 Open Access Article
Open Access ArticleMechanochemically-mediated dynamic imine bond conjugation for drug delivery using carbon dots
Gianluca
Fuoco
ab,
Gabrielle A.
Mandl
ab,
Carissa
De Mesa
ab,
John A.
Capobianco
 ab and
Rafik
Naccache
ab and
Rafik
Naccache
 *ab
*ab
aDepartment of Chemistry and Biochemistry and the Centre for NanoScience Research, Concordia University, Montreal H4B 1R6, Canada. E-mail: rafik.naccache@concordia.ca
bQuebec Centre for Advanced Materials, Concordia University, Montreal H4B 1R6, Canada
First published on 25th November 2025
Abstract
Efficient delivery of therapeutics remains a significant challenge, often hindered by solubility barriers encountered within aqueous biological environments. Carbon dots (CDs) offer a promising solution for drug delivery due to their aqueous dispersibility, generally low cytotoxicity and surfaces rich in functional groups which can enable facile conjugation of target payloads. However, traditional CD conjugation methods typically rely on robust covalent amide bonds that are difficult to cleave and require the use of hazardous coupling reagents. As an alternative covalent linkage, the solid-state mechanochemical formation of covalent imine bonds on the surfaces of CDs was achieved in this work. Vanillin (VAN), a model aldehyde molecule, was mechanochemically conjugated to the surface of CDs without the need for a solvent, or a catalyst. 1H-NMR analysis confirmed successful formation of an imine bond linkage due to the appearance of an imine proton at 8.1 ppm. Deconvolution of the N 1s spectrum revealed an increase in the relative N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C area at 398.5 eV from 38.9% to 61.9%, confirming the formation of new imine bonds. A drug loading capacity (DLC) of 5.02% was achieved and the pH-responsive drug release profiles were similar across all tested pH levels (5, 6, and 7.4) with release occurring over a period of 24 hours before reaching a plateau. In vitro cell viability assays showed that the CDs remained above 90% cell viability after 72 hours and exhibited low cytotoxicity in A549 model lung cancer cells, before and after mechanochemical conjugation with VAN.
C area at 398.5 eV from 38.9% to 61.9%, confirming the formation of new imine bonds. A drug loading capacity (DLC) of 5.02% was achieved and the pH-responsive drug release profiles were similar across all tested pH levels (5, 6, and 7.4) with release occurring over a period of 24 hours before reaching a plateau. In vitro cell viability assays showed that the CDs remained above 90% cell viability after 72 hours and exhibited low cytotoxicity in A549 model lung cancer cells, before and after mechanochemical conjugation with VAN.
1. Introduction
Nanomaterials have garnered significant attention over the last two decades, owing to their immense potential impact in the fields of chemistry, biology and physics.1–3 Over the past decade, nanomaterial-based targeted drug delivery has come to light.4,5 This interest stems from the materials’ ability to enhance the targeted and controlled release of therapeutic molecules, improvement of the circulation time and bioavailability, as well as an enhancement of the permeability across biological membranes.6–17 To date, a wide range of nanomaterials have been studied in drug delivery applications including inorganic (e.g. quantum dots and lanthanide-doped nanoparticles), polymeric (e.g. polyethylene glycol and polyethyleneimine nanoparticles), nanogels (e.g. chitosan and alginate nanogels) and lipid-based (e.g. liposomes and solid lipid nanoparticles) nanomaterials.18–25 While many show remarkable properties, some suffer from drawbacks such as poor cellular compatibility and uptake, requirements for post-synthetic surface modification to render them hydrophilic, as well as the lack of intrinsic traceable markers, to name a few examples. It is in this regard that carbon dots (CDs) offer a promising alternative, as they are readily dispersible in aqueous media, possess low cytotoxicity and exhibit intrinsic fluorescence with remarkable fluorescence quantum yields.26–35 CDs are described as quasi-spherical amorphous nanomaterials (∼10 nm) with a structure mostly comprising an sp2 and sp3 carbon framework with a surface rich in heteroatoms decorated with functional groups such as carboxylic acids, amines, amides, ketones and alcohols.36–43 These groups can be easily functionalized with a myriad of molecules, peptides or polymers of interest, making them valuable drug delivery vehicles.Current medical literature reports that over 40% of all drugs in clinical use are hydrophobic, and 90% or more of newly developed drugs are described as Class II (high permeability, low solubility), or Class IV (low permeability, low solubility).44 This inherent hydrophobicity often results in lower bioavailabilities due to limited dissolution of a drug within the aqueous bloodstreams. CDs, as hydrophilic drug delivery vehicles, offer a promising solution, however, some challenges persist. Most notably, functionalization of CDs typically relies on conjugating therapeutic agents through robust covalent amide linkages, which are highly stable, but difficult to cleave under physiological conditions, limiting release of the surface functionalized molecule and potentially affecting the pharmacophore.45 Additionally, amide bond formation often requires the need for expensive and hazardous coupling agents such as 1-ethyl-3-(3-dimethylaminoporpyl)carbodiimide (EDC), which is classified as toxic in contact with skin and very toxic to aquatic life with long lasting effects according to the safety data sheet.46,47 It is in this regard that imine bonds have been shown to be great alternatives, as they are covalent linkages which can form without the need for coupling agents and can cleave in response to slight changes in physiological pH.48–50 Acid labile bonds are often desirable, as nanocarriers are commonly uptaken through endocytosis and experience changes in pH ranging from a pH of 7.4 (blood stream) to a pH of 6.0 (early endosome) and finally a pH of 5.0 (late endosome/lysosome).51
