Open Access Article
Open Access ArticleType-dependent effects of nanoplastics on microglial activation and CXCR2-mediated chemotactic migration†
Jahong
Koo‡
ab,
Bohyeon
Jeong‡
a,
Jeong Yeob
Baek‡
 a,
Wang Sik
Lee
c,
Jiyoung
Gong
bc,
Subin
Park
ad,
Jiyeon
Hong
a,
Yugyeong
Sim
bc,
Dae Soo
Kim
be,
Sang Ryong
Kim
f,
Jinyoung
Jeong
a,
Wang Sik
Lee
c,
Jiyoung
Gong
bc,
Subin
Park
ad,
Jiyeon
Hong
a,
Yugyeong
Sim
bc,
Dae Soo
Kim
be,
Sang Ryong
Kim
f,
Jinyoung
Jeong
 *bc and
Da Yong
Lee
*bc and
Da Yong
Lee
 *ab
*ab
aBiotherapeutics Translational Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea. E-mail: daylee@kribb.re.kr
bKRIBB School, Korea University of Science and Technology (UST), Daejeon, South Korea
cEnvironmental Disease Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea. E-mail: jyjeong@kribb.re.kr
dDepartment of Biochemistry; Department of Medical Science, Chungnam National University School of Medicine, 266 Munhwa-ro, Jung-gu, Daejeon 35015, Korea
eDepartment of Digital Biotech Innovation center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea
fSchool of Life Sciences, BK21 FOUR KNU Creative BioResearch Group, Kyungpook National University, Daegu 41566, Korea
First published on 27th June 2025
Abstract
As plastic pollution continues to grow in various ecosystems, potential harmful effects of micro- and nanoplastics have become a great concern. Most studies on the biological effects of nanoplastics have been conducted using polystyrene nano- and microplastics. However, the majority of environmental plastic waste consists of a mixture of various types of plastics, such as polypropylene (PP), polyethylene, polystyrene (PS), polyvinyl chloride and polymethyl methacrylate (PMMA). In this study, we compared the biological effects of nanoplastics derived from three different types of plastics (PS, PP and PMMA) on the functions of microglia, which are the predominant immune cells with macrophage-like functions in the brain. Our experiments with cultured primary rat microglia revealed that the cells exposed to PMMA nanoplastic (PMMANP) exhibited the highest M1 phase activity. In addition, we found that PMMANP increased the migration ability of microglia by inducing the expression of chemokines, such as CXCL1 and CXCL2, in vitro and in vivo. These findings suggest that PMMANP-exposed brain microglia may accelerate neurological disorders by enhancing the recruitment of microglia and peripheral immune cells across the blood–brain barrier under neuropathological conditions.
Introduction
Recently, environmental nano- and microplastics (NMPs) have started being considered as potential environmental risk factors, as they are associated with various physiological malfunctions. These include metabolic disruption,1 ROS production,2 and defects in the activity of reproductive organs3 and the brain4 in animal models. Among various organs, the brain is particularly crucial as it regulates the activities of the other organs necessary for sustaining life. In this context, understanding the potential risks of NMPs to brain functions is crucial for assessing their impact on human health. We previously reported that maternal ingestion of polystyrene nanoplastics causes neurodevelopmental abnormalities and cognitive deficit in offspring.5 Similarly, NMPs can also cause defects in learning and memory, as well as various psychiatric abnormalities, such as autistic behaviors, anxiety, and depression-like symptoms.6–8Although the brain is more isolated from the vascular system than other organs due to the blood–brain barrier (BBB), NMP particles in the blood stream can still penetrate the BBB and infiltrate the brain parenchyma under various conditions.9 Once NMP particles infiltrate the brain, they can be recognized by microglia, the primary resident immune cells of the brain, which then initiate phagocytosis and immune responses. In fact, recent studies have shown that polystyrene NMPs significantly increase the permeability of the blood–brain barrier (BBB),10 leading to microglial activation,10,11 enhanced phagocytosis,12 and neuronal damage11 in the mouse brain. However, the molecular mechanisms underlying these effects depend on the size and surface properties of the particles. In addition, previous in vitro studies also showed nanoplastic uptake and M1 activation in microglia after polystyrene nanoplastic exposure.12,13 Microglial activation and the resulting neuroinflammation are closely associated with the pathophysiology of various brain diseases, including neurodegenerative diseases,14 depression,15 and developmental disorders.16 Therefore, NMPs could contribute to the onset or aggravation of brain diseases by enhancing microglial activation, which ultimately leads to the release of various proinflammatory cytokines (IL-1β, IL-6, and TNF-α) and cytotoxic molecules, including nitric oxide, reactive oxygen species and proteases.
Although there is increasing evidence of NMP-induced alterations in microglial functions in vitro and in vivo, most studies have used particles derived from a single type of plastic, particularly, polystyrene. However, the plastic pollutants in the environment consist of particles from various types of plastics, including polyethylene (PE), polypropylene (PP), polystyrene (PS) and polyvinyl chloride, and harmful plastic additives.17,18 Among these, PE, PP and PS are more abundant than the other plastic types in environmental plastic pollutants.19 Polymethyl methacrylate (PMMA) is widely used as a glass replacement in the manufacturing of various commercial lenses, monitors and biomaterials.20 Although environmental PMMA particles are less abundant than those from other plastics, they still have the potential to affect human organs. PMMA is used in biomaterials such as bone cements, contact lenses, screw fixation in bones, fillers for bone cavities and skull defects, and vertebrae stabilization in osteoporotic patients.20 Previous clinical data show that use of PMMA may cause significant neurotoxicity due to thermal damage and chemical toxicity during cranioplasty.21
In this study, we investigated the biological effects of 500 nm diameter nanoplastics derived from three different types of plastics, PS, PP, and PMMA, on brain microglia using cultured rat primary microglia. We found that particles from each type of plastic had differential effects on microglial activation and the expression of chemokines and cytokines. Among these nanoplastics, PMMA nanoplastics (PMMANPs) showed the most pronounced effects on the expression of chemokines, particularly CXCL1 and 2, and their receptor, CXCR2. Finally, our data show that PMMANP-treated microglia exhibited the highest migration ability in vitro and in vivo, suggesting that PMMANP has the greatest potential to recruit microglia and other immune cells under neuropathological conditions, thereby exacerbating neuroinflammation.
