 Open Access Article
Open Access ArticleA novel Janus nanomachine based on mesoporous silica nanoparticles anisotropically modified with PAMAM dendrimers for enzyme-controlled drug delivery†
Beatriz
Mayol
 a,
Esther
García-Díez
a,
Esther
García-Díez
 a,
Alexander
Hoppe
a,
Lucía
Espejo
a,
Miranda
Muñoz
a,
Marta
González
a,
Alexander
Hoppe
a,
Lucía
Espejo
a,
Miranda
Muñoz
a,
Marta
González
 a,
Anabel
Villalonga
a,
Teresa
Moreno
a,
Anabel
Villalonga
a,
Teresa
Moreno
 a,
Alfredo
Sánchez
a,
Diana
Vilela
a,
Narcisa
Martinez-Quiles
b,
Paloma
Martínez-Ruíz
a,
Alfredo
Sánchez
a,
Diana
Vilela
a,
Narcisa
Martinez-Quiles
b,
Paloma
Martínez-Ruíz
 a and
Reynaldo
Villalonga
a and
Reynaldo
Villalonga
 *a
*a
aNanosensors and Nanomachines Group, Faculty of Chemistry, Complutense University of Madrid, 28040 Madrid, Spain. E-mail: rvillalonga@quim.ucm.es
bDepartment of Immunology, Ophthalmology and ENT, School of Medicine, Complutense University, 28040 Madrid, Spain
First published on 3rd February 2025
Abstract
A masking/toposelective modification approach was employed to prepare a new organic–inorganic Janus nanomaterial by attaching ethylenediamine core polyamidoamine G-4.5 dendrimers to a defined face of mesoporous silica nanoparticles. The anisotropic colloid was then sequentially functionalized on the mesoporous face with (3-isocyanatopropyl)triethoxysilane, 1-(4-aminophenyl)-2-phenylethane-1,2-dione and β-cyclodextrin to assemble a novel H2O2-sensitive gating mechanism. The Janus nanomachine was finally constructed by immobilizing glucose oxidase on the dendrimeric face. The smart nanodevice released the encapsulated payload in the presence of H2O2 and glucose, and was successfully evaluated for the enzyme-controlled delivery of the antitumoral drug doxorubicin into HeLa cancer cells.
1. Introduction
During the last decades, increased attention has been paid to designing advanced drug delivery systems aimed at improving the pharmacological, pharmacokinetics and therapeutic properties of new or conventional pharmaceutical compounds.1 These formulated systems can improve drug stability and solubility, reduce toxicity, and enhance efficacy through on-demand time-controlled release mechanisms.2In this context, nanomaterials engineering constitutes an emergent and interdisciplinary research area that has contributed significantly to the development of new and efficient drug release systems.1,3–5 In this sense, a large assortment of nanomaterials, some of them appropriately functionalized with organic ligands or active biomolecules, have been employed as building elements to construct advanced nanocarriers able to release drugs at specific sites, times and/or under demand in response to specific chemical or physical stimuli.
Mesoporous silica nanoparticles (MSNs) are inorganic nanomaterials largely employed to develop new and sophisticated drug delivery systems due to their exceptional properties, such as low toxicity, high biocompatibility and load capacity, and tunability of pore size and shape through standardized synthetic protocols.6 MSNs can be also easily functionalized on their outer pore surface via silane coupling reagents, providing the nanomaterial with tailored functionalities. In particular, mechanization of MSNs with stimuli-responsive gate-like ensembles allows on-demand delivery of the encapsulated payload with precise temporal control.7,8
With the aim to design more sophisticated MSN-based smart nanocarriers for drug delivery, we previously introduced two new concepts: (i) the preparation of Janus colloids with metal nanoparticles (Au, Pt, Ir, etc.) and MSNs as opposite faces and their use as anisotropic “hardware” for the sequential and site-selective assembly of the encapsulation, control and release mechanisms, and (ii) the employment of enzymes as sensing and control units in the resulting device.9,10 Such approaches allowed easy and controlled preparation of highly selective nanomachines for smart delivery by using different metal nanoparticles, enzymes and enzyme combinations.11–13
Taking into account the large variety of redox enzymes producing H2O2 after catalytic transformation of their substrates,14 the construction of enzyme-powered mesoporous nanomachines mechanized with H2O2-sensitive gate-like ensembles stimulated our interest. However, mesoporous Janus nanoparticles bearing catalytically active metal nanoparticles cannot be employed to construct such nanocarriers due to the fast decomposition of H2O2 on such metallic surfaces.15 This limitation can be overcome by preparing new mesoporous Janus colloids provided with organic nanoparticles as opposite faces. In this sense, dendrimers constitute a good choice due to the unique properties of these synthetic and hyperbranched “soft” nanomaterials, such as nanometric size, structural uniformity with globular or ellipsoidal shape, monodispersity and high permeability of their internal cavities.16,17 In addition, several dendrimers are synthesized with high densities of functional groups at their surface, allowing their use as supports for the multipoint covalent immobilization of enzymes and other biomolecules.
