Unveiling the inhibition mechanism of host-defense peptide cathelicidin LL-37 on the amyloid aggregation of the human islet amyloid polypeptide†
Huayuan
Tang
 *ab
*ab
aDepartment of Engineering Mechanics, Hohai University, Nanjing 211100, P.R. China. E-mail: htang@hhu.edu.cn
bState Key Laboratory of Structural Analysis, Optimization and CAE Software for Industrial Equipment, Dalian University of Technology, Dalian 116024, P.R. China
First published on 22nd January 2025
Abstract
The aberrant aggregation of the human islet amyloid polypeptide (hIAPP) is a hallmark of type II diabetes. LL37, the only cathelicidin host-defense peptide in humans, plays essential roles in antimicrobial and immunomodulatory activities. Mounting evidence indicates that LL37 can inhibit the amyloid aggregation of hIAPP, suggesting possible interplays between infections and amyloid diseases while the mechanism remains unclear. In this paper, we explored the molecular interactions between hIAPP and LL37 using all-atom discrete molecular dynamics (DMD), a novel and predictive molecular dynamics engine with improved sampling efficiencies. We found that the LL37 peptides can effectively interact with hIAPP in monomer, oligomer, and fibril states driven by hydrophobic associations and pi–pi interactions. Specifically, the hydrophobic residues in the N- and C-termini of LL37 peptides can firmly bind with the monomeric and oligomeric hIAPP, especially in the amyloidogenic regions, to prevent the self-interactions of amyloidogenic regions and thus hinder the formation of amyloid fibrils. Furthermore, LL37 can bind to the elongation surfaces of the hIAPP fibril seeds with geometric incompatibility for monomer addition to block the fibril growth. Together, we identified the crucial residues and key driving forces for the interactions between LL37 and hIAPP peptides and revealed the related dynamics and conformational changes. The uncovered mechanism can contribute to a better understanding of the pathological links between microbial infections and amyloid diseases and guide the designs of novel therapies combining antimicrobial and anti-amyloid functions.
Introduction
The aberrant accumulation of deposits formed by the human islet amyloid polypeptide (hIAPP) is a hallmark in more than 90% of patients with type II diabetes (T2D) affecting hundreds of millions of people worldwide.1,2 The hIAPP is a 37-residue peptide hormone that is synthesized and stored in pancreatic β-cell islets, playing essential roles in regulating glucose homeostasis in the physiological monomeric conformation.3,4 As a highly amyloidogenic protein, the intrinsically disordered hIAPP will misfold and undergo amyloid aggregation under T2D conditions.5 The amyloid aggregation of hIAPP can be described as a nucleation-and-growth process that involves the self-assembly of the hIAPP peptides into cytotoxic oligomers and amyloid fibrils with in-register β-sheet structures via a series of conformational changes.6,7 Increasing evidence suggests that the amyloid aggregation of hIAPP that leads to the formation of soluble prefibrillar hIAPP oligomers and insoluble hIAPP fibrils in pancreatic islets is related to the degeneration and dysfunction of β cells to produce insulin and regulate the blood glucose.8,9Antimicrobial peptides are the key components of the innate immune system of humans defending against invading microorganisms, which attracts growing attention considering the urgent demands to develop a new generation of antibiotics against multi-drug-resistant pathogens.10,11 The 37-residue polypeptide LL37 is the only known cathelicidin-derived antimicrobial peptide expressed in humans.12,13 Featured by its broad-spectrum activity, LL37 serves as the first line of defense against local infections and systemic pathogen invasions, playing a vital role in antimicrobial and immunomodulatory activities.14,15 By forming the cytotoxic fibrils in the presence of acidic phospholipids, LL37 can disrupt the membrane of microbes and achieve antimicrobial effects against infections caused by various pathogenic bacteria, viruses, fungi, and parasites.16 As an antimicrobial peptide, LL37 features the ability to treat diseases caused by microorganisms without developing resistance, making it a promising alternative to traditional antibiotics. Importantly, LL37 is found to prevent the formation of bacterial biofilms in vitro, thus destroying the protective layers of the bacterium.17 LL37 can also address microbial infections by attracting immune cells including neutrophils, which creates a positive feedback loop by producing more LL37 peptides.18 Moreover, LL37 can be expressed by a wide variety of immune and non-immune cells throughout the body, including the β cells of the pancreas, making LL37 a promising antibacterial and immunomodulatory candidate.19
Intriguingly, recent studies revealed that LL37 exerts several physiological functions in addition to killing pathogenic organisms. Specifically, accumulating experimental evidence indicates that LL37 can effectively inhibit the amyloid aggregation of hIAPP. Daily treatment with the LL37 peptide in mouse models can promote insulin and glucagon secretion from isolated rat islets, enhance β-cell neogenesis, and suppress the inflammation of pancreatic β cells of type 1 diabetes.20,21 The thioflavin T (ThT) binding assay showed that LL37 interfered with the aggregation of hIAPP in a dose-dependent manner, which was confirmed by the transmission electron microscopy (TEM) images that the incubation of hIAPP with equimolar LL37 led to amorphous aggregates.22 Hetero-assemblies with random coil structures were identified in the mixture of hIAPP and LL37.22 LL37 was further found to interact with hIAPP with a high binding affinity, prevent the amyloid aggregation of hIAPP in vitro and mitigate the pancreatic damage, suggesting a protective role of LL37 in β cells.22 Besides, the seeding effect of preformed hIAPP fibrils was also found to be suppressed by LL37.22 Since LL37 and hIAPP peptides coexist in the pancreas, LL37 is proposed to serve as a natural protector against T2D in humans. Besides, LL37 was found to effectively inhibit the amyloid aggregation of amyloid beta (Aβ) implicated in Alzheimer's disease by interacting with Aβ peptides with nanomolar affinity.23,24 In parallel, recent studies have found that various amyloid peptides, including Aβ, hIAPP, and α-synuclein, possess antibacterial capacities against common bacteria and fungi by disrupting the cell membranes of pathogens, and LL37 peptides exhibit amyloid-like properties as well.25,26 Together, the amyloid peptides hIAPP and antimicrobial peptides LL37, which are traditionally considered separate peptide families, are now recognized to be interconnected with each other.27,28