Experimental
Nanoplastics
Three different types of nanoplastics were obtained as follows: yellow-green fluorescent and transparent polystyrene nanoplastics (PSNPs; 500 nm) were purchased from Polyscience Inc. (Warrington, PA, USA). Pink fluorescent and transparent polymethyl methacrylate nanoplastics (PMMANPs) were purchased from PolyAn (Berlin, Germany). Polypropylene nanoplastics (PPNPs) were manually prepared as described in a previous study.22 PP pellets (0.13 g, Sigma-Aldrich) were dissolved in 20 mL of xylene by heating at 110 °C for 20 minutes with vigorous stirring. After heating was stopped, ethyl alcohol (100 mL) was gently added to the PP solution, causing precipitates to form. The solution was stirred for 4 hours, and then the precipitates were vacuum filtered using a 0.2 μm PTFE membrane filter and dried at room temperature. The resulting precipitates were collected as a powder and stored until use. All nanoplastics are negatively charged, stabilized colloidal particles with plane surfaces and no surface modification. The biological effects of these nanoplastics on the function of microglia were examined at 0–100 μg ml−1. Tween-20 (0.01%) solution in distilled water, without nanoplastics, was used as the control. Rhodamine B isothiocyanate, an organic fluorescent dye, was conjugated with PPNP to determine its localization.Characterization of nanoplastics
The morphologies and sizes of the three different nanoplastics were measured using a field-emission scanning electron microscope (FE-SEM, Quanta 250 FEG, FEI). The nanoplastics, suspended in aqueous solution, were deposited on a silicon wafer. After drying, the nanoplastics were gold coated using a sputter coater (Polaron SC7640, Quorum Technology, Ltd). SEM images were obtained at an acceleration voltage of 5 kV.Animals
One-day-old Sprague-Dawley (SD) rats were purchased from Daehan Biolink Co. (Eumseong-gun, Korea). All animal experiments complied with the National Institutes of Health Guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) at the Korea Research Institute of Bioscience & Biotechnology (KRIBB) (permit number: KRIBB-AEC-24191).Culture of rat primary cortical microglia
Microglia were cultured from the cerebral cortices as previously described, with modifications.23 Briefly, cerebral cortices were removed from the neonatal rat brain and the tissues were dissociated into single cells using 0.1% trypsin. The cells were plated in 75 cm2 T-flasks (one hemisphere/two flasks) and maintained in a growth medium (DMEM containing 10% FBS and 1% penicillin/streptomycin) (Gibco, Grand island, NY, USA) for 2–3 weeks. Microglia on the surface were then detached by mild shaking and filtered through a 40 μm cell strainer to remove astrocytes and cell clumps. Isolated microglia were plated on 24-well plates (4 × 105 cells per well) for the cell viability assay, nanoplastic uptake studies, immunofluorescence staining and cytokine/chemokine array. For quantitative real-time PCR (qRT-PCR), cells were plated in 6-well plates (1.2 × 106 cells per well). One hour later, the plates were washed with the medium to remove unattached cells. After 1 day, the culture medium was replaced with a pure microglial medium (DMEM containing 2% FBS and penicillin/streptomycin). After 24 hours, the cells were treated with nanoplastics. As positive controls, 100 ng ml−1 lipopolysaccharide (LPS) from Escherichia coli O111:B4 (Sigma-Aldrich, St Louis, MO, USA) and 10 U ml−1 thrombin (Sigma-Aldrich) were used.Optical diffraction tomographic analysis
To analyze the uptake and internalization patterns of the three different nanoplastics in microglia, cells were subcultured in tissue culture dishes (FluoroDish™, WPI Inc., USA). After 3 hours of exposure to 50 μg mL−1 concentration of each nanoplastic, the cells were analyzed using a holotomographic microscope equipped with a fluorescence module (3D Cell Explorer-Fluo, Nanolive, Lausanne, Switzerland). Fluorescence images of nanoplastic uptake by microglia were obtained through Z-stacking of 96 slices of fluorescence images, with excitation and emission settings specific to each nanoplastic. Holotomographic images were captured using 520 nm light and a ×60 magnification lens. Refractive index (RI) values were analyzed using STEVE software from Nanolive to digitally stain and distinguish nanoplastics from the cytoplasm inside the cells.Immunofluorescence staining
Immunofluorescence staining was performed as previously described.5,24 For the detection of microglia and M1-activated immune cells, Iba1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, FUJIFILM Wako Chemical, Osaka, Japan) and CD68 (1
1000, FUJIFILM Wako Chemical, Osaka, Japan) and CD68 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, Bio-Rad, Hercules, CA, USA) antibodies were used, respectively. As a marker for neurons, neurofilament antibody (1:1000, Biolegend, San Diego, CA, USA) was used. The signals were visualized using appropriate fluorescence conjugated secondary antibodies. Images were obtained using a fluorescence microscope (Olympus, Tokyo, Japan) for 2D images and a confocal microscope (Zeiss, Oberkochen, Germany) for Z-stack images.
100, Bio-Rad, Hercules, CA, USA) antibodies were used, respectively. As a marker for neurons, neurofilament antibody (1:1000, Biolegend, San Diego, CA, USA) was used. The signals were visualized using appropriate fluorescence conjugated secondary antibodies. Images were obtained using a fluorescence microscope (Olympus, Tokyo, Japan) for 2D images and a confocal microscope (Zeiss, Oberkochen, Germany) for Z-stack images.
Measurement of nanoplastic uptake
The uptake velocity of nanoplastics was analyzed by measuring the fluorescence intensity of dye-conjugated nanoplastics in Iba1-positive microglia at various time points (5 to 180 minutes). To quantify the uptake rate, the fluorescence intensity of nanoplastics in individual Iba1-positive microglial cells was measured. Each intensity value was first normalized to the corresponding cell size. Finally, the percentage of nanoplastic uptake at each time point was calculated relative to the final time point, with the 180-minute value set as 100%.Quantitative real-time PCR (qRT-PCR) analysis
Total RNA from microglia was extracted using a PURY RNA Plus (GenDEPOT, Baker, TX) according to the manufacturer's instruction. RNA samples were reverse transcribed into complementary DNA (cDNA) using a TOPscript™ RT Dry MIX kit (dT18 plus) (Enzynomics, Daejeon, Republic of Korea) following the standard protocol. The cDNA was then used for quantitative real-time PCR (qRT-PCR) with a StepOnePlus™ real-time PCR system (Applied Biosystems, Waltham, MA, USA). The thermocycling conditions were as follows: initial denaturation at 95 °C for 3 minutes, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. Each reaction was performed in triplicate and gene expression was quantified using a FAST SYBR™ Green Master Mix kit (Applied Biosystems). Cycle threshold (Ct) values were calculated using the 2−ΔΔCT method, with GAPDH as the internal reference. Primer pairs used are listed in Table S1† and were purchased from Xenotech (Daejeon, Republic of Korea).mRNA sequencing (RNA-seq) and analysis
RNA samples were analyzed using an Agilent 2100 Bioanalyzer system (Agilent Biotechnologies, Santa Clara, CA, USA). Only samples of high-quality RNA (RNA integrity number ≥ 7.5) were used in the following mRNA sample preparation for sequencing. Using the TruSeq RNA Sample Preparation Kit V2, purification and library construction were carried out with total RNA, and Illumina NextSeq 1000 machines (Illumina, San Diego, CA, USA) were used for sequencing with a read length of 2 × 100 bases. A filtered read set was created using the Cutadapt v1.18 (https://cutadapt.readthedocs.io/en/stable/) command line parameters ‘-a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC-A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTA -m 50 -O 5’ and Sickle v1.33 (https://github.com/najoshi/sickle) was used to remove the low quality sequence (Phred score < 20) to a minimum length of 50 bp.25,26 We assessed the quality of the paired-end reads using FastQC version 0.11.4. In addition, duplicate sequences were examined through the application of the FASTQC tool. The trimmed data containing low-quality reads and the poly-N sequences were processed using NGSQCToolkit v2.3.3 (https://github.com/mjain-lab/NGSQCToolkit).27 Then reads were aligned to Rat genome version GRCr8. Transcripts obtained were quantified in the format of fragments per kilobase million (FPKM) using StringTie v2.2.1 (https://github.com/gpertea/stringtie) to calculate expression values and obtain normalized counts.28 Among total genes (n = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 558), protein-coding genes (n = 19
558), protein-coding genes (n = 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 957) were selected according to the Ensemble database (https://ensembl.org/Homo_sapiens/). The entire data set was transformed using log2 (FPKM + 1) for data normalization. We used multiple statistical tests to obtain differentially expressed genes (DEGs), including a t-test, Wilcoxon rank-sum test, and fold change.
