Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Green genes from blue greens: challenges and solutions to unlocking the potential of cyanobacteria in drug discovery
Benjamin
Philmus
 *a,
Nicole E.
Avalon
*a,
Nicole E.
Avalon
 b,
Yousong
Ding
b,
Yousong
Ding
 c,
Drew T.
Doering
c,
Drew T.
Doering
 d,
Alessandra S.
Eustáquio
d,
Alessandra S.
Eustáquio
 e,
William H.
Gerwick
e,
William H.
Gerwick
 f,
Hendrik
Luesch
f,
Hendrik
Luesch
 cg,
Jimmy
Orjala
cg,
Jimmy
Orjala
 e,
Shaz
Sutherland
e,
Shaz
Sutherland
 a,
Arnaud
Taton
a,
Arnaud
Taton
 h and
Daniel
Udwary
h and
Daniel
Udwary
 d
d
aDepartment of Pharmaceutical Sciences, Oregon State University, Corvallis, OR 97333, USA. E-mail: benjamin.philmus@oregonstate.edu
bDepartment of Pharmaceutical Sciences, School of Pharmacy & Pharmaceutical Sciences, University of California, Irvine, Irvine, CA 92617, USA
cDepartment of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, University of Florida, Gainesville, FL 32610, USA
dU.S. Department of Energy, Joint Genome Institute, Lawrence Berkeley National Laboratory, One Cyclotron Road, Berkeley, CA 94720, USA
eDepartment of Pharmaceutical Sciences, Retzky College of Pharmacy, University of Illinois, Chicago, Chicago, IL 60612, USA
fCenter for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92092, USA
gProgram in Cancer and Stem Cell Biology, Duke-NUS Medical School, Singapore, 169857, Singapore
hSchool of Biological Sciences, University of California, San Diego, La Jolla, CA 92093, USA
First published on 15th July 2025
Abstract
Cyanobacteria are prolific producers of biologically active compounds that are important in influencing ecology, behavior of interacting organisms, and as leads in drug discovery efforts. Here we discuss the challenges faced by all natural product researchers, especially those that focus on cyanobacteria, and then describe progress that has been made in these areas. We also propose some solutions, paths forward, and thoughts for consideration on these challenges.
1. Background
Cyanobacteria, also known as ‘blue-green algae’, emerged approximately 3 billion years ago and have since developed a remarkable array of metabolic traits through extensive evolutionary adaptation. These include oxygenic photosynthesis, nitrogen fixation, UV and desiccation tolerance, and the production of a diverse repertoire of secondary metabolites. Notably, their ability to perform oxygenic photosynthesis is credited with transforming Earth's atmosphere into its current oxygen-rich state and continues to have profound implications for future climate scenarios. Since the 1970s, cyanobacteria have been acknowledged as an exceptional source of bioactive and structurally diverse secondary metabolites. More recently, cyanobacteria have been recognized as a carbon-negative chassis for synthetic biology applications, including production of biofuels, sunscreens, and drug leads.1Cyanobacterial natural products play significant environmental and ecological roles.2–4 In aquatic environments ranging from freshwater to marine, species such as Microcystis and Nodularia produce cyclic peptides, including microcystins and nodularins, which exhibit potent protein phosphatase inhibition.5,6 When ingested, these compounds are highly toxic to the liver of mammalian species. These toxigenic cyanobacteria can grow to high cell densities and lead to harmful cyanobacterial blooms, posing serious ecological and public health challenges.7
Cyanobacteria have also been pivotal in Earth's evolutionary history, predating the presence of atmospheric oxygen and the protective ozone layer that shields against UV-A and UV-B radiation.8 In response to these harsh conditions, they evolved the ability to produce natural products that protect against UV damage. For example, the mycosporine-like amino acids (MAAs), such as shinorine, are small molecules featuring an aminocyclohexenone ring that provide effective protection against UV-B radiation.9,10 Similarly, scytonemin, an indole alkaloid dimer produced by many cyanobacterial species, offers protection against UV-A wavelengths. These compounds highlight the adaptive versatility of cyanobacteria in diverse and challenging environments.11
Cyanobacterial metabolites exhibit distinctive features, such as nitrogen-rich frameworks, covalent incorporation of halogen atoms, and hybrid structures integrating components from different biosynthetic pathways.12 Biologically, cyanobacterial secondary metabolites target a variety of molecular processes, including proteases, ion channels, tubulin, actin, and other critical cellular features of potential eukaryotic competitors. This specificity has particularly positioned cyanobacterial compounds as promising candidates for anticancer and antiproliferative therapies.13
Prominent examples of marine cyanobacterial natural products with anticancer or potential anticancer activity include dolastatin 10,14 curacin A,15 and apratoxin A,16 each notable for their architectural complexity and unique mechanisms of action. These compounds derive their distinctive chemical structures from the integrated activities of nonribosomal peptide synthetases (NRPS) and polyketide synthases (PKS), yet their structural diversity results in markedly different biochemical properties.