A complete understanding of the mechanism between hIAPP and LL37 is essential to deciphering the communication between the pathogenesis of amyloid diseases and the host defense against microbial infections, which remains unclear. In this study, we investigated the mechanisms of interactions between hIAPP and LL37 at the molecular level in silico using all-atom discrete molecular dynamics (DMD) with microsecond-long simulations. DMD is a novel and predictive molecular dynamics algorithm with improved computational capability and sampling efficiency that is used by us and many other groups to study the structure and amyloid aggregation of intrinsically disordered proteins.29–31 A sufficient number of independent simulations with varying initial positions and velocities were performed to guarantee adequate sampling of the structures, dynamics, and interactions of hIAPP and LL37. LL37 was found to be a dynamic and disordered peptide with two helical regions. The self-assembly of LL37 peptides was contributed by the helix-to-helix contacts with the helical structures of the peptides enhanced during the aggregation. Driven by the hydrophobic association and pi–pi interactions, the LL37 peptides can effectively interact with hIAPP in the monomer, oligomer, and fibril states. Specifically, the N-terminus and C-terminus of the LL37 peptides can firmly bind with the monomeric and oligomeric hIAPP, especially in the amyloidogenic regions, to prevent the self-interactions of amyloidogenic regions and thus hinder the formation of amyloid fibrils. Electrostatic interactions between charged residues, instead, played a minor role in the crosstalk between hIAPP and LL37. Furthermore, LL37 can bind to the elongation surface of the hIAPP fibril seeds with geometric incompatibility for monomer addition to block the growth of hIAPP fibrils. Together, our simulations identified the crucial residues and critical driving forces for the interactions between LL37 and hIAPP peptides and clarified the related dynamics and conformational changes, thus giving a comprehensive picture of the intermolecular interactions driving the inhibition of LL37 on the amyloid aggregation of hIAPP.
Materials and methods
Discrete molecular dynamics simulations
The computer simulations were conducted with the united-atom discrete molecular dynamics (DMD). DMD is a novel simulation engine that offers rapid and predictive capabilities with improved sampling efficiency. Compared with the classic molecular dynamics with continuous interaction potentials, DMD describes the molecular interactions by optimized, stepwise discrete functions. The potential energy is constant at each energy step in DMD. Thus, each atom moves with a constant velocity until its distance to the neighbouring atoms is equal to a potential step where the potential energy will change. Collisions occurred when atoms crossed the energy steps, where the velocities were updated by solving the ballistic equations considering the conservation of energy, momentum, and angular momentum.32 Thus, the trajectory of the system in DMD can be determined by calculating a series of consecutive collision events. To reduce the computational cost, the positions and velocities were updated only for the two colliding atoms during each collision event. Furthermore, the potential collisions with their neighboring atoms were determined by advanced sort algorithms in DMD, thus achieving enhanced sampling and computational efficiencies compared with classic molecular dynamics.Medusa force field that has been extensively benchmarked was used in our DMD simulations,33,34 which considered both the bonded interactions (i.e., covalent bonds, bond angles, and dihedrals) and non-bonded interactions (i.e., van der Waals, solvation, hydrogen bond, and electrostatic terms) between atoms. The classic CHARMM force field was used to assign the parameters for van der Waals, covalent bonds, bond angles, and dihedrals.35 The implicit solvent model based on the effective energy function 1 (EEF 1) was adopted to account for the solvation effect,36 whereas an explicit reaction-like model considering the distance and angle dependences was utilized to describe hydrogen bonds between peptides.37 The Debye–Huckel approximation was applied to compute the screened electrostatic interactions between charged atoms, assuming a Debye length of ∼10 Å corresponding to a physiological monovalent salt concentration of 100 mM. In combination with the Medusa force field, DMD has widely been used to model protein folding, amyloid aggregation, and bio-nano interactions. The obtained results are in excellent agreement with the experiments.38–40
Simulation setups
The initial atomic structures of hIAPP and LL37 peptides were taken from the Protein Data Bank (PDB) with IDs 2L86 and 2K6O,41 respectively. The sequences of the hIAPP and LL37 peptides are shown in Fig. 1A. A disulfide bond is formed between Cys2 and Cys7 of the hIAPP peptide. Arg and Lys residues were positively charged with +1e, whereas Asp and Glu residues were negatively charged with −1e. The charge neutrality of the system was preserved by adding counter ions. The structures and dynamics of monomeric hIAPP and LL37 peptides were simulated in a cubic box of 7.0 nm. To explore the molecular interaction mechanisms between hIAPP and LL37, the interactions of a monomeric hIAPP with a monomeric LL37 peptide (hIAPP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LL37 = 1
LL37 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and two hIAPP peptides with two LL37 peptides (hIAPP
1), and two hIAPP peptides with two LL37 peptides (hIAPP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LL37 = 2
LL37 = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were modelled. The box sizes were adjusted to maintain the same peptide concentration of hIAPP at 4.7 mM. To investigate the interaction of the LL37 peptide with the hIAPP fibril seeds, a preformed hIAPP fibril (PDB ID: 6ZRF) was adopted to interact with the monomeric LL37 peptide. The hIAPP fibril was kept fixed to reduce the computational cost, while the LL37 peptide can move freely to explore the possible binding with the fibril. The peptides were initially randomly positioned in the simulation box with minimal atomic distances larger than 1.5 nm. Thirty independent simulations were conducted for each system, starting from different initial positions and velocities to ensure sufficient samplings. Each independent simulation ran for 500 ns, accumulating 15 μs simulation time in total for each system. The temperature of the system was maintained at 300 K using the Andersen thermostat in DMD simulations.
