Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Post-SELEX modification of quinine aptamers through neoacetalization†‡
Heidi
Kähkölä
 ,
Muditha
Herath
,
Pasi
Virta
,
Muditha
Herath
,
Pasi
Virta
 and
Tuomas
Lönnberg
and
Tuomas
Lönnberg
 *
*
Department of Chemistry, University of Turku, Henrikinkatu 2, 20500 Turku, Finland. E-mail: tuanlo@utu.fi
First published on 6th January 2025
Abstract
In this article, a neoacetalization-based method for post-SELEX modification of aptamers is introduced. Three modified quinine binding aptamer scaffolds were synthesized by replacing three different nucleosides of the binding site with a (2R,3S)-4-(methoxyamino)butane-1,2,3-triol residue. These aptamer scaffolds were incubated in different aldehyde mixtures with and without quinine, allowing the reversible formation of N-methoxy-1,3-oxazinane (MOANA) nucleoside analogues through dynamic combinatorial chemistry. UHPLC-MS analysis identified two aldehydes, namely methyl 4-formylbenzoate and 3-nitrobenzaldehyde, with significantly different tendency to react with one of the aptamer scaffolds in the presence and absence of quinine. The quinine binding affinity of these two modified aptamers was determined by isothermal titration calorimetry (ITC). Unexpectedly, the 3-nitrobenzaldehyde derivatized aptamer dimerized on binding quinine at the relatively high concentration of the ITC. In addition, we discovered that with another modified aptamer, quinine binding caused cleavage of the N–O bond of the (2R,3S)-4-(methoxyamino)butane-1,2,3-triol residue.
1. Introduction
Aptamers are single-stranded DNA or RNA oligonucleotides that often have high affinity and specificity for their target molecule. They can be identified for virtually any target, including small molecules,1 proteins,2–4 cells5,6 and even tissues.7,8 Especially in diagnostic and therapeutic applications, aptamers are widely used due to their smaller size, lower immunogenicity, higher stability, chemical synthesis and cheaper production compared to antibodies.9–11However, natural nucleic acids have a poor chemical diversity provided by only five canonical nucleobases, and their ability to bind more demanding targets, such as single enantiomers of small organic molecules or glycosylated proteins, is limited. They could benefit from chemical modifications,10,12,13 likely resulting in aptamers with superior affinities compared to current ones.
Aptamers are obtained by a process known as Systematic Evolution of Ligands by EXponential enrichment (SELEX), which involves the directed selection of oligonucleotides from combinatorial oligonucleotide libraries in vitro. A target molecule is incubated with an oligonucleotide library, and oligonucleotides bound to the target molecule are amplified by a polymerase chain reaction (PCR). The cycle is repeated multiple times, gradually enriching the library with sequences expressing an increased affinity for the target molecule.14,15 Due to the limited tolerance of polymerase enzymes to chemical modifications, amplification of modified nucleic acids in the selection phase of SELEX is difficult. To circumvent this problem, post-SELEX optimization approaches have been developed over the recent years.16–22 Dynamic combinatorial chemistry (DCC) is an attractive approach for introducing chemical modifications to aptamers after SELEX. DCC enables the simultaneous screening of diverse modifications from large pools. Through ligand-driven selection, the most effective modifications for optimizing target binding can be identified. Previously, dynamic and pH-responsive N-methoxyoxazolidine formation has been employed for the reversible assembly of split aptamers, using quinine as a template.23 Under acidic conditions, this reaction is in dynamic equilibrium, but it can be freezed by increasing the pH above 7.24,25 Inspired by the neoglycosylation,26 this kind of N,O-acetalization of N-methoxyamines can be referred to as neoacetalization.
Here, we present the post-SELEX modification of aptamers through reversible formation of neoacetals. The quinine-binding cocaine aptamer MN4 was used in this study, as it is known to have the highest binding affinity for quinine of cocaine-binding aptamers.27 The binding pocket of the MN4 is located at the three-way junction formed by its three stems. In the free form, the aptamer is already fully folded to its secondary structure and no significant structural change takes place due to ligand binding.1,28–31 One of the three nucleosides within the binding pocket of MN4 – T19, C20 or A21 – was replaced by the previously reported (2R,3S)-4-(methoxyamino)butane-1,2,3-triol32 (MABT) residue. Each of these modified aptamer scaffolds was incubated with several pools of aldehydes at pH 5.5, both in the presence and absence of quinine, allowing the formation of new modified aptamers by DCC through formation of N-methoxy-1,3-oxazinane (MOANA) nucleotide analogues (Scheme 1). The affinity constant and other thermodynamic parameters of quinine binding were determined for the most promising modified aptamer candidates by isothermal titration calorimetry (ITC).