957) were selected according to the Ensemble database (https://ensembl.org/Homo_sapiens/). The entire data set was transformed using log2 (FPKM + 1) for data normalization. We used multiple statistical tests to obtain differentially expressed genes (DEGs), including a t-test, Wilcoxon rank-sum test, and fold change.
Cytokine/chemokine array
Secreted cytokines and chemokines were measured using a Rat Cytokine Array Panel A kit (ARY008, Proteome Profiler™ Array) from R&D Systems (Minneapolis, MN, USA), following the manufacturer's instructions. Culture supernatants from primary microglia were used for the array. The targeted cytokines and chemokines included the following: GM-CSF, ICAM-1, IFN-γ, IL-1α, IL-1β, IL-1ra, IL-2, IL-3, IL-4, IL-6, IL-10, IL-13, IL-17, LIX, L-selectin, CCL3, CCL5, CCL20, CINC-1, CINC-2α/β, CINC-3, CNTF, CX3CL1, CXCL9, CXCL10, CXCL7, TIMP-1, TNF-α, and VEGF.Enzyme-linked immunosorbent assay (ELISA)
Culture supernatants from primary microglia were collected at 24 hours after nanoplastic treatment and centrifuged to remove floating debris. The levels of CXCL1 and CXCL2 in the supernatants were measured using ELISA kits (DY515, Rat CXCL1/CINC-1 DuoSet ELISA; DY525, Rat CXCL2/CINC-3 DuoSet ELISA) from R&D Systems, following the manufacturer's protocols. CXCL1 and CXCL2 levels were calculated and expressed as pg mL−1.Measurement of cell migration ability
Cell migration ability was measured as described previously,29,30 with partial modification. Briefly, microglia (4 × 105 cells per well) were seeded into the lower wells of 24-well plates. After 24 hours, microglia (1 × 105 cells per 100 μL) were added to the upper wells of Corning® Transwell® polycarbonate membrane cell culture inserts (Corning, Corning, NY, USA), which have an uncoated 8 μm filter on the bottom. Microglia in the lower wells were treated with nanoplastics in the presence or absence of 200 nM SB225002, an inhibitor of CXCR2 (Millipore Sigma, Burlington, MA, USA). The cell-bearing filters from the upper wells were fixed with 70% ethanol for 10 minutes and rinsed with distilled water, and the microglial cells remaining on the upper side of each filter were removed with a cotton swab. The lower side of the filters was then stained with 0.2% crystal violet in 20% methanol (Sigma-Aldrich) for 5 minutes, followed by rinsing with distilled water. Cells that migrated to the lower side of the filter were counted in three random fields per filter using a light microscope (Zeiss, Oberkochen, Germany).Stereotaxic injection and tissue processing
The process of stereotaxic injection was performed as previously described,24 with minor modification to suit our conditions. Mice were injected with 1 μl of PMMANP (1 mg ml−1 in PBS) and phosphate-buffered saline (PBS) into the striatum (anteroposterior, −0.3 mm; mediolateral, ±2.0 mm; dorsoventral, −3.5 mm from bregma). After the injection, the needle was left in place for 10 minutes before being slowly withdrawn. The mice were sacrificed and their brains were collected 4 days post surgery. The collected brains were fixed in 4% paraformaldehyde (PFA) for 2 days, followed by dehydration in 30% sucrose. The tissues were embedded in FSC22 frozen section compound (Leica, Wetzlar, Germany) and cryosectioned at a thickness of 35 μm using a CM1520 cryotome (Leica).Statistical analysis
All data were obtained from at least three independent replicates and analyzed using either a two-tailed or one-tailed unpaired t-test, or one-way ANOVA with post-hoc testing using Dunnett's method. Statistical analyses were performed with GraphPad Prism (GraphPad Software version 5.0, La Jolla, CA, USA), and p-values less than 0.05 were considered statistically significant.Results and discussion
Physicochemical characterization of nanoplastics
We chose nanoplastics derived from three different types of plastics (PS, PP, and PMMA) to investigate their effects on brain microglia. SEM imaging revealed that the particles had a spherical morphology, with size distributions of 443.28 ± 16.41 nm for PSNP, 480.68 ± 77.54 nm for PPNP, and 419.16 ± 35.24 nm for PMMANP (Fig. 1A and B). In addition, we measured the hydrodynamic diameter and zeta potential values of these nanoplastics. The hydrodynamic sizes were 488.7 ± 13.11 nm for PSNP, 719.97 ± 35.36 nm for PPNP, and 482.77 ± 10.85 nm for PMMANP. PSNP and PMMANP exhibited high homogeneity with narrow size distributions, while PPNP exhibited a relatively broad distribution. The zeta potential values were negative: −63.7 ± 1.17 mV for PS, −55.1 ± 0.17 mV for PP, and −73.83 ± 28.74 mV for PMMA. The physicochemical characteristics of these nanoplastics are detailed in Table S2.† To confirm their chemical characteristics, we performed FTIR analysis (Fig. S1†). The FTIR spectra showed the characteristic peaks for PSNP at 695 cm−1 (aromatic CH out-of-plane bend), 1026 cm−1 (aromatic CH bend), 1491 and 1600 cm−1 (C–C aromatic ring stretch), 2918 cm−1 (CH alkyl stretch), and 3023 cm−1 (aromatic CH stretch); for PPNP at 808 and 839 cm−1 (CH2 rock, CC stretch), 1373 cm−1 (CH3 bend), 1455 cm−1 (CH2 bend), and 2838 and 2917 cm−1 (CH stretch); and for PMMANP at 1154 cm−1 (CO stretch), 1192 cm−1 (CH3 rock), 1389 cm−1 (CH3 bend), 1731 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch), and 2950 and 2994 cm−1 (CH stretch), which are consistent with a previous study.31
O stretch), and 2950 and 2994 cm−1 (CH stretch), which are consistent with a previous study.31
 | ||
| Fig. 1 Characterization of nanoplastics. (A) SEM images and (B) size distribution of three different types of nanoplastics (PSNP (PS), PPNP (PP) and PMMANP (PMMA)). | ||
Nanoplastic types differ in microglial absorption
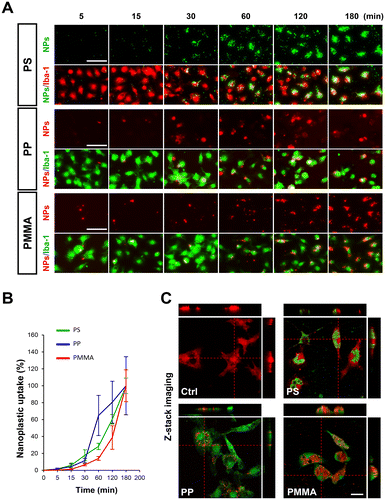
