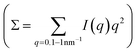Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An albumin unfolding and refolding cycle induced by a time-controlled pH jump†
Alessandra
Del Giudice‡
 *,
Daniele
Del Giudice§‡
,
Emanuele
Spatola¶
,
Valentina
Alemanno
,
Luciano
Galantini
*,
Daniele
Del Giudice§‡
,
Emanuele
Spatola¶
,
Valentina
Alemanno
,
Luciano
Galantini
 and
Stefano
Di Stefano
and
Stefano
Di Stefano

Department of Chemistry, Sapienza University of Rome, P.le A. Moro 5, 00185 Rome, Italy. E-mail: alessandra.delgiudice@uniroma1.it
First published on 26th September 2024
Abstract
Given the intimate connection between the structure and function of biological macromolecules, the ability to temporally control their unfolding–refolding process enables temporal regulation over specific functionalities, potentially applicable in innovative domains, including the construction of protein-based actuators or programmable catalysis and drug release in complex biotechnological processes. We show here how a temporally controlled protein unfolding–refolding cycle can be coupled in time with programmed pH sequences achieved through the spontaneous decomposition of an activated carboxylic acid. Specifically, we illustrate this process at the molecular level using albumin, the most prevalent protein found in plasma, for which a temporary shift from native to unfolded forms is promoted using nitroacetic acid, able to undergo base-catalysed decarboxylation when solubilized in water solution. As detected by small angle X-ray scattering and intrinsic tryptophan fluorescence, starting from the protein in its native form, the acid addition triggers unfolding to a partially denatured state and a subsequent time-tunable pH rise with gradual refolding that recapitulates the intermediate steps detected at the same pH values by static acidification, fitting within a framework of full reversibility of the structural changes as a function of the protein protonation state. At the end of the pH jump, the native structure is fully recovered, making this method a chemical tool to achieve a complete protein conformational sequence programmed in the timeframe of minutes without further intervention.
Introduction
Temporal control of supramolecular processes, such as self-assembly, host–guest interactions, operation of molecular machines or phase separation by means of chemical stimuli,1–7 is one of the hottest topics in the recent literature. The main reason for this can be found in the aspiration to reproduce in the laboratory the most typical feature of every biosystem, which consumes a chemical species to maintain a functional state different from the resting one. Control over the stimulus supply (amount and procedure) translates into temporal control of the system properties. We demonstrate here that a temporally controlled protein unfolding–refolding cycle can be accomplished by exploiting a decarboxylation-based method to control the pH. In the light of the close relationship between the structure and properties of biological macromolecules, the method would allow temporal control over specific functionalities exploitable in possible applications from protein-based actuators8 to programmable biocatalytic systems. The process was demonstrated for albumin, the most abundant protein in plasma. Knowing the ability of this protein to reversibly switch among different conformations by varying pH, a system was designed which was able to perform a temporary transition from native to unfolded forms and gradually refold through a time-tuneable pH cycle.We and others have indeed shown that simple reagents can be employed to finely control the pH of a water solution over time.10,11 The consumption of such reagents allows complementary pHlow–pHhigh–pHlow12–14 or pHhigh–pHlow–pHhigh15–19 sequences, whose amplitude and duration can be regulated at will. Since the properties of many biotic and abiotic systems in water solution depend on pH, the above programmable pH sequences can be used to have time control over such properties. In other words, the above reagents allow the system under investigation to pass from the resting state A to a different state B during the first pH variation and to come back from state B to state A during the second pH variation. Such a process is affected by the reactivity of the stimulus employed and its concentration and can be often modulated with the addition of enzymes13,20,21 or other additives. Generally, the system persists in state B as long as the stimulus is present and finally comes back to its resting state A when the stimulus is exhausted.
In particular, we used nitroacetic acid (NAA, 1), an activated carboxylic acid22–24 able to undergo base-catalysed decarboxylation, as a stimulus to achieve pHhigh–pHlow–pHhigh sequences. The latter were exploited to drive the operation of host–guest systems15 and DNA based molecular devices.16 The addition of acid 1 to a water solution results in an instantaneous lowering of the pH due to the dissociation of 1 to 1− and H+. Then, the resulting carboxylate 1− slowly loses carbon dioxide, giving the nitronate anion 2− which eventually acts as a base, increasing the pH to the final value (Fig. 1a).
 | ||
| Fig. 1 (a) Transient variation of the pH of a water solution due to the decarboxylation of acid 1. (b) Different conformations assumed by HSA in response to pH variation as a function of the protonation state (titration curve adapted from the original work of Tanford9). (c) The transient pH variation due to the addition of 1 causes programmed HSA conformational changes (unfolding–refolding). | ||
Among biomolecules, proteins are the most sensitive ones to pH variations due to the presence of acidic and basic functions in the lateral chains of the aminoacidic core, which affect the protonation state of the protein and hence its conformation.