In this paper, we describe the preparation of novel Janus nanoparticles with ethylenediamine core polyamidoamine G-4.5 dendrimers (PAMAM) and MSN opposite faces. These colloids were mechanized on the mesoporous face with a novel H2O2-sensitive gate-like ensemble based on 1-(4-aminophenyl)-2-phenylethane-1,2-dione moieties supramolecularly capped with β-cyclodextrin. The anisotropic nanomaterial was also functionalized with glucose oxidase (GOx, EC 1.1.3.4) covalently immobilized on the dendritic face to act as a signaling redox enzyme. During the last years, GOx has attracted increasing interest in the biomedical field due to its inherent biocompatibility, non-toxicity, and unique catalysis against β-D-glucose.18,19 As a proof-of-concept, this engineered nanomachine with enzymatic control was evaluated as a smart nanocarrier for the on-command and autonomous release of tris(2,2′-bipyridyl)dichlororuthenium(II) (Ru(bipy)3Cl2) and doxorubicin (Doxo) as model payloads.
2. Experimental
2.1. Materials
Ethylenediamine core polyamidoamine G-4.5 dendrimer was acquired from Dendritic Nanotechnologies Inc. (USA). Glucose oxidase from A. niger, (3-aminopropyl)triethoxysilane (APTES), tetraethoxysilane (TEOS), cetyltrimethylammonium bromide (CTAB), (3-isocyanatopropyl)triethoxysilane (ICPTES), N-hydroxysuccinimide (NHS), tris(2,2′-bipyridyl)dichlororuthenium(II) hexahydrate, β-cyclodextrin (βCD), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC) and PBS were purchased from Sigma-Aldrich (USA). Iscove's modified Dulbecco's medium (IMDM), fetal bovine serum (FBS), WGA (wheat germ agglutinin) conjugated to Alexa Flour-488 and Hoechst 333442 were purchased from Thermo Fisher. Doxorubicin (Doxo) was acquired from Sequoia Research Products Ltd (USA). Solvents were provided by Scharlau (Spain). All other reagents were of analytical grade.2.2. General techniques
Spectrophotometric analyses were performed with an Ultrospec™ 8000 Dual Beam UV/VIS spectrophotometer (Biochrom, UK). 1H NMR and 13C NMR measurements were performed with Bruker DPX 300 MHz equipment (USA). FT-IR spectra were measured on a PerkinElmer Spectrum 400 Series spectrometer (PerkinElmer, USA). Transmission electron microscopy (TEM) was performed with a JEOL JEM-2100 microscope (JEOL Ltd, Japan). Powder X-ray diffraction (XRD) was performed with an X'Pert MRD diffractometer (PANanalytical B.V., The Netherlands). Nitrogen adsorption/desorption isotherms and pore size distributions were determined with an ASAP 2020 Physisorption Analyzer (Micromeritics, USA). Thermal analysis was performed with TA Instruments SDT-Q600 apparatus (USA).2.3. Synthesis of 1-(4-aminophenyl)-2-phenylethane-1,2-dione (APPD)20
Acetophenone (484 mg, 4.0 mmol), CuCl2·2H2O (683 mg, 4 mmol) and I2 (303 mg, 1.2 mmol) were dissolved in 20 mL of anhydrous DMSO under an Ar atmosphere. The reaction mixture was stirred at 100 °C until complete reaction of acetophenone, as determined by TLC. The mixture was then cooled to room temperature, and 448 mg (4.8 mmol) of aniline was added. The temperature was adjusted to 100 °C and the mixture was stirred for 12 h under an Ar atmosphere. After cooling to room temperature, 30 mL of water was added, and the synthesized compound was extracted with EtOAc (3 × 20 mL). The combined EtOAc extracts were dried with Na2SO4 and the solvent eliminated using a rotoevaporator. The crude product was then purified by chromatography under isocratic conditions on a silica column using n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (5
EtOAc (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as eluent. Yield: 60% (553 mg, bright yellow powder). 1H NMR (300 MHz, CDCl3) δ/ppm: 7.89 (d, J = 7.00 Hz, 2 H), 7.71 (d, J = 8.69 Hz, 2 H), 7.55 (d, J = 7.36 Hz, 1 H), 7.41 (t, J = 8.20 Hz, 2 H), 6.56 (d, J = 8.77 Hz, 2 H). 13C NMR (75 MHz, CDCl3) δ/ppm: 195.5, 192.7, 152.9, 134.5, 133.5, 132.7, 129.9, 128.9, 123.2, 114.0. FT-IR: 3505 cm−1, 3376 cm−1, 2918 cm−1, 1630 cm−1, 1578 cm−1. MP: 128–129 °C.
1) as eluent. Yield: 60% (553 mg, bright yellow powder). 1H NMR (300 MHz, CDCl3) δ/ppm: 7.89 (d, J = 7.00 Hz, 2 H), 7.71 (d, J = 8.69 Hz, 2 H), 7.55 (d, J = 7.36 Hz, 1 H), 7.41 (t, J = 8.20 Hz, 2 H), 6.56 (d, J = 8.77 Hz, 2 H). 13C NMR (75 MHz, CDCl3) δ/ppm: 195.5, 192.7, 152.9, 134.5, 133.5, 132.7, 129.9, 128.9, 123.2, 114.0. FT-IR: 3505 cm−1, 3376 cm−1, 2918 cm−1, 1630 cm−1, 1578 cm−1. MP: 128–129 °C.