2) were modelled. The box sizes were adjusted to maintain the same peptide concentration of hIAPP at 4.7 mM. To investigate the interaction of the LL37 peptide with the hIAPP fibril seeds, a preformed hIAPP fibril (PDB ID: 6ZRF) was adopted to interact with the monomeric LL37 peptide. The hIAPP fibril was kept fixed to reduce the computational cost, while the LL37 peptide can move freely to explore the possible binding with the fibril. The peptides were initially randomly positioned in the simulation box with minimal atomic distances larger than 1.5 nm. Thirty independent simulations were conducted for each system, starting from different initial positions and velocities to ensure sufficient samplings. Each independent simulation ran for 500 ns, accumulating 15 μs simulation time in total for each system. The temperature of the system was maintained at 300 K using the Andersen thermostat in DMD simulations.
Computational analysis
The secondary structures of peptides were calculated based on the dictionary secondary structure of protein (DSSP) program.42 Two atoms were considered to be in contact if the atomic distance was within 0.65 nm. Two residues were regarded to form residue contact when there was any atomic contact between the heavy atoms of the residues. The two-dimensional potential of mean force (PMF) was calculated using the probability distribution function, i.e., −kBT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P(x,y), where kB is the Boltzmann constant, T is the temperature, and P(x,y) represents the probability of a conformation with the coordinates of x and y.
P(x,y), where kB is the Boltzmann constant, T is the temperature, and P(x,y) represents the probability of a conformation with the coordinates of x and y.
Results and discussion
Structural dynamics of the LL37 peptide
The structural dynamics of LL37 were first investigated based on the DMD simulations of the LL37 monomer. The thirty independent simulations, starting with different initial positions and velocities, reached equilibrium states after 200 ns (Fig. S1†). The overall secondary structure contents (Fig. S2†) and the ensemble-averaged secondary structure propensities of each residue (Fig. 1B) were calculated based on the equilibrated simulations. LL37 was intrinsically disordered with coil and bend as the dominant structures with contents around 50%, especially at the N-terminus, C-terminus, and residues 15–20. Besides, LL37 exhibited a relatively high content of the helical structure, with propensities higher than 35% to form two separate α-helices at residues 5–14 and 21–31. The β-sheet content was low in the monomeric LL37 peptide, which was mainly formed at residues 4–9 and 13–21.To better understand the LL37 peptide structure, the residue-wise intra-peptide contact frequency map was calculated (Fig. 1C). The high contact frequencies at the diagonal region confirmed the two helical structures at residues 5–14 and 21–31. The anti-diagonal region of the contact frequency map indicated the formation of anti-parallel β-sheet structures between residues 4–9 and 13–21, consistent with the secondary structure propensities of each residue. The remaining regions of the intra-peptide contact frequency map had low values and no obvious patterns, suggesting that the LL37 peptide was intrinsically disordered with residues frequently changing the contact states. The obtained structure of LL37 agreed well with the results of three-dimensional triple-resonance NMR spectroscopy, which showed that the LL37 peptide adopted a helix-bend-helix motif spanning residues 2–31 with the helical bend located between residues Gly-14 and Glu-16, and the C-terminus is disordered.41 The high consistency between the simulation results and experiments demonstrated the accuracy of DMD in predicting the structures of intrinsically disordered proteins.
The structure of the LL37 peptide was further described by the PMF with respect to the number of helical residues and radius of gyration (Rg) (Fig. 1D). The free energy landscape of the LL37 peptide spanned a broad region with the Rg ranging from 1.0 nm to 2.0 nm and the number of helical residues ranging from 5 to 30, further confirming the intrinsically disordered feature of the LL37 peptide. Two main basins can be found in the free energy landscape. The first basin (A in Fig. 1D) had a high helical residue number and a small Rg value, corresponding to the compact conformations with two helices in contact. The second basin (B in Fig. 1D) was of low helical residues and slightly larger Rg, where only the residues 21–30 formed a helical structure. The N terminus adopted a rand coil structure with flexible conformations, which enabled basin B to exhibit a broader range of Rg than basin A. Together, the LL37 was a dynamic and disordered peptide with residues 5–14 and 21–31 of high propensities to form helical structures.