Before investigating the effects of nanoplastics on microglia, we first performed dynamic light scattering (DLS) analysis on the three types of nanoplastics (PSNP, PPNP, and PMMANP) to determine whether each particle type remained well-dispersed and free from aggregation under our culture conditions. We found that all three types of nanoplastics remained well dispersed and free from aggregation in both distilled water (DW) and our culture medium (DMEM) for up to three days in vitro (Fig. S2†). To determine the optimal dosage for nanoplastic uptake studies, we assessed cytotoxicity using the MTT assay at various doses (0–200 μg ml−1) after 24 hours of treatment. Microglia exhibited decreased viability at doses of 100 μg ml−1 and above for PMMANP (Fig. S3†). Based on these findings, we treated microglia with 50 μg ml−1 nanoplastics, the highest non-toxic dosage, for our nanoplastic uptake analysis. Three hours after treatment, we used optical diffraction tomography (ODT) to examine the intracellular localization of the nanoplastics. We found that all three types of nanoplastics were predominantly localized in the cytoplasm, with no detectable presence in the nucleus (Fig. 2A and Fig. S4†). Our phase-contrast and fluorescence imaging data further confirmed that fluorescence signals of dye-conjugated nanoplastics were clearly distributed only in the cytoplasm and not in the nucleus of microglia (Fig. 2C). We then compared the uptake ratios of each type of nanoplastic based on our ODT data and observed the highest uptake in microglia treated with PMMANP, in comparison with those treated with PSNP and PPNP (Fig. 2B). We also analyzed the nanoplastic uptake velocity by measuring the intensity of fluorescence signals of dye-conjugated nanoplastics in microglia, positively stained with the Iba-1 marker, at various time points (5–180 minutes). Although the majority of the nanoplastic signals were detected inside the microglia, a few fluorescent particles were not absorbed and remained outside the cells up to 3 hours after exposure to nanoplastics. The relative uptake velocity of each nanoplastic is presented as normalized values, with the highest intensity value (100%) at 180 minutes (Fig. 3A and B). We found that PPNP had the highest uptake velocity at early time points (30–60 minutes), despite its lower overall absorption ratio than that of other nanoplastics. These data may suggest that PPNP could be involved in acute microglial activation at early time points because of its higher absorption velocity. Conversely, PMMANP exhibited the lowest uptake velocity, although it had the highest absorption ratio (Fig. 2B and 3B). When the uptake of these nanoplastics was examined through z-stack imaging, it was confirmed that they are located inside the microglia, rather than on the surface (Fig. 3C). These results indicate that different types of nanoplastics exhibit varying absorption rates in microglia.Microglia exposed to PMMANP exhibit M1-phase phenotypes and increased expression of genes related to chemotaxis
Since the highest uptake ratio was observed in microglia treated with PMMANP (Fig. 2), we first investigated the phenotypes of these microglia. We measured the mRNA expression of markers for proinflammatory M1 microglia and anti-inflammatory/neural protective M2 microglia using qRT-PCR. We found that M1 microglial markers, including TNF-α (1.49-fold increase at 3 hours and 1.43-fold increase at 6 hours) and CD86 (1.74-fold increase at 3 hours and 1.87-fold increase at 6 hours), were significantly elevated in microglia at 3 and 6 hours after PMMANP treatment (Fig. 4A). Although proinflammatory cytokines such as IL-1β and TNF-α were also increased in microglia treated with PSNP (<1.5-fold) and PPNP (around 1.8-fold) at 24 hours after treatment, the fold changes were lower than those of the PMMANP group (2–3-fold) (Fig. 4B). The mRNA expression of Iba1, a known marker of microglia, was not altered by PMMANP (Fig. S5†). In contrast, the expression of M2 microglial markers, including ARG1, IGF1, CD206, and TGFβ, showed no significant changes at 3 to 6 hours after PMMANP treatment (Fig. 4A). To understand the molecular mechanisms underlying PMMANP-induced microglial activation, we analyzed the gene expression profile by mRNA sequencing in microglia after PMMANP treatment. To reduce potential errors, we performed two independent analyses and selected common genes. Our DEG analysis revealed that 71 genes were up-regulated and 45 genes were down-regulated (≥1.7 fold change) in microglia following PMMANP treatment (Fig. 4C and Table S3†). Although PMMANP exposure induced M1 microglial activation (Fig. 4A), the increase in proinflammatory cytokines in PMMANP-treated microglia was relatively modest (≤1.3 fold change) compared to that in LPS-treated microglia (Table S4†). Instead of cytokines, PMMANP exposure notably elevated the expression of chemokines associated with chemotaxis in microglia (Table S4†). KEGG pathway enrichment analysis indicated an enhancement of CXCR2-mediated chemokine signaling in PMMANP-exposed microglia (Fig. 4D). This was corroborated by qRT-PCR, which confirmed increased CXCR2 mRNA expression in PMMANP-exposed microglia. However, CXCL11—despite its elevation in RNA sequencing data from the PMMANP group—and its receptor, CXCR3, did not show changes in our qRT-PCR analysis (Fig. 4E). In addition, we examined the microglial morphology and surface marker expression following nanoplastic exposure but observed no significant differences among the groups (data not shown). However, functional changes varied depending on the type of nanoplastic, as indicated by nitric oxide (NO) secretion, which was increased by PSNP and PMMANP, but not by PPNP (Fig. S6†). Collectively, these findings suggest that PMMANP exposure may facilitate CXCR2-mediated microglial chemotactic migration.To measure the protein levels of cytokines and chemokines secreted by microglia, we performed a cytokine/chemokine array using culture supernatants from microglia exposed to nanoplastics. Consistent with the mRNA sequencing results, microglia treated with PMMANP exhibited substantial increases in chemokine secretion compared to cytokine secretion (Fig. 5A). Among the groups exposed to the three types of nanoplastics, PMMANP-treated microglia showed the highest increases in chemokine secretion, including CXCL1 (2.25-fold), CXCL2 (3.63-fold), CCL5 (1.23-fold), CXCL10 (1.51-fold), and CCL3 (1.74-fold), compared to the control (Fig. 5A). This increase in CXCL1, CXCL2, and CCL3 was also confirmed in microglia treated with lower dosages of PMMANP (1–25 μg ml−1) (Fig. S7†). In contrast, PSNP only caused a slight increase in CCL3, while PPNP increased CXCL1, CXCL2, and CCL3 by 1.04-fold, 1.44-fold, and 1.81-fold, respectively. Among the various chemokines that were increased in PMMANP-exposed microglia, CXCL1 and CXCL2 showed higher increases than others (Fig. 5B). Our ELISA results showed a marked increase in the secretion of CXCL1 and CXCL2 from microglia upon exposure to PMMANPs, with CXCL1 levels increasing by 41.57 ± 17.59-fold and CXCL2 by 6.45 ± 2.27-fold compared to the control. In contrast, PSNP exposure did not induce significant secretion changes (CXCL1: 4.88 ± 0.08-fold; CXCL2: 1.18 ± 0.8-fold). PPNP exposure led to a notable increase in CXCL1 secretion (22.59 ± 9.95-fold), but the elevation in CXCL2 (2.09 ± 1.05-fold) was not statistically significant (Fig. 5C). Consistently, the mRNA expression levels of CXCL1 and CXCL2 were only modestly elevated in PSNP- and PPNP-treated microglia, and remained substantially lower than those observed in PMMANP-treated cells (Fig. 5D).