2.4. Preparation of Janus nanoparticles (N1)
MSNs (N0) were first prepared by dissolving 1.0 g of CTAB in 0.48 L of water and further adding 3.5 mL of 2.0 mol L−1 NaOH solution. The mixture was heated to 80 °C and 5.0 mL of TEOS was added dropwise under vigorous magnetic stirring and constant temperature. The mixture was stirred for 2 h under the same conditions and, after cooling, the resulting white solid was filtered, exhaustively washed with water and methanol, and dried at 60 °C. The solid was finally calcined at 550 °C for 5 h.To prepare the PAMAM–MSN Janus nanoparticles, 200 mg of solid N0 was dispersed in 10 mL of 1.0 μM CTAB in 6.7% EtOH aqueous solution. The mixture was heated to 75 °C and 1.0 g of paraffin wax was then added. After melting the paraffin wax, the mixture was stirred at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and at 75 °C by using an Ultra Turrax T-10 homogenizer (IKA, Germany). The resulting emulsion was then magnetically stirred at 4000 rpm for 1 h at 75 °C, further cooled to room temperature, and mixed with 10 mL of MeOH. The unmasked faces of the MSNs were then provided with primary amino groups by adding 200 μL of APTES, and stirring the mixture for 3 h at room temperature. The mixture was further filtered, and the resulting solid was exhaustively washed with MeOH and then dispersed in 25 mL of methanol.
000 rpm for 10 min and at 75 °C by using an Ultra Turrax T-10 homogenizer (IKA, Germany). The resulting emulsion was then magnetically stirred at 4000 rpm for 1 h at 75 °C, further cooled to room temperature, and mixed with 10 mL of MeOH. The unmasked faces of the MSNs were then provided with primary amino groups by adding 200 μL of APTES, and stirring the mixture for 3 h at room temperature. The mixture was further filtered, and the resulting solid was exhaustively washed with MeOH and then dispersed in 25 mL of methanol.
In parallel, 10 μL of PAMAM was first dissolved in 4.81 mL of 0.1 mol L−1 sodium phosphate buffer, pH 6.0, and then 100 μL of 0.2 μg mL−1 EDAC and 80 μL of 0.2 μg mL−1 NHS were added. The mixture was stirred at 4 °C for 1 h, then diluted up to 120 mL with 0.1 mol L−1 sodium phosphate buffer, pH 7.5, and mixed with the methanolic dispersion of paraffin-masked amino-activated MSNs. The mixture was stirred at room temperature for 18 h, and further centrifuged. The resulting solid was sequentially washed with MeOH, mixtures of MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHCl3 2
CHCl3 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and finally with CHCl3 until total elimination of the paraffin wax. The PAMAM–MSN Janus nanoparticles (N1) were kept in a desiccator until use.
2, and finally with CHCl3 until total elimination of the paraffin wax. The PAMAM–MSN Janus nanoparticles (N1) were kept in a desiccator until use.
2.5. Preparation of gated and Ru(bipy)3Cl2-loaded Janus nanoparticles (N2)
A dispersion of 50 mg of solid N1 in 3.0 mL of anhydrous toluene was mixed with 50 μL of ICPTES and stirred at room temperature for 18 h. The mixture was centrifuged, and the solid was 3-times washed with toluene and further redispersed in 1.0 mL of the same solvent. The synthesized APPD (45 mg) was dissolved in 2.0 mL of toluene, and mixed with the nanoparticle suspension. The mixture was stirred at room temperature for 18 h, centrifuged and the solid was 3-times washed with toluene. The 1-(4-aminophenyl)-2-phenylethane-1,2-dione-branched MSNs were dried at 60 °C for 12 h, and kept in a desiccator until use.To prepare the dye-loaded nanoparticles, the MSNs previously modified were dispersed in 3.0 mL of 0.1 mol L−1 sodium phosphate buffer, pH 7.5, containing 25 mg of Ru(bipy)3Cl2. The mixture was stirred overnight at room temperature, and then 60 mg of βCD was added. The mixture was kept under continuous stirring for 18 h at room temperature, then centrifuged and the resulting solid N2 was exhaustively washed with the same phosphate buffer, dried at 60 °C, and kept in a desiccator until use.