LL37 peptides self-assembled by helix-to-helix contacts
The self-assembly of LL37 is a crucial part of the interactions with hIAPP peptides and is essential for its functional role in antimicrobial activities.43 The molecular mechanisms for the aggregation of the LL37 peptides were investigated by the dimerization simulations. The residue-wise inter-peptide contact frequency map was calculated after the simulations reached the equilibrium states (Fig. 2A). As indicated by the contact frequency map and the accumulative contact frequency of each residue, the self-assembly of the LL37 peptide was mainly contributed by the residues 1–6 and 20–32. Both the self-interactions and the cross-interactions between these two regions were found to exhibit high contact frequencies, especially the self-interactions between residues 20–32. Residues 1–6 and 20–32 were identified to form α-helices (Fig. 1B), suggesting that the self-assembly of LL37 peptides was contributed by the helix-to-helix contacts, as shown by the typical conformations of LL37 dimers (Fig. 2D). This is confirmed by that the residues with high contact frequencies were intervened with residues of low contact frequencies in the residue-wise inter-peptide contact frequency map (Fig. 2A), which is a typical feature of helix-to-helix contacts due to the molecular structure of an α-helix. The revealed helix-to-helix contacts during the self-assembly of LL37 are consistent with the experimental results that the fibrils formed by the LL37 active core consisted of densely packed helices.44We next investigated the structural change of the LL37 peptide upon the self-assembly. The secondary structural propensities of each residue in LL37 dimers and the residue-wise intra-peptide contact frequency map were calculated (Fig. 2B and Fig. S3†). The helix propensities of residues 15–31, the second helical region, were significantly enhanced by converting the coil and bend structures into helices (Fig. S3†). In contrast, the helix propensities of residues 5–10 in the first helical region were slightly reduced. The overall contents and the corresponding propensities at each residue of the turn and β-sheet structures remained almost unchanged during the dimerization of LL37 (Fig. 2B and Fig. S3†). The changes of the secondary structures were consistent with the variations of the intra-peptide contact frequency of the LL37 peptide in dimers compared with the monomeric LL37 (Fig. 2B). Specifically, the intra-peptide contact frequency in the diagonal region of residues 15–31 increased, whereas the diagonal region of residues 5–10 reduced, reflecting the changes of helical structures. The contact frequencies in the off-diagonal regions were reduced mainly due to the rigidification of the peptides with enhanced helical structures that eliminated the contacts between the N- and C-terminus. Therefore, the self-assembly of LL37 peptides was driven by the helix-to-helix contacts and can further enhance the helical structures of the peptides.
LL37 inhibited the amyloid aggregation of hIAPP by interfering with the contact between amyloidogenic regions
We next investigated the interactions between the LL37 and hIAPP peptides to uncover the molecular of their crosstalk. The system with an LL37 peptide interacting with a hIAPP in a monomeric state (LL37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hIAPP = 1
hIAPP = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was considered. The residue-wise inter-peptide contact frequencies between each hIAPP residue and LL37 residue were calculated (Fig. 3A). As indicated by the contact frequency map and the accumulative contact frequencies of each hIAPP residue, the LL37 peptide mainly interacted with the residues 22–29 and 13–19 of hIAPP, which are the primary and second amyloidogenic core regions for the hIAPP aggregation (Fig. S3†). On the other hand, the binding of the LL37 with hIAPP peptide was mainly contributed by two regions, e.g., residues 1–15 and 18–34, which was consistent with the experimental results obtained using consecutive peptide arrays of LL37 with ten residues that covered the full-length LL37 peptide by positionally shifting one residue for each decamer peptide.22 Moreover, several peaks of contact frequency can be identified, including 1LL2, 5FF6, 13I, 17F, 20IV21, 24I, 27FL28, and 31LV32, which are all hydrophobic amino acids. In contrast, the negatively charged residues of the LL37 peptide, which is supposed to have high binding affinities with the positively charged residues 1K and 11R of the hIAPP peptide, exhibited low contact frequencies. This could be because the negatively charged residues of LL37 are surrounded by positively charged residues, such as 10KEK12, 15KEFK18, and 25KD26, the electrostatic repulsion of which prevented the binding of these regions. Mutagenesis of the identified residues or binding assays can be conducted in the future to corroborate the simulation results. Therefore, the LL37 peptide was found to interact with the amyloidogenic regions of the hIAPP peptide by hydrophobic interactions and pi–pi interactions.
1) was considered. The residue-wise inter-peptide contact frequencies between each hIAPP residue and LL37 residue were calculated (Fig. 3A). As indicated by the contact frequency map and the accumulative contact frequencies of each hIAPP residue, the LL37 peptide mainly interacted with the residues 22–29 and 13–19 of hIAPP, which are the primary and second amyloidogenic core regions for the hIAPP aggregation (Fig. S3†). On the other hand, the binding of the LL37 with hIAPP peptide was mainly contributed by two regions, e.g., residues 1–15 and 18–34, which was consistent with the experimental results obtained using consecutive peptide arrays of LL37 with ten residues that covered the full-length LL37 peptide by positionally shifting one residue for each decamer peptide.22 Moreover, several peaks of contact frequency can be identified, including 1LL2, 5FF6, 13I, 17F, 20IV21, 24I, 27FL28, and 31LV32, which are all hydrophobic amino acids. In contrast, the negatively charged residues of the LL37 peptide, which is supposed to have high binding affinities with the positively charged residues 1K and 11R of the hIAPP peptide, exhibited low contact frequencies. This could be because the negatively charged residues of LL37 are surrounded by positively charged residues, such as 10KEK12, 15KEFK18, and 25KD26, the electrostatic repulsion of which prevented the binding of these regions. Mutagenesis of the identified residues or binding assays can be conducted in the future to corroborate the simulation results. Therefore, the LL37 peptide was found to interact with the amyloidogenic regions of the hIAPP peptide by hydrophobic interactions and pi–pi interactions.
The interaction between the hIAPP and LL37 peptides induced the structural changes of each peptide. As indicated by the secondary structure propensities of each residue (Fig. 3C–F and Fig. S4†), the crosstalk between the hIAPP and LL37 peptides enhanced the helical propensities of residues 5–17 of hIAPP peptides. The β-sheet propensities of residues 13–19 and 22–29 were promoted by forming the inter-peptide hydrogen bonds with LL37. Meanwhile, the first helical region of the LL37 peptide (residues 5–14) was converted into a β-sheet structure, while the second helical region (residues 21–31) remained almost unchanged. The β-sheet structures of the LL37 peptide at residues 4–9 and 13–21 were significantly promoted by interacting with the hIAPP peptide. Together, the β-sheet structures of both hIAPP and LL37 peptides were elevated by forming inter-peptide hydrogen bonds and the helix of hIAPP was enhanced due to the crosstalk between hIAPP and LL37 peptides.