The potent microtubule depolymerizing agent dolastatin 10, a modified linear peptide–polyketide produced by the marine cyanobacterium Caldora penicillata, served as the starting point for the development of monomethyl auristatin E (MMAE), the cytotoxic payload of five currently approved antibody–drug conjugates (ADCs). The success of brentuximab vedotin, the first MMAE ADC for the treatment of Hodgkins lymphoma and anaplastic large cell lymphoma in 2011, and now a blockbuster drug with over $1B in annual sales, contributed to the ADC revolution and validated this enabling technology as a therapeutic modality.17 Dolastatin 10 (Fig. 1) is a linear hexapeptide featuring two amino acids that are PKS-extended by two carbons each via malonyl-CoA units. The C-terminal cysteine is cyclized and oxidatively decarboxylated to form a thiazole ring. Mechanistically, dolastatin 10 and its analogs inhibit microtubule assembly by binding to the vinca alkaloid site on the β-tubulin subunit, disrupting cellular division. Curacin A (Fig. 1), another structurally intriguing molecule, combines PKS-derived malonyl extensions with a single cysteine unit, producing a distinctive combination of cyclopropyl and thiazoline rings. As a potent antimitotic agent, curacin A binds to the colchicine site of microtubules, interfering with their dynamic behavior essential for mitosis. The gatorbulins (Fig. 1),18 modified cyclodepsipeptides targeting a new pharmacological site of tubulin, represent the most recent scaffold and mechanism that modulate tubulin dynamics.
Largazole (Fig. 1) is among the most potent histone deacetylase (HDAC) inhibitors discovered so far and has selectivity for class I isoforms, rivaling the approved drug romidepsin.19 A largazole analogue, bocodepsin, reached Phase I clinical trials. Carmaphycins (Fig. 1), linear modified peptides, are potent proteasome inhibitors and serve as a template for the development of organism-specific inhibitors.20 While the proteasome is a validated target for cancer, synthetic analogues of the carmaphycins possess selectivity for the parasite proteasome and are now in development to treat tropical parasite infections, including trypanosomiasis. The macrocyclic polyketide–peptide hybrids apratoxins and coibamides (Fig. 1) have been shown to perturb cotranslational translocation by targeting the translocon Sec61 in the endoplasmic reticulum, representing a new mechanism for inhibiting protein maturation.21,22 Elucidating the novel mechanism of these two agents has opened up opportunities for development for different indications linked to Sec61. A biosynthetic hybrid of three different moieties, a peptide–polyketide–glycoside termed iezoside was found to be a sarco/endoplasmic reticulum Ca2+ ATPase pump (SERCA) inhibitor, linking an unusual structure to function.23 Functionally, cyanobacterial natural products display a wide range of biological activities against diverse molecular targets. Synthetic structural modifications of their unique structures can lead to improved drug-like properties.
The remarkable structural diversity of cyanobacterial natural products stems from the unique reactivity and specificity of their biosynthetic enzymes. Over the past three decades, significant progress has been made in understanding the function of some of these enzymes, the pathways they catalyze, and the gene clusters that serve as the ultimate repository of this biosynthetic information.24–26 A natural extension of these discoveries has been the effort to harness and transfer this biosynthetic potential into other microbial systems that are more amenable to cultivation and genetic engineering. A summary of these efforts can be found in recent reviews by Dhakal et al.27 and Baunach et al.28 These efforts include the use of various traditional genetically tractable hosts such as E. coli, various Actinobacteria, and yeast as well as various model cyanobacterial species. The use of a cyanobacterial chassis allows for the environmentally friendly production of these high-value chemicals, as cyanobacteria are considered a sustainable platform for biotechnological applications due to their photoautotrophic and nitrogen-fixing capacities. But some technical barriers in working with cyanobacteria include low titers of their natural products (typically 0.1–0.2% dry weight), slow growth rates, and difficulty removing associated bacteria,29,30 which has led to them being underrepresented in drug discovery efforts.
This perspective aims to provide an overview of recent advancements in the use of cyanobacterial natural product pathway activation in native producers and capture, harness and expression of natural product BGCs in heterologous hosts, challenges faced by all researchers involved in natural product research, and issues specifically facing researchers working on cyanobacterial natural product pathways, while highlighting critical challenges that continue to impede progress in this promising area.