2.6. Preparation of the dye-loaded enzyme-controlled nanomachine (N3)
The nanomachine N3 was prepared by dispersing 10 mg of N2 nanoparticles in 1.0 mL of cold 0.1 mol L−1 sodium phosphate buffer, pH 6.0, containing 5 mg of EDAC and 5 mg of NHS. The mixture was stirred for 30 min at 4 °C, centrifuged and 3-times washed with cold 0.1 mol L−1 sodium phosphate buffer, pH 7.5, and finally redispersed in 1.0 mL of the same cold buffer. Glucose oxidase (1.2 mg) was added, and the mixture was stirred overnight at 4 °C. The solid was purified by centrifugation, 3-times washed and redispersed in 1.0 mL of the same cold buffer. The dispersion of the nanomachine N3 was stored in a refrigerator until use.2.7. Preparation of the drug-loaded enzyme-controlled nanomachine (N4)
The Doxo-loaded nanomachine was prepared as previously described in Sections 2.5 and 2.6, but by dispersing 10 mg of solid N2 in 4 mL of 1.88 mg mL−1 Doxo solution in 0.1 M sodium phosphate buffer, pH 7.5, instead of using the model dye. The dispersion of the resulting nanomachine N4, at 10 mg mL−1 final concentration in 0.1 mol L−1 sodium phosphate buffer, pH 7.5, was kept in a refrigerator until use. Similar nanomachines without the Doxo payload (N4A) and without Doxo and GOx (N4B) were also prepared as controls for cell viability assays.2.8. Release assays
Solid N3 was dispersed in 100 mmol L−1 sodium phosphate buffer, pH 7.5, at 3.0 mg mL−1 final concentration. For release assays, samples of 2.5 mL were employed in each batch experiment. Glucose at 0.05–0.1 mol L−1 or H2O2 at 0.1–1.0 mol L−1 final concentration was added under continuous stirring to trigger the gate-opening mechanism, and the released dye was monitored spectrophotometrically at 454 nm in time intervals after centrifugation. In parallel, control experiments were performed by using N3 dispersions without addition of glucose.2.9. Cell culture
HeLa human cervix adenocarcinoma cells (ATCC) were grown in Iscove's modified Dulbecco's medium (IMDM) (25 mM D-glucose) supplemented with 10% fetal bovine serum (FBS) and antibiotics (penicillin 100 U mL−1 and streptomycin 100 μg mL−1, Invitrogen, Fisher Scientific). The cells were cultured at 37 °C in a humidified 5% CO2 atmosphere and subcultured twice a week by trypsinization.2.10. MTT cell viability assay
Cell viability was determined using the standard tetrazolium salt MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay.21 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 HeLa cells were seeded per well in a 96-well plate in a volume of 160 μL of medium.
000 HeLa cells were seeded per well in a 96-well plate in a volume of 160 μL of medium.
The following day, the cells were incubated at 4 °C for 15 min to synchronize them for the endocytosis. Then, the medium was removed and the nanoparticles N4 (100 μg mL−1) were added in D-PBS (Dulbecco's phosphate solution with calcium and magnesium) during a 30 min incubation. Control wells without nanoparticles contained D-PBS. After the cells were washed twice with D-PBS, different media containing 5% FBS and antibiotics (penicillin 100 U mL−1 and streptomycin 100 μg mL−1) were added: DMEM with 0, 25, 50, and 100 mM glucose and incubated during 24 h.
The next day, 20 μL of MTT (5 mg mL−1 in PBS) was added to the cells and incubated for 3 hours at 37 °C. Then, the medium was replaced with 150 μL of DMSO and incubated for 1 hour. Finally, the absorbance was measured at 490 nm using a Biotek ELX800 absorbance microplate reader.
2.11. Confocal microscopy
Internalization and cargo delivery studies using N4 were performed on HeLa cells. For this purpose, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 HeLa cells were seeded in 24-well plates, with each well covered by a 12 mm diameter glass coverslip, and placed in a CO2 cell incubator at 37 °C and allowed to attach for 24 h. Next, we proceeded as above for 0, 25, and 100 mM glucose-supplemented DMEM.
000 HeLa cells were seeded in 24-well plates, with each well covered by a 12 mm diameter glass coverslip, and placed in a CO2 cell incubator at 37 °C and allowed to attach for 24 h. Next, we proceeded as above for 0, 25, and 100 mM glucose-supplemented DMEM.
After 4, 6, and 24 hours of incubation, the cells were fixed with 10% formalin solution (Sigma) in PBS at room temperature and permeabilized with 0.1% Triton X-100 for 5 min. After 3 washes with PBS, cells were blocked with 2% BSA in PBS for 10 min. Then the cells were stained with 2 μg mL−1 WGA (wheat germ agglutinin) conjugated to Alexa Flour-488 for 10 min and secondly with 1 μg mL−1 Hoechst 333442 for 5 min. Both incubation periods were protected from light and immediately followed by 3 washes with PBS.
The coverslips were mounted on microscope slides and examined under an Olympus FluoView-1200 confocal laser scanning microscope (Olympus Life Science, UCM microscope facility), using a 60× oil immersion objective. Images at a resolution of 2048 × 2048 pixels were processed with Fiji Image J software (https://imagej.net/software/fiji/).
2.12. Fluorescence microscopy
Cell morphology and internalization of nanoparticles were observed with a Zeiss Axio Imager A1 fluorescence microscope coupled to a Zeiss Axiocam 202 mono-camera while using ZEN 3.6 Blue Edition software. For this purpose, 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded onto 12-well plates, treated with N4 and glucose and further stained as previously described.
000 cells per well were seeded onto 12-well plates, treated with N4 and glucose and further stained as previously described.