It is known that the tertiary structure of human serum albumin (HSA), a protein that under physiological conditions (pH = 7.1–7.5) is found in its folded native (N) form, is highly affected by pH variations.25 According to the widely accepted Foster's model,26,27 lowering the pH to 4.3 induces the protein transition to a partially open conformation identified as the F (“fast migrating”) form, stable up to a pH value as low as 2.7. Moreover, the F form can experience a second transition to the E (“expanded”) form at even lower pH (Fig. 1b), in which the protein further loses secondary and tertiary structures and expands to a highly flexible chain of its subdomains, which locally maintain a high degree of native folding, together with the covalent disulfide bonds.
Taking advantage of the in situ production of acid due to the hydrolysis of glucono-δ-lactone, we have previously studied the unfolding process of HSA by means of fluorescence spectroscopy and small angle X-ray scattering (SAXS), enabling continuous monitoring of the protein structure in solution as a function of a smooth pH decrease.28 This approach has allowed the deconvolution of structural models for the main conformational components of the protein under acidic conditions, highlighting the intermediately unfolded conformation represented by the F-form, which never reached 100% at low ionic strength and in the absence of ligands. It has also provided interesting insights into the role of fatty acid binding within the high affinity sites in the stabilization of the native tertiary structure, so that lower pH values are required to trigger the acid transition towards partially unfolded conformations.29
Here we show a complementary approach for easily monitoring over time the conformational changes of HSA. Starting from the protein in its native form, we employed nitroacetic acid 1 to achieve a fast acidification of the solution upon dissolution, with the consequent denaturation of the protein, followed by a slow increase in the pH without further intervention, which enables full protein refolding with a predictable time dependence. This method allows for a whole protein conformational cycle to be realized by the addition of a single reagent, prompting us to study in detail the refolding process by SAXS (Fig. 1c), verifying the reversibility and identifying the main structural components.
Results and discussion
pH vs. time profiles of albumin solutions induced by nitroacetic acid
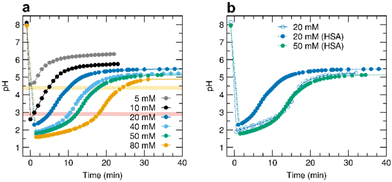
We initially carried out a series of experiments to assess the extent to which it is possible to control the duration and entity of a pHhigh–pHlow–pHhigh cycle induced by the addition of acid 1 in albumin solutions (0.055 mM HSA), prepared in 5.0 mM Tris HCl and 150 mM NaCl at 25 °C. The initial pH is always set at 8.1, and then different amounts of 1 are added while monitoring the pH variation over time (Fig. 2a). Increasing the initial acid concentration has three effects on the pH profile. First, the larger the amount of 1 added, the larger the initial pH jump and the lower the pHlow value reached at the end of the first jump. Second, the lower the value of pHlow, the slower the following pH ramp, which translates into a longer duration of the pH cycle. Third, the value of pHhigh finally reached by the system slightly decreases with increased initial concentration of 1 since a larger amount of carbon dioxide, in equilibrium with carbonic acid, is liberated during the reaction.15 On increasing the concentration of 1 from 5.0 to 80 mM, we indeed observed a decrease of both the pHlow and pHhigh values and a temporal expansion (longer duration) of the acidic transient state.By comparing the pH versus time profiles obtained with the same concentration of 1 and under the same buffer conditions, with and without HSA, it is evident that the presence of the protein affects the pH cycle, neutralizing part of the added 1 and consequently causing a less pronounced acid excess, resulting in an earlier pHlow–pHhigh jump. For example, the pH versus time trend obtained with 20 mM of 1 reasonably agrees with the profile obtained with 50 mM added acid and 0.055 mM of protein (Fig. 2b). Assuming that a similar pH rise over time implies a similar initial molar ratio between 1 and the base, 68 extra base equivalents per protein molecule can be estimated. This is a reasonable number considering that the HSA sequence has 98 total aspartate and glutamate residues that act as weak bases.