2. Challenges that impact all natural product researchers
2.1. General challenge #1: effective prioritization of BGCs
Previous research has identified up to 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 984 gene cluster families (GCFs) in 16
984 gene cluster families (GCFs) in 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 004 complete bacterial genomes with the majority (97%) being considered orphan biosynthetic gene clusters (BGCs).31 Hence, prioritization of BGCs to pursue for in-depth studies becomes essential. For example, assuming 20 kb for each cluster and a price of commercial synthesis of $0.09 per base, the cost of synthesizing one BGC from each GCF would be $30
004 complete bacterial genomes with the majority (97%) being considered orphan biosynthetic gene clusters (BGCs).31 Hence, prioritization of BGCs to pursue for in-depth studies becomes essential. For example, assuming 20 kb for each cluster and a price of commercial synthesis of $0.09 per base, the cost of synthesizing one BGC from each GCF would be $30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 571
571![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. This is without considering the salary of researchers to assemble the synthesized fragments into BGCs, transformation into appropriate heterologous production hosts, and chemical analysis. Similarly, cultivation of 16
200. This is without considering the salary of researchers to assemble the synthesized fragments into BGCs, transformation into appropriate heterologous production hosts, and chemical analysis. Similarly, cultivation of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 004 bacterial strains in 3–4 media followed by chemical extraction and analysis would be an enormous undertaking. It is apparent that the prioritization of BGCs to work on is essential in order to maximize research efforts to find new chemistry and biochemistry, and to ensure the highest return on investment of research funding. To this end, multiple research groups have been engaged in orthogonal efforts to prioritize BGCs in different bacteria, which are briefly reviewed below. Prioritization of BGCs for expression is typically guided by one of the following goals:
004 bacterial strains in 3–4 media followed by chemical extraction and analysis would be an enormous undertaking. It is apparent that the prioritization of BGCs to work on is essential in order to maximize research efforts to find new chemistry and biochemistry, and to ensure the highest return on investment of research funding. To this end, multiple research groups have been engaged in orthogonal efforts to prioritize BGCs in different bacteria, which are briefly reviewed below. Prioritization of BGCs for expression is typically guided by one of the following goals:
(1) linking a compound of interest to a putative BGC, (2) exploring BGCs based on enzymatic novelty, (3) identifying specific enzymes or enzyme groups that would result in structural novelty, or (4) assessing bioactivity based on accessory genes and, (5) artificial intelligence (AI)-based approaches.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 BGCs, while MIBiG only contains 146 experimentally verified BGCs linked to cyanobacterial compounds.41 Cross-referencing of biosynthetic repositories and compound-based repositories is underway and can be linked to omics datasets deposited in established databases.42 These linked datasets can be analyzed to improve the knowledge surrounding BGC-to-structure relationships and are critical to advancing the field. Tools such as NPLinker,43 IsoAnalyst,44 NPOmix,45 and strategies such as inverse stable isotopic labeling46,47 all use muti-omics techniques developed to address this gap. As technological and computational advances are made, natural product scientists can explore the unknown biosynthetic spaces through these emerging strategies and techniques. Although none of these tools are exclusive to cyanobacterial natural product discovery, all can be applied.
000 BGCs, while MIBiG only contains 146 experimentally verified BGCs linked to cyanobacterial compounds.41 Cross-referencing of biosynthetic repositories and compound-based repositories is underway and can be linked to omics datasets deposited in established databases.42 These linked datasets can be analyzed to improve the knowledge surrounding BGC-to-structure relationships and are critical to advancing the field. Tools such as NPLinker,43 IsoAnalyst,44 NPOmix,45 and strategies such as inverse stable isotopic labeling46,47 all use muti-omics techniques developed to address this gap. As technological and computational advances are made, natural product scientists can explore the unknown biosynthetic spaces through these emerging strategies and techniques. Although none of these tools are exclusive to cyanobacterial natural product discovery, all can be applied.
One of the bottlenecks in linking BGCs to their encoded natural products is the innate need for unambiguous structure elucidation of a natural product to then allow for retrobiosynthesis and linking of a congruent BGC. NMR-based approaches such as SMART2.0,60 SMART-Miner,61 and DeepSAT62 are accelerating structure elucidation through AI recognition of HSQC spectra as interpreted in the algorithm as either multidimensional vectors or Morgan Fingerprints and then matched with structural features of known NPs. The output provides the structures of known compounds with similar structural features, providing a jumpstart to the elucidation process. Mass spectrometry-based approaches such as Spec2Vec,63 MS2DeepScore,64 and COSMIC65 that mine the metabolome enhance fragment identification and compound and analogue annotation, thus also accelerating the structure elucidation process. Other tools such as NPOMix45 utilize AI and ML to link BGCs to mass spectral data. These tools are quite dependent upon paired, high-quality training sets, which are often pulled from public repositories, though at the current time are limited in number. Repositories and analysis tools designed specifically for paired datasets such as NPLinker,43 GraphOmics,66 and Anvi'o67 exist and are expanding to meet the growing needs within the community.
Since the final goal for many NP discovery efforts is targeting human disease, predicting bioactivity from structure and/or BGCs is an appealing application of AI and an important consideration for prioritization of BGCs for heterologous expression. Building upon the ideas for targeting resistance, regulation, and key evolutionary genes, AI can be used to identify new markers of resistance, identify key genes or chemical substructures associated with bioactivity, and apply this knowledge to prioritization schemes for BGCs producing biologically active molecules. For example, chemical structures have been used to predict bioactivity in tools such as PECAN,58 while nucleotide and amino acid sequences from BGCs have also been used to predict bioactivities,57 allowing for another level of BGC prioritization for expression, testing, and potentially development of natural products. Future advancements of new AI and ML models would be beneficial to accelerate structure determination, and linking BGCs with compound production and their resulting biological properties.
2.2. General challenge #2: many natural product pathways are large and complex
Natural product BGCs within bacterial genomes range in size from 5 kbp to 200 kbp with the total number of discrete BGCs correlating with genome size.48 In addition, BGC size and the number of genes often relates directly to the structural complexity of the associated natural product. However, cloning and successful expression of large BGCs takes both financial and temporal resources.One of the issues facing synthetic biologists is the lack of large-scale studies that detail successes and failures of natural product BGCs, particularly the failures. As is typical in science, only the successes are reported in publications leaving the failures hidden in laboratory notebooks. As Samuel Smiles wrote, “We learn wisdom from failure much more than from success. We often discover what will do, by finding out what will not do; and probably he who never made a mistake never made a discovery”.68 As noted by Kadjo and Eustáquio,69 success rates for the four large scale heterologous expression studies reported range between 11–32% suggesting that optimization of hosts, development of new hosts in addition to large scale studies, potentially collaborative across laboratories, would be beneficial to the community as a whole. In some of these large-scale studies both positive and negative results are reported to provide data so that guidelines can be applied to maximize success in heterologous expression experiments. These data can also be used to populate machine learning or artificial intelligence data sets for training.