2. Results and discussion
2.1. Oligonucleotide synthesis
Three modified aptamer scaffolds were synthesized by an automated oligonucleotide synthesizer. In each scaffold, the previously reported MOANA phosphoramidite building block33 replaced a different base: either T19, C20 or A21 (Table 1). While the other nucleotides showed near quantitative yields for all couplings, the coupling efficiency of MOANA phosphoramidite building block was ca. 90% for the aptamers MN4-T19, MN4-C20 and MN4-A21. After the synthesis, the solid support and the protecting groups were removed by a conventional ammonolysis. Since purification was challenging due to the co-elution of the desired products and side products lacking the MOANA residue, MN4-T19, MN4-C20 and MN4-A21 were derivatized to slower eluting analogues through reaction with the hydrophobic cyclohexanecarboxaldehyde. The crude products were purified by RP-HPLC (Fig. S1–S3 in the ESI‡). Aptamer scaffolds were characterized by UHPLC-MS (Fig. S4–S9 in the ESI‡) and quantified by UV/Vis spectrophotometry.| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| MN4-T19 | GGCGACAAGGAAAATCCTXCAACGAAGTGGGTCGCC |
| MN4-C20 | GGCGACAAGGAAAATCCTTXAACGAAGTGGGTCGCC |
| MN4-A21 | GGCGACAAGGAAAATCCTTCXACGAAGTGGGTCGCC |
2.2. Derivatization of modified aptamers with aldehydes
MN4-T19, MN4-C20 and MN4-A21 were derivatized with various aldehydes both in the presence and absence of quinine to investigate the impact of quinine on the product composition. Modifications favoring quinine binding would be expected to be overrepresented in the product mixtures in the presence of quinine, while those retarding quinine binding should be underrepresented or altogether absent. The aldehyde mixtures used in this study are summarized in Table 2, and their chemical structures are presented in the ESI (Table S1‡). Within each mixture, all aldehydes had unique molecular weights to allow unambiguous identification of the products by UHPLC-MS analysis. All three aptamer scaffolds were incubated separately with a large excess of aldehydes in aqueous cacodylate buffer (pH 5.5) at room temperature protected from light. The reactions were monitored by UHPLC-MS over 2 weeks at regular intervals, after which the relative intensities of the starting material and product peaks no longer changed. The relative intensities of naked aptamer scaffolds and their DCC products are reported in the ESI (Table S2‡), along with the mass spectra, UV spectra and extracted ion UPLC traces of the DCC reaction products (Fig. S10–S69‡). Quantification of the DCC products and naked aptamers was based on the integration of the extracted ion ([M − 5H]5−) chromatograms, assuming equal ionizability for closely related structures.| Aldehydes | Abbreviation | |
|---|---|---|
| Group 1 | 2-(Adenin-9-yl) acetaldehyde | a |
| 2-(Cytosin-1-yl) acetaldehyde | b | |
| 2-(Guanin-9-yl) acetaldehyde | c | |
| 2-(Thymin-1-yl) acetaldehyde | d | |
| 2-(Imidazol-4-yl) acetaldehyde | e | |
| 2-(2-Methylbenzimidazol-1-yl) acetaldehyde | f | |
| Group 2 | 3-Benzyloxypropionaldehyde | g |
| Cyclohexanecarboxaldehyde | h | |
| D-Ribose | i | |
| D-Ribose-5-phosphate | j | |
| Glyoxylic acid | k | |
| Tribromoacetaldehyde | l | |
| Group 3.1 | 4-Carboxybenzaldehyde | m |
| Benzaldehyde | n | |
| Group 3.2 | 4-Hydroxybenzaldehyde | o |
| 4-Methoxybenzaldehyde | p | |
| Group 3.3 | Methyl 4-formylbenzoate | q |
| 3-Nitrobenzaldehyde | r |
Aldehyde group 1 included acetaldehyde derivatives of nucleobases34 and other heterocycles. With MN4-T19 and MN4-C20, 2-(adenin-9-yl) acetaldehyde derivatized aptamer scaffolds were the main products both in the absence and presence of quinine. With MN4-A21, on the other hand, the main components were the unreacted and formaldehyde derivatized aptamer scaffolds regardless of the presence of quinine, even though the 2-(adenin-9-yl) acetaldehyde derivatized aptamer was also formed. Also, 2-(guanin-9-yl) acetaldehyde and 2-(imidazol-4-yl) acetaldehyde derivatized aptamers could be detected with all three aptamer scaffolds in both reaction mixtures.