It has been shown that mechanochemistry is an attractive approach to imine bond formation.52 The mechanochemical approach is particularly interesting because it avoids the use of solvents, which can sometimes lead to solubility issues during the conjugation procedure. Mechanochemistry involves the use of mechanical energy, typically in the form of grinding or milling in order to drive reactions forward.53 This field of chemistry has gained significant traction in recent years due to its ability to minimize or eliminate the use of solvents and thus overcoming any solubility barriers and rendering syntheses more unstainable in the process.54,55 Mechanochemistry can also significantly reduce reaction times, and avoids the use of harsh catalysts and toxic solvents. While solution-based approaches have been successful in the formation of imine bonds employing CDs, the use of mechanochemistry to form imine bonds on the surfaces of CDs remains unexplored, to our knowledge.56
Herein, mechanochemistry was leveraged to efficiently attach vanillin (VAN), a model drug to the surfaces of nitrogen-doped CDs, forming stable yet cleavable imine bonds. Furthermore, the mechanochemical functionalization process, kinetics of drug release, and the potential in vitro effects of this novel functionalization strategy were also explored. The undertaken approach in this work can translate beyond the conjugation of a model molecule to the conjugation of drugs containing aldehyde functional groups such as the antiviral medication Bofutrelvir, potentially improving therapeutic efficacy.57 The mechanochemical approach to CD surface conjugation provides a promising solution to challenges such as solubility mismatches between therapeutic molecules and CDs, as it eliminates the need for solvents.58 Furthermore, conjugation times may also be improved which can otherwise last upwards of 24 hours. This approach can accelerate the translation of CD systems beyond proof-of-concept studies.
2. Materials and methods
2.1. Chemicals and reagents
Citric acid, pentaethylenehexamine (PH6), vanillin, methanol, chloroform, methanol-d4 and methanol-d4 (0.03% v/v TMS) were all purchased from Sigma Aldrich. Acetone was purchased from Caledon Laboratories Ltd. Sodium phosphate dibasic heptahydrate and uranyl acetate were purchased from Fisher Scientific. 3.5–5 kDa regenerated cellulose membranes were purchased from Cole-Parmer Canada. Milli-Q water was produced in-house, and Dulbecco modified eagle medium (DMEM, 319-065 CL) and Fetal bovine serum (Heat Inactivated, US origin, cat. no. 080-150) and Phosphate Buffered Saline 10× (311-410-CL) were purchased from Wisent Bioproducts. Sterile 96 well culture plates were obtained from Sarstedt (cat. no. 83.3924.005).2.2. Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. After centrifugation the supernatant is discarded and acetone is reintroduced, repeating the process for a total of three times. The pelleted, purified CDs were dispersed in water, frozen at −80 °C and then lyophilized overnight. The solid material was then crushed into a fine powder using a mortar and pestle resulting in purified PH6-CDs with an approximate yield of 10% (Fig. 1).
000g for 10 min. After centrifugation the supernatant is discarded and acetone is reintroduced, repeating the process for a total of three times. The pelleted, purified CDs were dispersed in water, frozen at −80 °C and then lyophilized overnight. The solid material was then crushed into a fine powder using a mortar and pestle resulting in purified PH6-CDs with an approximate yield of 10% (Fig. 1).
2.3. Characterization
3. Results and discussion
Nitrogen doped pentaethylenehexamine (PH6)-CDs, known for their high fluorescence quantum yield (20%), are efficiently uptaken in cells and show no signs of toxicity in HeLa & HFF cells beyond 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 µg mL−1, which is desirable for biological applications and, more specifically, for drug delivery.27 Additionally these CDs have been shown to localize to lysosomes and are uptaken primarily through endocytosis.27 PH6-CDs have many amine groups decorating their surface, which are necessary for imine bond formation, a bond typically chosen for drug delivery due to its dynamic nature.59 These CDs were synthesized via a microwave-assisted reaction using citric acid as a carbon source and PH6 as a nitrogen source.
000 µg mL−1, which is desirable for biological applications and, more specifically, for drug delivery.27 Additionally these CDs have been shown to localize to lysosomes and are uptaken primarily through endocytosis.27 PH6-CDs have many amine groups decorating their surface, which are necessary for imine bond formation, a bond typically chosen for drug delivery due to its dynamic nature.59 These CDs were synthesized via a microwave-assisted reaction using citric acid as a carbon source and PH6 as a nitrogen source.