Shift of albumin fluorescence spectra during the pH jump
Upon evaluating the influence of HSA on pH cycles triggered by 1, we focused on the protein conformational transitions upon pH variations over time. With this aim, we initially resorted to fluorescence spectroscopy, monitoring the emission of the only tryptophan residue present in the protein (Trp214) by using excitation at λexc = 295 nm. Indeed, this is considered a diagnostic probe to study the tertiary structure of HSA since its emission is sensitive to the local polarity around the cleft where the fluorescent residue is located (Fig. S1†). In its native state (pH > 4.3), the emission shows a maximum (λmax) at 340 nm. After the addition of 20 mM 1, the emission spectra are blue shifted (λmax = 330 nm), as expected when the protein is in its acid form and the fluorophore is found in a less polar local environment.30,31 As the decarboxylation of 1 takes place, the fluorescence emission starts to move back to the original position, strongly suggesting that the protein recovers the native form at less acidic pH (Fig. 3b). Notably, following λmax as a function of pH, a midpoint around pH 4.4 is observed, above which the transition to the native conformation is known to occur (Fig. 3c). Therefore, the presence of nitroacetic acid 1 and nitromethane 2 does not seem to affect the conformational stability of HSA and its intrinsic pH refolding behaviour.Following albumin refolding over time using small angle X-ray scattering
To provide more insights into the conformational cycles experienced by HSA under the action of 1, we used SAXS, which is sensitive to electron density inhomogeneities at the 1–100 nm scale, and for dilute samples of biological macromolecules, SAXS provides information about their average size, shape, and folding state directly in solution.32–36Since the conformation of HSA well and reversibly responds to such pH variations, a clear change of the SAXS profiles as a function of time was detected (Fig. S3† and Fig. 4a, b and c). The clarity of the collected data is due to the simple protocol allowing continuous monitoring of the same sample, not needing preparation of a series of samples at different pH values and thus removing a source of variability. The transition is more clearly observed when showing the data in the Kratky plot representation (I·q2vs. q),|| which emphasizes the difference between a fully folded state, whose scattering profile follows in the high q regime the law expected for sharp interfaces (1/q4), and flexible states, which rather tend to follow the polymer statistics (1/q2). In addition, the presence of maxima in this representation is easily detected and their position provides information about the size of folded portions of the studied macromolecules, shifting to larger q values when smaller compact entities prevail within a flexible state, over a larger fully globular structure.37,38 In this case, we used the subtended area under the Kratky plot as an index of protein refolding. This quantity, if calculated up to the isosbestic point among the curves and normalized by the native value, grows towards 1.0 when the protein refolds (Fig. 4c bottom panel). A clear indication of reversibility is also denoted by the overlap between the data collected for the refolded samples at the end of the pHlow–pHhigh jump and those of the initial stock solutions (Fig. S3†).
The radius of gyration (Rg) obtained from the initial slope of the SAXS profile, according to a Guinier fit, is an immediate indication of the overall size of the protein molecules and is observed to decrease as a function of time (Fig. 4c top panel).
Both parameters indicate the increasing presence of partially unfolded and extended states upon increasing the concentration of 1 and that the recovery to the native size and folding state occurs with different time delays, as expected from the pH measurements. Therefore, using different acid amounts allows for tuning, in the range of 1 to 20 minutes, the time after which the protein begins to refold from an initially non-native state.
Reversibility and structural components of albumin acid denaturation
By converting the time scale to a pH scale, we would expect to observe superimposable trends of the protein size and folding index for different acid concentrations, if they are representative of an “instantaneous” equilibrium state, according to the slow rate of pH variation – minutes – compared to the fast acid unfolding and refolding – below seconds – reported for albumin.39 This assumption is found to be approximately verified, except for a deviation observed at the highest 1 concentration (Fig. 4d, yellow trace). This is likely due to an incorrect evaluation of the pH value corresponding to the SAXS data. Off-line pH measurements indeed have limitations to deduce what occurs at the sample position during SAXS collection, especially at high 1 concentration and therefore high CO2 release. Possibly, due to a faster nucleation of bubbles favoured by flow, moving meniscus and capillary interfaces (Fig. S4†), the pH can rise faster, and the protein refolds accordingly. With knowledge of the reversible nature of albumin acid transitions,26 we conclude that the protein folding state seems a better indicator of the local pH than the off-line electrode measurements.The apparent molecular mass obtained by assuming a fixed partial specific volume of 0.735 cm3 g−1 (ref. 40) shows a maximum during the pH rise towards neutrality around pH 3–3.5 (Fig. 5a, middle panel), possibly indicating that upon moving towards the isoelectric point,41 when electrostatic repulsions decrease, an increasing population of weak oligomers could transiently exist.** To further test whether the protein conformation detected by SAXS measurements during the time-dependent refolding is indeed an “instantaneous” equilibrium corresponding to a certain average protonation state, we prepared control samples by static acidification of the same HSA solution via the addition of known amounts of a strong acid (HCl, as reported in ESI Table S2†). The comparison between the SAXS profiles of the control samples brought to constant acidic pH values and those accessed at the same pH values in the time-dependent pH jump show that they substantially overlap (Fig. S5†), suggesting that the 1 decarboxylation-coupled refolding recapitulates the same equilibrium average conformations that take place in static samples.