The main bottlenecks in heterologous expression include building expression vectors, cultivating genetically engineered organisms and performing chemical extractions, compound isolation, and full structure elucidation. The automation of these steps could enhance workflows and throughput. One additional hurdle is the lack of institutional knowledge and training programs for using non-model organisms. While vectors and strains are frequently shared, the tips and tricks with non-model organisms are sometimes lost when a trainee leaves a lab. Additionally, depending on project priorities, the successes and failures of industry researchers may be left unpublished.
To overcome some of these barriers, the formation of consortiums to pool resources and results can amplify efficiency and promote success in heterologous expression. Funded centers where trainees could come and learn techniques, protocols, and interact with experts will facilitate the adoption of heterologous expression in more laboratories. Initial studies with both model and non-model organisms could focus on which clusters are successfully expressed including metadata such as original organism, BGC size, plasmid assembly methods, heterologous host, among others. This could be expanded in the future to include some of the nuances important in expression, such as regulators present in the BGC and the need for precursor pathways. The formation of Biofoundries by the Department of Energy is a promising beginning in this direction.
3. Challenges faced by natural products researchers focusing on cyanobacteria
3.1. Richness and diversity of cyanobacterial BGCs: reanalysis of high-quality genomes reveals the enormous potential and opportunity cyanobacteria provide
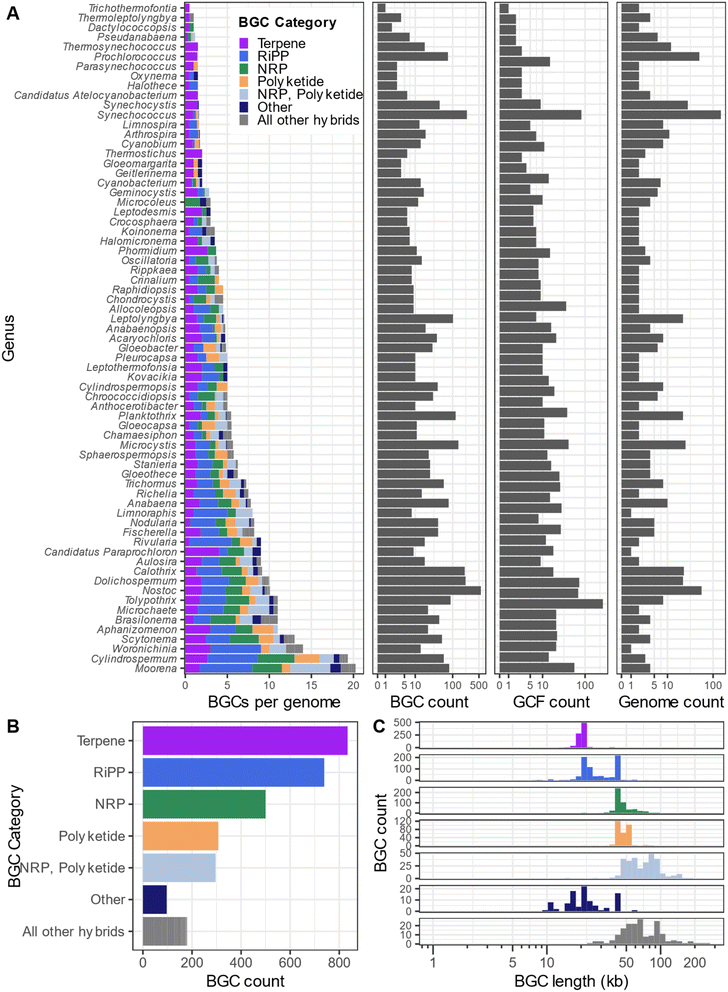
We surveyed the Secondary Metabolite Collaboratory (SMC) to provide an overview of BGCs found in cyanobacterial genomes (Fig. 2). We identified 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 112 BGCs in 4090 cyanobacterial genomes (data accessed 2/26/2025). Since fragmented and incompletely-assembled genomes often have BGCs split across contigs, we filtered our dataset to genomes of “chromosome” or “complete” assembly quality, as listed on NCBI. After filtering, our dataset consisted of 2953 BGCs in 1881 GCFs across 326 cyanobacterial genomes (302 genomes did not have a BGC detected by AntiSMASH). Of these annotated BGCs, 834 are classified as terpene, 739 are classified as ribosomally synthesized and post-translationally modified peptides (RiPPs), 500 are classified as NRPSs, 307 are classified as PKS, 296 are classified as NRPS–PKS hybrids, 97 are classified as “Other” and 180 are classified as combinations of BGC categories. In contrast, MIBiG 4.0 (ref. 41) contains 146 linked natural product/BGC pairs from cyanobacteria demonstrating the potential of new compounds and pathways that remain to be discovered from cyanobacteria.
112 BGCs in 4090 cyanobacterial genomes (data accessed 2/26/2025). Since fragmented and incompletely-assembled genomes often have BGCs split across contigs, we filtered our dataset to genomes of “chromosome” or “complete” assembly quality, as listed on NCBI. After filtering, our dataset consisted of 2953 BGCs in 1881 GCFs across 326 cyanobacterial genomes (302 genomes did not have a BGC detected by AntiSMASH). Of these annotated BGCs, 834 are classified as terpene, 739 are classified as ribosomally synthesized and post-translationally modified peptides (RiPPs), 500 are classified as NRPSs, 307 are classified as PKS, 296 are classified as NRPS–PKS hybrids, 97 are classified as “Other” and 180 are classified as combinations of BGC categories. In contrast, MIBiG 4.0 (ref. 41) contains 146 linked natural product/BGC pairs from cyanobacteria demonstrating the potential of new compounds and pathways that remain to be discovered from cyanobacteria.