The CDs were first characterized to assess their physicochemical and optical properties. UV-Vis and Fluorescence spectroscopy were first carried out to determine if a material with its own unique optical signatures is produced in comparison to the starting materials. The absorption spectrum of PH6-CDs depicts a π → π* transition centered at 240 nm and an n → π* transition centered at 350 nm, which are ascribed to the sp2 domains found within the structure of the CDs, and the carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) functional groups decorating the surfaces, respectively (Fig. 2a). The bands are clearly different than those present in the precursors, providing strong evidence of CD formation. A characteristic blue fluorescence27 with an emission maximum centred at 450 nm was observed following 350 nm excitation, as shown in Fig. 2b. Once again, this fluorescence is different than both precursors, with citric acid and pentaethylenehexamine having no intrinsic fluorescence.
O) functional groups decorating the surfaces, respectively (Fig. 2a). The bands are clearly different than those present in the precursors, providing strong evidence of CD formation. A characteristic blue fluorescence27 with an emission maximum centred at 450 nm was observed following 350 nm excitation, as shown in Fig. 2b. Once again, this fluorescence is different than both precursors, with citric acid and pentaethylenehexamine having no intrinsic fluorescence.
PXRD analysis was carried out do determine if the synthesized material depicts any Bragg's reflections; the lack of reflections would point towards the formation of an amorphous material rather than a crystalline material. Crystallinity of the CDs was assessed using PXRD and they were found to be amorphous in nature as demonstrated by an amorphous halo observed from 10–30° 2θ (Fig. 2c), which is consistent with current CD literature.60,61 Notably, the citric acid precursor shows clear Braggs peaks indicating the shift from a crystalline precursor to an amorphous CD. The crystallinity of the PH6 precursors could not be assessed as it is a liquid. TEM analysis (Fig. S1) was also carried out to determine the size and morphology of the dots. The CDs were quasi spherical in shape with a particle size of 2.5 ± 0.7 nm and assize distribution spanning 1–4 nm.
FT-IR was carried out to determine the chemical composition of the synthesized material. This analysis allows for a direct comparison to the chemical composition of starting materials. FT-IR analysis (Fig. 2d) shows that the surface of the CDs is rich in amine and hydroxyl groups (3268 cm−1), sp3 alkane groups (2938 cm−1, 2821 cm−1), carbonyl functional groups originating from ketones, or acids (1695 cm−1), carbonyl groups originating from amide functionalities (1645 cm−1) and alkene groups (1540 cm−1). Citric acid also shows similar carboxylic acid vibrations, particularly the hydroxyl stretching vibration at 3276 cm−1 and the associated carbonyl stretching vibration at 1691 cm−1. PH6 shows clear amine vibrations with an N–H stretch at 3273 cm−1 and the corresponding bending vibration at 1601 cm−1. All stretches and bends are in accordance with other similar CD systems discussed in the literature.62,63 These results highlight that the functional groups present in the precursors translate effectively to the functional groups found on the surfaces of the CDs while also showing some expected differences when undergoing the transformation to form a nanomaterial.
Lastly, important functionalities relating to the different proton environments of the CDs were also assessed using 1H-NMR spectroscopy (Fig. 2e). 1H-NMR was carried out to support the findings of FT-IR and ensure that the functional groups of the CDs are unique and comparable to that of the precursors. Protons relating to sp2 domains were confirmed following the presence of a multiplet spanning 5.5–6.5 ppm. Many signals can be observed spanning the region of 2.5–4.25 ppm, depicting the presence of protons directly bonded to electronegative atoms such as nitrogen (amine groups), or protons bonded to carbon atoms, which in turn are bonded to electronegative functionalities (amide groups). Many multiplets can also be observed from 0–2 ppm, which are representative of protons bonded to carbon atoms that are not neighbouring any electronegative functionalities. The 1H-NMR spectrum of citric acid clearly shows a quartet at 3 ppm related to the proton attached to the alpha carbon. The 1H-NMR spectrum of PH6 also evidences a multiplet at 3 ppm ascribed to the protons of the central alkyl groups. It's interesting to note that the 1H-NMR spectrum of the CDs in this region is drastically more complex compared to the precursors, indicating the formation of a higher order structure with a surface likely decorated by many different types of carboxylic acid and amine bearing functional groups.
VAN, a model aldehyde molecule exhibiting therapeutic effects, was conjugated to PH6-CDs utilizing mechanochemistry as a novel approach for the formation of this type of linkage. Conjugation of VAN to PH6-CDs was first assessed by FT-IR to evaluate if a change in chemical composition can be observed. The most intense stretching vibration of VAN is that of the aldehyde functional group at 1658 cm−1 which seems to be absent post-conjugation, as observed in the FT-IR spectrum of VAN-CDs, suggesting conversion of the aldehyde group into an imine bond (Fig. S2). However, the FT-IR spectra before and after conjugation are quite similar and therefore drawing a conclusion solely from this technique would not be possible. 1H-NMR was then carried out to further assess the formation of imine bonds and evaluate if a change in the 1H-NMR spectrum can be observed. A singlet associated with the aldehyde proton of VAN can be seen downfield at 9.80 ppm and is no longer present after conjugation, providing strong evidence for its transformation (Fig. S3). Focusing on the downfield region, it is quite clear that a new broad singlet is observed at 8.10 ppm, associated with the proton of a newly-formed imine bond between VAN and the CDs (Fig. 3a) A newly-formed multiplet spanning from 6.50–7.50 ppm can also be observed due to the aromatic protons of VAN. It is important to note that the chemical shifts of these aromatic protons are also shifted upfield in comparison to native VAN indicating formation of a covalent bond. The broad nature of the singlet and multiplet associated with the imine and aromatic protons, respectively, is also important. Every primary amine on the surfaces of PH6-CDs may exhibit different local chemical environments resulting in the formation of imine bonds, which are slightly chemically different and thus broad signals are observed rather than the highly resolved aromatic protons of native VAN. It should be noted that PH6-CDs do exhibit a very weak and broad singlet at 8.1 ppm prior to conjugation due to the fact that some imine content is already incorporated into the structure of the CDs following their synthesis. A control spectrum of only ball milled CDs (Fig. 3b) was also obtained to ensure that the observed chemical shifts do not arise from a reaction only involving CDs. A broad singlet at 8.10 ppm is observed with no aromatic protons, similar to native PH6-CDs, indicating that the CDs do not react with each other during ball milling. A second control involving ball milling VAN was also carried out and we note that no differences were observed relative to native VAN (Fig. 3b).