LL37 hinders the oligomerization of hIAPP by reducing the inter-peptide interactions
To reveal the interactions of LL37 peptides with hIAPP in the oligomerization process, we investigated the dynamic coaggregation process of two hIAPP peptides incubated with two LL37 peptides (e.g., LL37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hIAPP = 2
hIAPP = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
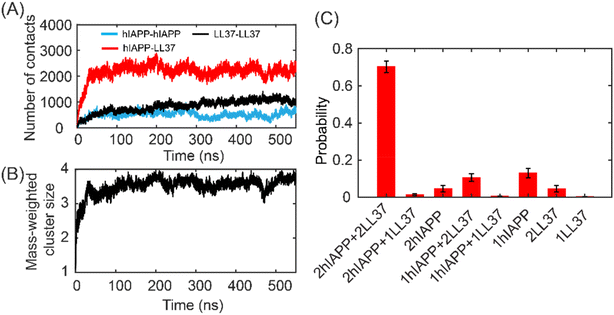
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). The number of atomic contacts between peptides gradually increased and reached equilibrium states after 200 ns (Fig. 4A). Extensive contacts between hIAPP and LL37 peptides can be observed. Besides, the atomic contacts of LL37–LL37 were higher than those of hIAPP–hIAPP. The dimerization propensity of the system with LL37–LL37 was higher than that with hIAPP–hIAPP (Fig. S5†), suggesting that the binding affinity of LL37–LL37 was stronger than that of hIAPP–hIAPP. The mass-weighted cluster size of the oligomer formed by the LL37 and hIAPP peptides showed that most of the peptides aggregated into a tetramer (Fig. 4B). The distributions of the oligomer species confirmed that over 70% of the oligomers were hetero tetramers (Fig. 4C). The hIAPP in the monomeric state was 13% and the monomeric LL37 peptide was only 0.1%, suggesting that LL37 is more aggregation-prone than hIAPP (Fig. S5†). Taken together, the LL37 peptides can strongly interact with hIAPP to form hetero-oligomers.
2). The number of atomic contacts between peptides gradually increased and reached equilibrium states after 200 ns (Fig. 4A). Extensive contacts between hIAPP and LL37 peptides can be observed. Besides, the atomic contacts of LL37–LL37 were higher than those of hIAPP–hIAPP. The dimerization propensity of the system with LL37–LL37 was higher than that with hIAPP–hIAPP (Fig. S5†), suggesting that the binding affinity of LL37–LL37 was stronger than that of hIAPP–hIAPP. The mass-weighted cluster size of the oligomer formed by the LL37 and hIAPP peptides showed that most of the peptides aggregated into a tetramer (Fig. 4B). The distributions of the oligomer species confirmed that over 70% of the oligomers were hetero tetramers (Fig. 4C). The hIAPP in the monomeric state was 13% and the monomeric LL37 peptide was only 0.1%, suggesting that LL37 is more aggregation-prone than hIAPP (Fig. S5†). Taken together, the LL37 peptides can strongly interact with hIAPP to form hetero-oligomers.
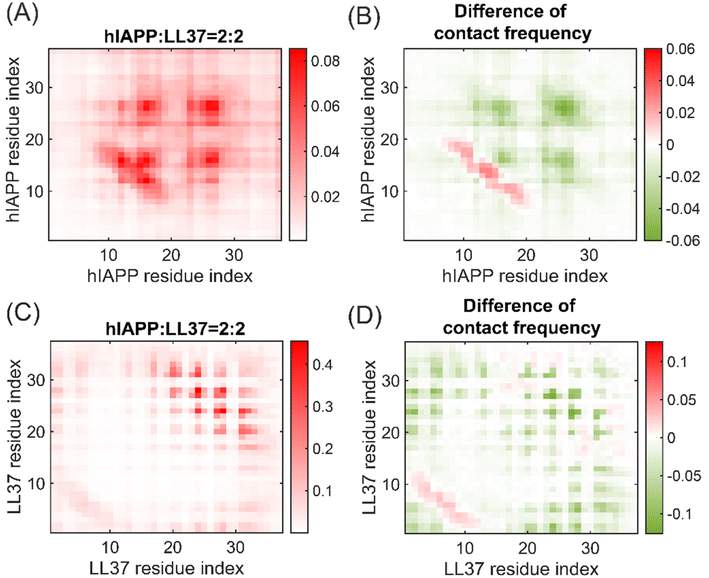
The influence of LL37 on the oligomerization of hIAPP was investigated by the contacts between each residue. The dimerization of hIAPP is driven by the contacts between the amyloidogenic regions at residues 13–19 and 22–29 in the presence of LL37 peptides (Fig. 5A), similar to the self-assembly of hIAPP peptides (Fig. S6†). Notably, the presence of LL37 peptides significantly reduced the inter-peptide contact frequencies, especially at the amyloidogenic regions 13–19 and 22–29 (Fig. 5B). Therefore, LL37 inhibited the oligomerization of hIAPP peptides by binding to the core regions and prevented the contacts between the core regions. The interactions between LL37 peptides in the hetero-tetramer remained to be mediated by the helix-to-helix contacts between residues 1–6 and 20–32 (Fig. 5C). Moreover, the helix-to-helix contacts between the LL37 peptides were also hindered by the hIAPP peptides, suggesting that hIAPP peptides can inhibit the aggregation of LL37. Therefore, the formation of hetero-oligomers of hIAPP and LL37 effectively impeded the amyloid aggregation of hIAPP by reducing the interactions between amyloidogenic regions of hIAPP peptides.