3. Results and discussion
The novel organic–inorganic PAMAM–MSN Janus nanoparticles were prepared according to the experimental protocol represented in Scheme 1. To assemble the anisotropic nanomaterial, a mask-protecting assisted site-selective modification approach was employed by partially confining the N0 mesoporous nanoparticles at the interface of a Pickering emulsion, using paraffin wax as oil phase.9 The exposed nanoparticle surface was then enriched with reactive primary amino groups by modification with APTES, allowing further attachment of carboxylic acid-terminated PAMAM G-4.5 molecules through a carbodiimide-mediated reaction.To characterize the resulting Janus nanoparticles, which was obtained in 43% yield, TEM analysis was performed and representative images are shown in Fig. 1. The anisotropic colloid N1 showed a similar quasi-spherical morphology to that of the initial mesoporous silica nanoparticles N0, with an average particle diameter of 102 ± 7 nm and an MCM-41 type channel-like mesoporous structure. To further visualize the attached PAMAM dendron molecules, the nanomaterial was previously stained with aqueous sodium phosphotungstate.22 No significant staining was observed for N0, but TEM images of N1 confirmed the anisotropic morphology of the modified nanoparticles, showing stained regions corresponding to the dendritic moieties covalently attached to the mesoporous nanomaterial.
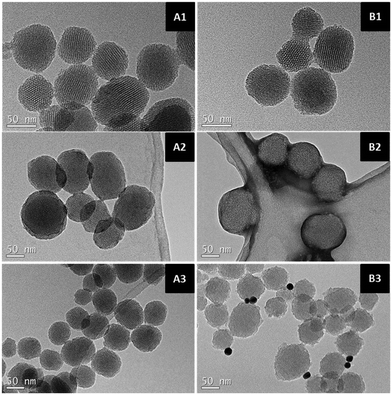 | ||
| Fig. 1 Representative TEM images of solids N0 (A) and N1 (B) before (1) and after staining with sodium phosphotungstate (2) or covalent attachment of Au nanoparticles (3). | ||
To reinforce our hypothesis on the toposelective attachment of PAMAM molecules on a preferential face of N0, the carboxylic acid residues at the surface of the dendrimer in the N1 Janus colloids were modified with cystamine, and Au nanoparticles were further attached as nanosized markers. For comparison, a similar protocol was followed with raw N0 nanoparticles. TEM images revealed that Au nanoparticles were not attached to N0, but metal colloids were clearly associated with a preferential face of the PAMAM–MSN Janus nanoparticles. An additional experiment was also performed to demonstrate the attachment of PAMAM moieties to mesoporous silica nanoparticles in the anisotropic colloid by the covalent immobilization of the enzyme horseradish peroxidase on the carboxylic acid-enriched dendrimer surface. A colorimetric enzymatic assay revealed that only 30 mU of peroxidase per milligram of nanoparticles was immobilized on raw N0, probably associated with the non-covalent adsorption of the enzyme on the highly porous nanomaterial surface. In contrast, the enzymatic activity immobilized on the Janus nanomaterials reached 400 mU mg−1, due to the presence of the PAMAM molecules acting as excellent supports for the multipoint attachment of the catalytically active protein.
As is represented in Scheme 2, N1 nanoparticles were further provided on the mesoporous face with a novel H2O2-sensitive ligand by sequential attachment of (3-isocyanatopropyl)triethoxysilane (ICPTES) and 1-(4-aminophenyl)-2-phenylethane-1,2-dione (APPD). Loading the pores of nanoparticles with the reporter dye and capping the dione derivative with β-cyclodextrin yielded the solid N2. Fig. 1S in the ESI† shows the FT-IR spectra of these nanomaterials. All nanoparticles reveal the characteristics peaks of mesoporous silica nanoparticles with main bands at 3430 cm−1, 1090 cm−1, 800 cm−1 and 460 cm−1 attributed to O–H, Si–OH, SiO4 and Si–O vibrations.10–13 The spectra of N1 and N2 nanoparticles show broad peaks at 3430 cm−1 corresponding to the stretching of the O–H and N–H groups in the dendrimer. These spectra also show peaks at 2925 cm−1 and 1550 cm−1 ascribed to the stretching and bending of C–H groups, respectively, suggesting the presence of PAMAM and APTES in the nanomaterial. The last peak could also be ascribed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching in the aromatic rings of APPD residues. Small bands at 1480–1340 cm−1 spectra proved the presence of the encapsulated ruthenium complex in N2,23 while the shoulder peak at 1160 cm−1 could be ascribed to the C–OH stretching in the βCD moieties.
C stretching in the aromatic rings of APPD residues. Small bands at 1480–1340 cm−1 spectra proved the presence of the encapsulated ruthenium complex in N2,23 while the shoulder peak at 1160 cm−1 could be ascribed to the C–OH stretching in the βCD moieties.
 | ||
| Scheme 2 Preparation of the enzyme-powered Janus nanomachine N3 for controlled release. N1: PAMAM–MSN Janus nanoparticles; N2: Ru(bipy)3Cl2-loaded N1. | ||
Thermogravimetric analysis revealed that all nanoparticles experienced a primary weight loss at temperatures up to 92 °C due to the thermal-induced desorption of physically adsorbed H2O molecules (Fig. 2S in the ESI†). At higher temperatures up to 1000 °C, N0 exhibited a slight weight loss of only 3% due to the thermal decomposition of pendant hydroxyl groups on the nanoparticle surface. In contrast, N1 showed a second transformation with a maximum rate of thermal-induced decomposition at 567 °C, which could be ascribed to the loss of pendant APTES groups and PAMAM molecules. A more complex pattern was observed for N2, showing three consecutive and more pronounced thermal-induced transformation processes with maximum rate of decomposition at 324 °C, 458 °C and 915 °C. These processes could be ascribed to the transformation of the organic molecules composing the gating mechanism, the silane and dendritic residues attached to the nanoparticle surface and the encapsulated ruthenium complex.