As no sensitivity to the presence of quinine was detected with the relatively similar aldehydes of group 1, the ones for group 2 were selected to represent a much wider range of physicochemical properties. Disappointingly, only 3-benzyloxypropionaldehyde was incorporated with a reasonable efficiency to any of the aptamer scaffolds and in all cases much less in the presence of quinine than in the absence thereof. With MN4-T19 and MN4-C20, some 3-benzyloxypropionaldehyde adduct was formed in the presence of quinine, but the naked aptamers and their formaldehyde derivatives were the main components. In contrast, MN4-A21 was derivatized with 3-benzyloxypropionaldehyde only in the absence of quinine and even then, naked, formaldehyde derivatized and acetaldehyde derivatized MN4-A21 dominated the equilibrium mixture.
Since quinine did not affect the DCC product composition with aldehyde groups 1 and 2, aldehydes of the group 3 were divided into small subgroups, each consisting of only two benzaldehydes with different substituents but similar electronic properties. The rate of aldehyde exchange at the MABT scaffold is strongly dependent on electron density at the carbonyl carbon.33 Furthermore, given the previously reported equilibrium constants for N-methoxyoxazolidines,24 it seems likely that benzaldehydes with electron-withdrawing substituents would form inherently more stable MOANA nucleosides than those with electron-donating substituents, regardless of the presence of quinine. The Hammett substituent constants (σ) for the aldehydes chosen for group 3 are presented in Table 3.
| Aldehyde | Hammett constant value (σ) | |
|---|---|---|
| Group 3.1 | 4-Carboxybenzaldehyde | −0.05 |
| Benzaldehyde | 0 | |
| Group 3.2 | 4-Hydroxybenzaldehyde | −0.38 |
| 4-Methoxybenzaldehyde | −0.28 | |
| Group 3.3 | Methyl 4-formylbenzoate | +0.39 |
| 3-Nitrobenzaldehyde | +0.55 |
Subgroup 3.1 contained 4-carboxybenzaldehyde (σ = −0.05) and benzaldehyde (σ = 0). In the absence of quinine, only MN4-C20 formed derivatized aptamers with 4-carboxybenzaldehyde and benzaldehyde and equilibrium still favored the starting material. In the presence of quinine, none of the three aptamer scaffolds were derivatized with aldehydes. However, it was discovered that the N-methoxy group of (2R,3S)-4-(methoxyamino)butane-1,2,3-triol residue was cleaved from MN4-T19 in the presence of quinine. The same reaction was also observed with MN4-C20 and MN4-A21, but to a lesser extent.
Subgroup 3.2 consisted of 4-hydroxybenzaldehyde (σ = −0.38) and 4-methoxybenzaldehyde (σ = −0.28). In the absence of quinine, the naked MN4-T19, MN4-C20 and MN4-A21 were the main components, along with their formaldehyde and acetaldehyde derivatives. In the presence of quinine, the N-methoxy group was cleaved from every aptamer scaffold to some extent but again much more with MN4-T19 than with MN4-C20 and MN4-A21. Small traces of MN4-T19 without the N-methoxy group was also found in the absence of quinine.
Subgroup 3.3 contained methyl 4-formylbenzoate (σ = 0.39) and 3-nitrobenzaldehyde (σ = 0.55). Without quinine, MN4-T19 was derivatized with both methyl 4-formylbenzoate and 3-nitrobenzaldehyde, although the naked aptamer scaffold was still the main component. Like with previous aldehyde groups, loss of the N-methoxy group was the main reaction in the presence of quinine, the aldehyde derivatives remaining as minor components. Quinine affected most on the product composition of MN4-C20. In the absence of quinine, the main products were 3-nitrobenzaldehyde- and methyl 4-formylbenzoate-derivatized aptamers (MN4-C20r and MN4-C20q, respectively) with similar relative intensities. However, with quinine, the product composition was changed, and methyl 4-formylbenzoate-derivatized aptamer became a clear main product. The relative intensity of the 3-nitrobenzaldehyde derivative had decreased compared to the methyl 4-formylbenzoate derivative and was only half of that of the latter. Additionally, the relative amount of formaldehyde derivative increased upon the addition of quinine and some loss of the N-methoxy group was also observed. Mass spectra of MN4-C20 in the absence and presence of quinine are presented in Fig. 1. With MN4-A21, the main component was the naked aptamer scaffold along with the formaldehyde and acetaldehyde derivatives, regardless of whether quinine was present or not. Methyl 4-formylbenzoate- and 3-nitrobenzaldehyde-derivatized MN4-A21 were also formed but only as minor products. In the presence of quinine, a small amount of MN4-A21 without the N-methoxy group was also found.