XPS was also carried out to assess the elemental composition of CDs. Deconvolution of the XPS spectra would allow for the direct observation in changes to surface functional groups confirming the formation of imine bonds (Fig. 4). The survey scan of PH6-CDs (Fig. 4a) shows that the surfaces are mostly composed of carbon (67.8%), nitrogen (19.0%) and oxygen (12.4%). These same elements are also present in similar proportions (Fig. 4b) following drug loading due to elemental similarities between PH6-CDs and VAN.
In order to observe differences between PH6-CDs and VAN-CDs, high resolution scans of the three main elements (C, N and O) were carried out. The deconvoluted high resolution C 1s spectrum of PH6-CDs shows two main binding energies at around 285 and 287.5 eV respectively (Fig. 4c). These binding energies can be associated with C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds found on the surface of the dots.62 Similar relative areas of both these binding energies is also shown in the deconvoluted high resolution C 1s spectrum of VAN-CDs (Fig. 4f), confirming that they do not play a role in the conjugation of VAN. The deconvoluted high resolution N 1s spectrum of PH6-CDs shows two main binding energies at around 398.5 and 399.5 eV respectively (Fig. 4d) and can be attributed to the presence of N
O bonds found on the surface of the dots.62 Similar relative areas of both these binding energies is also shown in the deconvoluted high resolution C 1s spectrum of VAN-CDs (Fig. 4f), confirming that they do not play a role in the conjugation of VAN. The deconvoluted high resolution N 1s spectrum of PH6-CDs shows two main binding energies at around 398.5 and 399.5 eV respectively (Fig. 4d) and can be attributed to the presence of N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and N–C bonds on the surface.62,64 The relative areas under these peaks are 38.9% and 61.1%, respectively. Following conjugation with VAN (Fig. 4g), a notable shift in these proportions is observed, with the relative areas changing to 61.9% (N
C and N–C bonds on the surface.62,64 The relative areas under these peaks are 38.9% and 61.1%, respectively. Following conjugation with VAN (Fig. 4g), a notable shift in these proportions is observed, with the relative areas changing to 61.9% (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) and 38.1% (N–C). This inversion indicates a transformation of amine (C–N) groups into imine (C
C) and 38.1% (N–C). This inversion indicates a transformation of amine (C–N) groups into imine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) groups as a result of mechanochemical conjugation with VAN. The deconvoluted high resolution O 1s spectrum of PH6-CDs shows two main binding energies at around 530.5 and 532 eV owing to the presence of C
N) groups as a result of mechanochemical conjugation with VAN. The deconvoluted high resolution O 1s spectrum of PH6-CDs shows two main binding energies at around 530.5 and 532 eV owing to the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds originating from amide and carboxylic acid functional groups respectively.62 Once again the relative area of both these binding energies are very similar in the deconvoluted high resolution O 1s spectrum of VAN-CDs (Fig. 4h) confirming that they do not play a role in the conjugation of VAN and further highlighting that amine groups are the only affected functional groups during the conjugation process.
O bonds originating from amide and carboxylic acid functional groups respectively.62 Once again the relative area of both these binding energies are very similar in the deconvoluted high resolution O 1s spectrum of VAN-CDs (Fig. 4h) confirming that they do not play a role in the conjugation of VAN and further highlighting that amine groups are the only affected functional groups during the conjugation process.
To confirm that VAN was bonding to CDs as individual molecules through imine bonds rather than electrostatically associating to the surfaces as crystals PXRD was carried out. A loss of crystallinity was observed as VAN is conjugated to the CD surface (Fig. S4). This demonstrates that individual molecules of VAN are taken apart from the bulk crystal during the conjugation process facilitated by the mechanochemical reaction between the aldehyde group of VAN and the amines on the surfaces of PH6-CDs.