Nitrogen adsorption/desorption isotherms were measured for all nanoparticles to determine their specific surface area, pore diameter and morphology by applying the BET and BJH models. As is illustrated in Fig. 3S in the ESI,† N0 and N1 showed type IV nitrogen adsorption/desorption isotherms, typical of mesoporous supports,24 with BET specific surface areas of 1053 m2 g−1 and 482 m2 g−1, respectively. The large specific surface area and the absence of hysteresis loops in the corresponding isotherm suggested that all pores are highly accessible in N0. However, the small hysteresis loops observed at high relative pressure values in the N1 isotherm, as well as the noticeable reduction in its specific surface area, suggested that some pores were partially blocked in this Janus material. This fact could be ascribed to the toposelective modification of the mesoporous nanoparticle with APTES and further attachment of PAMAM molecules. This hypothesis was also supported by the pore size distribution values obtained by the BJH model. As can be appreciated from Fig. 4S (ESI†), the average pore diameter of the mesoporous nanoparticles changed from 2.2 nm to 2.0 nm after covalent attachment of the dendritic molecules.
A completely different nitrogen adsorption/desorption behavior was observed for N2, showing the characteristic isotherm pattern of mesoporous materials with filled pores, with low BET specific surface area of 98 m2 g−1 and no appreciable porosity, suggesting all pores were filled with the reporter dye. Finally, XRD analysis for N0–N2 (Fig. 5S in the ESI†) revealed three diffraction peaks, assigned to the (100), (110) and (200) Bragg reflections, which are consistent with the characteristic diffraction pattern of MCM-41 type mesoporous silica nanoparticles with hexagonal pore structures.25 This fact was previously confirmed by TEM analysis for solids N0 and N1 as shown in Fig. 1.
Solid N2 was then employed as support for the assembly of the enzyme-controlled nanomachine N3 by covalent immobilization of glucose oxidase on the dendritic molecules. Enzymatic and protein assays revealed that 0.1 U and 0.15 μg were immobilized in each milligram of N3, representing an immobilized specific activity of 0.7 mU mg−1 enzyme. In addition, alkaline hydrolysis and spectrophotometric determination of the reporter dye revealed an amount of 3.2 mg of ruthenium complex encapsulated in each gram of N3.
Scheme 3 presents the proposed functional mechanism for the nanomachine N3. Upon addition of glucose as the INPUT chemical signal, the GOx-mediated catalytic transformation of this monosaccharide yields gluconic acid and H2O2, and this oxidant should break the 1,2-dione linkage at the gate-like ensemble on the mesoporous face, thus releasing the reported dye as the OUTPUT signal. To demonstrate this hypothesis, the novel H2O2-sensitive gating mechanism in N3 was first evaluated. Fig. 2A shows the time-course of Ru(bipy)3Cl2 release from N3 in the presence and absence of H2O2. No significant dye release was noticed for the control experiment without the addition of the trigger. However, the concentration of dye delivered to the solution increased progressively with the time of incubation for the mixtures upon addition of H2O2. It was also noticed that the release was higher for higher concentrations of the trigger. These facts suggest that H2O2 disrupts the gating ensemble on the nanomachine pore surface, leading to on-command release of the encapsulated dye.
 | ||
| Scheme 3 Proposed mechanism for the enzyme-powered Janus nanomachine N3 for on-command controlled release. | ||
In a further experiment, glucose was employed as the trigger, and the results of this kinetics release assay are shown in Fig. 2B. Similarly to the previous experiment, no significant increase in the absorbance measured at 454 nm was observed for the control solution without glucose, suggesting that the nanomachine N3 was tightly capped, and the dye was not released. However, a noticeable cargo release was achieved over time after the addition of glucose. This fact suggests that glucose is rapidly transformed by the integrated enzyme, leading to the production of H2O2 with consequent disruption of the 1-(4-aminophenyl)-2-phenylethane-1,2-dione based gate-like ensemble at the mesoporous face thus promoting the release of the encapsulated Ru(bipy)3Cl2 complex, as represented in Scheme 3. It was also demonstrated that this autonomous release was highly selective, as illustrated in Fig. 6S in the ESI.† In fact, negligible dye release from N3 was observed after 120 min of incubation in the presence of lactose, sucrose, galactose or fructose at 100 mmol L−1 final concentration. However, maximum release capacity was exhibited for N3 incubated with glucose under similar experimental conditions.
Once the H2O2-sensitive gating mechanism and nanomachine operation were confirmed, a similar enzyme-powered smart nanocarrier N4 was prepared but using the antitumoral drug Doxo as cargo. Nanoparticle hydrolysis and spectrophotometric determination revealed that N4 contained 33 μg of Doxo per milligram of nanocarrier, representing 4.4% of encapsulation efficiency. An immobilized specific activity for GOx of 0.71 mU mg−1 enzyme was also confirmed.