 | ||
| Fig. 1 Mass spectra of MN4-C20 with aldehyde mixture 3.3 in the absence (a) and presence (b) of quinine. | ||
The aromatic aldehydes incorporated most efficiently into the aptamer scaffolds. The formation of the oxazinane ring with these aldehydes creates structures, which resemble the canonical nucleobases. Incorporation of aldehydes to MN4-T19 and MN4-C20 was more efficient than to the MN4-A21 regardless of the presence of quinine. It has previously been reported that in a double helix, incorporation of canonical nucleobase acetaldehyde derivatives to MABT residue is strongly favored by base stacking and Watson–Crick base pairing.35 However, in this case A21 is not located in a double helix, but instead in a folded three-dimensional structure, probably lowering the effect of base stacking. Also, A21 forms a non-canonical base pair with G29 in the binding pocket of the aptamer, indicating that Watson–Crick base pairing is unfavored at this site. Since T19 and C20 are unpaired at the three-way junction,30 base pairing likely has no impact on which aldehydes are incorporated, but base stacking might. Overall, the addition of quinine retarded the incorporation of aldehydes into the aptamer scaffolds, presumably due to a steric clash at the binding site.
2.3. Isothermal titration calorimetry (ITC)
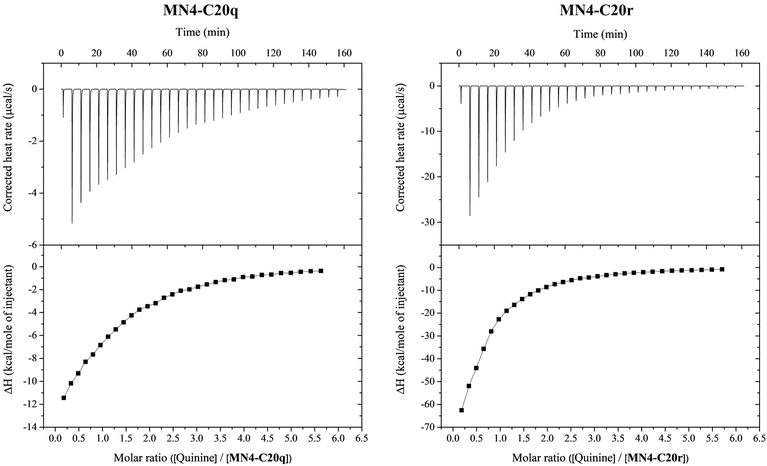
After identifying the dynamic combinatorial library formed by MN4-C20, methyl 4-formylbenzoate and 3-nitrobenzaldehyde as the most responsive to the presence of quinine, the two aldehyde derivatives (MN4-C20q and MN4-C20r, Fig. 2) were synthesized on a scale sufficient for isothermal titration calorimetry studies. The RP-HPLC traces of crude products as well as UV and mass spectra of purified products are presented in the ESI (Fig. S70–S73‡). The affinity and other thermodynamic parameters of quinine binding of the aldehyde derivatized aptamers MN4-C20q and MN4-C20r were determined by ITC. MN4-C20q and MN4-C20r were dissolved in 20 mM Tris·HCl buffer (pH 7.4) containing 140 mM NaCl and 5 mM KCl to make 207 and 204 μM aptamer solutions, respectively. The aptamers were titrated with 2.6 mM quinine in the same buffer. The calorimetric titrations consisted of 32 injections. The titrant was injected every 300 s, the first injection being 0.5 μl and the others 2.4 μl. In all experiments, the reference power was 11 μCal s−1, initial delay 60 s and stirring speed 1000 rpm. All experiments were conducted at 25 °C. Control measurements were performed by titrating quinine into buffer. Data were fitted to a one-site binding model and corrected for the heat of dilution. The first injection (0.5 μl) was excluded from each data set to remove the effect of titrant diffusion during the equilibration. The obtained binding isotherms are presented in Fig. 3 and the values for stoichiometry (N), affinity constant (Ka), enthalpy (ΔH) and entropy (ΔS) in Table 4.| Aptamer | N | K a (M−1) | ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) |
|---|---|---|---|---|
| MN4-C20q | 1.20 ± 0.04 | 6500 ± 400 | −19.4 ± 0.8 | −48 ± 2 |
| MN4-C20r | 0.50 ± 0.03 | 8900 ± 400 | −148 ± 8 | −480 ± 30 |
Due to the low affinity of the modified aptamers for quinine, the titrations yielded non-sigmoidal binding isotherms and attained c values were relatively low (1.6 and 0.92 for MN4-C20q and MN4-C20r, respectively). The obtained ΔH° values had good reproducibility for a given c value. Even better approach to determine ΔH° at low c conditions is by performing binding experiments at different temperatures and deriving ΔH° through a van't Hoff analysis.36 However, due to the limited availability of material, the binding experiments were conducted only at one temperature. Also, the impact of statistical errors increases with non-sigmoidal isotherms, making the determination of not only ΔH but also Ka less reliable.37,38
Instead of the expected 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (N = 1), the obtained N values for MN4-C20q and MN4-C20r were 1.2 and 0.50, respectively. The slight deviation from 1 observed in the case of MN4-C20q is probably due to errors in determining the aptamer concentration rather than indicating a real deviation from 1