After fully characterizing VAN-CDs, the drug loading capacity (DLC) was investigated. Quantitative NMR (qNMR) analysis was carried out (Fig. 5a) as there is clear resolution between protons arising from the conjugated drug and the native CDs. qNMR is a well-established method for determining the DLC of different vehicles.65 The protons attributed to VAN and TMS were integrated and a DLC of 5.02% was determined according to the following formulas.66,67
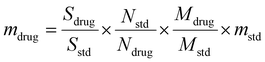 | (1) |
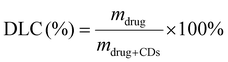 | (2) |
Drug release experiments were conducted at 37 °C at different pH to simulate naturally occurring cellular environments. Prior to carrying out the release studies, a standard curve was generated according to the absorbance readings at a λmax of 275 nm for different concentrations of VAN (Fig. S4). The obtained calibration equation corresponds to a linear trendline confirmed by an r2 value >0.99. Cumulative release was observed at pH 5.0, 6.0 and 7.4 with an increasing trend over a period of 24 hours until a plateau was eventually reached (Fig. 5b). We observed that the successful loading and release was indeed achieved. The release occurred over several hours, which can be advantageous in terms of dose reduction since a longer release period can allow for drug concentrations to be within the therapeutic concentration range for a longer period of time, reducing the frequency of administration. Moreover, drug release from an imine covalent bond offers an alternate covalent linkage in comparison to covalent amide bonds which are commonly used and difficult to cleave under physiological conditions. Effect of pH on drug release was also investigated and showed no difference in release profile. Imine bonds, known for their sensitivity to acidic environments are highly influenced by their local chemical environment.68 CDs as complex materials have many functional groups on their surfaces, many of which are electron withdrawing, potentially destabilizing local imine bonds and allowing for hydrolysis at a neutral pH. Nonetheless, insensitivity to pH can be advantageous for drug delivery applications such as wound healing where pH fluctuations are common between healthy and damaged/infected tissue.69 A drug delivery system whose release profile does not change in relation to pH allows for consistent and predictable release, leading to greater efficiency in wound treatment. This finding confirms the aqueous cleavage of imine bonds.
Lastly, In Vitro studies were conducted to investigate if the mechanochemical procedure induces any cytotoxicity (Fig. 5c). Human A549 lung cancer cells were used as a model system due to fast replication times, facile culture conditions and accessibility. Furthermore, VAN does not induce cytotoxicity in A549 cells as opposed to other cell lines.70 This ensures that any observable cytotoxicity stems solely from the mechanochemical surface modification of PH6-CDs rather than from VAN itself. In all cases, 50 µg CDs were administered, corresponding to the upper limit of the most common dosage range (10–60 µg), ensuring that a lack of toxicity is not due to minimal dosing.71,72 A 72 hours incubation time was used, corresponding to three cell division cycles, again ensuring any adverse effects would be observed beyond a single mitotic cycle. As expected, the metabolic activity observed for all treatment conditions indicates negligible acute cytotoxicity. It is worth mentioning that treatment with VAN-CDs did result in a small but statistically significant decrease in metabolic activity to 91.8%. In-depth mechanistic studies are required to elucidate the precise cause for this small but significant difference in viability. Regardless, metabolic activity is still well above the commonly accepted cytotoxicity thresholds of 70%.73,74 As such we can conclude that the mechanochemical conjugation process does not render the CDs cytotoxic, verifying the viability of this new approach.
4. Conclusions
In conclusion, our findings have brought forward a novel approach for functionalizing therapeutic agents onto the surfaces of hydrophilic CDs via a mechanochemically assisted route. This approach can allow for the formation of new covalent bonds without the need for expensive or toxic coupling agents, furthermore, this approach circumvents solubility challenges, which can be encountered during traditional solution-based conjugation methods. Our approach also introduces a sustainable, solvent-free method that maintains many of the desirable properties of CDs, such as low cytotoxicity and intrinsic fluorescence, while enhancing their scope as nanocarriers. It also shows that it is possible to have successful drug conjugation through covalent imine bonds with a DLC of 5.02%. Given the lack of dependence on pH, the delivery system can be advantageous for applications in drug delivery such as wound healing. The lack of cytotoxicity in A549 cells before and after mechanochemical conjugation confirms that our approach is safe and effective in terms of development of a drug delivery vehicle. Ultimately, our findings not only broaden the applicability of CDs but also lay the groundwork for greener drug conjugation strategies in nanomedicine, paving the way for the expanded use of CD-based delivery systems across a wide range of therapeutic agents. Future research could focus on optimizing the stability of surface imine bonds by tailoring the types and ratios of CD precursors to modify the surface chemistry. The goal would be to render the formed imine bonds stable at a neutral pH and labile in slightly acidic environments for controlled and targeted drug release applications, enhancing the efficacy and versatility of this delivery system.Author contributions
G. F. and R. N.; methodology, G. F., G. A. M.; software, G. F. and G. A. M.; validation, G. F., G. A. M., R. N.; formal analysis, G. F., G. A. M., C. M.; investigation, G. F., G. A. M., C. M.; resources, R. N., J. A. C.; data curation, G. F., G. A. M., C. M.; writing—original draft preparation, G. F.; writing—review and editing, R. N., G. A. M., J. A. C.; visualization, G. F.; supervision, R. N.; project administration, G. F. and R. N.; funding acquisition, R. N., J. A. C. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflicts of interest regarding publication of this research.Data availability
The data presented is contained within the article or supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5nr03355a.Any additional information concerning this body of work are available upon request from the corresponding author.