As is reported in Fig. 3A, N4 was absolutely capped when incubated in 100 mmol L−1 sodium phosphate buffer, pH 7.5, and consequently, no Doxo release was noticed. However, a time-dependent drug release was observed upon addition of glucose at 100 mmol L−1 final concentration, suggesting that this monosaccharide triggers the cargo delivery through a similar mechanism proposed for N3. Accordingly, N4 was evaluated as a smart nanocarrier for Doxo delivery in in vitro experiments using HeLa cancer cells as the test model.
Cell viability studies were initially performed by incubating HeLa cells for 24 h with suspensions of nanoparticles at 100 μg mL−1 final concentration, and viable cells were further determined by using the MTT assay.21 As is illustrated in Fig. 3B, viability of HeLa cells is lower in the absence of glucose because this sugar is needed as a carbon source for cell growth and maintenance. HeLa cells remain viable in the absence of the nanoparticles, but these nanomaterials seem to be toxic for the cells, causing them significant death when applied to the incubation medium. When incubated in a medium with 25 mM glucose, cell viability was not significantly affected suggesting this sugar concentration acts as nutrient for the cells and is not enough to trigger the drug release mechanism in N4. However, higher glucose concentrations of 100 mM lead to significant cell death in the presence of N4, suggesting the nanomachine successfully releases the drug under these conditions through the enzyme-mediated mechanism previously proposed.
With the aims being to demonstrate the cell internalization of N4 and furthermore the enzyme-mediated autonomous drug release, confocal microscopy experiments were performed. Fig. 4 shows representative confocal images of Alexa Flour-488-labeled WGA (green), Hoescht (blue), doxorubicin (red) and merged staining (green, red and blue) for untreated HeLa cells, as well as for HeLa cells first treated with N4 and further incubated with glucose. Two incubation times (4 h and 24 h) and different glucose concentrations (0, 25 and 100 mmol L−1) were employed for this experiment.
As expected, in the absence of glucose, due to the absence of the carbon source required for cellular homeostasis, the morphology of control cells without nanoparticles seem to be affected, and additionally a significant decrease in cell density was observed, which was more accentuated after 24 h. This result agrees with those previously achieved in cell viability assays. In the presence of N4, a noticeable red fluorescence was observed, suggesting the Doxo-loaded nanoparticles were internalized by HeLa cells. The intensity and dispersion of the Doxo fluorescence was noticeably higher, and the morphology of HeLa cells was significantly affected when glucose was added to the incubation media. Such effects increased when both longer incubation times and higher glucose concentrations were employed. These facts suggest that the internalized nanomachines were able to release the drug in the presence of the trigger according to the mechanism previously proposed, thus stimulating cell death.
To confirm this hypothesis, HeLa cells were first treated with N4, then incubated with glucose and finally stained as previously described. As control, cells non-treated with the nanoparticles were employed. The resulting living cells were observed by fluorescence microscopy, and representative images are shown in Fig. 7S–9S in the ESI.†
As is illustrated in Fig. 7S,† Doxo fluorescence was not observed in control experiments based on cells non-treated with nanoparticles. However, a clear red fluorescence was detected in those incubated with N4, demonstrating cell internalization of these nanomaterials (Fig. 8S†). In addition, the intensity of Doxo fluorescence was higher at high glucose concentrations, suggesting major release of the drug from N4 according to the proposed mechanism.
Fig. 9S† illustrates two HeLa cells in the same preparation but with different responses toward N4. On the right is a cell that internalized the nanoparticles, while on the left is another that has not internalized them. Clear red and blue fluorescence can be observed for the cell on the right, suggesting Doxo release has caused morphological changes including high membrane permeability. Accordingly, Hoechst penetrates more easily into the cells staining the nuclei. In contrast, only Alexa Flour-488-labeled WGA green staining was observed in the HeLa cell on the left. This fact suggests that N4 was not endocytosed, and the cell membrane was also less permeable to Hoechst staining.
4. Conclusions
An original organic–inorganic Janus nanoparticle based on mesoporous silica nanoparticles toposelectively modified with PAMAM dendrimers is herein reported. These anisotropic colloids were used to assemble an autonomous nanomachine mechanized with novel H2O2-sensitive gate-like ensembles on the mesoporous face and facilitated glucose oxidase-based enzymatic control on the dendritic face. This nanodevice was able to release the encapsulated cargo when glucose was used as the trigger, and was successfully employed to deliver the anticancer drug doxorubicin into HeLa cells. We envision that the present results can open new avenues for the further development of a large variety of autonomous nanodevices controlled by H2O2 producing redox enzymes for the smart and on-command delivery of drugs and other compounds.Author contributions
BM: investigation, visualization, writing – original draft. EG, AH, LE, MM, MG, AV, TM: investigation. AS, DV: supervision. NMQ, PMR: methodology and supervision. RV: conceptualization, methodology, supervision, funding acquisition, writing – original draft.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from the Spanish Ministry of Economy and Competitiveness (projects CTQ2017-87954-P and PID2021-125723NB-I00), Complutense University of Madrid (project PR27/21-015), and the Spanish Ministry of Science and Innovation (project CNS2022-135255) is gratefully acknowledged. E.G. and M.G. also thank the Public Employment Service (SEPE) and NextGenerationEU funds for financial support through the Investigo Program (CT19/23-INVM-129 and CT19/23-INVM-56).References
- X. Cai, M. Jin, L. Yao, B. He, S. Ahmed, W. Safdar, I. Ahmad, D.-B. Cheng, Z. Lei and T. Sun, J. Mater. Chem. B, 2023, 11, 716–733 Search PubMed.