To gain a more detailed understanding of the inhibition mechanism of LL37 peptides on the oligomerization of hIAPP, the 2D PMF with respect to the number of residue contacts between hIAPP peptides and the center-to-center distance of hIAPP peptides in the oligomers were calculated (Fig. 6). Peptides with a large distance and a low residue contact were loosely connected whereas peptides with a small distance and a high residue contact were densely packed. As shown in the figures, the free energy landscape covered a wild spectrum of residue contacts and peptide distances, suggesting that the oligomers are highly dynamic with varying internal connections. In the self-assembly of hIAPP peptides, the free energy landscape featured a broad basin with peptide distance varying from 0.8 nm to 2.1 nm and residue contacts ranging from 9 to 26 (Fig. 6A). In the presence of LL37 peptides, however, the basin shifted to the region with low residue contacts and large peptide distances (Fig. 6B). The typical conformations showed that the hIAPP peptides were separated away by the LL37 peptides in the basin region. Thus, the LL37 peptides altered the structure of hIAPP oligomers by enlarging the peptide distance and hindering the residue contacts.
LL37 disrupted the fibril growth of hIAPP by caping on the elongation surface of fibrils
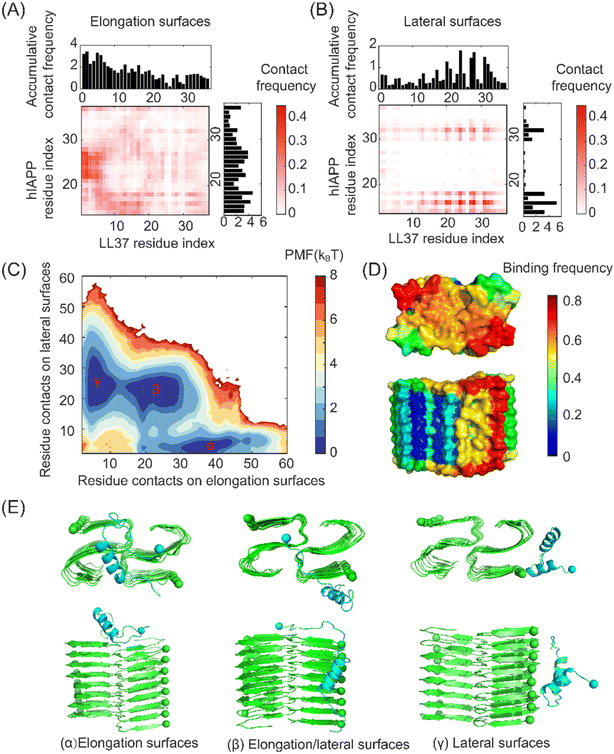
The fibril seeds can be nucleated during the amyloid aggregation of hIAPP. We further investigated the interactions between the LL37 peptide and the hIAPP fibril seeds. The high-resolution atomic structure of the hIAPP fibril obtained by cryo-EM was adopted.45 The N-terminal residues 1–13 were highly disordered and flexible without clear density in cryo-EM, and only the residues 14–37 were considered the fibril structure. The residue-wise contact frequencies between the LL37 peptide and the peptides on the elongation and lateral surfaces of the hIAPP fibrils were calculated (Fig. 7A&B). As indicated by the figures, the LL37 peptide exhibited distinct binding properties on the lateral and elongation surfaces of the hIAPP fibrils. Specifically, the residues 1–20 of LL37 peptides had high binding affinities with the residues 14–19 and 22–29 of hIAPP peptides on the elongation surfaces. In contrast, the regions 14–19 and 29–31 of hIAPP peptides on the lateral surfaces preferred to bind with the residues 19–32 of the LL37 peptides. The amyloidogenic residues 22–29 were buried in the concave region on the lateral surfaces and thus exhibited low binding affinities with LL37 peptides (Fig. 7D).To gain a better understanding of the interactions between the LL37 peptide and hIAPP fibrils, the 2D PMF with respect to the residue contacts on elongation surfaces and lateral surfaces was calculated (Fig. 7C). Three basins on the free energy landscape can be identified, corresponding to binding with the elongation surface, lateral surfaces, and elongation/lateral surface (Fig. 7E). The three basins had similar free energy values, suggesting that the LL37 peptides can adopt the three binding conformations with similar probabilities. Importantly, by binding on the amyloidogenic region of the elongation surfaces of hIAPP fibrils, LL37 peptides induced the geometrical incompatibility for the adding of hIAPP peptides to form in-register β-sheet structures, thus blocking the growth of fibrils. At the same time, the LL37 peptides stuck at the lateral surfaces can impede the secondary nucleation of hIAPP peptides by hindering the binding and diffusion of the hIAPP peptides. Together, LL37 inhibited the growth of hIAPP fibrils by blocking the fibril elongation and hindering the secondary nucleation process.