Acknowledgements
The authors would like to acknowledge funding sources for financial support for this research. RN and JAC are grateful to NSERC for funding the Discovery program and the Quebec Centre for Advanced Materials for financial support. RN also acknowledges Concordia University for financial support through the Concordia University Research Chair Program. GF acknowledges the NSERC CGS-M program while GM is thankful to the NSERC PDF program for financial support. The authors are also grateful to Mr Luis Alfonso Páramo Serrano for their assistance with TEM analyses and Mrs Victoria Lapointe for their assistance with XRD analyses.References
- G. A. Ozin and A. Arsenault, Nanochemistry: a chemical approach to nanomaterials, Royal Society of Chemistry, 2015 Search PubMed.
- P. Bergese and K. Hamad-Schifferli, Nanomaterial Interfaces in Biology, Springer, 2013, vol. 1025 Search PubMed.
- J. Yao, Y. Sun, M. Yang and Y. Duan, J. Mater. Chem., 2012, 22, 14313–14329 RSC.
- X. Cheng, Q. Xie and Y. Sun, Front. Bioeng. Biotechnol., 2023, 11, 1177151 CrossRef PubMed.
- B. Dutta, K. C. Barick and P. A. Hassan, Adv. Colloid Interface Sci., 2021, 296, 102509 CrossRef CAS PubMed.
- Q. Wang, X. Huang, Y. Long, X. Wang, H. Zhang, R. Zhu, L. Liang, P. Teng and H. Zheng, Carbon, 2013, 59, 192–199 CrossRef CAS.
- Q. Zeng, D. Shao, X. He, Z. Ren, W. Ji, C. Shan, S. Qu, J. Li, L. Chen and Q. Li, J. Mater. Chem. B, 2016, 4, 5119–5126 RSC.
- G. Calabrese, G. De Luca, G. Nocito, M. G. Rizzo, S. P. Lombardo, G. Chisari, S. Forte, E. L. Sciuto and S. Conoci, Int. J. Mol. Sci., 2021, 22, 11783 CrossRef CAS PubMed.
- W. Li, Y. Ma, L. Ou, C. Xu, Y. Wei, K. Yang and B. Yuan, J. Hazard. Mater., 2024, 465, 133382 CrossRef CAS PubMed.
- C. Kong, K. Wang, L. Sun, H. Zhao, T. Wang, W. Zhou, D. Wu and F. Xu, Int. J. Nanomed., 2024, 3611–3622 CrossRef CAS PubMed.
- D. M. Arvapalli, A. T. Sheardy, K. Allado, H. Chevva, Z. Yin and J. Wei, ACS Appl. Bio Mater., 2020, 3, 8776–8785 CrossRef CAS PubMed.
- W. Luo, L. Zhang, X. Li, J. Zheng, Q. Chen, Z. Yang, M. Cheng, Y. Chen, Y. Wu and W. Zhang, Nano Res., 2022, 15, 9274–9285 CrossRef CAS.
- J. Li, Q. Li, Q. Yang, Q. Tang, X. Hu, Q. Liu and L. Zhang, Mater. Today Nano, 2024, 100532 CrossRef CAS.
- H. Zhao, J. Duan, Y. Xiao, G. Tang, C. Wu, Y. Zhang, Z. Liu and W. Xue, Chem. Mater., 2018, 30, 3438–3453 CrossRef CAS.
- T. Feng, X. Ai, G. An, P. Yang and Y. Zhao, ACS Nano, 2016, 10, 4410–4420 CrossRef CAS PubMed.
- F. Yan, Y. Jiang, X. Sun, Z. Bai, Y. Zhang and X. Zhou, Microchim. Acta, 2018, 185, 1–34 CrossRef CAS PubMed.
- Z. Wang, Z. Wang and F. Wu, ChemMedChem, 2022, 17, e202200003 CrossRef CAS PubMed.
- C. T. Matea, T. Mocan, F. Tabaran, T. Pop, O. Mosteanu, C. Puia, C. Iancu and L. Mocan, Int. J. Nanomed., 2017, 5421–5431 CrossRef CAS.
- A. Bagheri, H. Arandiyan, C. Boyer and M. Lim, Adv. Sci., 2016, 3, 1500437 CrossRef PubMed.
- X. Wang, D. Niu, C. Hu and P. Li, Curr. Pharm. Des., 2015, 21, 6140–6156 CrossRef CAS PubMed.
- Y. Tian, Z. Gao, N. Wang, M. Hu, Y. Ju, Q. Li, F. Caruso, J. Hao and J. Cui, J. Am. Chem. Soc., 2022, 144, 18419–18428 CrossRef CAS PubMed.
- L. He, Z. Shang, H. Liu and Z. Yuan, BioMed Res. Int., 2020, 2020, 1487259 CrossRef PubMed.
- L. Xing, Y.-T. Fan, L.-J. Shen, C.-X. Yang, X.-Y. Liu, Y.-N. Ma, L.-Y. Qi, K.-H. Cho, C.-S. Cho and H.-L. Jiang, Int. J. Biol. Macromol., 2019, 141, 85–97 CrossRef CAS PubMed.