1 stoichiometry (N = 1), the obtained N values for MN4-C20q and MN4-C20r were 1.2 and 0.50, respectively. The slight deviation from 1 observed in the case of MN4-C20q is probably due to errors in determining the aptamer concentration rather than indicating a real deviation from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. However, the N value of 0.5 for the titration of MN4-C20r could not be attributed to inaccuracies in the aptamer concentration but instead suggested simultaneous binding of two aptamers to one quinine molecule. This interpretation is further supported by the entropy change of quinine binding, which was with MN4-C20r ten times more negative than with MN4-C20q. It has been previously reported that one MN4 aptamer has two different binding sites for its substrates and depending of the NaCl concentration, one or two quinine molecules can bind to it at the same time.39 However, to our knowledge, two MN4 aptamers have not been observed to bind one quinine molecule at the same time. This kind of spontaneous binding of two aptamers to one target molecule has previously been reported with some protein-binding aptamers.40,41
1 stoichiometry. However, the N value of 0.5 for the titration of MN4-C20r could not be attributed to inaccuracies in the aptamer concentration but instead suggested simultaneous binding of two aptamers to one quinine molecule. This interpretation is further supported by the entropy change of quinine binding, which was with MN4-C20r ten times more negative than with MN4-C20q. It has been previously reported that one MN4 aptamer has two different binding sites for its substrates and depending of the NaCl concentration, one or two quinine molecules can bind to it at the same time.39 However, to our knowledge, two MN4 aptamers have not been observed to bind one quinine molecule at the same time. This kind of spontaneous binding of two aptamers to one target molecule has previously been reported with some protein-binding aptamers.40,41
Based on DCC results, both MN4-C20q and MN4-C20r formed in the presence of quinine, but the equilibrium yield of MN4-C20r was only about a half of that of MN4-C20q. In contrast, the affinity constants determined by ITC were 8900 M−1 for MN4-C20r and 6500 M−1 for MN4-C20q, indicating that the quinine is bound to MN4-C20r with almost 40% greater affinity. However, the obtained results by DCC and ITC are not exactly comparable since buffer, pH and ionic strength vary between analyses. Perhaps even more importantly, concentrations of the aptamers as well as quinine varied drastically between the DCC and ITC experiments. While only a 0.9 μM aptamer concentration was used in DCC, in ITC it was 204–207 μM. The higher concentration would favor quinine induced dimerization of MN4-C20r, perhaps to the point that the binding stoichiometry of MN4-C20r is different under the DCC and ITC conditions.
Based on ITC results, only MN4-C20r forms a dimer in the presence of quinine under the ITC experiment conditions. MN4-C20q and MN4-C20r differ from each other only by the aldehydes used for the derivatization. Although both methyl 4-formylbenzoate and 3-nitrobenzaldehyde are aromatic and have a hydrogen bond acceptor attached to the ring, they differ in the position of the substituents – methyl 4-formylbenzoate has an ester group at the para position and 3-nitrobenzaldehyde a nitro group at the meta position. However, the mechanism of the dimerization is unknown, and it is uncertain whether two aptamer molecules hybridize with each other or bind to different regions of the quinine. Therefore, the effect of the positions of the substituents on dimerization is unclear and should be studied further with other para and meta substituted aldehydes.
Binding of both MN4-C20q and MN4-C20r to quinine is driven by negative enthalpy compensated by unfavorable binding entropy as previously reported for MN4.27,42,43 Both MN4-C20q and MN4-C20r bind to quinine with reasonable affinity but still much weaker than MN4.27 However, the unexpected dimerization of MN4-C20r precludes a meaningful comparison of the binding affinities of MN4-C20q and MN4-C20r. For reference, the binding of the naked MN4-C20 to quinine was also investigated by ITC, but only very weak interaction was observed. Therefore, the thermodynamic parameters could not be determined. The corrected heat rate and the binding isotherm obtained for the naked MN4-C20 are presented in the ESI (Fig. S76‡).