We calculated the distribution and average size of cyanobacterial BGCs (Fig. 2). The majority of NRPS and PKS BGCs are within the range of 45–60 kb, while the majority of NRPS–PKS hybrids are larger (50–110 kb), terpene and RiPP BGCs are generally smaller in size (ca. 20 kb). The majority of NRPS, PKS, and NRPS–PKS hybrid BGCs should be accessible by established methodologies developed for cloning large DNA fragments.70 As previously mentioned, 2142 compounds are annotated in NPAtlas as deriving from cyanobacteria whereas 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833 BGCs (not including terpene BGCs) were identified in SMC from this phylum; therefore, a maximum of only 9.4% of the chemical diversity of cyanobacteria has been discovered to date. To our knowledge, no lanthipeptides have been isolated from cyanobacteria, except the prochlorosins, despite the presence of LanM homologs in multiple genomes, suggesting an untapped resource for the discovery of novel lanthipeptides. We note that the estimated terpene and RiPP BGC sizes of ca. 20 kb is most likely larger than the true lengths of these BGCs. These sizes are an artifact of the definition of BGC boundaries by AntiSMASH which identifies a biosynthetic core gene (e.g. terpene cyclase) and extends 10 kb in either direction and then examines the genomic DNA for additional core biosynthetic genes. This procedure most likely inflates the size of terpene and RiPP BGCs.
833 BGCs (not including terpene BGCs) were identified in SMC from this phylum; therefore, a maximum of only 9.4% of the chemical diversity of cyanobacteria has been discovered to date. To our knowledge, no lanthipeptides have been isolated from cyanobacteria, except the prochlorosins, despite the presence of LanM homologs in multiple genomes, suggesting an untapped resource for the discovery of novel lanthipeptides. We note that the estimated terpene and RiPP BGC sizes of ca. 20 kb is most likely larger than the true lengths of these BGCs. These sizes are an artifact of the definition of BGC boundaries by AntiSMASH which identifies a biosynthetic core gene (e.g. terpene cyclase) and extends 10 kb in either direction and then examines the genomic DNA for additional core biosynthetic genes. This procedure most likely inflates the size of terpene and RiPP BGCs.
3.2. Cyanobacterial specific challenge #1: accessing NP biosynthesis by native producers
Other abiotic factors may also impact the regulatory networks governing NP biosynthesis. For example, the production of scytonemin and mycosporine-like amino acids (MAAs) is stimulated by UV and other stresses.73 The production of different jamaicamide analogs occurs at different times of the day,74 suggesting an alignment with the circadian clock or an effect of light conditions. Siderophores such as cyanochelins are stimulated by iron deprivation,75 while the eagle-killing toxin aetokthonotoxin (AETX) requires the accumulation of potassium bromide from a plant partner.76
Despite the advances described above, a vast knowledge gap exists for the roles of specialized metabolomes and their diverse inducing cues in comparison to the rich diversity of known compounds and identified cyanobacterial BGCs. Systematically evaluating culture conditions in conjunction with multi-omics approaches, integrating genomics, transcriptomics, proteomics, and metabolomics data, can provide a comprehensive understanding of how these factors influence growth and BGC expression. In addition, HD cultivation approaches could be deployed at different scales for NP research. Furthermore, genetic engineering is increasingly becoming a viable solution for accessing cyanobacterial NPs from native producers, which we discuss below.
Despite the extensive defense mechanisms of cyanobacteria, innovative approaches can overcome these barriers. For example, foreign DNA can be protected by methylation in vivo or in vitro prior to its introduction into the strains of interest such as Anabaena and N. punctiforme.82 A few strains of Arthrospira platensis (Spirulina), grown industrially for its nutritional value, but refractory to genetic manipulations, gain natural competence for genetic transformation when co-cultured with heterotrophic bacteria. This discovery has enabled the genetic engineering of this organism for biotechnological applications.83 Natural competence in Synechococcus elongatus is controlled by the circadian clock and is maximal at dusk and early night in darkness.84 These findings along with progress in molecular biology and more affordable sequencing and gene synthesis, can substantially help in developing new tools and methods to facilitate genetic manipulation of cyanobacteria, particularly those with multiple predicted BGCs.
To study and engineer microbial communities under native environmental conditions, in situ manipulation approaches of complex microbial cultures are emerging.85 Microbes amenable to genetic manipulation are identified by exposing the microbial community to a randomly integrating mobile genetic element (a transposon). Then the community is sequenced to map and quantify transposon insertions. Once genetically amenable organisms are identified, genetic material can be inserted at a specific locus using approaches that leverage RNA-guided CRISPR-associated transposase systems. Such approaches can be used for the prospection of new genetically tractable strains and opens new avenues for studying cyanobacteria under conditions similar to their native environment.