Inhibition mechanism of the LL37 peptides on the amyloid aggregation of hIAPP
Based on the above results, the molecular mechanism for the inhibition of hIAPP aggregation by the LL37 peptides can be proposed (Fig. 8). The amyloid aggregation of hIAPP involves the nucleation of soluble oligomers with dynamic structures, which further forms the fibril seeds with in-register β-sheet structures via conformational changes. The fibrils further grow by elongation where fibril lengths increase by monomer addition on the elongation surface. Driven by the hydrophobic associations and pi–pi interactions, the N- and C-terminal regions of the LL37 peptide can bind with the amyloidogenic regions of hIAPP to form heterooligomers. The strong binding affinities between the LL37 and hIAPP peptides can effectively prevent the interactions between the amyloidogenic regions of hIAPP peptides, hindering the conversion into amyloid fibrils. Furthermore, LL37 peptides can bind on the elongation surfaces of the fibrillar seeds, inducing the geometric incompatibility to block the fibril growth via monomer addition. Taken together, the host-defense peptide cathelicidin LL37 can interact with the hIAPP peptides and interfere with the amyloid aggregation at multiple stages. | ||
| Fig. 8 Schematic plot of the inhibition mechanism of the LL37 peptides on the amyloid aggregation of hIAPP. | ||
Conclusions
Mounting evidence suggests that the hIAPP related to T2D and the host-defense peptide cathelicidin LL37 exhibit biological connections and LL37 can effectively inhibit the amyloid aggregation of hIAPP with the mechanism remaining elusive. In this paper, we investigated the molecular interactions underlying the inhibition effect of LL37 peptides on the aggregation of hIAPP using all-atom DMD simulations. By conducting multiple independent simulations with different initial positions and velocities to ensure sufficient sampling, we showed that the LL37 peptide was an intrinsically disordered protein with two helices formed at residues 5–14 and 21–31. The helix-to-helix contacts between LL37 peptides mediated the self-assembly of LL37 peptides, which further enhanced the helical structures. The hydrophobic residues of LL37 peptides had strong binding affinities with the hIAPP, especially in the amyloidogenic regions, driven by hydrophobic associations and pi–pi interactions. The binding of the LL37 peptides with the hIAPP prevented the self-interactions of amyloidogenic regions, hindering the formation of amyloid fibrils. Furthermore, the elongation of hIAPP fibril seeds was blocked by the binding of LL37 peptides to the fibril elongation surfaces with geometric incompatibility for monomer addition. Together, our simulation gave a comprehensive picture of the intermolecular interactions that are responsible for the inhibition of LL37 on the amyloid aggregation of hIAPP at various stages. The efficient inhibition of the amyloid aggregation of hIAPP by LL37 indicated potential therapeutic opportunities by utilizing the functional properties of antimicrobial peptides. The mechanism revealed by our study can contribute to a better understanding of the pathological links between microbial infections and amyloid diseases and guide the design of novel therapies combining antimicrobial and anti-amyloid functions. Further research can be conducted to investigate the amyloid inhibition effects of LL37 derivatives and analogs, such as KR-12 (residues 18–29 of LL37), which was proposed to be the smallest peptide of LL37 exhibiting antimicrobial activity, thus facilitating the design of therapeutic candidates with improved efficiencies.Author contributions
H. T. conceived the project, conducted the simulations, analyzed the data, and wrote the manuscript.Data availability
The data supporting this article have been included as part of the ESI.† Typical trajectories of the interactions between LL37 and hIAPP are available at Zenodo at https://doi.org/10.5281/zenodo.14260346Conflicts of interest
The author declares no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 12402237), the Natural Science Foundation of Jiangsu Province (No. BK20241505), and the Research Foundation of the State Key Laboratory of Structural Analysis, Optimization and CAE Software for Industrial Equipment, Dalian University of Technology (No. GZ24106).References
- E. Ahmad, S. Lim, R. Lamptey, D. R. Webb and M. J. Davies, Lancet, 2022, 400, 1803–1820 CrossRef.
- N. Louros, J. Schymkowitz and F. Rousseau, Nat. Rev. Mol. Cell Biol., 2023, 24, 912–933 CrossRef PubMed.
- P. Westermark, A. Andersson and G. T. Westermark, Physiol. Rev., 2011, 91, 795–826 CrossRef PubMed.
- D. Milardi, E. Gazit, S. E. Radford, Y. Xu, R. U. Gallardo, A. Caflisch, G. T. Westermark, P. Westermark, C. L. Rosa and A. Ramamoorthy, Chem. Rev., 2021, 121, 1845–1893 CrossRef.
- P. Cao, A. Abedini and D. P. Raleigh, Curr. Opin. Struct. Biol., 2013, 23, 82–89 CrossRef.
- T. P. J. Knowles, M. Vendruscolo and C. M. Dobson, Nat. Rev. Mol. Cell Biol., 2014, 15, 384–396 CrossRef.
- T. C. T. Michaels, D. Qian, A. Šarić, M. Vendruscolo, S. Linse and T. P. J. Knowles, Nat. Rev. Phys., 2023, 5, 379–397 CrossRef.
- M. G. Iadanza, M. P. Jackson, E. W. Hewitt, N. A. Ranson and S. E. Radford, Nat. Rev. Mol. Cell Biol., 2018, 19, 755–773 CrossRef.
- Q. Cao, D. R. Boyer, M. R. Sawaya, P. Ge and D. S. Eisenberg, Nat. Struct. Mol. Biol., 2020, 27, 653–659 CrossRef.
- B. P. Lazzaro, M. Zasloff and J. Rolff, Science, 2020, 368, eaau5480 CrossRef PubMed.
- Y. Huan, Q. Kong, H. Mou and H. Yi, Front. Microbiol., 2020, 11, 582779 CrossRef PubMed.
- D. Vandamme, B. Landuyt, W. Luyten and L. Schoofs, Cell. Immunol., 2012, 280, 22–35 CrossRef.
- U. H. N. Dürr, U. S. Sudheendra and A. Ramamoorthy, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 1408–1425 CrossRef.
- M. A. Alford, B. Baquir, F. L. Santana, E. F. Haney and R. E. W. Hancock, Front. Microbiol., 2020, 11, 1902 CrossRef PubMed.
- S. Bhattacharjya, Z. Zhang and A. Ramamoorthy, Biomolecules, 2024, 14, 320 CrossRef PubMed.
- F. Neville, M. Cahuzac, O. Konovalov, Y. Ishitsuka, K. Y. C. Lee, I. Kuzmenko, G. M. Kale and D. Gidalevitz, Biophys. J., 2006, 90, 1275–1287 CrossRef CAS PubMed.
- J. Overhage, A. Campisano, M. Bains, E. C. W. Torfs, B. H. A. Rehm and R. E. W. Hancock, Infect. Immun., 2008, 76, 4176–4182 CrossRef CAS.
- B. Yang, D. Good, T. Mosaiab, W. Liu, G. Ni, J. Kaur, X. Liu, C. Jessop, L. Yang, R. Fadhil, Z. Yi and M. Q. Wei, BioMed Res. Int., 2020, 2020, 8349712 CrossRef PubMed.