- A. Samad, Y. Sultana and M. Aqil, Curr. Drug Delivery, 2007, 4, 297–305 CrossRef CAS.
- R. H. Müller, K. Mäder and S. Gohla, Eur. J. Pharm. Biopharm., 2000, 50, 161–177 CrossRef PubMed.
- F. Yarur, J. R. Macairan and R. Naccache, Environ. Sci.: Nano, 2019, 6, 1121–1130 RSC.
- A. Clermont-Paquette, K. Larocque, A. Piekny and R. Naccache, Mater. Adv., 2024, 5, 3662–3674 RSC.
- Y.-Y. Yao, G. Gedda, W. M. Girma, C.-L. Yen, Y.-C. Ling and J.-Y. Chang, ACS Appl. Mater. Interfaces, 2017, 9, 13887–13899 CrossRef CAS PubMed.
- Z. Wang, H. Liao, H. Wu, B. Wang, H. Zhao and M. Tan, Anal. Methods, 2015, 7, 8911–8917 RSC.
- T. Feng, X. Ai, H. Ong and Y. Zhao, ACS Appl. Mater. Interfaces, 2016, 8, 18732–18740 CrossRef CAS PubMed.
- J. Qi, R. Zhang, X. Liu, Y. Liu, Q. Zhang, H. Cheng, R. Li, L. Wang, X. Wu and B. Li, ACS Appl. Nano Mater., 2023, 6, 9071–9084 CrossRef CAS.
- T. Feng, X. Ai, G. An, P. Yang and Y. Zhao, ACS Nano, 2016, 10, 4410–4420 CrossRef CAS PubMed.
- W. Gao, S. Xiang, M. Bai, Y. Ruan, J. Zheng, X. Cao, Y. Xu, Y. Chen and W. Weng, Polymer, 2022, 257, 125278 CrossRef CAS.
- M. Maruthapandi, A. Saravanan, P. Das, J. H. T. Luong and A. Gedanken, Biotechnol. Adv., 2021, 53, 107843 CrossRef CAS PubMed.
- L. Jiang, W. Cao, Z. Li, C. Wang and H. Bi, Small, 2024, 20, 2402827 CrossRef CAS PubMed.
- S. Chahal, J. R. Macairan, N. Yousefi, N. Tufenkji and R. Naccache, RSC Adv., 2021, 11, 25354–25363 RSC.
- F. Yan, Y. Jiang, X. Sun, Z. Bai, Y. Zhang and X. Zhou, Microchim. Acta, 2018, 185, 1–34 CrossRef CAS PubMed.
- K. Dimos, Curr. Org. Chem., 2016, 20, 682–695 CrossRef CAS.
- K. J. Mintz, M. Bartoli, M. Rovere, Y. Zhou, S. D. Hettiarachchi, S. Paudyal, J. Chen, J. B. Domena, P. Y. Liyanage and R. Sampson, Carbon, 2021, 173, 433–447 CrossRef CAS.
- S. Li, L. Li, H. Tu, H. Zhang, D. S. Silvester, C. E. Banks, G. Zou, H. Hou and X. Ji, Mater. Today, 2021, 51, 188–207 CrossRef CAS.
- N. Papaioannou, M.-M. Titirici and A. Sapelkin, ACS Omega, 2019, 4, 21658–21665 CrossRef CAS PubMed.
- T. V. de Medeiros, J. Manioudakis, F. Noun, J.-R. Macairan, F. Victoria and R. Naccache, J. Mater. Chem. C, 2019, 7, 7175–7195 RSC.
- H. Shabbir, E. Csapó and M. Wojnicki, Inorganics, 2023, 11, 262 CrossRef CAS.
- C. Hogarth, K. Arnold, A. McLauchlin, S. P. Rannard, M. Siccardi and T. O. McDonald, J. Mater. Chem. B, 2021, 9, 9874–9884 RSC.
- S. D. Hettiarachchi, E. K. Cilingir, H. Maklouf, E. S. Seven, S. Paudyal, S. Vanni, R. M. Graham and R. M. Leblanc, Nanoscale, 2021, 13, 5507–5518 RSC.
- 1-Ethyl-3-(3-Dimethylaminopropyl)carbodiimide, Hydrochloride, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/E2247, (accessed 23 April 2025).
- 1-Ethyl-3-(3-Dimethylaminopropyl)carbodiimide, Hydrochloride, SDS Cat No. E2247, Thermo Fisher Scientific, https://assets.thermofisher.com/TFS-Assets/LSG/SDS/E2247_MTR-NALT_EN.pdf, (accessed 23 April 2025).
- X. Qu and Z. Yang, Chem. – Asian J., 2016, 11, 2633–2641 CrossRef CAS PubMed.
- Y. Tao, S. Liu, Y. Zhang, Z. Chi and J. Xu, Polym. Chem., 2018, 9, 878–884 RSC.
- H. Ding, P. Tan, S. Fu, X. Tian, H. Zhang, X. Ma, Z. Gu and K. Luo, J. Controlled Release, 2022, 348, 206–238 CrossRef CAS PubMed.