Similarly, using transparent nanoplastics that are not conjugated with a fluorescent dye, we also observed a significantly greater increase in the mRNA expression of CXCL1 and CXCL2 following PMMANP exposure (Fig. S8†). These findings are consistent with our mRNA sequencing results, which showed that PMMANP induced increases in CXCL1, CXCL2 (Table S4†) and CXCR2 (Fig. 4D and E), the common receptor for both CXCL1 and CXCL2.32
Exposure to PMMANPs induces chemotactic migration of microglia mediated by CXCR2
Since both CXCL1 and CXCL2, which are increased by PMMANP exposure, are chemokines that promote the recruitment of CXCR2-expressing peripheral immune cells and microglia,33–36 we investigated whether the elevated secretion levels of CXCL1 and CXCL2 enhance microglial migration. Our migration assay data obtained using a transwell system (Fig. 6A) clearly show that PPNP exposure induced a 2.1-fold increase in the number of migrated cells, whereas PMMANP exposure led to a relatively high 3.9-fold increase in microglial migration. However, no increase in migration ability was observed in microglia exposed to PSNPs (Fig. 6B). Although there was a significant increase in CCL3 in microglia treated with PPNP (Fig. 5A), this increase in PPNP-induced microglial migration may not be mediated by CCL3. We did not observe any change in the expression of CCR5, the receptor for CCL3, following PPNP treatment (data not shown). Our migration assay results are consistent with the data showing a higher increase in secreted CXCL1 and CXCL2 in microglia exposed to PMMANPs than those exposed to PPNP, and no increase in microglia exposed to PSNPs (Fig. 4). To investigate the mechanism underlying PMMANP-induced microglial migration, we co-treated microglia with SB225002, a potent competitive antagonist of CXCR2, a G-protein coupled receptor.37,38 We found that SB225002 completely inhibited PMMANP-induced microglial migration, indicating that this migration is mediated by CXCL1/CXCL2–CXCR2 signaling pathways in PMMANP-exposed microglia (Fig. 6B). Moreover, our in vivo data also show that PMMANP increases the activation and chemotactic attraction of immune cells in the mouse brain 4 days after stereotaxic injection of PMMANPs (1 μg) into the striatum. Our immunofluorescence staining data clearly show that CD68 signals, a marker for activated microglia and macrophages, were significantly higher in the PMMANP-injected group than in the PBS-treated control (Fig. 6C–E). Although Iba-1-positive signals were not significantly increased, the PMMANP group showed a strong increasing tendency compared to the control (Fig. 6C–E). To examine whether these immune cell activation and migration induced by PMMANPs affect neurons, tissues were stained with a marker for M- and H-type neurofilaments. We found a decrease in neurofilament signals in the PMMANP group compared to the control (Fig. 6C–E). It should be noted that the decrease of the neurofilament observed in the PMMANP group could, in part, be due to the direct effects of the nanoplastic particles, rather than immune cell activation alone. Collectively, these data demonstrate that PMMANPs have a greater impact on M1 microglial activation and chemokine expression than PSNPs and PPNPs, resulting in the highest increase in chemotactic migration of microglia.Discussion
In this study, we systematically compared the biological effects of three distinct nanoplastics—PSNP, PPNP, and PMMANP—on brain microglia. We found that each type of nanoplastic exhibited a unique uptake pattern in microglia and induced differential alterations in microglial functions, including M1-phase activation, chemokine secretion, and migration. These findings suggest that the polymer composition of nanoplastics plays a critical role in shaping their biological effects. Indeed, previous studies have reported differential biological outcomes depending on the type of plastic from which nanoplastics are derived. For example, PSNP has been shown to impair learning and memory,5,8 and to induce anxiety- and depression-like behaviors,6 whereas PENPs have been associated with social deficits and repetitive behaviors, potentially linking them to autism spectrum disorders.7 Such type-specific nanoplastic effects have also been observed in peripheral organs, including the liver39,40 and the lungs.41 Consistent with these previous studies, our data support the idea that nanoplastics of different polymer types elicit distinct microglial neuroinflammatory responses through divergent molecular mechanisms in the brain.In our study, PMMANP showed the highest biological effects on microglia although the environmental prevalence is relatively low compared to the other types of nanoplastics. In this regard, investigating the potential harmfulness of PMMANP is crucial for human health. In this study, we have used spherical particles rather than fibers, fragments, or pellets, even though spherical nanoplastics are less abundant than other shapes in the environment. It is well known that non-spherical nanoplastics exhibit varying cytotoxicity due to differences in their surface roughness,42,43 and their shapes can vary from one instance to another. This suggests that even when other forms of nanoplastics are synthesized using the same method, they may differ in length and thickness. Taken together, the use of spherical nanoplastics, which have a uniform surface and consistent size, appears to be an ideal choice for maintaining experimental consistency. Our data clearly indicate that PMMANP can induce immune cell recruitment, which may lead to exacerbation of brain inflammation and, consequently, brain damage. We observed distinct biological effects of different types of nanoparticles on brain microglial function. These findings are consistent with previous reports, which suggest that the biological effects and underlying mechanisms of nanoparticles can vary depending on their type and surface modifications and that these variations can influence their effects within cells and organisms.44–47 Our results show the greatest upregulation of CXCL1/2-induced chemotaxis in microglia exposed to PMMANP. This upregulation might be due to the high capability of PMMANP to infiltrate microglia. In addition, from a physicochemical perspective, PMMANP includes ester bonds and a fibrous molecular structure, which could contribute to increased duration, resistance to abrasion, and mechanical strength.