3.3. Cyanobacterial specific challenge #2: improving heterologous expression of cyanobacterial NP pathways
Although there have been numerous cases of successful expression of cyanobacterial NP pathways, there are significant improvements to be made to increase the probability of successful heterologous expression, and increase NP titers. Here we describe the more traditional approaches and then we identify strategies to further enhance these markers of success (Fig. 3).Only about a dozen other strains of cyanobacteria have been reported to be genetically tractable.107 The filamentous strain Leptolyngbya BL0902 is suitable for large-scale cultivation. Anabaena ATCC 33047 shows rapid growth, nitrogen fixation, and high salt tolerance, making it a promising alternative to PCC 7120. The symbiotic strain Nostoc punctiforme PCC 73102 harbors several BGCs, which may help understand the regulation mechanisms for specialized metabolites and provide clues to improve the expression of BGCs.
Typically, genetic engineering in cyanobacteria has relied on antibiotic markers and the selection of recombinant clones under antibiotic pressure. For more advanced engineering, several approaches have been developed to construct markerless mutant strains. Recent methods rely on CRISPR/Cas systems. Cas9 and Cas12a have been successfully used in various cyanobacterial strains, with Cas12a being preferred due to its minimal impact on cell growth.108,109 Additionally, the Cas12a system allows for multiple guide RNAs to be expressed from a single array, facilitating the editing of multiple loci simultaneously. For metabolic engineering, CRISPR systems greatly simplify the construction of complex strains that would otherwise require multiple antibiotic genes and enable genetic modifications in native loci without polar effects, including small modifications. CRISPR interference (CRISPRi) technologies, using inactivated Cas nucleases, have also been developed to regulate gene expression at transcriptional and translational levels.108
Moreover, metabolic engineering for improved growth and increased compound yield can benefit from Genome-Scale Models (GSMs). GSMs can predict metabolome and flux changes to identify potential targets for higher production and have been developed for a few model strains like S. elongatus PCC 7942, UTEX 2973 and Synechocystis PCC 6803.107 Tools like OptFlux and OptForce help identify engineering targets to improve specific metabolite production, such as n-butanol in PCC 6803.110 These useful models are continuously being improved to detect key metabolic differences between strains and identify bottlenecks to improve growth and compound production.
To improve fitness and productivity, Adaptive Laboratory Evolution (ALE) experiments can be deployed. This involves selecting mutant strains that outperform the original population through repeated culture transfers under selective conditions or various stresses. ALE experiments, generally accelerated by mutagenic agents or hypermutation systems, have been used in Synechocystis PCC 6803 and S. elongatus PCC 7942 and resulted in strains with better tolerance to high light, temperature, and salt stress, as well as strains with increased production of phenylpropanoids or better ethanol tolerance.111–114
3.4. Cyanobacterial specific challenge #3: increasing our fundamental understanding of cyanobacterial physiology, biosynthesis, and regulation to help guide host development
Environmental conditions including abiotic factors, discussed above, and biotic factors can impact growth and the regulation of pathways encoding for the specialized metabolomes. Better knowledge of the specific conditions that induce expression of different NP BGCs and a deeper understanding of the regulation mechanisms that govern NP pathways can be used in the laboratory to elicit the production of NPs (Fig. 3). However, while the regulation of gene expression in cyanobacteria has been extensively studied for fundamental processes like photosynthesis and nitrogen fixation, research on the regulation of specialized metabolomes in cyanobacteria is limited and largely focused on freshwater toxin pathways.71,115 Moreover, beside physical and chemical parameters, biotic factors and the endogenous circadian clock may affect the regulation of the specialized metabolome but little is known about their effect on the production of NPs.In natural habitats, cyanobacteria live in association or compete with various microorganisms. These interactions affect the growth of cyanobacteria and likely their secondary metabolism. Several NPs have antimicrobial properties,116 while others could mediate communication between cyanobacteria and their symbiotic partners.117 However, the impact of other organisms on cyanobacterial metabolism is largely unknown. Different studies have shown varied effects on the specialized metabolome. The co-culture of heterotrophic bacteria with Microcystis led to degradation of compounds released in the media while negligible intracellular changes were found.118 In contrast, significant differences in the specialized metabolome of N. punctiforme PCC 73102 were found when the strain was grown associated with its host in comparison to free-living.81
Cyanobacteria rely heavily on light for their metabolism and have an endogenous circadian clock to predict day/night cycles and adjust expression of most genes, accordingly.119,120 The cyanobacterial clock has been extensively studied in S. elongatus PCC 7942 and functional clocks have also been described in Anabaena PCC 7120, Cyanothece ATCC 51142, and Synechocystis PCC 6803.121–123 Despite the pervasive control of gene expression by the clock in cyanobacteria, only a very few studies have investigated how day/night cycles may affect specialized metabolomes and have led to mixed results.124 Interestingly, the production of different jamaicamides in Moorena producens (formerly Lyngbya majuscula) occurs at different times of the day,24 suggesting that it could be regulated by the clock. In addition to collecting samples at different times of day or growing cultures under different light regimens, genetic approaches that manipulate the clock could be used to maximize yields.125
High-throughput methods to grow and monitor cultures under controlled conditions could help assess more comprehensively the role of abiotic and biotic factors, as well as endogenous mechanisms (such as the circadian clock) governing BGC expression.