- C. C. Scott, F. Vacca and J. Gruenberg, in Seminars in cell & developmental biology, Elsevier, 2014, vol. 31, pp. 2–10 Search PubMed.
- S. Han, J. Lee, E. Jung, S. Park, A. Sagawa, Y. Shibasaki, D. Lee and B.-S. Kim, ACS Appl. Bio Mater., 2021, 4, 2465–2474 CrossRef CAS PubMed.
- S. Pagola, Crystals, 2023, 13, 124 CrossRef CAS.
- J.-L. Do and T. Friščić, ACS Cent. Sci., 2017, 3, 13–19 CrossRef CAS PubMed.
- X. Liu, Y. Li, L. Zeng, X. Li, N. Chen, S. Bai, H. He, Q. Wang and C. Zhang, Adv. Mater., 2022, 34, 2108327 CrossRef CAS PubMed.
- B.-B. Chen, S. Chang, L. Jiang, J. Lv, Y.-T. Gao, Y. Wang, R.-C. Qian, D.-W. Li and M. E. Hafez, J. Colloid Interface Sci., 2022, 621, 464–469 CrossRef CAS PubMed.
- Frontier Biotechnologies, Bofutrelvir, https://adisinsight.springer.com/drugs/800062531, (accessed 11 October 2025).
- A. Saravanan, M. Maruthapandi, P. Das, S. Ganguly, S. Margel, J. H. T. Luong and A. Gedanken, ACS Appl. Bio Mater., 2020, 3, 8023–8031 CrossRef CAS PubMed.
- Y. Tao, S. Liu, Y. Zhang, Z. Chi and J. Xu, Polym. Chem., 2018, 9, 878–884 RSC.
- J. Macairan, I. Zhang, A. Clermont-Paquette, R. Naccache and D. Maysinger, Part. Part. Syst. Charact., 2020, 37, 1900430 CrossRef CAS.
- P. Das, S. Ganguly, M. Bose, S. Mondal, S. Choudhary, S. Gangopadhyay, A. K. Das, S. Banerjee and N. C. Das, Mater. Sci. Eng., C, 2018, 88, 115–129 CrossRef CAS PubMed.
- J. Manioudakis, F. Victoria, C. A. Thompson, L. Brown, M. Movsum, R. Lucifero and R. Naccache, J. Mater. Chem. C, 2019, 7, 853–862 RSC.
- T. V. de Medeiros, A. Macina, J. P. de Mesquita and R. Naccache, RSC Appl. Interfaces, 2024, 1, 86–97 RSC.
- E. Gala, G. C. D. Bandomo, M. Vettori, S. Royuela, M. Martínez-Fernández, J. I. Martínez, E. Salagre, E. G. Michel, F. Zamora and J. Lloret-Fillol, J. Mater. Chem. A, 2025, 13, 1142–1152 RSC.
- D. Guimarães, J. Noro, A. Loureiro, A. Cavaco-Paulo and E. Nogueira, Colloids Surf., B, 2019, 179, 414–420 CrossRef PubMed.
- W. Xin, W. Yumei, G. Xianmou, G. Kaijun, L. Jing, L. Sanming, Z. Lin and H. Li, Int. J. Nanomed., 2020, 15, 7451–7468 CrossRef CAS PubMed.
- T. Miura, N. Sugimoto, S. Bhavaraju, T. Yamazaki, Y. Nishizaki, Y. Liu, A. Bzhelyansky, C. Amezcua, J. Ray and E. Zailer, Chem. Pharm. Bull., 2020, 68, 868–878 CrossRef CAS PubMed.
- G. Ahmad, N. Rasool, K. Rizwan, A. A. Altaf, U. Rashid, M. Z. Hussein, T. Mahmood and K. Ayub, Molecules, 2019, 24, 2609 CrossRef CAS PubMed.
- S. Saghazadeh, C. Rinoldi, M. Schot, S. S. Kashaf, F. Sharifi, E. Jalilian, K. Nuutila, G. Giatsidis, P. Mostafalu and H. Derakhshandeh, Adv. Drug Delivery Rev., 2018, 127, 138–166 CrossRef CAS PubMed.
- K. Lirdprapamongkol, H. Sakurai, S. Suzuki, K. Koizumi, O. Prangsaengtong, A. Viriyaroj, S. Ruchirawat, J. Svasti and I. Saiki, In Vivo, 2010, 24, 501–506 CAS.
- J. Ortenzio, L. Degn, A. Goldstein-Plesser, J. K. McGee, J. Navratilova, K. Rogers, R. M. Zucker and W. K. Boyes, NanoImpact, 2019, 14, 100156 CrossRef PubMed.
- H. Sahin, O. Yucel, P. Holloway, E. Yildirim, S. Emik, G. Gurdag, G. Tanriverdi and G. Erkanli Senturk, Pharmaceuticals, 2024, 17, 1567 CrossRef CAS PubMed.
- A. Azqueta, H. Stopper, B. Zegura, M. Dusinska and P. Møller, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2022, 881, 503520 CrossRef CAS PubMed.
- M. R. Romano, C. Gatto, L. Giurgola, E. Ragazzi and J. D. Tóthová, Transl. Vis. Sci. Technol., 2021, 10, 24 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2025 |