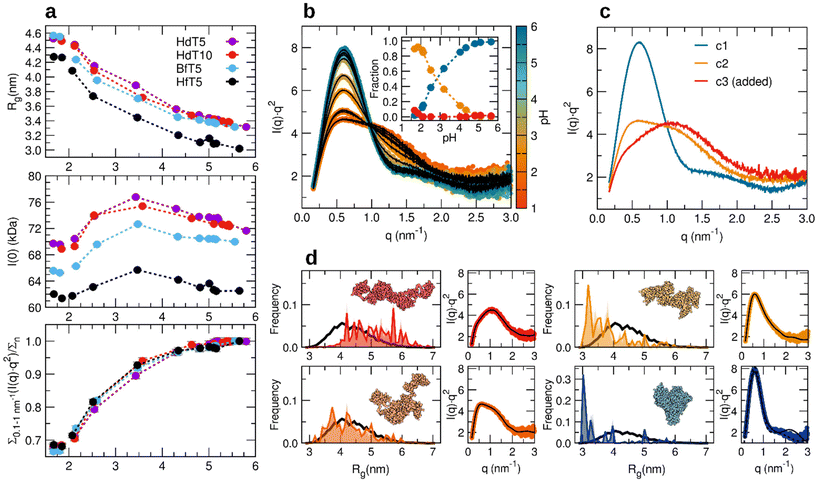 | ||
| Fig. 5 (a) Refolding trends as detected using SAXS parameters (Rg, I(0) and the index of refolding (Σ/Σn)) for different albumin preparations acidified with 40 mM and 60 or 80 mM (data points for pH < 2) acid 1: defatted HSA (purple), defatted HSA with double concentration of Tris HCl buffer (red), fatted BSA (cyan), and fatted HSA (black). All solutions contain NaCl 0.15 M. (b) Collection of SAXS data in Kratky plot representation for the fatted HSA sample shown as coloured dots according to the assigned pH, and data fit (black lines) as a linear combination of three components, shown in (c), weighted by the volume fractions reported in the inset. The first two components come from applying a component analysis to the dataset, while the third component was added to consider a fully expanded conformation, as measured in the absence of added NaCl and at pH < 1.6 (HCl > 40 mM). (d) Description of the selected SAXS profiles of fatted HSA (red is component 3 of panel (c), dark orange is 5 minutes after the addition of 80 mM acid 1, and light orange and blue are respectively 7.5 min and 5 h after the addition of 40 mM acid 1) according to an ensemble of models obtained by assuming that the unfolded albumin structure is composed of structured helical loops connected by random linkers. In the distribution of Rg values, the black line represents the overall pool of generated conformers, while the coloured areas show the optimized selection of conformers to fit the SAXS data as shown in a lateral panel. A representative conformer is shown for the non-native states, while for the refolded albumin, a fit of the data with the crystal structure46 is shown as a dashed line. | ||
We also followed time-dependent pH jumps by SAXS carried out with different albumin preparations from commercially purified products (Fig. 5, defatted HSA in Tris HCl 5 mM (HdT5), defatted HSA with double the concentration of Tris HCl buffer (HdT10), fatted bovine serum albumin BSA (BfT5), and fatted HSA (HfT5)). The general refolding trends as a function of pH are essentially unchanged for the human protein, except for a systematic shift of Rg to smaller values for the preparation not deprived of bound fatty acid molecules (“fatted”), probably due to an intrinsically smaller number of dimers and oligomers in the lyophilized starting product.†† For the HfT5 preparation indeed, the starting stock solution of native protein can be satisfyingly described by means of the monomeric albumin crystal structure (Fig. 5d). We therefore focus on this dataset to reach a more detailed structural interpretation of the refolding process, in terms of the minimum possible conformational states involved and previously proposed structural components.
The set of SAXS data collected during the 1-triggered unfolding and time-dependent refolding could be described reasonably well in terms of just two SAXS profiles, a fully folded component and a partially unfolded one (Fig. S6b†), while a third, further expanded component could marginally contribute at pH < 2 (Fig. 5b and c).‡‡ The optimized concentration profiles of these three structural components suggest that the mid-point of the transition between the partially unfolded and the native form as observed in these SAXS refolding experiments is located close to pH 3 (Fig. 5b). This should be compared to the component-based interpretation of previously collected unfolding trends with glucono-δ-lactone time-dependent acidification (in the absence of added salts), where a similar intermediately unfolded component, ascribed to the “F form” of the literature, seemed to reach a maximum around pH 4, while already coexisting with further expanded conformations clearly detected for pH lower than 3. Such a difference could be ascribed to the low ionic strength in the unfolding experiments, implying a reduced screening of the intra-chain electrostatic repulsions and an emphasized population of extended conformations, despite the nominal pH value measured with the electrode being the same.42
The overall folding state of albumin could be visualized by applying an ensemble optimization method to fit selected SAXS profiles and by considering a particular conformer found to be highly contributing and recurrent in repeated optimization experiments (Fig. 5d). The further extended component, not detected in these pH jumps due to the requirement for stronger inter-subdomain repulsions, can be described by including a relevant portion of models with Rg values above 5 nm, and shows an even higher level of flexibility (wider Rg distribution) compared to the reference random pool generated using the compact helical loop-flexible linker assumption. The SAXS profile with the lowest degree of folding accessed with the highest concentration of 1 (nominal pH 1.9) suggests an albumin conformation with a larger globular core involving the close-packed central subdomains and higher orientation flexibility for the N-terminal and C-terminal loops. Such a low-pH state substantially coincides with the main non-native component detected along the refolding path, which, together with an increasing fraction of native structures, allows for the simplest interpretation as a coexistence of the two main conformations in varying amounts (native and partially unfolded). The inspection of models that fit the SAXS data at higher pH upon refolding (e.g. at pH 3) could give an alternative view to this simplified assumption and suggest the involvement of possible conformers with a lower degree of separation of the terminal loops.
When correlating the results of SAXS and fluorescence from separate measurements using the unified pH scale (Fig. S8†), a clear asynchrony is detected between the two probes. The sharp shift of the tryptophan fluorescence spectrum back to the native position (accompanied by a more gradual change of the emission intensity) is completed only when the protein fully recovers the SAXS profile of the original native solution (pH > 4.7) and the mid-point of the transition observed through fluorescence corresponds to a global protein folding, which is already more than 90% similar to that of the native state. The discrepancy has to do with the multi-domain nature of albumin and the different sensitivity of the two techniques to global and local changes of the protein structure: while the SAXS signal mainly detects the earlier refolding and terminal domain recompaction taking place around pH 3, it is much less sensitive to the local re-adjustments within the central protein domain, where tryptophan is located. Here the fluorescent probe experiences a less polar environment compared to the native state irrespective of the variable degree of unfolding of the terminal domains, until a final local rearrangement restores the native polarity of its site in the central domain cleft characteristic of the native conformation. Such asynchronicity had already been observed more robustly with simultaneous detection of SAXS and fluorescence emission during time-dependent acid-induced unfolding (rather than refolding as realized here) of the same sample,28 and it contributes to a clear understanding of the multi-step nature of albumin's isomerisations at acidic pH.31
Conclusions
We showed that an activated carboxylic acid, namely nitroacetic acid 1, can be used as a chemical stimulus to control the conformation of human serum albumin. Fluorescence and SAXS data allow us to monitor over time the structural transitions experienced by the protein in response to the transient pH change triggered by acid 1. This method enables the pH to change in situ with predictable kinetics, without the addition of further external reactants or salts and without the need for dialysis or dilution protocols, allowing us to simply monitor the initial unfolding and time-dependent refolding event of the same protein sample in an automated experimental modality, well suited for synchrotron SAXS setups.Starting from a native protein sample, the acid 1 addition triggers the unfolding to a partially denatured state of albumin ascribed mainly to the “F” form, and the gradual time-dependent refolding can be described as an increase of the native fraction, which reaches 100% when the pH climbs back to 5, allowing for a full protein conformational cycle. The final state at the end of the pHhigh–pHlow–pHhigh cycle is indistinguishable from the initial starting state, and the intermediate steps of the refolding kinetics recapitulate both the inverse process of unfolding upon acidification28 and the average protein conformations detected at the same pH values by static acidification with HCl, showing that neither acid 1 nor its byproduct affects the conformational transition or the stability of the protein, fitting within a framework of full reversibility of the structural changes as a function of the protein protonation state. Such reversibility, appreciated through various methods since very early works (e.g. ref. 26), has now been clearly shown in time-programmed refolding experiments of the same sample without further intervention.
A whole conformational cycle (native–unfolded–native) can therefore be driven and temporally controlled by the single addition of a simple reagent. Together with other systems used to transiently control the pH, our protocol based on the use of acid 1 can be helpful in the study of pH responsive structural transitions of macromolecules. The biological origin of proteins, their high structural sensitivity towards pH and the intimate link between their structure and functions make these results particularly interesting for the development of pioneering and sustainable applications, from protein-driven actuators in robotics to customizable catalysis and drug delivery in advanced biotechnologies.
Data availability
Data supporting this article have been included as part of the ESI.† Raw data from synchrotron SAXS measurements at the BM29 beamline of ESRF are accessible on the ESRF data portal at https://data.esrf.fr/doi/10.15151/ESRF-ES-514136926 and https://data.esrf.fr/doi/10.15151/ESRF-ES-61665128.Final data shown in this article, including the spectroscopic and SAXS data plotted in all the figures, are available at Zenodo at https://doi.org/10.5281/zenodo.13169259.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
SDS thanks Ateneo 2022 Sapienza (RG1221815C85AF91). CNR-ISB is thanked for the use of the spectrofluorometer. The ESRF synchrotron, Grenoble, France, is acknowledged for providing the SAXS beamtime at BM29 under BAG proposals MX2363, MX2291, and MX1949 and Mark Tully is thanked for assistance. The Partnership for Soft Condensed Matter (PSCM) at ESRF is acknowledged for providing lab support. ADG acknowledges co-financing of the Sapienza University of Rome and the European Union—FSE-REACT-EU, PON Research and Innovation 2014–2020 DM1062/2021 for the RTD-A contract. We deeply thank the late Professor Nicolae Viorel Pavel for his valuable suggestions and help in synchrotron experiments, with everlasting gratitude for his scientific and human contribution to our group.References
- K. Das, L. Gabrielli and L. J. Prins, Angew. Chem., Int. Ed., 2021, 60, 20120–20143 CrossRef CAS PubMed.
- L. S. Kariyawasam, M. M. Hossain and C. S. Hartley, Angew. Chem., Int. Ed., 2021, 60, 12648–12658 CrossRef CAS.
- S. Borsley, D. A. Leigh and B. M. W. Roberts, Nat. Chem., 2022, 14, 728–738 CrossRef CAS PubMed.
- E. Del Grosso, E. Franco, L. J. Prins and F. Ricci, Nat. Chem., 2022, 14, 600–613 CrossRef CAS PubMed.
- Z. Li, J. Wang and I. Willner, Adv. Funct. Mater., 2022, 32, 2200799 CrossRef CAS.
- P. S. Schwarz, M. Tena-Solsona, K. Dai and J. Boekhoven, Chem. Commun., 2022, 58, 1284–1297 RSC.
- E. Shandilya, A. S. Bains and S. Maiti, J. Phys. Chem. B, 2023, 127, 10508–10517 CrossRef CAS PubMed.
- L. R. Khoury and I. Popa, Nat. Commun., 2019, 10, 1–9 CrossRef CAS PubMed.
- C. Tanford, J. Am. Chem. Soc., 1950, 72, 441–451 CrossRef CAS.
- D. Del Giudice, F. Frateloreto, C. Sappino and S. Di Stefano, Eur. J. Org. Chem., 2022, 2022, e202200407 CrossRef CAS.
- C. Sharma, I. Maity and A. Walther, Chem. Commun., 2023, 59, 1125–1144 RSC.
- T. Heuser, A. K. Steppert, C. Molano Lopez, B. Zhu and A. Walther, Nano Lett., 2015, 15, 2213–2219 CrossRef CAS PubMed.
- L. Heinen, T. Heuser, A. Steinschulte and A. Walther, Nano Lett., 2017, 17, 4989–4995 CrossRef CAS PubMed.