For successful heterologous expression, the host machinery and BGC promoters must be compatible. This is achievable through pathway refactoring or promoter swapping. Several inducible and constitutive promoters from cyanobacteria and E. coli, as well as synthetic promoters126 and fully orthogonal systems, using the T7 RNA polymerase and its cognate promoter, have been characterized in several strains of cyanobacteria.98 While refactoring BGCs and exchanging promoters has been effective for NP biosynthesis in E. coli, S. elongatus and Anabaena PCC 7120, these strategies become challenging with large BGCs.89,105,127 Alternatively, transforming a BGC with its native regulatory sequences into a compatible host is feasible. Anabaena PCC 7120 appears to be a suitable host for several unmodified cyanobacterial BGCs.99,100,128,129 Additionally, several other BGC promoters from marine cyanobacteria have appeared compatible with its transcription machinery.128 An interesting approach has been to evaluate whether the 12 sigma factors of Anabaena PCC 7120 could enable the expression of cyanobacterial promoters in E. coli. Although no individual sigma factor could drive the expression of an entire BGC, several sigma factors could drive the expression of individual promoters.130
Combining genetic approaches with controlled culture methods for a better understanding of the specialized metabolome regulatory networks can help develop broadly applicable approaches for NP expression in both native and heterologous hosts.
4. Conclusions
A common theme among cyanobacterial natural products is their biological potency, target diversity and opportunity to discover new mechanisms of action. Their structures can lead to first-in-class or best-in-class drug candidates or at least serve as tool compounds to discover new cell biology. The application of synthetic biology has lagged behind in cyanobacteria compared to Actinobacteria (particularly Streptomyces) and Pseudomonadota, but with new tools such as CRISPR and a focus on a sustainable green bioeconomy, this is poised to change. Here we have outlined some challenges and some of the resources needed to push forward the synthetic biology of cyanobacteria with a special emphasis on NRPS, PKS, and NRPS–PKS derived natural products/specialized metabolites. To advance synthetic biology in cyanobacteria and thus accelerate drug discovery and green chemistry efforts, we recommend a greater investment in the study of their BGCs, investigation of more cyanobacterial strains, establishment of an open data sharing platform that includes both positive and negative results, and the creation of collaborative research teams or centers.5. Data availability
The data and analytical pipeline used in the generation of Fig. 2 is available at our GitHub repository https://github.com/dtdoering/2025-cyano_bgc_perspective.6. Conflicts of interest
There are no conflicts of interest to declare.7. Acknowledgements
The authors would like to thank the following funding sources. National Institutes of Health [R15GM117541 to B. P., 5R01GM107550 to W. H. G., R35GM128742 and RM1GM145426 to Y. D., RM1GM145426] and the Debbie and Sylvia DeSantis Chair Professorship to H. L., P01 CA125066 to J. O. S. S was supported by T32AT010131 (to Drs Mahmud and van Breemen, Oregon State University). A. T. was supported by R35GM118290 (to Susan S. Golden, UC San Diego). A portion of these data were produced by the US Department of Energy Joint Genome Institute (https://ror.org/04xm1d337; operated under Contract No. DE-AC02-05CH11231) in collaboration with the user community.8. References
- The Cyanobacteria: Molecular Biology, Genomics and Evolution, ed. A. Herrero and E. Flores, Caister Academic Press, Norfolk, 2008 Search PubMed.
- P. N. Leão, N. Engene, A. Antunes, W. H. Gerwick and V. Vasconcelos, Nat. Prod. Rep., 2013, 29, 372–391 RSC.
- S. Mazard, A. Penesyan, M. Ostrowski, I. T. Paulsen and S. Egan, Mar. Drugs, 2016, 14, 97 CrossRef PubMed.
- Ecology of Cyanobacteria II: Their Diversity in Space and Time, ed. S. R. Whitton, E. H. C. McKenzie and K. D. Hyde, Springer Science & Business Media, New York, 2012 Search PubMed.
- C. MacKintosh, K. A. Beattie, S. Klumpp, P. Cohen and G. A. Codd, FEBS Lett., 1990, 264, 187–192 CrossRef CAS.
- S. Yoshizawa, R. Matsushima, M. F. Watanabe, K. Harada, A. Ichihara, W. W. Carmichael and H. Fujiki, J. Cancer Res. Clin. Oncol., 1990, 116, 609–614 CrossRef CAS PubMed.
- R. P. Rastogi, D. Madamwar and A. Incharoensakdi, Front. Microbiol., 2015, 6, 1254 Search PubMed.
- F. Garcia-Pichel, J. Lombard, T. Soule, S. Dunaj, S. H. Wu and M. F. Wojciechowski, mBio, 2019, 10, e00561 CrossRef CAS PubMed.
- V. Geraldes and E. Pinto, Pharmaceuticals, 2021, 14, 63 CrossRef CAS.
- P. M. D'Agostino, V. S. Javalkote, R. Mazmouz, R. Pickford, P. R. Puranik and B. A. Neilan, Appl. Environ. Microbiol., 2016, 82, 5951–5959 CrossRef.
- S. Sen and N. Mallick, Algal Res., 2022, 64, 102678 CrossRef.
- C. R. Pye, M. J. Bertin, R. S. Lokey, W. H. Gerwick and R. G. Linington, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5601–5606 CrossRef CAS.
- H. Luesch, E. K. Ellis, Q. Y. Chen and R. Ratnayake, Nat. Prod. Rep., 2025, 42, 208–256 RSC.
- H. Luesch, R. E. Moore, V. J. Paul, S. L. Mooberry and T. H. Corbett, J. Nat. Prod., 2001, 64, 907–910 CrossRef CAS PubMed.
- W. H. Gerwick, P. J. Proteau, D. G. Nagle, E. Hamel, A. Blokhin and D. L. Slate, J. Org. Chem., 1994, 59, 1243–1245 CrossRef CAS.