2.4. UV melting temperature and PAGE analyses
The quinine induced dimerization of MN4-C20r was also investigated by melting temperature studies and polyacrylamide gel electrophoresis (PAGE). The samples in both experiments were dissolved in the same buffer as used in ITC studies. Melting temperature studies were conducted with MN4-C20q and MN4-C20r samples both with and without quinine. In the absence of quinine, biphasic UV melting curves were obtained for both aptamers with melting temperatures for MN4-C20q 17.4 ± 1.0 °C and 54.4 ± 0.2 °C, and for MN4-C20r 14.1 ± 0.6 °C and 55.2 ± 0.1 °C (Fig. S77 and S78 in the ESI‡). However, no effect of dimerization could be seen in melting curves, since the concentrations of aptamers were low, similar to those used in the DCC experiments.MN4-C20q and MN4-C20r were also analyzed by PAGE in the presence of varying quinine concentrations to find out whether the mobility of MN4-C20r would change, indicating quinine-induced dimerization. No such effect was observed, again probably due to insufficient concentration of MN4-C20r (Fig. S79 in the ESI‡).
2.5. Cleavage of the N–O bond
In principle, the observed loss of approximately 30 units in the molecular weight of MN4-T19 in the presence of quinine could result from either cleavage of the N–O bond or oxidation of the methoxyamino group to the corresponding oxime, followed by hydrolysis to an aldehyde. To distinguish between these alternatives, decomposed MN4-T19 was digested with nuclease P1. Nuclease P1 was added to the neutralized reaction mixture containing mainly MN4-T19 without the N-methoxy group in the presence of quinine. The reaction was allowed to proceed at room temperature and the next day monomeric nucleotides were almost exclusively detected by UHPLC-MS analysis (Fig. S84 in the ESI‡). The sole exception was a trimer consisting of phosphodiester-linked (2R,3S)-4-aminobutane-1,2,3-triol, thymidine and deoxycytidine (Fig. 4). The molecular weight of this trimer could be determined with sufficient precision to allow it to be unambiguously assigned as the amine, rather than the aldehyde, product (Fig. S85 in the ESI‡). | ||
| Fig. 4 The structure of the trimer identified by UHPLC-MS analysis after digestion of the decomposed MN4-T19 with nuclease P1. | ||
The N–O cleavage was also investigated with small molecules by NMR to evaluate the role of the tertiary structure of the aptamer. (2R,3S)-4-(Methoxyamino)butane-1,2,3-triol and quinine were incubated in deuterated acetate buffer (pH 5.5) at room temperature for several days. For reference, (2R,3S)-4-(methoxyamino)butane-1,2,3-triol was also incubated in the absence of quinine in the same buffer. The reactions were monitored by NMR at regular intervals. Even after two weeks, no change in the NMR spectra was observed (Fig. S80 and S81 in the ESI‡). These results confirm that the tertiary structure of the aptamer is needed to bring quinine and the MABT residue sufficiently close to each other to cause cleavage of the N–O bond. Cleavage of the N–O bond in hydroxylamines can be induced by multiple reducing agents44,45 and bases46 but to our best knowledge, quinine induced N–O bond cleavage has not been previously reported. The quinuclidine substituent and the hydroxyl group at C9 of quinine are reported to activate both nucleophile and electrophile, respectively, allowing quinine to act as bifunctional organic catalyst.47 However, the reaction mechanism of quinine induced N–O cleavage in modified MN4 aptamers is unknown and should be studied further before any conclusions can be drawn about the role of quinine.
The N-methoxy group cleavage from the aptamer scaffolds was observed mostly with group 3 aldehydes, even though closer observation revealed that it occurred also with aldehydes of the groups 1 and 2 at some extent in the presence of quinine. However, it occurred preferentially with aromatic aldehydes and in the presence of quinine. The cleavage of the N-methoxy group was investigated also without aldehydes and N–O cleavage was observed even when only quinine and MN4-T19 were present (Fig. S82 and S83 in the ESI‡). Based on previous reports, the T19 is known to be the most critical base for cocaine binding, indicating that it occupies a crucial location at the binding site. It is also possible that quinine is in direct contact with T19.30 It has been previously proposed, that T19 is slightly changing its position due to the binding of the ligand.28 Presumably, in the present case, such a change could orient the N-methoxy group favorably for cleavage og the N–O bond.
3. Conclusions
We have developed a post-SELEX modification method of aptamers exploiting reversible formation of neoacetals. A single (2R,3S)-4-(methoxyamino)butane-1,2,3-triol residue was incorporated at three different positions at the three-way junction binding site of a quinine aptamer. In most cases, the dynamic combinatorial libraries formed on incubating the aptamer scaffolds in mixtures of aldehydes were insensitive to the presence of quinine. However, with some benzaldehydes clearly different profiles were obtained in the presence and absence of quinine. Determination of the thermodynamic parameters of quinine binding of the corresponding modified aptamers by isothermal titration calorimetry revealed unexpected dimerization of one, while the other bound with the usual 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. Binding affinities of both were rather modest and the different binding modes prevented a meaningful comparison. Finally, we discovered a rapid quinine-promoted cleavage of the N–O bond in some of the modified aptamers. This finding might prove valuable in further development of quinine aptasensors.