- G. Panzarasa, A. L. Torzynski, T. Sai, K. Smith-Mannschott and E. R. Dufresne, Soft Matter, 2020, 16, 591–594 RSC.
- D. Del Giudice, E. Spatola, M. Valentini, C. Bombelli, G. Ercolani and S. Di Stefano, Chem. Sci., 2021, 12, 7460–7466 RSC.
- D. Mariottini, D. Del Giudice, G. Ercolani, S. Di Stefano and F. Ricci, Chem. Sci., 2021, 12, 11735–11739 RSC.
- G. Fusi, D. Del Giudice, O. Skarsetz, S. Di Stefano and A. Walther, Adv. Mater., 2023, 35, 2209870 CrossRef CAS.
- J. Li, Y. Cui, Y. L. Lu, Y. Zhang, K. Zhang, C. Gu, K. Wang, Y. Liang and C.-S. Liu, Nat. Commun., 2023, 14, 1–14 CAS.
- G. Capocasa, F. Frateloreto, S. Correale Cavallari, M. Valentini, O. Lanzalunga and S. Di Stefano, Chem. – Eur. J., 2024, 30, e202303897 CrossRef CAS PubMed.
- T. Heuser, E. Weyandt and A. Walther, Angew. Chem., Int. Ed., 2015, 54, 13258–13262 CrossRef CAS.
- H. Che, S. Cao and J. C. M. Van Hest, J. Am. Chem. Soc., 2018, 140, 5356–5359 CrossRef CAS PubMed.
- C. Biagini and S. Di Stefano, Angew. Chem., Int. Ed., 2020, 59, 8344–8354 CrossRef CAS PubMed.
- E. Olivieri and A. Quintard, ACS Org. Inorg. Au, 2023, 3, 4–12 CrossRef CAS.
- D. Del Giudice and S. Di Stefano, Acc. Chem. Res., 2023, 56, 889–899 CrossRef CAS.
- T. J. Peters, All About Albumin, Elsevier, 1995 Search PubMed.
- C. Tanford, J. G. Buzzell, D. G. Rands and S. A. Swanson, J. Am. Chem. Soc., 1955, 77, 6421–6428 CrossRef CAS.
- D. C. Carter and J. X. Ho, Adv. Protein Chem., 1994, 45, 153–203 CrossRef CAS.
- A. Del Giudice, C. Dicko, L. Galantini and N. V. Pavel, J. Phys. Chem. B, 2017, 121, 4388–4399 CrossRef CAS PubMed.
- A. Del Giudice, L. Galantini, C. Dicko and N. V. Pavel, Colloids Surf., B, 2018, 168, 109–116 CrossRef CAS PubMed.
- R. F. Chen, Biochim. Biophys. Acta, Biophys. Incl. Photosynth., 1966, 120, 169–171 CrossRef CAS.
- M. Dockal, D. Carter and F. Rüker, J. Biol. Chem., 2000, 275, 3042–3050 CrossRef CAS PubMed.
- S. Doniach, Chem. Rev., 2001, 101, 1763–1778 CrossRef CAS.
- G. L. Hura, A. L. Menon, M. Hammel, R. P. Rambo, F. L. Poole, S. E. Tsutakawa, F. E. Jenney, S. Classen, K. A. Frankel, R. C. Hopkins, S. J. Yang, J. W. Scott, B. D. Dillard, M. W. W. Adams and J. A. Tainer, Nat. Methods, 2009, 6, 606–612 CrossRef CAS.
- S. Skou, R. E. Gillilan and N. Ando, Nat. Protoc., 2014, 9, 1727–1739 CrossRef CAS PubMed.
- S. Da Vela and D. I. Svergun, Curr. Res. Struct. Biol., 2020, 2, 164–170 CrossRef PubMed.
- T. N. Cordeiro, F. Herranz-Trillo, A. Urbanek, A. Estaña, J. Cortés, N. Sibille and P. Bernadó, in Advances in Experimental Medicine and Biology, Springer New York LLC, 2017, vol. 1009, pp. 107–129 Search PubMed.
- E. E. Lattman, K. M. Fiebig and K. A. Dill, Biochemistry, 1994, 33, 6158–6166 CrossRef CAS PubMed.
- P. Bernadó, Eur. Biophys. J., 2010, 39, 769–780 CrossRef PubMed.