Although microglia exposed to PMMANP show increased expression of M1 microglial markers, the majority of upregulated genes in these microglia are chemokines and chemotaxis-related genes. These gene expression patterns in PMMANP-exposed microglia differ significantly from those exposed to infectious pathogens, such as bacteria, LPS, a bacterial cell wall component, and virus, where a substantial increase of inflammatory cytokines is generally observed.48–50 The differences in the responses between microglia treated with nanoplastics and those exposed to pathogens may arise from distinct mechanisms of microglial activation. In fact, bacterial infection increases the expression of various receptors on microglia, such as toll-like receptors (TLRs), which can recognize specific peptidoglycans and proteins like flagellin found on bacterial cell walls.51,52 Similarly, viral infection also induces the expression of the purinergic receptor P2RY12, which senses adenosine triphosphate (ATP) and adenosine diphosphate (ADP) released from virus-infected neurons.48,53 In addition, several receptors in the TLR family also induce microglial activation by recognizing viral single-stranded and double-stranded RNA, which ultimately results in anti-viral effects.49,50 However, nanoplastics are composed of various polymers that do not possess specific binding affinity for any receptors on the surface of immune cells. Instead of receptor-mediated signaling, nanoplastics may bind to the plasma membrane through adsorption and influence microglial functions through mechanisms that do not involve surface receptors on microglia. In addition, unlike general pathogen-induced microglial activation, PMMANP-induced microglial activation was not accompanied by an increase in Iba1 expression. These findings are consistent with previous studies showing microglial activation without an increase in Iba1 expression in Alzheimer's disease.54
While previous studies have primarily focused on the uptake of nanoplastics, inflammatory cytokine secretion, and apoptosis of microglia,12,13,55 the current study identifies the critical impact of nanoplastics on chemotactic attraction induced by brain microglia, which plays a key role in immune cell recruitment. Although the increase in the expression of inflammatory cytokines was not very pronounced in PMMANP-exposed microglia, the upregulation of CXCL1/2 induced by PMMANP could still contribute to brain damage by enhancing the recruitment of CXCR2-expressing immune cells, which may accelerate inflammation in the brain. In fact, previous reports have shown that the CXCL1/2–CXCR2 axis plays a crucial role in the recruitment of peripheral leukocytes, including neutrophils, macrophages, and monocytes, into the brain parenchyma under various disease conditions, such as viral encephalitis, Alzheimer's disease, and brain tumors.34,56–58 Considering these previous data, our current study demonstrates the potential mechanisms by which inflammation and resulting damage could occur in the brain exposed to PMMANPs. This is supported by a series of experiments showing the upregulation of CXCL1/2–CXCR2 signaling and enhanced microglial migration following PMMANP treatment.
In addition to CXCR2-mediated migration, the observed increase in microglia following PMMANP injection may also result from local proliferation in response to inflammatory signals. Indeed, studies in disease models—such as status epilepticus—have shown that resident microglia proliferate locally via CSF-1R signaling, contributing significantly to microgliosis.59 Moreover, repeated peripheral LPS exposure induces hypertrophic microglial states and transient proliferation across brain regions, supporting the role of inflammatory stimuli in driving microglial expansion.60 These data suggest that microglial proliferation, potentially induced by microgliosis, may partially contribute to the PMMANP-induced increase in brain microglia.
Conclusions
This study demonstrates that PMMANP exposure induces M1 microglial activation, ultimately leading to CXCL1/2–CXCR2 pathway-mediated chemotactic migration of brain microglia. Given that the CXCL1/2–CXCR2 signaling axis also plays a role in the recruitment of peripheral immune cells that may exacerbate neuroinflammation, our findings offer important insights for future investigations into the biosafety of PMMA-derived biomaterials and consumer products. Although the biological effects of PSNPs and PPNPs on microglia were lower than those of PMMANPs, their substantially higher environmental prevalence suggests that they may still pose a potential risk to brain health. These results underscore the importance of considering both the toxic potential and real-world abundance of different nanoplastic types. Collectively, this study raises broader concerns regarding the neuroinflammatory risks posed by nanoplastics.Author contributions
Da Yong Lee: conceptualization, funding acquisition, investigation, methodology, project administration, supervision, and writing – review and editing. Jinyoung Jeong: data curation, funding acquisition, methodology, project administration, resources, supervision, and writing – original draft. Jahong Koo: data curation, formal analysis, investigation, validation, visualization, and writing – original draft. Bohyeon Jeong: data curation, investigation, and visualization. Jeong Yeob Baek: formal analysis, investigation, and visualization. Wang Sik Lee: formal analysis. Jiyeon Hong: formal analysis. Subin Park: formal analysis. Yugyeong Sim: data curation. Jiyoung Gong: data curation. Dae Soo Kim: methodology. Sang Ryong Kim: resources and visualization.Conflicts of interest
There are no conflicts to declare.Data availability
The data that support the conclusions of this study are available upon reasonable request from the corresponding author. Most of the experimental procedures and data have been included in the main text and the ESI.†Acknowledgements
This work was supported by the KRIBB Initiative Research Program (KGM5362521 and KGM5322523), the Brain Research Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (NRF-2019M3C7A1031534 (D. Y. L.)), the Basic Science Research Program of the NRF funded by the Korean government (Ministry of Education) (RS-2022-NR075598 (D. Y. L.)), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the MSIT (RS-2025-02305013), the Basic Science Research Program (RS-2024-00344622 (J. J.)) of the NRF and the National Research Council of Science & Technology (NST) (CAP20024-002 (J. J.)) funded by the MSIT of the Republic of Korea.References
- Y. Jin, L. Lu, W. Tu, T. Luo and Z. Fu, Sci. Total Environ., 2019, 649, 308–317 CrossRef CAS PubMed.
- R. Sun, M. Liu, F. Xiong, K. Xu, J. Huang, J. Liu, D. Wang and Y. Pu, Sci. Total Environ., 2024, 912, 169228 CrossRef CAS PubMed.