1 stoichiometry. Binding affinities of both were rather modest and the different binding modes prevented a meaningful comparison. Finally, we discovered a rapid quinine-promoted cleavage of the N–O bond in some of the modified aptamers. This finding might prove valuable in further development of quinine aptasensors.
Author contributions
H. K.: data curation, formal analysis, funding acquisition, investigation, supervision, validation, visualization, writing – original draft, writing – review and editing; M. H.: data curation, investigation; P. V.: methodology, supervision, writing – review and editing; T. L.: formal analysis, methodology, project administration, supervision, validation, writing – review and editing.Data availability
The data supporting this article have been included as part of the ESI.‡Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
Access to the MicroCal ITC200 isothermal titration calorimeter was provided by Turku Protein Core. The financial support from the Finnish Cultural Foundation (decision number 00230689) is gratefully acknowledged.References
- M. N. Stojanovic, P. De Prada and D. W. Landry, J. Am. Chem. Soc., 2001, 123, 4928–4931 CrossRef CAS.
- R. A. Potyrailo, R. C. Conrad, A. D. Ellington and G. M. Hieftje, Anal. Chem., 1998, 70, 3419–3425 CrossRef CAS PubMed.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564–566 CrossRef CAS PubMed.
- S. Centi, G. Messina, S. Tombelli, I. Palchetti and M. Mascini, Biosens. Bioelectron., 2008, 23, 1602–1609 CrossRef CAS PubMed.
- C. D. Medley, J. E. Smith, Z. Tang, Y. Wu, S. Bamrungsap and W. Tan, Anal. Chem., 2008, 80, 1067–1072 CrossRef CAS.
- J. Sam Lee, M. Kim, H. Jin, M. Kwak, E. Cho, K. S. Kim and D. E. Kim, Int. J. Pharm., 2024, 662, 124519 CrossRef CAS.
- K. Y. Wong, Y. Liu, M. S. Wong and J. Liu, Exploration, 2024, 4, 20230008 CrossRef CAS PubMed.
- L. Xu, J. Zhu, L. Rong, H. Yang, B. Wang, S. Lu, L. Zhang, F. Li, S. Yang, Z. Wang, C. Li, X. Hu, R. Liu, L. Zheng, H. Liu, H. Zhang, Y. Liu, D. Zhao, S. Zhao, L. Zhang, Y. Jia, S. Liang, Z. Guo, X. Xie, R. Liu and L. Zhang, Theranostics, 2024, 14, 3945–3962 CrossRef CAS.
- V. Thiviyanathan and D. G. Gorenstein, Proteomics:Clin. Appl., 2012, 6, 563–573 CAS.
- P. Röthlisberger and M. Hollenstein, Adv. Drug Delivery Rev., 2018, 134, 3–21 CrossRef PubMed.
- P. R. Bouchard, R. M. Hutabarat and K. M. Thompson, Annu. Rev. Pharmacol. Toxicol., 2010, 50, 237–257 CrossRef CAS.
- A. Shoji, M. Kuwahara, H. Ozaki and H. Sawai, J. Am. Chem. Soc., 2007, 129, 1456–1464 CrossRef CAS.
- A. M. Yoshikawa, A. Rangel, T. Feagin, E. M. Chun, L. Wan, A. Li, L. Moeckl, D. Wu, M. Eisenstein, S. Pitteri and H. T. Soh, Nat. Commun., 2021, 12, 1–12 CrossRef.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- S. Gao, X. Zheng, B. Jiao and L. Wang, Anal. Bioanal. Chem., 2016, 408, 4567–4573 CrossRef CAS PubMed.
- E. Campos-Fernández, L. S. Barcelos, A. G. Souza, L. R. Goulart and V. Alonso-Goulart, ACS Omega, 2020, 5, 3533–3541 CrossRef.
- A. Bashir, Q. Yang, J. Wang, S. Hoyer, W. Chou, C. McLean, G. Davis, Q. Gong, Z. Armstrong, J. Jang, H. Kang, A. Pawlosky, A. Scott, G. E. Dahl, M. Berndl, M. Dimon and B. S. Ferguson, Nat. Commun., 2021, 12, 2366 CrossRef CAS PubMed.
- L. Zhao, Q. Wang, Y. Yin, Y. Yang, H. Cui and Y. Dong, Molecules, 2022, 27, 5725 CrossRef CAS PubMed.
- K. Y. Lee, H. Kang, S. H. Ryu, D. S. Lee, J. H. Lee and S. Kim, J. Biomed. Biotechnol., 2010, 2010, 168306 Search PubMed.