- R. Bals, X. Wang, M. Zasloff and J. M. Wilson, Proc Natl Acad. Sci. U. S. A., 1998, 95, 9541–9546 CrossRef CAS.
- L. D. Pound, C. Patrick, C. E. Eberhard, W. Mottawea, G.-S. Wang, T. Abujamel, R. Vandenbeek, A. Stintzi and F. W. Scott, Diabetes, 2015, 64, 4135–4147 CrossRef CAS.
- J. Sun, L. Furio, R. Mecheri, A. M. van der Does, E. Lundeberg, L. Saveanu, Y. Chen, P. van Endert, B. Agerberth and J. Diana, Immunity, 2015, 43, 304–317 CrossRef CAS.
- V. Armiento, K. Hille, D. Naltsas, J. S. Lin, A. E. Barron and A. Kapurniotu, Angew. Chem., Int. Ed., 2020, 59, 12837–12841 CrossRef CAS.
- R. Mani, S. D. Cady, M. Tang, A. J. Waring, R. I. Lehrer and M. Hong, Proc Natl Acad. Sci. U. S. A., 2006, 103, 16242–16247 CrossRef CAS PubMed.
- S. J. Soscia, J. E. Kirby, K. J. Washicosky, S. M. Tucker, M. Ingelsson, B. Hyman, M. A. Burton, L. E. Goldstein, S. Duong, R. E. Tanzi and R. D. Moir, PLoS One, 2010, 5, e9505 CrossRef.
- L. Wang, Q. Liu, J.-C. Chen, Y.-X. Cui, B. Zhou, Y.-X. Chen, Y.-F. Zhao and Y.-M. Li, Biol. Chem., 2012, 393, 641–646 CrossRef CAS PubMed.
- N. B. Last and A. D. Miranker, Proc Natl Acad. Sci. U. S. A., 2013, 110, 6382–6387 CrossRef CAS.
- B. L. Kagan, Biophys. J., 2011, 100, 1597–1598 CrossRef CAS PubMed.
- Y. Tang, Y. Zhang, D. Zhang, Y. Liu, R. Nussinov and J. Zheng, Chem. Soc. Rev., 2024, 53, 8713–8763 RSC.
- Y. Xing, A. Nandakumar, A. Kakinen, Y. Sun, T. P. Davis, P. C. Ke and F. Ding, J. Phys. Chem. Lett., 2021, 12, 368–378 CrossRef.
- H. Tang, Y. Sun and F. Ding, J. Chem. Inf. Model., 2022, 62, 1760–1770 CrossRef.
- X. Liang, N. Andrikopoulos, H. Tang, Y. Wang, F. Ding and P. C. Ke, Small, 2024, 20, 2308753 CrossRef.
- S. Peng, F. Ding, B. Urbanc, S. V. Buldyrev, L. Cruz, H. E. Stanley and N. V. Dokholyan, Phys. Rev. E:Stat., Nonlinear, Soft Matter Phys., 2004, 69, 041908 CrossRef PubMed.
- S. Yin, F. Ding and N. V. Dokholyan, Nat. Methods, 2007, 4, 466–467 CrossRef.
- S. Yin, L. Biedermannova, J. Vondrasek and N. V. Dokholyan, J. Chem. Inf. Model., 2008, 48, 1656–1662 CrossRef PubMed.
- B. R. Brooks, R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan and M. Karplus, J. Comput. Chem., 1983, 4, 187–217 CrossRef.
- T. Lazaridis, Curr. Opin. Struct. Biol., 2000, 10, 139–145 CrossRef PubMed.
- F. Ding, J. M. Borreguero, S. V. Buldyrey, H. E. Stanley and N. V. Dokholyan, Proteins: Struct., Funct., Genet., 2003, 53, 220–228 CrossRef.
- Y. Li, H. Tang, H. Zhu, A. Kakinen, D. Wang, N. Andrikopoulos, Y. Sun, A. Nandakumar, E. Kwak, T. P. Davis, D. T. Leong, F. Ding and P. C. Ke, ACS Appl. Mater. Interfaces, 2021, 13, 29936–29948 Search PubMed.
- S. J. Bunce, Y. Wang, K. L. Stewart, A. E. Ashcroft, S. E. Radford, C. K. Hall and A. J. Wilson, Sci. Adv., 2019, 5, eaav8216 Search PubMed.
- Y. Sun, A. Kakinen, X. Wan, N. Moriarty, C. P. J. Hunt, Y. Li, N. Andrikopoulos, A. Nandakumar, T. P. Davis, C. L. Parish, Y. Song, P. C. Ke and F. Ding, Nano Today, 2021, 38, 101125 CrossRef PubMed.
- G. Wang, J. Biol. Chem., 2008, 283, 32637–32643 CrossRef CAS PubMed.
- W. Kabsch and C. Sander, Biopolymers, 1983, 22, 2577–2637 CrossRef CAS.
- E. Y. Lee, C. Zhang, J. Di Domizio, F. Jin, W. Connell, M. Hung, N. Malkoff, V. Veksler, M. Gilliet, P. Ren and G. C. L. Wong, Nat. Commun., 2019, 10, 1012 CrossRef PubMed.
- Y. Engelberg and M. Landau, Nat. Commun., 2020, 11, 3894 CrossRef CAS PubMed.
- C. Röder, T. Kupreichyk, L. Gremer, L. U. Schäfer, K. R. Pothula, R. B. G. Ravelli, D. Willbold, W. Hoyer and G. F. Schröder, Nat. Struct. Mol. Biol., 2020, 27, 660–667 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr05075d |
| This journal is © The Royal Society of Chemistry 2025 |