- L. Zeng, C. Zhou, W. Xu, Y. Huang, W. Wang, Z. Ma, J. Huang, J. Li, L. Hu, Y. Xue, T. Luo and L. Zheng, Ecotoxicol. Environ. Saf., 2023, 257, 114941 CrossRef CAS PubMed.
- W. Zhou, D. Tong, D. Tian, Y. Yu, L. Huang, W. Zhang, Y. Yu, L. Lu, X. Zhang, W. Pan, J. Shen, W. Shi and G. Liu, Adv. Healthc. Mater., 2023, 12, e2301799 CrossRef PubMed.
- B. Jeong, J. Y. Baek, J. Koo, S. Park, Y. K. Ryu, K. S. Kim, S. Zhang, C. Chung, R. Dogan, H. S. Choi, D. Um, T. K. Kim, W. S. Lee, J. Jeong, W. H. Shin, J. R. Lee, N. S. Kim and D. Y. Lee, J. Hazard. Mater., 2022, 426, 127815 CrossRef CAS PubMed.
- H. S. Shin, S. H. Lee, H. J. Moon, Y. H. So, H. R. Lee, E. H. Lee and E. M. Jung, J. Hazard. Mater., 2023, 454, 131465 CrossRef CAS PubMed.
- J. Zaheer, H. Kim, I. O. Ko, E. K. Jo, E. J. Choi, H. J. Lee, I. Shim, H. J. Woo, J. Choi, G. H. Kim and J. S. Kim, Environ. Int., 2022, 161, 107121 CrossRef CAS PubMed.
- C. W. Lee, L. F. Hsu, I. L. Wu, Y. L. Wang, W. C. Chen, Y. J. Liu, L. T. Yang, C. L. Tan, Y. H. Luo, C. C. Wang, H. W. Chiu, T. C. Yang, Y. Y. Lin, H. A. Chang, Y. C. Chiang, C. H. Chen, M. H. Lee, K. T. Peng and C. C. Huang, J. Hazard. Mater., 2022, 430, 128431 CrossRef CAS PubMed.
- D. J. Mc Carthy, M. Malhotra, A. M. O'Mahony, J. F. Cryan and C. M. O'Driscoll, Pharm. Res., 2015, 32, 1161–1185 CrossRef CAS PubMed.
- S. Shan, Y. Zhang, H. Zhao, T. Zeng and X. Zhao, Chemosphere, 2022, 298, 134261 CrossRef CAS PubMed.
- S. Shan, D. Cheng, H. Li, W. Yao, R. Kou, J. Ji, N. Liu, T. Zeng and X. Zhao, J. Hazard. Mater., 2025, 489, 137615 CrossRef CAS PubMed.
- Y. M. M. Paing, Y. Eom, G. B. Song, B. Kim, M. G. Choi, S. Hong and S. H. Lee, Sci. Total Environ., 2024, 924, 171681 CrossRef CAS PubMed.
- W. Kwon, D. Kim, H. Y. Kim, S. W. Jeong, S. G. Lee, H. C. Kim, Y. J. Lee, M. K. Kwon, J. S. Hwang, J. E. Han, J. K. Park, S. J. Lee and S. K. Choi, Sci. Total Environ., 2022, 807, 150817 CrossRef CAS PubMed.
- H. S. Kwon and S. H. Koh, Transl. Neurodegener., 2020, 9, 42 CrossRef PubMed.
- H. Wang, Y. He, Z. Sun, S. Ren, M. Liu, G. Wang and J. Yang, J. Neuroinflammation, 2022, 19, 132 CrossRef PubMed.
- X. Liao, J. Yang, H. Wang and Y. Li, J. Psychiatr. Res., 2020, 130, 167–176 CrossRef PubMed.
- L. Schroter and N. Ventura, Small, 2022, 18, e2201680 CrossRef PubMed.
- J. Bhagat, N. Nishimura and Y. Shimada, J. Hazard. Mater., 2021, 405, 123913 CrossRef CAS PubMed.
- E. D. Okoffo and K. V. Thomas, J. Hazard. Mater., 2024, 464, 133013 CrossRef CAS PubMed.
- R. Q. Frazer, R. T. Byron, P. B. Osborne and K. P. West, J. Long-Term Eff. Med. Implants, 2005, 15, 629–639 CrossRef CAS PubMed.
- S. Pikis, J. Goldstein and S. Spektor, J. Clin. Neurosci., 2015, 22, 139–143 CrossRef CAS PubMed.
- W. S. Lee, H. Kim, Y. Sim, T. Kang and J. Jeong, ACS Omega, 2022, 7, 2467–2473 CrossRef CAS PubMed.
- H. Li, Y. Xiang, Z. Zhu, W. Wang, Z. Jiang, M. Zhao, S. Cheng, F. Pan, D. Liu, R. C. M. Ho and C. S. H. Ho, J. Neuroinflammation, 2021, 18, 254 CrossRef CAS PubMed.
- E. Nam, Y. Lin, J. Park, H. Do, J. Han, B. Jeong, S. Park, D. Y. Lee, M. Kim, J. Han, M. H. Baik, Y. H. Lee and M. H. Lim, Adv. Sci., 2024, 11, e2307182 CrossRef PubMed.
- M. Martin, EMBnet. J., 2011, 17, 3 Search PubMed.
- N. Joshi and J. Fass, Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33), 2011 Search PubMed.
- R. K. Patel and M. Jain, PLoS One, 2012, 7, e30619 CrossRef CAS PubMed.
- M. Pertea, G. M. Pertea, C. M. Antonescu, T. C. Chang, J. T. Mendell and S. L. Salzberg, Nat. Biotechnol., 2015, 33, 290–295 CrossRef CAS PubMed.
- C. R. Justus, N. Leffler, M. Ruiz-Echevarria and L. V. Yang, J. Visualized Exp., 2014, 1, 51046 Search PubMed.
- J. Pijuan, C. Barcelo, D. F. Moreno, O. Maiques, P. Siso, R. M. Marti, A. Macia and A. Panosa, Front. Cell Dev. Biol., 2019, 7, 107 CrossRef PubMed.
- S. S. Suresh, S. Mohanty and S. K. Nayak, J. Remanufacturing, 2017, 7, 217–233 CrossRef.
- Y. Cheng, X. L. Ma, Y. Q. Wei and X. W. Wei, Biochim. Biophys. Acta, Rev. Cancer, 2019, 1871, 289–312 CrossRef CAS PubMed.
- X. Jin, S. H. Kim, H. M. Jeon, S. Beck, Y. W. Sohn, J. Yin, J. K. Kim, Y. C. Lim, J. H. Lee, S. H. Kim, S. H. Kang, X. Pian, M. S. Song, J. B. Park, Y. S. Chae, Y. G. Chung, S. H. Lee, Y. J. Choi, D. H. Nam, Y. K. Choi and H. Kim, Brain, 2012, 135, 1055–1069 CrossRef PubMed.