- R. P. Taylor, V. Chau, M. J. Zenkowich and L. H. Leake, Biophys. Chem., 1978, 7, 293–299 CrossRef CAS PubMed.
- E. Mylonas and D. I. Svergun, J. Appl. Crystallogr., 2007, 40, s245–s249 CrossRef CAS.
- A. Salis, M. Boström, L. Medda, F. Cugia, B. Barse, D. F. Parsons, B. W. Ninham and M. Monduzzi, Langmuir, 2011, 27, 11597–11604 CrossRef CAS PubMed.
- L. Medda, B. Barse, F. Cugia, M. Boström, D. F. Parsons, B. W. Ninham, M. Monduzzi and A. Salis, Langmuir, 2012, 28, 16355–16363 CrossRef CAS PubMed.
- Y.-Q. Yeh, K.-F. Liao, O. Shih, Y.-J. Shiu, W.-R. Wu, C.-J. Su, P.-C. Lin and U.-S. Jeng, J. Phys. Chem. Lett., 2017, 8, 470–477 CrossRef CAS.
- A. Del Giudice, C. Dicko, L. Galantini and N. V. Pavel, J. Phys. Chem. B, 2016, 120, 12261–12271 CrossRef CAS.
- C. Leggio, L. Galantini and N. V. Pavel, Phys. Chem. Chem. Phys., 2008, 10, 6741–6750 RSC.
- S. Sugio, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi, Protein Eng., 1999, 12, 439–446 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ob01289e |
| ‡ Co-first authors with equal contribution. |
| § Present address: Institute for Complex Molecular Systems and the Laboratory of Macromolecular and Organic Chemistry, Eindhoven University of Technology, P.O. Box 513, 5600 MB Eindhoven, The Netherlands. |
| ¶ Present address: Institute of Organic Chemistry, Ulm University Albert-Einstein-Allee 11, 89081 Ulm, Germany. |
|| I is the background subtracted scattered intensity and q is the reciprocal space scattering vector modulus q = 4π![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ/λ, where 2θ is the scattering angle and λ is the X-ray wavelength. θ/λ, where 2θ is the scattering angle and λ is the X-ray wavelength. |
** According to the relationships ISAXS(0)(cm−1) = MM(kDa)c(mg cm−3)(Δρm(cm g−1))2/NA and Δρm(cm g−1) = (ρm(el. g−1) − ρsolv(el. cm−3) ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) (cm3 g−1))rTh(cm el.−1), complementary explanations for the observed behaviour of ISAXS(0) could be related to a non-monotonic variation of the X-ray scattering contrast (Δρm (cm g−1)) as a function of pH due to changes in protein specific volume ( (cm3 g−1))rTh(cm el.−1), complementary explanations for the observed behaviour of ISAXS(0) could be related to a non-monotonic variation of the X-ray scattering contrast (Δρm (cm g−1)) as a function of pH due to changes in protein specific volume (![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) ), ion or water binding (impacting the actual chemical composition and therefore the electrons per mass (ρm)). ), ion or water binding (impacting the actual chemical composition and therefore the electrons per mass (ρm)). |
| †† The presence of such oligomeric species in an amount usually below 20% of the overall protein mass affects the average SAXS profile especially in the lower q region, and the estimated Rg (when above 2.8 nm under native conditions) and maximum size (when above 9 nm) are particularly sensitive parameters signalling their existence. However, the overall refolding trend as a function of pH is not affected by the co-existence of a small fraction of albumin dimers,43 which can undergo comparable conformational transitions to monomers, as seen for example for the structural response of albumin upon oxidation using sodium hypochlorite.44 |
| ‡‡ Considering the previous knowledge of the maximum degree of albumin expansion detected by SAXS upon acidification below pH 2.7, with Rg values above 4.5 nm and Kratky plots skewed to larger q values with a lower subtended area in the q range <1 nm−1,28,29,43,45 we tested if such an expanded component could be included to describe the refolding process in combination with the native form. This was not the case (Fig. S6c†), and to reach satisfactory agreement, the second component could not be assigned to a state in which the protein is fully expanded as allowed by its local disulfide bonds but had to provide for an intermediate state with the bimodal Kratky plot and an Rg value of around 3.8 nm, which could be rather ascribed to the putative F-form. If the SAXS profile of a further expanded conformation is artificially included as an additional third component, it can just help in describing the data collected at the lowest pH, but it is not appreciably contributing, as already pointed out by the goodness of fit of a two-component model. |
| This journal is © The Royal Society of Chemistry 2025 |