- H. Yan, P. Xu, H. Cong, B. Yu and Y. Shen, Mater. Today Chem., 2024, 37, 101997 Search PubMed.
- A. Jayakumar, S. Mathew, S. Radoor, J. T. Kim, J.-W. Rhim and S. Siengchin, Mater. Today Chem., 2023, 30, 101492 CrossRef CAS.
- B. Q. G. Le and T. L. H. Doan, Wiley Interdiscip. Rev.:Nanomed. Nanobiotechnol., 2023, 15, e1874 Search PubMed.
- V. Jadhav, A. Roy, K. Kaur, A. K. Rai and S. Rustagi, Nano-Struct. Nano-Objects, 2024, 37, 101103 Search PubMed.
- J. Gupta, M. Quadros and M. Momin, J. Drug Delivery Sci. Technol., 2023, 81, 104305 CrossRef CAS.
- S.-N. Zhao, R.-H. Tang, S.-J. Deng, G.-E. Chen, J. Ye and Z.-L. Xu, J. Mater. Sci., 2023, 58, 17610–17622 CrossRef CAS.
- B. Mayol, V. Dato, M. Rodriguez, E. Lucena, A. Villalonga, P. Díez, S. Jimenez-Falcao, F. Sancenón, A. Sánchez, D. Vilela, P. Martínez-Ruiz, R. Martínez-Máñez and R. Villalonga, J. Mater. Chem. B, 2022, 10, 6983–6990 RSC.
- R. Villalonga, P. Díez, A. Sánchez, E. Aznar, R. Martínez-Máñez and J. M. Pingarrón, Chem. – Eur. J., 2013, 19, 7889–7894 CrossRef CAS PubMed.
- B. Mayol, S. Pradana-López, A. García, C. De La Torre, P. Díez, A. Villalonga, C. Anillo, D. Vilela, A. Sánchez, P. Martínez-Ruiz, R. Martínez-Máñez and R. Villalonga, J. Colloid Interface Sci., 2024, 671, 294–302 CrossRef CAS PubMed.
- B. Mayol, A. Rodríguez, A. Villalonga, C. Anillo, D. Vilela, A. Sánchez, P. Martínez-Ruiz and R. Villalonga, J. Mater. Chem. B, 2023, 11, 7190–7196 RSC.
- S. Jimenez-Falcao, D. Torres, P. Martínez-Ruiz, D. Vilela, R. Martínez-Máñez and R. Villalonga, Nanomaterials, 2021, 11, 2492 CrossRef CAS PubMed.
- P. Díez, A. Sánchez, M. Gamella, P. Martínez-Ruíz, E. Aznar, C. De La Torre, J. R. Murguía, R. Martínez-Máñez, R. Villalonga and J. M. Pingarrón, J. Am. Chem. Soc., 2014, 136, 9116–9123 CrossRef PubMed.
- A. J. C. Wahart, J. Staniland, G. J. Miller and S. C. Cosgrove, R. Soc. Open Sci., 2022, 9, 211572 CrossRef CAS PubMed.
- K. Liu, S. He, L. Li, Y. Liu, Z. Huang, T. Liu, H. Wu, X. Jiang, K. Liu and F. Tian, Sci. Rep., 2021, 11, 9709 CrossRef CAS PubMed.
- A. Sánchez, A. Villalonga, G. Martínez-García, C. Parrado and R. Villalonga, Nanomaterials, 2019, 9, 1745 CrossRef PubMed.
- M. A. Mintzer and M. W. Grinstaff, Chem. Soc. Rev., 2011, 40, 173–190 RSC.
- L. H. Fu, C. Qi, Y. R. Hu, J. Lin and P. Huang, Adv. Mater., 2019, 31, 1808325 CrossRef PubMed.
- L. H. Fu, C. Qi, J. Lin and P. Huang, Chem. Soc. Rev., 2018, 47, 6454–6472 RSC.
- H. Zhang, X. Ren, W. Zhao, X. Tang and G. Wang, Adv. Synth. Catal., 2017, 359, 395–401 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- A. Villalonga, A. Sánchez, D. Vilela, B. Mayol, P. Martínez-Ruíz and R. Villalonga, Microchim. Acta, 2022, 189, 309 CrossRef CAS PubMed.
- K. M. Omberg, J. R. Schoonover, J. A. Treadway, R. M. Leasure, R. B. Dyer and T. J. Meyer, J. Am. Chem. Soc., 1997, 119, 7013–7018 Search PubMed.
- A. J. Di Pasqua, K. K. Sharma, Y.-L. Shi, B. B. Toms, W. Ouellette, J. C. Dabrowiak and T. Asefa, J. Inorg. Biochem., 2008, 102, 1416–1423 Search PubMed.
- S. Schacht, M. Janicke and F. Schüth, Microporous Mesoporous Mater., 1998, 22, 485–493 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03740e |
| This journal is © The Royal Society of Chemistry 2025 |