- Q. Zhang, J. Wang, X. Yao, S. Wu, W. Tian, C. Gan, X. Wan, C. You, F. Hu, S. Zhang, H. Zhang, K. Zhao, K. Shu and T. Lei, Front. Immunol., 2021, 12, 637053 CrossRef CAS PubMed.
- J. Wojcieszak, K. Kuczynska and J. B. Zawilska, J. Mol. Neurosci., 2022, 72, 1929–1951 CrossRef CAS PubMed.
- A. I. Rojo, G. McBean, M. Cindric, J. Egea, M. G. Lopez, P. Rada, N. Zarkovic and A. Cuadrado, Antioxid. Redox Signal., 2014, 21, 1766–1801 CrossRef CAS PubMed.
- P. Bakshi, C. Jin, P. Broutin, B. Berhane, J. Reed and M. Mullan, Bioorg. Med. Chem., 2009, 17, 8102–8112 CrossRef CAS PubMed.
- J. R. White, J. M. Lee, P. R. Young, R. P. Hertzberg, A. J. Jurewicz, M. A. Chaikin, K. Widdowson, J. J. Foley, L. D. Martin, D. E. Griswold and H. M. Sarau, J. Biol. Chem., 1998, 273, 10095–10098 CrossRef CAS PubMed.
- Y. Wen, S. Deng, B. Wang, F. Zhang, T. Luo, H. Kuang, X. Kuang, Y. Yuan, J. Huang and D. Zhang, Ecotoxicol. Environ. Saf., 2024, 278, 116439 CrossRef CAS PubMed.
- J. Hua, T. Zhang, X. Chen, B. Zhu, M. Zhao, K. Fu, Y. Zhang, H. Tang, H. Pang, Y. Guo, J. Han, L. Yang and B. Zhou, Sci. Total Environ., 2024, 947, 174541 CrossRef CAS PubMed.
- I. Kwabena Danso, J. H. Woo, S. Hoon Baek, K. Kim and K. Lee, Toxicol. Res., 2024, 40, 313–323 CrossRef CAS PubMed.
- D. Choi, J. Bang, T. Kim, Y. Oh, Y. Hwang and J. Hong, J. Hazard. Mater., 2020, 400, 123308 CrossRef CAS PubMed.
- D. Choi, J. Hwang, J. Bang, S. Han, T. Kim, Y. Oh, Y. Hwang, J. Choi and J. Hong, Sci. Total Environ., 2021, 752, 142242 CrossRef CAS PubMed.
- G. F. Schirinzi, I. Perez-Pomeda, J. Sanchis, C. Rossini, M. Farre and D. Barcelo, Environ. Res., 2017, 159, 579–587 CrossRef CAS PubMed.
- I. K. Danso, J. H. Woo and K. Lee, Molecules, 2022, 27, 313–323 CrossRef PubMed.
- J. Chen, Z. Xu, Y. Liu, A. Mei, X. Wang and Q. Shi, Ecotoxicol. Environ. Saf., 2023, 252, 114574 CrossRef CAS PubMed.
- C. M. Wolff, D. Singer, A. Schmidt and S. Bekeschus, J. Hazard. Mater., 2023, 459, 132194 CrossRef CAS PubMed.
- R. Fekete, C. Cserep, N. Lenart, K. Toth, B. Orsolits, B. Martinecz, E. Mehes, B. Szabo, V. Nemeth, B. Gonci, B. Sperlagh, Z. Boldogkoi, A. Kittel, M. Baranyi, S. Ferenczi, K. Kovacs, G. Szalay, B. Rozsa, C. Webb, G. G. Kovacs, T. Hortobagyi, B. L. West, Z. Kornyei and A. Denes, Acta Neuropathol., 2018, 136, 461–482 CrossRef CAS PubMed.
- L. Alexopoulou, A. C. Holt, R. Medzhitov and R. A. Flavell, Nature, 2001, 413, 732–738 CrossRef CAS PubMed.
- Y. L. Lin, M. Y. Lu, C. F. Chuang, Y. Kuo, H. E. Lin, F. A. Li, J. R. Wang, Y. P. Hsueh and F. Liao, Front. Immunol., 2020, 11, 614743 CrossRef CAS PubMed.
- J. Spielbauer, E. Glotfelty, H. Sarlus, R. A. Harris, R. D. Heijtz and T. E. Karlsson, Brain, Behav., Immun., 2024, 121, 43–55 CrossRef CAS PubMed.
- M. Ifuku, L. Hinkelmann, L. D. Kuhrt, I. E. Efe, V. Kumbol, A. Buonfiglioli, C. Kruger, P. Jordan, M. Fulde, M. Noda, H. Kettenmann and S. Lehnardt, Acta Neuropathol. Commun., 2020, 8, 159 CrossRef CAS PubMed.
- S. E. Haynes, G. Hollopeter, G. Yang, D. Kurpius, M. E. Dailey, W. B. Gan and D. Julius, Nat. Neurosci., 2006, 9, 1512–1519 CrossRef CAS PubMed.
- K. E. Hopperton, D. Mohammad, M. O. Trepanier, V. Giuliano and R. P. Bazinet, Mol. Psychiatry, 2018, 23, 177–198 CrossRef CAS PubMed.
- J. Antunes, P. Sobral, M. Martins and V. Branco, Environ. Toxicol. Pharmacol., 2023, 104, 104298 CrossRef CAS PubMed.
- F. Wu, Y. Zhao, T. Jiao, D. Shi, X. Zhu, M. Zhang, M. Shi and H. Zhou, J. Neuroinflammation, 2015, 12, 98 CrossRef PubMed.
- B. D. Michael, L. Bricio-Moreno, E. W. Sorensen, Y. Miyabe, J. Lian, T. Solomon, E. A. Kurt-Jones and A. D. Luster, Cell Rep., 2020, 32, 108150 CrossRef CAS PubMed.
- K. Zhang, L. Tian, L. Liu, Y. Feng, Y. B. Dong, B. Li, D. S. Shang, W. G. Fang, Y. P. Cao and Y. H. Chen, PLoS One, 2013, 8, e72744 CrossRef CAS PubMed.
- L. Feng, M. Murugan, D. B. Bosco, Y. Liu, J. Peng, G. A. Worrell, H. L. Wang, L. E. Ta, J. R. Richardson, Y. Shen and L. J. Wu, Glia, 2019, 67, 1434–1448 CrossRef PubMed.
- J. Kim, O. Sullivan, K. Lee, J. Jao, J. Tamayo, A. M. Madany, B. Wong, P. Ashwood and A. V. Ciernia, J. Neuroinflammation, 2024, 21, 233 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5nr00638d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |