 Open Access Article
Open Access ArticleRecent advances in discovery and biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPP)-derived lipopeptides
Shumpei
Asamizu
 *
*
Engineering Biology Research Center, Kobe University, 1-1 Rokkodai, Nada, Kobe 657-8501, Japan. E-mail: shumpei.asamizu@port.kobe-u.ac.jp
First published on 16th July 2025
Abstract
Covering: This review summarizes recent advances in the discovery, biosynthesis, and bioactivity of RiPP-derived lipopeptides, covering studies published up to 2024.
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a diverse superfamily of natural products unified by a common biosynthetic logic: The peptide backbone is genetically encoded, and the translated precursor peptide undergoes a series of post-translational modifications catalyzed by maturase enzymes to produce the final bioactive compound. Despite their structural complexity, RiPPs are encoded by relatively small biosynthesis gene clusters. RiPP maturase enzymes are diverse and often promiscuous, offering significant biotechnological potential. However, their lack of conserved features makes genome-based discovery of novel RiPPs challenging. Recent advances in biosynthetic understanding and genome mining techniques have led to the identification of numerous uncharacterized RiPP biosynthetic gene clusters, often flanked by genes encoding non-RiPP moieties, in microbial genomes. Leveraging this information, a new class of natural products, hybrids of RiPPs and non-RiPP elements, has recently been discovered. Among them, RiPPs bearing fatty acyl groups, referred to as RiPP-derived lipopeptides, represent a newly emerging class of lipopeptide natural products with significant antimicrobial activity.
1. Introduction
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a diverse superfamily of natural products unified by a common biosynthetic logic.1–3 Their biosynthesis typically begins with a ribosomally synthesized precursor peptide, which often comprises a leader and a core region, although alternative architectures also exist. The core peptide undergoes various post-translational modifications catalyzed by maturase enzymes and is subsequently proteolytically cleaved to yield the final bioactive mature product.Advances in high-throughput gene sequencing technologies have ushered in a new era of natural product discovery, with RiPPs3 emerging as one of the fastest-growing fields, supported by the development of specialized bioinformatic tools.4,5 Despite the structural complexity and large molecular size of RiPPs, their biosynthesis gene clusters (BGCs) are relatively small. The maturase enzymes involved in RiPP biosynthesis are remarkably diverse and often promiscuous in substrate recognition, making them highly attractive for biotechnological applications. However, the discovery of novel RiPPs is hindered by their limited structural predictability in nature and the frequent absence of characteristic features in their maturase enzymes, which restricts their straightforward identification as RiPP-associated enzymes during genome mining.
Growing interest in RiPP-based therapeutics has been fueled by the discovery of compounds exhibiting diverse bioactivities, including antibacterial, antifungal, antiviral, antinociceptive, and antitumor effects.6 These wide-ranging activities underscore how post-translational modifications drive the diversity of RiPPs, enabling them to engage a broad array of biological targets across different domains of life.
RiPPs exhibit unique modes of action.6 For example, nisin—a class I lanthipeptide produced by Lactococcus lactis—is the most extensively studied RiPP and shows potent antimicrobial activity against many non-proteobacterial species.6 For over 50 years, nisin has been widely used in the food industry to combat foodborne pathogens. Its mechanism involves binding to the cell wall precursor lipid II and forming stable pores in the bacterial membrane, composed of both nisin and lipid II, ultimately leading to bacterial cell death.7,8
Thiostrepton, a thiopeptide antibiotic originally isolated in the 1950s from Streptomyces azureus, is used in veterinary medicine and features a characteristic 26-membered macrocyclic core.6 It exhibits potent growth–inhibitory activity against Firmicutes, including hemolytic streptococci. Thiostrepton exerts its antimicrobial effect by binding to the 50S ribosomal subunit, thereby inhibiting protein synthesis through two mechanisms: it blocks mRNA–tRNA translocation mediated by the GTPase elongation factor G (EF-G), and it interferes with EF-Tu-catalyzed delivery of aminoacyl-tRNA.9–11
A more recent finding is darobactin, isolated from Photorhabdus khanii HGB1456, which selectively kills Gram-negative pathogens by targeting BamA, the central component of the essential BAM complex that is responsible for folding and inserting outer membrane proteins,12 mimicking the recognition signal of native substrates and displacing a lipid molecule.13
Alongside the discovery of RiPP natural products, novel modification patterns have also been revealed,1,2,14 including fatty acylation of the RiPP backbone. Lipopeptide natural products (acylated peptides) include clinically valuable antibiotics, such as the antibacterial agents colistin/polymyxin B15 and daptomycin (DAP),16 as well as the antifungal agent echinocandin,17 all of which are produced by nonribosomal peptide synthetase (NRPS) pathways.18,19 Advances in genome mining have revealed numerous uncharacterized RiPP BGCs, often flanked by genes encoding non-RiPP elements, in microbial genomes. This has led to the discovery of hybrid natural products, including a newly emerging class of RiPP-derived lipopeptides bearing fatty acyl groups, which exhibit notable antimicrobial activity. In addition to the N-acylated RiPPs modified by short-chain acyl groups, this review summarizes recent advances in the discovery, biosynthesis, and bioactivity of RiPP-derived lipopeptides bearing medium- and long-chain acyl modifications, covering reports up to 2024 in this rapidly developing field. Although RiPPs with short-chain acyl modifications are not typically classified as lipopeptides, the term “lipopeptide” is used in a broader sense in this review to encompass such modifications for the sake of comprehensive coverage.
2. Polar functionalized fatty acylated RiPPs
2.1. Discovery of RiPP-derived lipopeptides
RiPP-derived lipopeptides with basic functional groups at the fatty acid terminus are designated “polar functionalized fatty acylated RiPPs” (PFARs). The first report of the isolation of RiPP-derived lipopeptides was the discovery of albopeptins A and B from Streptomyces albofaciens JC-82-120 by Isono et al. in 1986 (ref. 20) (Fig. 1A). The structure of albopeptin B was later fully characterized by Oikawa et al. in 2022,21 informed by insights from the biosynthesis genes identified in S. albofaciens JC-82-120 (details described in Section 2.4). | ||
| Fig. 1 Chemical structures of type I lipolanthines from Actinobacteria. (A) Representative compounds include albopeptin B from Streptomyces albofaciens and S. nigrescens HEK616;21,31 microvionin from Microbacterium arborescens;22 nocavionin from Nocardia terpenica;22 nocaviogua from Nocardia sp. XZ19_369;25 and solabiomycins A (R = CH3) and B (R = H) from S. lydicus.31 Deoxysolabiomycins A (R = CH3) and B (R = H) were obtained from a S. lydicus ΔsolS mutant.34 (B) Structural features of avionin, labionin, and solabionin.31 (C) Reaction scheme for avionin formation catalyzed by the enzyme LanKC.24 | ||
2.2. Lipolanthines: microvionin and nocavionin
In 2018, with the advancement of genome mining techniques for discovering RiPP natural products, the Süssmuth group reported the isolation and full structural characterization (except for the stereochemistry of a quaternary carbon) of a new class of antibiotics, named lipolanthines. These are hybrids of an N,N′-dimethylguanidyl fatty acid and an avionin-type RiPP (see below). The identified lipolanthines include microvionin, produced by Microbacterium arborescens 5913, and nocavionin, from Nocardia terpenica DSM44935 (ref. 22) (Fig. 1A). A putative BGC was identified for microvionin (the mic BGC) and nocavionin (the noc BGC); these BGCs were classified as type I lipolanthine BGCs. They contrast with type II lipolanthine BGCs, which additionally contain type I polyketide synthase (PKS)-encoding gene(s). Microvionin exhibited potent growth–inhibitory activity toward both methicillin-susceptible Staphylococcus aureus (MSSA) and methicillin-resistant S. aureus, with minimum inhibitory concentration (MIC) values of 0.1 and 0.46 μg mL−1, respectively (Table 1).| Isolated product | Group | Original strain | Heterologous expression host | Bioactivity | Reference | |
|---|---|---|---|---|---|---|
| Microvionin | Lipolanthine/avionin | Microbacterium arborescens 5913 | Methicillin-resistant S. aureus (MRSA) | MIC: 0.46 μg mL | 22 | |
| Methicillin-susceptible S. aureus (MSSA) | MIC: 0.1 μg mL−1 | |||||
| Nocavionin | Lipolanthine/avionin | Nocardia terpenica DSM44935 | 22 | |||
| Nocaviogua A | Lipolanthine/avionin | Nocardia sp. XZ19_369 | Weak cytotoxic | 25 | ||
| Nocaviogua B | ||||||
| Goadvionin A1 | Polyketide/RiPP hybrid lipopeptide/avionin | Streptomyces sp. TP-A0584 | Streptomyces lividans TK23 | S. aureus | MIC: 6.4 μg mL−1 | 26 |
| Goadvionin B2 | ||||||
| Albopeptin B | Lipolanthine/solabionin | Streptomyces albofaciens JC-82-120 | Antifungal | MIC: 12.5 μg mL−1 | 20 and 21 | |
| Streptomyces nigrescens HEK616 | Gram-positive bacteria | MIC: 12.5 μg mL−1 | 31 | |||
| Solabiomycin A | Lipolanthine/solabionin | Streptomyces lydicus NBRC 13058 | M. tuberculosis H37Rv | MIC95: 3.125 μg mL−1 | 31 | |
| M. smegmatis | MIC: 6.4 μM | |||||
| Solabiomycin B | M. smegmatis | MIC: 25.6 μM | ||||
| Deoxysolabiomycin A | Lipolanthine/labionin | Streptomyces lydicus NBRC 13058 ΔsolS | M. smegmatis | MIC: 25.6 μM | 34 | |
| Deoxysolabiomycin B | M. smegmatis | MIC: 51.2 μM | ||||
| Kamptornamide | Selidamide/AviCys | Kamptonema sp. PCC 6506 | Escherichia coli | 35 | ||
| Phaeornamide | Selidamide/AviCys | Pseudophaeobacter arcticus DSM 23566 | Escherichia coli | 35 | ||
| Prethioviridamide | Thioamide/AviCys | Streptomyces olivoviridis NA05001 | Mitochondrial respiratory chain complex V inhibitor | 38 | ||
| Lipoavitide | Lipoavitide/AviMeCys | Streptomyces sp. NRRL S-1521 | Streptomyces albus J1074 | Hemolytic activity | 44 | |
| Polytheonamide | Candidatus Entotheonella spp. | P388 murine leukemia cells | IC50: 68 pg mL−1 | 46 | ||
| Goadsporin | LAP | Streptomyces sp. TP-A0584 | Anti-Streptomyces | 55 and 56 |
The RiPP moiety of lipolanthines contains a bicyclic octapeptide featuring a central quaternary carbon atom, referred to as avionin (Fig. 1B), which is generated through the cooperative action of a class III lanthipeptide synthetase (LanKC)23 and a cysteine decarboxylase (LanD).24 LanD catalyzes the decarboxylation of a C-terminal cysteine residue to generate 2-aminovinyl-cysteine (AviCys). Meanwhile, the kinase domain of LanKC phosphorylates the hydroxyl groups of two serine residues of the core peptide in an ATP-dependent manner and subsequently eliminates the phosphate to generate dehydroalanine residues. The formation of the avionin structure proceeds via a thiol attack on the exomethylene of one dehydroalanine, followed by a nucleophilic attack by the resulting enol on the second dehydroalanine. This sequence of 1,4-additions forms the characteristic bicyclic structure of the avionin-containing peptide (Fig. 1C).
The fatty acyl unit of lipolanthines features an N-terminal dimethyl guanidino group. Interestingly, a comparison of BGCs for putative lipolanthines suggests that the terminal guanidine group in microvionin and nocavionin is installed by the combined action of an aldehyde dehydrogenase (MicY or NocY) and a thiamine diphosphate (ThDP)-dependent acetolactone synthase (MicJ or NocJ).22 In contrast, in a type II lipolanthine BGC, which contains a type I PKS, the guanidine group appears to be introduced by an arginine:glycine amidinotransferase, which putatively catalyzes the transfer of a guanidino group from arginine to glycine.22 However, the product of this type II lipolanthine BGC remains uncharacterized.
In addition to the discovery of nocavionin, two analogs, nocaviogua A and B, with slight differences in the amino acid sequence, have also been isolated from Nocardia sp. XZ19_369 (ref. 25) (Fig. 1A). Nocaviogua A exhibited weak cytotoxic effects against tested cell lines [e.g., non-small cell lung cancer (NCI-H2170) and breast cancer (MDA-MB-231) cell lines], while nocavioguas A and B showed no significant antibacterial or antifungal activity (Table 1). Comparing micorovinin and nacavionin analogs, how differences in chemical modifications—such as the presence of trans olefins in the fatty acid moiety or variations in the amino acid residues in the RiPP core—affect bioactivity remains unknown.
2.3. Polyketide/RiPP-hybrid lipopeptides: goadvionins
Hiroyasu Onaka and colleagues performed genome mining for RiPP BGCs, which led to the identification of the gdv BGC for goadvionin in Streptomyces sp. TP-A0584. Heterologous expression of the gdv BGC in S. lividans TK23 resulted in the production of eight congeners and successful isolation of polyketide/RiPP-hybrid lipopeptides, goadvionins A1 and B2 (ref. 26) (Fig. 2A). The RiPP moiety of the goadvionins features a bicyclic octapeptide with an avionin structure, similar to those found in microvionin and nocavionin, but differing in the amino acid residues. | ||
| Fig. 2 Biosynthesis and chemical structures of goadvionin and synthetic analogs. (A) Chemical structures of goadvionins A1 and B2, isolated from Streptomyces sp. TP-A0584.26 (B) Proposed reaction scheme of the in vivo acyl carrier protein (ACP)-dependent acyl transferase reaction catalyzed by GdvG during goadvionin biosynthesis.26 The C29 fatty acyl group synthesized through the sequential actions of GdvFB (KS), FG (KR), FZ (DH), and FI (ER) is predicted to be loaded onto the N-terminal ACP domain of GdvPKS by GdvL (see details in Fig. 3). A final extension with a building block supplied from methylmalonyl-CoA subsequently generates the C31 very long-chain fatty acid (VLCFA) unit on the C-terminal ACP* domain of GdvPKS. Although a DH domain is present in GdvPKS, it is likely non-functional, as the final product retains a hydroxyl group. Abbreviations: KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; ER, enoylreductase. (C) Chemical structure of stearoylgoadpeptin B, generated by in vitro synthesis.26 (D) Reaction scheme of the in vitro ACP*-dependent non-natural acyl transferase reaction catalyzed by GdvG for the synthesis of stearoylgoadpeptin B.26 ACP* refers to a truncated version of the C-terminal ACP domain from GdvPKS. | ||
The fatty acyl moiety of goadvionins is highly distinctive, featuring an N-terminal trimethylammonium-containing a linear C31 very-long-chain fatty acid (VLCFA) with multiple hydroxyl groups. Conventional collision-induced dissociation MS/MS was insufficient to elucidate its structure because of limited C–C bond cleavage and low resolution. To address this, radical-induced dissociation techniques—specifically hydrogen attachment/abstraction dissociation (HAD)–MS/MS—were employed.27 This method uses interactions between precursor ions and neutral hydrogen radicals (H˙), enabling efficient C–C bond cleavage in mild conditions with high mass resolution.28 Application of HAD–MS/MS allowed precise localization of all hydroxyl groups within the VLCFA moiety of goadvionins.26
In the biosynthesis of the fatty acyl group of goadvionin, it is proposed that, first, L-lysine is converted to trimethyllysine by a methyltransferase (GdvMT), and then the α-amine is oxidized by an NADH-dependent dehydrogenase (GdvY). A ThDP-dependent enzyme, GdvJ, is assumed to catalyze the decarboxylation of the substrate, forming an aldehyde product. After carboxylation of the aldehyde by unknown enzyme, acyl CoA ligase (GdvI) generates the CoA form, which can be loaded onto the acyl carrier protein (ACP) for chain elongation (Fig. 3). This pathway was suggested by genetic work and incorporation studies of 13C6-lysine, which showed a 5 Da mass shift compared with incorporation of 12C6-lysine; were the full 13C6-lysine molecule to be incorporated, there would be a mass shift of 6 Da. The observation of a 5 Da shift indicates that one carbon is lost by decarboxylation.26 This mechanism is likely conserved in the synthesis of NRP-derived lipopeptides, e.g., fusaricidin A.29 Interestingly, the incorporation pattern of L-arginine is different in biosynthesis of type I polyketides, e.g., azalomycins from S. violaceusniger DSM4137. Azalomycins consist of a 34-membered macrolactone ring and a guanidine-containing side-chain. Arginine monooxygenase and 4-guanidinobutyramide hydrolase are predicted to synthesize 4-guanidinylbutyryl-CoA as the starter unit for the polyketide synthesis.30
 | ||
| Fig. 3 Proposed biosynthesis pathway of the C31 very-long-chain fatty acid (VLCFA) in goadvionin biosynthesis. L-lysine undergoes modification to serve as the trimethylamine-containing C5 starter unit for fatty acid synthesis.26 In the biosynthesis of the intermediate C29 VLCFA moiety in goadvionins, dehydration and enoyl reduction steps are skipped during the first five elongation cycles. The final chain extension to generate a C31 VLCFA is catalyzed by Gdv polyketide synthase (PKS) using methylmalonyl-CoA as the substrate. | ||
The VLCFA structure of goadvionin is predicted to be assembled through extension with 12 malonyl-CoA units, beginning with the loading of a trimethyllysine-derived C5 starter unit on the GdvACP, a dedicated ACP in goadvionin biosynthesis, via enzymes associated with the fatty acid synthesis (FAS) system (GdvFB-FG-FZ-FI). In the final step of the fatty acyl synthesis, following the transfer of a fatty acyl group onto an N-terminal ACP domain of GdvPKS, a dedicated type I PKS in goadvionin biosynthesis, a C3 unit derived from methylmalonyl-CoA, is incorporated (Fig. 3). This dedicated PKS system for polyketide/RiPP-hybrid lipopeptides is distinct from that for type I lipolanthines,22 which presumably uses fatty acyl chains generated solely by the FAS system. The dedicated GDV FAS system likely catalyzes only a ketoreduction step during the first five elongation cycles, resulting in the formation of five consecutive hydroxyl groups along the fatty acyl chain. Structural diversification is introduced during the tailoring steps: a desaturase, GdvC, installs a cis-olefin, and an oxygenase (GdvH) hydroxylates the C30′ position of the terminal trimethylamine moiety observed in goadvionin A1 (Fig. 2). These variations in the acyl moiety along with the two RiPP cores derived from different precursor peptide genes (gdvA and gdvB) give rise to eight congeners. The enzymatic condensation of the fatty acyl group to the N-terminus of the RiPP (goadpeptins) will be described later (please see Section 5). Goadvionin B2 exhibited growth-inhibitory activity toward Gram-positive bacteria, including MSSA, with an MIC value of 6.4 μg mL−1 (Table 1).
2.4. Lipolanthine: albopeptin B
Before the isolation of microvionin, nocavionin, and goadvionin, Isono et al. reported the isolation of albopeptins A and B from S. albofaciens JC-82-120.20 Structural elucidation based on spectral data proved challenging, presumably because of the poor solubility and the conformational flexibility of the compounds, which resulted in broadened and low-intensity NMR spectra. The discovery of lipolanthines prompted a revision of the structure of albopeptin B (Fig. 1A), which was later carefully recharacterized by Oikawa et al.21 The genome of S. albofaciens JC-82-120 was sequenced, leading to the identification of a putative alb BGC. The alb cluster contains a LanKC homologue responsible for labionin formation (Fig. 1B), but lacks a LanD homologue, which is typically required for AviCys formation and, consequently, for the formation of avionin. As indicated by the genes in the alb BGC, the RiPP moiety of albopeptins indeed lacked the AviCys structure. Notably, the sulfur atom in the labionin is oxygenated to form a sulfoxide, giving rise to a structure named “solabionin”31 (Fig. 1B).The presence of a free carboxyl group on the RiPP and an unmodified guanidino terminus on the fatty acyl chain results in a zwitterionic molecule. RiPP-derived lipopeptides often feature positively charged functional groups [e.g., a (dimethyl)guanidino or trimethylamine group] at the terminus of the fatty acyl moiety. The importance of such functional groups is demonstrated, for example, by the NRP-derived lipopeptide fusaricidin A; removal of the guanidino group from the fatty acyl chain of fusaricidin A significantly decreases its activity toward S. aureus.32 Albopeptins exhibited growth–inhibitory activity toward several fungi and Gram-positive bacteria, with MIC values of around 12.5 μg mL−120 (Table 1).
2.5. Lipolanthines: solabiomycins
Inspired by the discovery of type I lipolanthines and goadvionin, genome mining and stable isotope-guided comparative metabolomics were employed to discover further PFAR natural products, solabiomycins.Goadvionins were shown to incorporate L-lysine, as demonstrated by the incorporation of 13C6-lysine.26 In contrast, microvionin and nocavionin were predicted to incorporate L-arginine as the starter unit for their fatty acyl moieties.22 Genome mining was guided by the presence of acetolactate synthase homologues (MicJ, NocJ, and GdvJ), and putative BGCs for PFARs were identified. Strains harboring such putative BGCs were cultured with stable isotope-labeled amino acids such as 13C6,15N4-L-arginine or 13C6,15N2-L-lysine, and incorporation was monitored through comparative secondary metabolomic analysis. This stable isotope labeling method resulted in the discovery of solabiomycins A and B from S. lydicus NBRC 13058, along with albopeptin B produced by S. nigrescens HEK616 (Fig. 1A).31 However, when 13C6,15N4-arginine was incorporated, an unexpected mass shift of 9 Da (rather than 8 Da) was observed for both solabiomycins and albopeptins, suggesting that no decarboxylation occurred during biosynthesis of the guanidino fatty acid chain.31 This result made ambiguous the function of acetolactate synthase homologs (GdvJ, MicJ, NocJ, SolJ, and NapJ) in the biosynthesis.
The involvement of the identified BGCs in the production of solabiomycins and albopeptins was confirmed through gene knockout experiments: disruption of solG, encoding a putative Gcn5-related N-acetyltransferase (GNAT) family acyltransferase in the sol BGC, abolished solabiomycin production in S. lydicus,31 while deletion of napKC, encoding a putative type III lanthionine synthase in the nap BGC, resulted in the loss of albopeptin biosynthesis in S. nigrescens.31
The antituberculosis activity of solabiomycins was evaluated by assessing the viability of Mycobacterium tuberculosis H37Rv through ATP quantification using the BacTiter assay system. Both solabiomycins exhibited a 95% inhibitory concentration (MIC95) of 3.125 μg mL−131 (Table 1), indicating their potential as antituberculosis agents. However, the mode of action of RiPP-type lipopeptides, as well as the mechanisms by which producer strains resist their own compounds (“self-resistance”), remains largely unknown (see also Section 6). RNA-Seq analysis was performed on the nap BGC responsible for albopeptin B biosynthesis in S. nigrescens HEK616.31 Transcription of the genes encoding a putative ABC transporter permease, NapT, and two hypothetical proteins—NapV, which contains a conserved domain associated with cell wall-active antibiotic response proteins,33 and NapW—was observed alongside the expression of other biosynthesis enzymes. Although further investigation is needed, characterization of these gene products may shed light on the self-resistance mechanism and reveal potential target molecules of RiPP-derived lipopeptides.
2.6. Lipolanthines: deoxysolabiomycins
Similar to albopeptins, solabiomycins feature a characteristic sulfoxide group within the labionin moiety, a structural motif referred to as solabionin in the RiPP portion of the molecule (Fig. 1B). A comparative analysis of the chemical structures of albopeptins and solabiomycins, and of the respective nap and sol BGCs, revealed that the sol BGC contains a single putative oxidase gene, solS, whereas the nap BGC harbors two oxidase-like genes, napS and napN. Given that albopeptins also possess a unique N-hydroxyl group, the conserved oxidase genes solS and napS were predicted to be involved in the sulfoxide formation. Gene knockout experiments showed that the solS gene, encoding a putative flavin adenine dinucleotide (FAD)–nicotinamide adenine dinucleotide phosphate (NAD(P))-binding protein, is involved in the sulfoxidation of an alkyl sulfide in solabiomycin.31 Deoxysolabiomycins A and B were isolated from the S. lydicus ΔsolS mutant.34 The catalytic activity of SolS was demonstrated in vitro using recombinant protein expressed in Escherichia coli. When incubated with the putative substrates (i.e., deoxysolabiomycins) and the cofactors FAD and NADPH, SolS catalyzed sulfoxidation, converting deoxysolabiomycins into solabiomycins.34The bioactivity of deoxysolabiomycins against Gram-positive bacteria, including Mycolicibacterium smegmatis, was evaluated. The results indicated that the sulfoxide group is critical for antibacterial activity, because deoxysolabiomycins exhibited significantly decreased growth inhibition compared with the equivalent solabiomycins. Specifically, the MIC of deoxysolabiomycin A increased from 6.4 μM to 25.6 μM upon removal of the sulfoxide moiety34 (Table 1).
3. Medium-chain fatty acylated RiPPs
3.1. Selidamides: kamptornamide and phaeornamide
In addition to N-terminal acylation of RiPPs by polar functionalized fatty acyl chains, as described above, a RiPP-derived lipopeptide featuring N-acylation on an amino acid side-chain was discovered following heterologous expression of cyanobacterial genes. Jörn Piel and colleagues undertook genome mining and discovered a unique class of RiPP-derived lipopeptides via heterologous gene expression in E. coli. The identified BGCs from cyanobacteria contained LanM-type class II lanthipeptide synthases23 along with GNAT family acyltransferases.35 Putative gene clusters from Kamptonema sp. PCC 6506 (kspA–kspRMN) and Pseudophaeobacter arcticus DSM 23566 (phaA–phaNRIM) were heterologously expressed in E. coli BL21 (DE3). N-terminal His-tagged precursor peptides (KspA or PhaA) were co-expressed with the associated modification enzymes, and the resulting products—named kamptornamide and phaeornamide respectively—were obtained after cleavage of the leader peptide by the endoproteinase LahT150, which specifically cleaves peptide bonds between consecutive glycine residues36 (Fig. 4). | ||
| Fig. 4 Chemical structures of selidamides of cyanobacterial origin. Kamptornamide was identified following the heterologous expression of ksp genes from Kamptonema sp. in Escherichia coli.35 Phaeornamide was obtained following the heterologous expression of pha genes from Pseudophaeobacter arcticus in E. coli.35 | ||
These compounds were not detected in the original strains; it thus remains unclear whether they are true natural products or artifacts of heterologous expression. Nonetheless, the products exhibited unique acylation features. Both compounds were N-acylated at the side-chain of an ornithine residue derived from arginine, presumably generated by arginase activity (of KspR or PhaR). Kamptornamide was N-acylated with dodecanoic acid (C12), and phaeornamide was modified with 3-(S)-hydroxydecanoic acid (C10) at the side-chain of 4-(S)-hydroxyornithine. Because the BGCs did not encode enzymes for fatty acid biosynthesis, it is likely that the fatty acids were supplied by the primary metabolism of E. coli. However, E. coli does not typically accumulate medium-chain fatty acids such as C10 and C12 forms; so, how, specifically, the fatty acid modifications take place is interesting. A possible explanation is that the culture temperature (isopropyl β-D-1-thiogalactopyranoside induction at 18 °C) may have affected the composition of the E. coli fatty acids.37 Additionally, because the BGCs do not encode a dedicated ACP, this observation raises the possibility that the acylation steps catalyzed by KspN and PhaN, which have a conserved CoA-binding pocket predicted in their primary structures, are likely to use acyl-CoA as substrates rather than acyl-ACP (which is observed in the biosynthesis of goadvionin). However, this hypothesis requires experimental validation. Because of their structural features, in which N-acylation occurs on the amine group of an ornithine side-chain, this compound class was named selidamides (“selida” means “side” in ancient Greek). The bioactivities of these compounds have not been elucidated.
4. Short-to-medium-chain acylation
4.1. AviCys-containing thioamide: prethioviridamide
Relatively short-chain acyl groups have been found in several RiPP natural products. Thioviridamide is an AviCys-containing cyclic peptide, with, uniquely, five thioamides in its side-chain. It was found to contain a 2-hydroxy-2-methyl-4-oxopentanoyl (HMOP) group at its N-terminus. Thioviridamide was isolated from S. olivoviridis NA05001.38 It was later revealed that the true metabolite is prethioviridamide,39 which contains a pyruvyl group at the N-terminus of the RiPP core (Fig. 5A). Investigation of the tva BGC in S. olivoviridis NA05001 showed that it lacks an acyltransferase homolog. It was therefore proposed that the pyruvyl moiety is formed from the N-terminal serine residue of the peptide via dehydration, enamine–imine tautomerization, and hydrolysis (Fig. 6A).40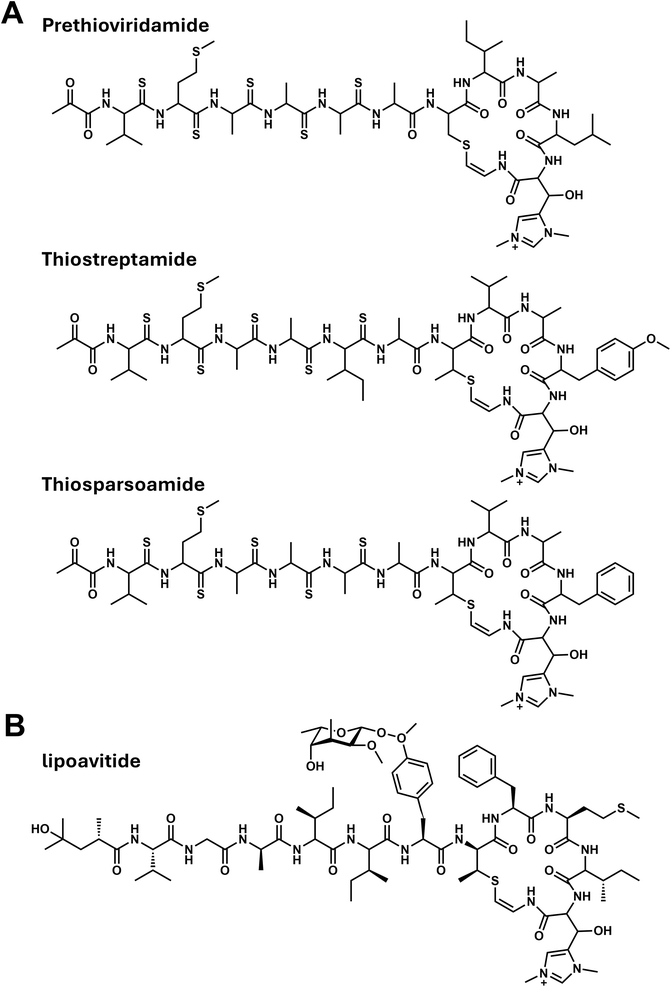 | ||
| Fig. 5 Chemical structures of 2-aminovinyl-cysteine (AviCys)-containing peptides from Actinobacteria. (A) Prethioviridamide from S. olivoviridis NA05001,38 thiostreptamide from Streptomyces sp. NRRL S-4,42 and thiosparsoamide from S. sparsogenes ATCC 25498.43 (B) Lipoavitide from Streptomyces sp. NRRL S-1521.44 | ||
 | ||
| Fig. 6 N-Terminal acyl modifications in ribosomally synthesized and post-translationally modified peptide (RiPP) biosynthesis. (A) Formation of the N-terminal pyruvyl moiety from a serine residue in prethioviridamide biosynthesis.40 (B) Formation of the N-terminal 5,5-dimethyl-2-oxohexanoyl group from a threonine residue in polytheonamide biosynthesis.50 (C) Proposed biosynthesis pathway of 4-hydroxy-2,4-dimethylpentanoyl-CoA (HMP-CoA) from isobutyryl-CoA and methylmalonyl-CoA in lipoavitide biosynthesis.44 (D) CoA-dependent N-acyltransferase reaction catalyzed by LpvE in lipoavitide biosynthesis.44 | ||
Prethioviridamide induces selective apoptosis of various cancer cells—particularly those expressing the adenoviral oncogene E1A—by binding to and inhibiting mitochondrial respiratory chain complex V (F1Fo-ATP synthase)41 (Table 1).
The formation of AviCys-containing macrocycles is a unique feature of these thioamide-containing RiPP natural products. In the biosynthesis of thiostreptamide S4 from Streptomyces sp. NRRL S-4 (Fig. 5A), it is predicted that a phosphotransferase (TsaC) and a HopA1-like protein (TsaD), a class V lanthipeptide synthetases, catalyze the formation of dehydroalanine (or dehydrobutyrine), while a LanD-like enzyme (TsaF) generates the AviCys residue. Notably, a canonical lanthionine synthetase (such as LanKC or LanM), which are typically involved in the macrocyclization steps in the formation of lipolanthines, goadvionins, and selidamides, is absent from the BGCs for biosynthesis of RiPPs with thioamide groups (i.e., tva and tsa for prethioviridamide and thiostreptamide, respectively).42 Interestingly, in the biosynthesis of thiosparsoamide in S. sparsogenes ATCC 25498, a LanKC homolog (SpaKC) located outside the spa cluster was found to catalyze the dehydration of a serine residue and was involved in the formation of the AviCys-containing macrocycle with LanD homologue SpaF.43 Further experimental validation is required to reveal the functions of the phosphotransferase and HopA1-like protein conserved in the thioamide BGC.
4.2. AviMeCys-containing lipopeptides: lipoavitides
In thioamide biosynthesis, the N-terminal pyruvyl moiety is predicted to be derived from a serine residue (Fig. 6A). A newly identified class of fatty acylated RiPPs features an AviCys-containing core with structural similarity to thioamides (Fig. 5B). Huimin Zhao and colleagues performed genome mining for RiPP BGCs encoding conserved AviCys (or AviMeCys) biosynthesis genes, such as tsaC, tsaD, and tsaF, known from thiostreptamide S4 biosynthesis. Using the CAPTURE method (Cas12a-assisted precise targeted cloning with in vivo Cre-lox recombination), they cloned the lpv BGC from Streptomyces sp. NRRL S-1521. Heterologous expression in S. albus J1074 led to the discovery of lipoavitides, a new class of AviMeCys-containing lipopeptides modified with a 4-hydroxy-2,4-dimethylpentanoyl (HMP) group (Fig. 5B).44 While lipoavitides lack thioamide groups,45 they feature unique modifications, including O-glycosylation (2-O-methyl-β-6-deoxygulose) of a tyrosine side-chain and incorporation of a D-alanine residue derived from serine.The biosynthesis pathway of 4-hydroxy-2,4-dimethylpentanoate was systematically investigated.44 It was found that LpvG, a 3-ketoacyl-ACP synthase III, catalyzes the initial condensation of isobutyryl-CoA and methylmalonyl-CoA to produce 3-oxo-2,4-dimethylpentanoyl-CoA (Fig. 6C). This intermediate undergoes sequential transformations: ketoreduction by LpvV, dehydration by LpvF, and enoyl reduction also by LpvH, yielding 2,4-dimethylpentanoyl-CoA. The P450 enzyme LpvS subsequently hydroxylates this intermediate to generate HMP-CoA. The enzymatic condensation of the fatty acyl group to the N-terminus of the RiPP is described later (please see Section 5). Notably, the acyltransferase LpvE displays substrate promiscuity, accepting various acyl-CoA derivatives to modify the RiPP scaffold.
The lipoavitides exhibit hemolytic activity, with the N-terminal HMP moiety playing a critical role in this bioactivity; removal of the HMP group, leaving only the RiPP core, led to a marked reduction in the hemolytic activity.44 While the fatty acyl moieties of other RiPP-derived lipopeptides, such as lipolanthins and goadvionins, have been identified, their precise contributions to antibacterial activity remain unclear. Elucidating the role of these lipid modifications will be an important direction for future research.
4.3. Polytheonamide
Polytheonamide, one of the most extensively modified RiPPs, is composed of 48 amino acid residues and features a remarkable array of modifications, including eight tert-leucines, three β-hydroxyvalines, six γ-N-methylasparagines, two γ-N-methyl-β-hydroxyasparagines, and a β,β-dimethylmethionine sulfoxide. Notably, 18 of its residues are D-amino acids. It was originally isolated from the marine sponge Theonella swinhoei by the Fusetani and Matsunaga groups.46 Uncultivated filamentous bacteria “Candidatus Entotheonella” spp. have been proposed as symbionts of marine sponge and a source of marine natural products.47 It was later found that polytheonamide is produced by symbiotic Entotheonella spp., members of the uncultivated candidate phylum “Tectomicrobia,” with genome sizes exceeding 9 megabases.48Because of its structural complexity and the presence of numerous nonproteinogenic and D-amino acids, polytheonamide was initially thought to be a nonribosomal peptide product. However, Piel and colleagues identified the ribosomal origin of polytheonamide through metagenomic analysis of T. swinhoei.49 The poy BGC responsible for polytheonamide biosynthesis was discovered in the assembled metagenomic data, revealing that only eight gene products are required to construct this highly complex molecule.50 The N-terminus of polytheonamide features a unique 5,5-dimethyl-2-oxohexanoyl group (Fig. 6B). Structural comparison with the core peptide suggests that a threonine residue is transformed into this unusual acyl moiety through a remarkable series of biosynthetic steps, including dehydration catalyzed by PoyF, formal tert-butylation at a nonactivated carbon by PoyC, and formation of the 2-oxo group via hydrolytic cleavage of the enamide intermediate by PoyH (Fig. 6B). These insights led to the successful biosynthesis of the polytheonamide-related compound, aeronamide A, in the culturable host Microvirgula aerodenitrificans, and further enabled the production of polygeonamides, whose biosynthetic gene cluster (BGC) originates from the uncultivated Rhodospirillaceae bacterium BRH-c57.51
Polytheonamide B exhibits potent cytotoxicity against P388 murine leukemia cells, with an IC50 value of 68 pg mL−1.46 Its structure is a right-handed β-helix approximately 45 Å long, which forms a hydrophilic pore with inner diameter ∼4 Å. This unique structure enables the formation of transmembrane channels that selectively permeate monovalent cations. The high cytotoxicity of polytheonamide B is attributed to its ability to form single-molecule ion channels in biological membranes.52 The Inoue group, in addition to accomplishing the total synthesis of polytheonamide,53 has extensively investigated its structure–activity relationship. Notably, they demonstrated that N-terminal modifications with hydrophobic groups tend to enhance cytotoxic activity.54 In particular, the palmitamide derivative bearing an N-terminal C16 palmitoyl group exhibited even greater potency than the natural polytheonamide. These findings underscore the critical role of N-terminal acylation in modulating the bioactivity of this peptide.
4.4. N-Acetylayed RiPPs
Goadsporin was first isolated from Streptomyces sp. TP-A0584 through a screen for secondary metabolites that induce pigment production and morphological differentiation in S. lividans.55,56 The god BGC was identified via heterologous expression, revealing that goadsporin is a ribosomally synthesized linear azole-containing peptide featuring dehydroamino acids and N-terminal acetylation.57 The GNAT family acetyltransferase GodH was shown to catalyze N-terminal acetylation using acetyl-CoA as a substrate.58 Whether GodH exhibits substrate promiscuity and can transfer alternative acyl groups—similar to LpvE in lipoavitide biosynthesis—remains to be investigated. Goadsporin likely targets the bacterial signal recognition particle (SRP), because overexpression of paralogous Ffh homologs (the product of the godI gene), a component of SRP, confers self-resistance (Table 1).57In addition to goadsporin, N-acetylated RiPPs have been identified in a variety of bacterial species. For example, members of the microviridin family—tricyclic depsipeptides produced by Microcystis spp. (cyanobacteria)—feature N-terminal acetylation.59 Albusnodin, a lasso peptide from Streptomyces albus, is also post-translationally modified with an acetyl group.60 Paenibacillin, a lantibiotic produced by the Firmicute Paenibacillus polymyxa, likewise exhibits N-terminal acetylation.61
5. Diversification of fatty acyl moieties
In vitro experiments revealed that, in goadvionin biosynthesis, the GNAT family acyltransferase GdvG recognizes the C-terminal ACP domain in GdvPKS and catalyzes the condensation reaction between a non-natural stearoyl (C18) unit loaded onto the ACP and the N-terminus of the RiPP moiety (Fig. 2B–D and Table 2).26 The transferase reaction was successfully performed using a stearoyl substrate, which is markedly different from the native substrate (Fig. 2B), demonstrating the substrate promiscuity of GdvG. This suggests the potential of the enzyme for generating analogs with non-natural acyl groups. In contrast, in lipoavitide biosynthesis, LpvE was shown to recognize acyl-CoA as a substrate,44 bypassing the need for a specific ACP (Fig. 6D and Table 2). LpvE appears to preferentially recognize shorter acyl chains (less than C6), but also accepts acyl-CoA with a branched chain, an aromatic group, and even an alkynyl group, likely determined by the size of the cavity for the acyl-CoA substrate.44| Isolated product | Fatty acyl moiety (putative donor substrate) | Acylation position in RiPPs | Putative acyl transferase homologs | Condensation mechanism | Experimental evidence | Reference |
|---|---|---|---|---|---|---|
| a N/A: not available. | ||||||
| Microvionin | Dimethyl guanidine terminal C15 fatty acyl group | RiPP N-terminal | MicB | Fatty acyl-ACP dependent (putative) | N/A | 22 |
| Nocavionin | Dimethyl guanidine terminal C13 (with 3 trans double bonds) fatty acyl group | RiPP N-terminal | NocB1/NocB2 | Fatty acyl-ACP dependent (putative) | N/A | 22 |
| Goadvionins | Trimethylamine terminal C31 fatty acyl group | RiPP N-terminal | GdvG | Fatty acyl-ACP dependent | In vivo (gene disruption): Detection of intermediates | 26 |
| Stearoyl group | RiPP N-terminal | GdvG | Fatty acyl-ACP dependent | In vitro reaction | 26 | |
| Solabiomycins | Guanidine terminal C13 fatty acyl group | RiPP N-terminal | SolG | Fatty acyl-ACP dependent (putative) | In vivo (gene disruption): Abolishment of production | 31 |
| Albopeptins | Guanidine terminal C13 fatty acyl group | RiPP N-terminal | AlbG/NapG | Fatty acyl-ACP dependent (putative) | N/A | 21 and 31 |
| Kamptornamide | C12 fatty acyl group | Side chain amine of N-terminal ornithine residue | KspN | Fatty acyl-CoA dependent (putative) | In vivo (heterologous biosynthesis) | 35 |
| Phaeornamide | 3-(S)-Hydroxydecanoyl (C10) group | Side chain amine of N-terminal ornithine residue | PhaN | Fatty acyl-CoA dependent (putative) | In vivo (heterologous biosynthesis) | 35 |
| Lipoavitide | 4-Hydroxy-2,4-dimethylpentanoyl group | RiPP N-terminal | LpvE | Fatty acyl-CoA dependent | In vitro reaction | 44 |
Based on comparison of BGCs with those for goadvionin,26 it appears that synthesis of microvionin from M. arborescens,22 nocavionin from N. terpenica,22 solabiomycins from S. lydicus,31 and albopeptins from S. albofaciens or S. nigrescens21,31 may use ACP-dependent transfer of acyl moieties by a GNAT family acyltransferase, because specific ACP homologues are present in their BGCs (Table 2). In contrast, similar to the BGCs for lipoavitide,44 the synthesis of kamptornamide from Kamptonema sp.35 and phaeornamide from P. arcticus35 may not require an ACP, instead directly using acyl-CoA as a substrate (Table 2).
The range of substrate tolerance of the acyltransferase enzymes enables the incorporation of diverse acyl groups in RiPP bioengineering. However, expanding structural diversity further will require overcoming the challenge of substrate tolerance toward the RiPP core. Detailed three dimensional structural characterization of relevant enzymes will be essential for more precise and rational protein engineering.
Fatty acyl moieties play a critical role in bioactivity, as demonstrated in NRPS-derived antibiotics such as DAP, where the acyl chain modulates both antibacterial efficacy62 and hemolytic toxicity.32 DAP, a semisynthetic derivative of the A21978C complex from S. roseosporus, is produced industrially by supplementing toxic decanoic acid.63 Recently, Kang and colleagues developed a synthetic biology strategy for selective biosynthesis of C10 decanoic acid, streamlining DAP production.64 Such control over fatty acid chain length could enable synthetic biology approaches to generate novel RiPP-derived lipopeptides.
6. Bioactivity – potential molecular targets of RiPP-type lipopeptides
RiPP-derived lipopeptides exhibit diverse bioactivities, including antibacterial, antifungal, and cytotoxic hemolytic effects (Table 1). However, their mechanisms of action remain largely uncharacterized, and further investigation will be essential to fully realize their potential as therapeutic agents. Nevertheless, insights into their modes of action may be drawn from studies of compounds with similar structural features to those found in RiPP-derived lipopeptides:Peptide antibiotics containing AviCys are known to interact with the bacterial cell wall precursor lipid II, forming pores in the membrane.65 This dual mechanism—lipid II sequestration combined with pore formation—leads to bacterial cell lysis and death, and has been described for several lanthipeptides, including mutacin-1140 and epidermin. Other lanthipeptides, such as mersacidin, are currently believed to function solely through lipid II sequestration without pore formation.
The NRPS-derived lipopeptide DAP acts by forming calcium-dependent micelles, enabling its insertion into phosphatidylglycerol-containing membranes. Once inserted, it oligomerizes and either forms cation-selective pores or disrupts membrane integrity by lipid extraction, leading to ion leakage and cell death.66 Another NRPS-derived lipopeptides, polymyxins (e.g., colistin) act by binding to lipid A in lipopolysaccharide (LPS), displacing divalent cations and destabilizing the outer membrane. The lipid tail inserts into the membrane, promoting self-mediated uptake into the periplasm, where colistin targets LPS precursors in the inner membrane. This disrupts membrane integrity, causes cytoplasmic leakage, and leads to bacterial cell death.67 These NRPS-derived lipopeptides may offer valuable insights into the mechanisms of action of RiPP-derived lipopeptides.
7. Concluding remarks
Recent genome mining efforts have uncovered a novel class of RiPP-based natural products that undergo fatty acylation during their biosynthesis, generating hybrid molecules that combine RiPP cores with fatty acid moieties. These RiPP-derived lipopeptides exhibit antibacterial and antifungal activities, and hold significant promise as therapeutic agents. The RiPP precursors are genetically encoded, but structural diversification can be achieved through in vitro post-translational enzymatic modifications.68–70 Efficient selection of functional variants can be facilitated using target-guided mRNA display screening,71 which may aid the development of this class of molecules for therapeutic uses.The molecular diversity revealed through genome mining, combined with rational design for the biological production of RiPP-derived lipopeptides, may advance drug discovery efforts in response to the growing challenge of antimicrobial resistance.72 Moreover, integrating in vitro engineering strategies with in vivo screening and production platforms for RiPPs73,74 holds great promise for the development of novel RiPP-derived lipopeptide therapeutics in the future.
8. Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.9. Conflicts of interest
The author declares no conflict of interest.10. Acknowledgments
This project was supported by a research grant from the Novozymes Japan Research Foundation. We thank Edanz (https://www.jp.edanz.com/ac) for editing a draft of this manuscript. This article was made open access with funding support from Kobe University.11. References
- D. Richter and J. Piel, Curr. Opin. Chem. Biol., 2024, 80, 102463 CrossRef CAS PubMed.
- Q. Guo and B. I. Morinaka, Curr. Opin. Chem. Biol., 2024, 81, 102483 CrossRef CAS PubMed.
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K. D. Entian, M. A. Fischbach, J. S. Garavelli, U. Goransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Muller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. Reaney, S. Rebuffat, R. P. Ross, H. G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Sussmuth, J. R. Tagg, G. L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey and W. A. van der Donk, Nat. Prod. Rep., 2013, 30, 108–160 RSC.
- M. M. Zdouc, J. J. J. van der Hooft and M. H. Medema, Trends Pharmacol. Sci., 2023, 44, 532–541 CrossRef CAS PubMed.
- A. M. Kloosterman, M. H. Medema and G. P. van Wezel, Curr. Opin. Biotechnol., 2021, 69, 60–67 CrossRef CAS PubMed.
- C. Ongpipattanakul, E. K. Desormeaux, A. DiCaprio, W. A. van der Donk, D. A. Mitchell and S. K. Nair, Chem. Rev., 2022, 122, 14722–14814 CrossRef CAS PubMed.
- E. Breukink, I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. G. Sahl and B. de Kruijff, Science, 1999, 286, 2361–2364 CrossRef CAS PubMed.
- S. T. Hsu, E. Breukink, E. Tischenko, M. A. Lutters, B. de Kruijff, R. Kaptein, A. M. Bonvin and N. A. van Nuland, Nat. Struct. Mol. Biol., 2004, 11, 963–967 CrossRef CAS PubMed.
- G. L. Conn, D. E. Draper, E. E. Lattman and A. G. Gittis, Science, 1999, 284, 1171–1174 CrossRef CAS PubMed.
- B. T. Wimberly, R. Guymon, J. P. McCutcheon, S. W. White and V. Ramakrishnan, Cell, 1999, 97, 491–502 CrossRef CAS PubMed.
- M. V. Rodnina, A. Savelsbergh, N. B. Matassova, V. I. Katunin, Y. P. Semenkov and W. Wintermeyer, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9586–9590 CrossRef CAS PubMed.
- Y. Imai, K. J. Meyer, A. Iinishi, Q. Favre-Godal, R. Green, S. Manuse, M. Caboni, M. Mori, S. Niles, M. Ghiglieri, C. Honrao, X. Ma, J. J. Guo, A. Makriyannis, L. Linares-Otoya, N. Bohringer, Z. G. Wuisan, H. Kaur, R. Wu, A. Mateus, A. Typas, M. M. Savitski, J. L. Espinoza, A. O'Rourke, K. E. Nelson, S. Hiller, N. Noinaj, T. F. Schaberle, A. D'Onofrio and K. Lewis, Nature, 2019, 576, 459–464 CrossRef CAS PubMed.
- H. Kaur, R. P. Jakob, J. K. Marzinek, R. Green, Y. Imai, J. R. Bolla, E. Agustoni, C. V. Robinson, P. J. Bond, K. Lewis, T. Maier and S. Hiller, Nature, 2021, 593, 125–129 CrossRef CAS PubMed.
- Y. Zheng, Y. Cong, E. W. Schmidt and S. K. Nair, Acc. Chem. Res., 2022, 55, 1313–1323 CrossRef CAS PubMed.
- K. D. Roberts, M. A. Azad, J. Wang, A. S. Horne, P. E. Thompson, R. L. Nation, T. Velkov and J. Li, ACS Infect. Dis., 2015, 1, 568–575 CrossRef CAS PubMed.
- L. Robbel and M. A. Marahiel, J. Biol. Chem., 2010, 285, 27501–27508 CrossRef CAS PubMed.
- Q. Yue, L. Chen, X. Zhang, K. Li, J. Sun, X. Liu, Z. An and G. F. Bills, Eukaryotic Cell, 2015, 14, 698–718 CrossRef CAS PubMed.
- R. D. Sussmuth and A. Mainz, Angew Chem. Int. Ed. Engl., 2017, 56, 3770–3821 CrossRef PubMed.
- M. Dell, K. L. Dunbar and C. Hertweck, Nat. Prod. Rep., 2022, 39, 453–459 RSC.
- K. Isono, K. Kobinata, H. Okawa, H. Kusakabe, M. Uramoto, K. Ko, T. Misato, S.-W. Tai, C.-T. Ni and Y.-C. Shen, Agric. Biol. Chem., 1986, 50, 2163–2165 CAS.
- H. Oikawa, Y. Mizunoue, T. Nakamura, E. Fukushi, J. Yulu, T. Ozaki and A. Minami, Biosci., Biotechnol., Biochem., 2022, 86, 717–723 CrossRef PubMed.
- V. Wiebach, A. Mainz, M. J. Siegert, N. A. Jungmann, G. Lesquame, S. Tirat, A. Dreux-Zigha, J. Aszodi, D. Le Beller and R. D. Sussmuth, Nat. Chem. Biol., 2018, 14, 652–654 CrossRef CAS PubMed.
- M. Montalban-Lopez, T. A. Scott, S. Ramesh, I. R. Rahman, A. J. van Heel, J. H. Viel, V. Bandarian, E. Dittmann, O. Genilloud, Y. Goto, M. J. Grande Burgos, C. Hill, S. Kim, J. Koehnke, J. A. Latham, A. J. Link, B. Martinez, S. K. Nair, Y. Nicolet, S. Rebuffat, H. G. Sahl, D. Sareen, E. W. Schmidt, L. Schmitt, K. Severinov, R. D. Sussmuth, A. W. Truman, H. Wang, J. K. Weng, G. P. van Wezel, Q. Zhang, J. Zhong, J. Piel, D. A. Mitchell, O. P. Kuipers and W. A. van der Donk, Nat. Prod. Rep., 2021, 38, 130–239 RSC.
- V. Wiebach, A. Mainz, R. Schnegotzki, M. J. Siegert, M. Hugelland, N. Pliszka and R. D. Sussmuth, Angew Chem. Int. Ed. Engl., 2020, 59, 16777–16785 CrossRef CAS PubMed.
- S. Chang, Y. Luo, N. He, X. Huang, M. Chen, L. Yuan and Y. Xie, Front. Chem., 2023, 11, 1233938 CrossRef CAS PubMed.
- R. Kozakai, T. Ono, S. Hoshino, H. Takahashi, Y. Katsuyama, Y. Sugai, T. Ozaki, K. Teramoto, K. Teramoto, K. Tanaka, I. Abe, S. Asamizu and H. Onaka, Nat. Chem., 2020, 12, 869–877 CrossRef CAS PubMed.
- H. Takahashi, S. Sekiya, T. Nishikaze, K. Kodera, S. Iwamoto, M. Wada and K. Tanaka, Anal. Chem., 2016, 88, 3810–3816 CrossRef CAS PubMed.
- H. Takahashi, Y. Shimabukuro, D. Asakawa, S. Yamauchi, S. Sekiya, S. Iwamoto, M. Wada and K. Tanaka, Anal. Chem., 2018, 90, 7230–7238 CrossRef CAS PubMed.
- J. Li and S. E. Jensen, Chem. Biol., 2008, 15, 118–127 CrossRef CAS PubMed.
- H. Hong, T. Fill and P. F. Leadlay, Angew Chem. Int. Ed. Engl., 2013, 52, 13096–13099 CrossRef CAS PubMed.
- S. Asamizu, S. Ijichi, S. Hoshino, H. Jo, H. Takahashi, Y. Itoh, S. Matsumoto and H. Onaka, ACS Chem. Biol., 2022, 17, 2936–2944 CrossRef CAS PubMed.
- N. Bionda, J. P. Pitteloud and P. Cudic, Future Med. Chem., 2013, 5, 1311–1330 CrossRef CAS PubMed.
- S. Boyle-Vavra, S. Yin, D. S. Jo, C. P. Montgomery and R. S. Daum, Antimicrob. Agents Chemother., 2013, 57, 83–95 CrossRef CAS PubMed.
- S. Ijichi, S. Hoshino, S. Asamizu and H. Onaka, Bioorg. Med. Chem. Lett., 2023, 89, 129323 CrossRef CAS PubMed.
- F. Hubrich, N. M. Bosch, C. Chepkirui, B. I. Morinaka, M. Rust, M. Gugger, S. L. Robinson, A. L. Vagstad and J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2113120119 CrossRef CAS PubMed.
- S. C. Bobeica, S. H. Dong, L. Huo, N. Mazo, M. I. McLaughlin, G. Jimenez-Oses, S. K. Nair and W. A. van der Donk, Elife, 2019, 8, e42305 CrossRef PubMed.
- L. Hoogerland, S. P. H. van den Berg, Y. Suo, Y. W. Moriuchi, A. Zoumaro-Djayoon, E. Geurken, F. Yang, F. Bruggeman, M. D. Burkart and G. Bokinsky, Nat. Commun., 2024, 15, 9386 CrossRef CAS PubMed.
- Y. Hayakawa, K. Sasaki, H. Adachi, K. Furihata, K. Nagai and K. Shin-ya, J. Antibiot., 2006, 59, 1–5 CrossRef CAS PubMed.
- M. Izawa, S. Nagamine, H. Aoki and Y. Hayakawa, J. Gen. Appl. Microbiol., 2018, 64, 50–53 CrossRef CAS PubMed.
- M. Izawa, T. Kawasaki and Y. Hayakawa, Appl. Environ. Microbiol., 2013, 79, 7110–7113 CrossRef CAS PubMed.
- S. Takase, R. Kurokawa, Y. Kondoh, K. Honda, T. Suzuki, T. Kawahara, H. Ikeda, N. Dohmae, H. Osada, K. Shin-Ya, T. Kushiro, M. Yoshida and K. Matsumoto, ACS Chem. Biol., 2019, 14, 1819–1828 CrossRef CAS PubMed.
- T. H. Eyles, N. M. Vior, R. Lacret and A. W. Truman, Chem. Sci., 2021, 12, 7138–7150 RSC.
- J. Lu, Y. Wu, Y. Li and H. Wang, Angew Chem. Int. Ed. Engl., 2021, 60, 1951–1958 CrossRef CAS PubMed.
- H. Ren, C. Huang, Y. Pan, S. R. Dommaraju, H. Cui, M. Li, M. G. Gadgil, D. A. Mitchell and H. Zhao, Nat. Chem., 2024, 16, 1320–1329 CrossRef CAS PubMed.
- A. Sikandar, M. Lopatniuk, A. Luzhetskyy, R. Muller and J. Koehnke, J. Am. Chem. Soc., 2022, 144, 5136–5144 CrossRef CAS PubMed.
- T. Hamada, S. Matsunaga, G. Yano and N. Fusetani, J. Am. Chem. Soc., 2005, 127, 110–118 CrossRef CAS PubMed.
- E. W. Schmidt, A. Y. Obraztsova, S. K. Davidson, D. J. Faulkner and M. G. Haygood, Mar. Biol., 2000, 136, 969–977 CrossRef CAS.
- M. C. Wilson, T. Mori, C. Ruckert, A. R. Uria, M. J. Helf, K. Takada, C. Gernert, U. A. Steffens, N. Heycke, S. Schmitt, C. Rinke, E. J. Helfrich, A. O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crusemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama and J. Piel, Nature, 2014, 506, 58–62 CrossRef CAS PubMed.
- M. F. Freeman, C. Gurgui, M. J. Helf, B. I. Morinaka, A. R. Uria, N. J. Oldham, H. G. Sahl, S. Matsunaga and J. Piel, Science, 2012, 338, 387–390 CrossRef CAS PubMed.
- M. F. Freeman, M. J. Helf, A. Bhushan, B. I. Morinaka and J. Piel, Nat. Chem., 2017, 9, 387–395 CrossRef CAS PubMed.
- A. Bhushan, P. J. Egli, E. E. Peters, M. F. Freeman and J. Piel, Nat. Chem., 2019, 11, 931–939 CrossRef CAS PubMed.
- T. Hamada, S. Matsunaga, M. Fujiwara, K. Fujita, H. Hirota, R. Schmucki, P. Güntert and N. Fusetani, J. Am. Chem. Soc., 2010, 132, 12941–12945 CrossRef CAS PubMed.
- M. Inoue, N. Shinohara, S. Tanabe, T. Takahashi, K. Okura, H. Itoh, Y. Mizoguchi, M. Iida, N. Lee and S. Matsuoka, Nat. Chem., 2010, 2, 280–285 CrossRef CAS PubMed.
- N. Shinohara, H. Itoh, S. Matsuoka and M. Inoue, ChemMedChem, 2012, 7, 1770–1773 CrossRef CAS PubMed.
- Y. Igarashi, Y. Kan, K. Fujii, T. Fujita, K. Harada, H. Naoki, H. Tabata, H. Onaka and T. Furumai, J. Antibiot., 2001, 54, 1045–1053 CrossRef CAS PubMed.
- H. Onaka, H. Tabata, Y. Igarashi, Y. Sato and T. Furumai, J. Antibiot., 2001, 54, 1036–1044 CrossRef CAS PubMed.
- H. Onaka, M. Nakaho, K. Hayashi, Y. Igarashi and T. Furumai, Microbiology, 2005, 151, 3923–3933 CrossRef CAS PubMed.
- T. Ozaki, K. Yamashita, Y. Goto, M. Shimomura, S. Hayashi, S. Asamizu, Y. Sugai, H. Ikeda, H. Suga and H. Onaka, Nat. Commun., 2017, 8, 14207 CrossRef CAS PubMed.
- N. Ziemert, K. Ishida, A. Liaimer, C. Hertweck and E. Dittmann, Angew Chem. Int. Ed. Engl., 2008, 47, 7756–7759 CrossRef CAS PubMed.
- C. Zong, W. L. Cheung-Lee, H. E. Elashal, M. Raj and A. J. Link, Chem. Commun., 2018, 54, 1339–1342 RSC.
- E. Huang and A. E. Yousef, Microbiol. Res., 2015, 181, 15–21 CrossRef CAS PubMed.
- J. H. Lakey and E. J. A. Lea, Biochim. Biophys. Acta, 1986, 859, 219–226 CrossRef CAS PubMed.
- F. M. Huber, R. L. Pieper and A. J. Tietz, J. Biotechnol., 1988, 7, 283–292 CrossRef CAS.
- C. H. Ji, S. Park, K. Lee, H. W. Je and H. S. Kang, J. Am. Chem. Soc., 2024, 146, 30434–30442 CrossRef CAS PubMed.
- E. S. Grant-Mackie, E. T. Williams, P. W. R. Harris and M. A. Brimble, JACS Au, 2021, 1, 1527–1540 CrossRef CAS PubMed.
- W. R. Miller, A. S. Bayer and C. A. Arias, Cold Spring Harbor Perspect. Med., 2016, 6, a026997 CrossRef PubMed.
- E. V. K. Ledger, A. Sabnis and A. M. Edwards, Microbiology, 2022, 168, 001136 CrossRef CAS PubMed.
- J. S. Chang, A. A. Vinogradov, Y. Zhang, Y. Goto and H. Suga, ACS Cent. Sci., 2023, 9, 2150–2160 CrossRef CAS PubMed.
- A. A. Vinogradov, M. Shimomura, Y. Goto, T. Ozaki, S. Asamizu, Y. Sugai, H. Suga and H. Onaka, Nat. Commun., 2020, 11, 2272 CrossRef CAS PubMed.
- Y. C. Zhang, K. Hamada, D. T. Nguyen, S. Inoue, M. Satake, S. Kobayashi, C. Okada, K. Ogata, M. Okada, T. Sengoku, Y. Goto and H. Suga, Nat. Catal., 2022, 5, 682 CrossRef CAS.
- S. E. Iskandar and A. A. Bowers, ACS Med. Chem. Lett., 2022, 13, 1379–1383 CrossRef CAS PubMed.
- C. E. Seyfert, C. Porten, B. Yuan, S. Deckarm, F. Panter, C. D. Bader, J. Coetzee, F. Deschner, K. Tehrani, P. G. Higgins, H. Seifert, T. C. Marlovits, J. Herrmann and R. Muller, Angew Chem. Int. Ed. Engl., 2023, 62, e202214094 CrossRef CAS PubMed.
- T. Si, Q. Tian, Y. Min, L. Zhang, J. V. Sweedler, W. A. van der Donk and H. Zhao, J. Am. Chem. Soc., 2018, 140, 11884–11888 CrossRef CAS PubMed.
- R. S. Ayikpoe, C. Shi, A. J. Battiste, S. M. Eslami, S. Ramesh, M. A. Simon, I. R. Bothwell, H. Lee, A. J. Rice, H. Ren, Q. Tian, L. A. Harris, R. Sarksian, L. Zhu, A. M. Frerk, T. W. Precord, W. A. van der Donk, D. A. Mitchell and H. Zhao, Nat. Commun., 2022, 13, 6135 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |
