2,4-triazoles: high affinity somatostatin receptor-4 agonist synthesis and structure-activity relationships, RSC Med. Chem., 2025, 16, 945–960 RSC
.
- C. R. Sondergaard, M. H. Olsson, M. Rostkowski and J. H. Jensen, Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values, J. Chem. Theory Comput., 2011, 7(7), 2284–2295 CrossRef CAS PubMed
.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, The Cambridge Structural Database, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72(Pt 2), 171–179 CrossRef CAS PubMed
.
- J.-P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, Numerical integration of cartesian equations of motion of a system with constraints - molecular dynamics of N-alkanes, J. Comput. Phys., 1977, 23(3), 327–341 CrossRef CAS
.
- R. B. Murphy, D. M. Philipp and R. A. Friesner, A Mixed Quantum Mechanics/Molecular Mechanics (QM/MM) Method for Large-Scale Modeling of Chemistry in Protein Environments, J. Comput. Chem., 2000, 21, 1442–1457 CrossRef CAS
.
- D. M. Philipp and R. A. Friesner, Mixed ab initio QM/MM modeling using frozen orbitals and tests with alanine dipeptide and tetrapeptide, J. Comput. Chem., 1999, 20, 1468–1494 CrossRef CAS
.
- J. J. Stewart, MOPAC: a semiempirical molecular orbital program, J. Comput. Aided Mol. Des., 1990, 4(1), 1–105 CrossRef CAS PubMed
.
-
J. J. P. Stewart, MOPAC2016, Stewart Computational Chemistry, Colorado Springs, 2016 Search PubMed
.
- A. D. Bochevarov, E. Harder, T. F. Hughes, J. R. Greenwood, D. A. Braden, D. M. Philipp, D. Rinaldo, M. D. Halls, J. Zhang and R. A. Friesner, Jaguar: A High-Performance Quantum Chemistry Software Program with Strengths in Life and Materials Sciences, Int. J. Quantum Chem., 2013, 113, 2110–2142 CrossRef CAS
.
- A. Anoop, S. Ranganathan, B. D. Dhaked, N. N. Jha, S. Pratihar, S. Ghosh, S. Sahay, S. Kumar, S. Das, M. Kombrabail, K. Agarwal, R. S. Jacob, P. Singru, P. Bhaumik, R. Padinhateeri, A. Kumar and S. K. Maji, Elucidating the role of disulfide bond on amyloid formation and fibril reversibility of somatostatin-14: relevance to its storage and secretion, J. Biol. Chem., 2014, 289(24), 16884–16903 CrossRef CAS PubMed
.
- F. Fogolari, A. Brigo and H. Molinari, The Poisson-Boltzmann equation for biomolecular electrostatics: a tool for structural biology, J. Mol. Recognit., 2002, 15(6), 377–392 CrossRef CAS PubMed
.
- J. J. Klicić, R. A. Friesner, S.-Y. Liu and W. C. Guida, Accurate prediction of acidity constants in aqueous solution via density functional theory and self-consistent reaction field methods, J. Phys. Chem. A, 2002, 106, 1327–1335 CrossRef
.
- A. D. Bochevarov, M. A. Watson, J. R. Greenwood and D. M. Philipp, Multiconformation, Density Functional Theory-Based pK(a) Prediction in Application to Large, Flexible Organic Molecules with Diverse Functional Groups, J. Chem. Theory Comput., 2016, 12(12), 6001–6019 CrossRef CAS PubMed
.
- P. Politzer and J. S. Murray, Computational prediction of condensed phase properties from statistical characterization of molecular surface electrostatic potentials, Fluid Phase Equilib., 2001, 185, 129–137 CrossRef CAS
.
- A. V. Onufriev and E. Alexov, Protonation and pK changes in protein-ligand binding, Q. Rev. Biophys., 2013, 46(2), 181–209 CrossRef CAS PubMed
.
-
R. Mazzaferro, M. Ferrara, R. Giovannini, I. Lingard and K. Rudolf, Morpholine and 1,4-oxazepane amides as somatostatin receptor subtype 4 (sstr4) agonists, WO2016075240A1, 2016
.
-
R. Giovannini, Y. Cui, H. Doods, M. Ferrara, S. Just, R. Kuelzer, I. Lingard, R. Mazzaferro and K. Rudolf, New somatostatin receptor subtype 4 (sstr4) agonists, WO2014184275A1, 2014
.
| This journal is © The Royal Society of Chemistry 2025 |