- K. S. Schmidt, S. Borkowski, J. Kurreck, A. W. Stephens, R. Bald, M. Hecht, M. Friebe, L. Dinkelberg and V. A. Erdmann, Nucleic Acids Res., 2004, 32, 5757–5765 CrossRef CAS PubMed.
- Y. Nonaka, W. Yoshida, K. Abe, S. Ferri, H. Schulze, T. T. Bachmann and K. Ikebukuro, Anal. Chem., 2013, 85, 1132–1137 CrossRef CAS PubMed.
- A. Aho and P. Virta, Chem. Commun., 2023, 59, 5689–5692 RSC.
- A. Aho, M. Sulkanen, H. Korhonen and P. Virta, Org. Lett., 2020, 22, 6714–6718 CrossRef CAS PubMed.
- A. Aho, A. Äärelä, H. Korhonen and P. Virta, Molecules, 2021, 26, 490 CrossRef CAS PubMed.
- F. Peri, P. Dumy and M. Mutter, Tetrahedron, 1998, 54, 12269–12278 CrossRef CAS.
- O. Reinstein, M. Yoo, C. Han, T. Palmo, S. A. Beckham, M. C. J. Wilce and P. E. Johnson, Biochemistry, 2013, 52, 8652–8662 CrossRef CAS PubMed.
- M. A. D. Neves, O. Reinstein and P. E. Johnson, Biochemistry, 2010, 49, 8478–8487 CrossRef CAS PubMed.
- E. Daems, D. Dewaele, K. Barylyuk, K. De Wael and F. Sobott, Talanta, 2021, 224, 121917 CrossRef CAS PubMed.
- M. A. D. Neves, O. Reinstein, M. Saad and P. E. Johnson, Biophys. Chem., 2010, 153, 9–16 CrossRef CAS PubMed.
- Z. R. Churcher, D. Garaev, H. N. Hunter and P. E. Johnson, Biophys. J., 2020, 119, 1147–1156 CrossRef CAS PubMed.
- M. N. K. Afari, P. Virta and T. Lönnberg, Org. Biomol. Chem., 2022, 20, 3480–3485 RSC.
- J. Wallin and T. Lönnberg, Eur. J. Org. Chem., 2022, e202200538 CrossRef CAS.
- B. P. Sutherland, P. J. LeValley, D. J. Bischoff, A. M. Kloxin and C. J. Kloxin, Chem. Commun., 2020, 56, 11263–11266 RSC.
- M. N. K. Afari, N. Heikinmäki, P. Virta and T. Lönnberg, ChemBioChem, 2024, e202400666 Search PubMed.
- J. Tellinghuisen, Anal. Biochem., 2008, 373, 395–397 CrossRef CAS PubMed.
- J. Tellinghuisen, Methods Enzymol., 2004, 383, 245–282 CAS.
- J. Tellinghuisen, Methods Cell Biol., 2008, 84, 739–780 CAS.
- M. A. D. Neves, S. Slavkovic, Z. R. Churcher and P. E. Johnson, Nucleic Acids Res., 2017, 45, 1041–1048 CAS.
- S. Manochehry, J. Gu, E. M. McConnell, B. J. Salena and Y. Li, ChemBioChem, 2020, 21, 2029–2036 CrossRef CAS.
- A. Pica, I. R. Krauss, V. Parente, H. Tateishi-Karimata, S. Nagatoishi, K. Tsumoto, N. Sugimoto and F. Sica, Nucleic Acids Res., 2017, 45, 461–469 CrossRef CAS.
- S. Slavkovic, M. Altunisik, O. Reinstein and P. E. Johnson, Bioorg. Med. Chem., 2015, 23, 2593–2597 CrossRef CAS.
- S. Slavkovic, Z. R. Churcher and P. E. Johnson, Bioorg. Med. Chem., 2018, 26, 5427–5434 CrossRef CAS PubMed.
- G. E. Keck, T. T. Wager and S. F. McHardy, Tetrahedron, 1999, 55, 11755–11772 CrossRef CAS.
- S. Scholz and B. Plietker, Org. Chem. Front., 2016, 3, 1295 RSC.
- K. V. Nikitin and N. P. Andryukhova, Mendeleev Commun., 2000, 10, 32–33 CrossRef.
- H. Hiemstra and H. Wynberg, J. Am. Chem. Soc., 1981, 103, 417–430 CrossRef CAS.
Footnotes |
| † The graphical abstract was created in BioRender. Salminen, J. (2024) https://BioRender.com/i45t158. |
| ‡ Electronic supplementary information (ESI) available: The experimental procedures and analytical data, including RP-HPLC, UHPLC-MS, ITC, UV melting temperatures, PAGE and 1H NMR spectroscopic data. See DOI: https://doi.org/10.1039/d4ob01973c |
| This journal is © The Royal Society of Chemistry 2025 |