- H. Luesch, W. Y. Yoshida, R. E. Moore, V. J. Paul and T. H. Corbett, J. Am. Chem. Soc., 2001, 123, 5418–5423 CrossRef CAS.
- Drugs R&D, 2011, 11, 85–95, https://pmc.ncbi.nlm.nih.gov/articles/PMC3585690/ Search PubMed.
- S. Matthew, Q.-Y. Chen, R. Ratnayake, C. S. Fermaintt, D. Lucena-Agell, F. Bonato, A. E. Prota, S. T. Lim, X. Wang, J. F. Díaz, A. L. Risinger, V. J. Paul, M. Á. Oliva and H. Luesch, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2021847118 CrossRef CAS PubMed.
- K. Taori, V. J. Paul and H. Luesch, J. Am. Chem. Soc., 2008, 130, 1806–1807 CrossRef CAS.
- A. R. Pereira, A. J. Kale, A. T. Fenley, T. Byrum, H. M. Debonsi, M. K. Gilson, F. A. Valeriote, B. S. Moore and W. H. Gerwick, ChemBioChem, 2012, 13, 810–817 CrossRef CAS.
- D. Tranter, A. O. Paatero, S. Kawaguchi, S. Kazemi, J. D. Serrill, J. Kellosalo, W. K. Vogel, U. Richter, D. R. Mattos, X. Wan, C. C. Thornburg, S. Oishi, K. L. McPhail, J. E. Ishmael and V. O. Paavilainen, ACS Chem. Biol., 2020, 15, 2125–2136 CrossRef CAS.
- A. O. Paatero, J. Kellosalo, B. M. Dunyak, J. Almaliti, J. E. Gestwicki, W. H. Gerwick, J. Taunton and V. O. Paavilainen, Cell Chem. Biol., 2016, 23, 561–566 CrossRef CAS PubMed.
- N. Kurisawa, A. Iwasaki, K. Teranuma, S. Dan, C. Toyoshima, M. Hashimoto and K. Suenaga, J. Am. Chem. Soc., 2022, 144, 11019–11032 CrossRef CAS.
- A. C. Jones, L. Gu, C. M. Sorrels, D. H. Sherman and W. H. Gerwick, Curr. Opin. Chem. Biol., 2009, 13, 216–223 CrossRef CAS.
- J.-C. Kehr, D. Gatte Picchi and E. Dittmann, Beilstein J. Org. Chem., 2011, 7, 1622–1635 CrossRef CAS PubMed.
- P. M. D'Agostino, Nat. Prod. Rep., 2023, 40, 1701–1717 RSC.
- D. Dhakal, M. Chen, H. Luesch and Y. Ding, J. Ind. Microbiol. Biotechnol., 2021, 48, kuab003 CrossRef CAS.
- M. Baunach, A. Guljamow, M. Miguel-Gordo and E. Dittmann, Nat. Prod. Rep., 2024, 41, 347–369 RSC.
- H. Shiraishi, Biosci., Biotechnol., Biochem., 2015, 79, 331–341 CrossRef CAS PubMed.
- R. A. Hughes, Y. Zhang, R. Zhang, P. G. Williams, J. S. Lindsey and E. S. Miller, Appl. Environ. Microbiol., 2017, 83, e01068–01017 CAS.
- A. Gavriilidou, S. A. Kautsar, N. Zaburannyi, D. Krug, R. Müller, M. H. Medema and N. Ziemert, Nat. Microbiol., 2022, 7, 726–735 CrossRef CAS PubMed.
- M. Elyashberg and D. Argyropoulos, Magn. Reson. Chem., 2021, 59, 669–690 CrossRef CAS.
- C. G. Jones, M. W. Martynowycz, J. Hattne, T. J. Fulton, B. M. Stoltz, J. A. Rodriguez, H. M. Nelson and T. Gonen, ACS Cent. Sci., 2018, 4, 1587–1592 CrossRef CAS PubMed.
- N. Zigon, V. Duplan, N. Wada and M. Fujita, Angew. Chem., Int. Ed., 2021, 60, 25204–25222 CrossRef CAS.
- J. A. van Santen, S. A. Kautsar, M. H. Medema and R. G. Linington, Nat. Prod. Rep., 2021, 38, 264–278 RSC.
- H. W. Zhang, C. Lv, L. J. Zhang, X. Guo, Y. W. Shen, D. G. Nagle, Y. D. Zhou, S. H. Liu, W. D. Zhang and X. Luan, Biomed. Pharmacother., 2021, 141, 111833 CrossRef CAS PubMed.
- J. A. van Santen, E. F. Poynton, D. Iskakova, E. McMann, T. A. Alsup, T. N. Clark, C. H. Fergusson, D. P. Fewer, A. H. Hughes, C. A. McCadden, J. Parra, S. Soldatou, J. D. Rudolf, E. M.-L. Janssen, K. R. Duncan and R. G. Linington, Nucleic Acids Res., 2021, 50, D1317–D1323 CrossRef.
- M. R. Jones, E. Pinto, M. A. Torres, F. Dörr, H. Mazur-Marzec, K. Szubert, L. Tartaglione, C. Dell'Aversano, C. O. Miles, D. G. Beach, P. McCarron, K. Sivonen, D. P. Fewer, J. Jokela and E. M.-L. Janssen, Water Res., 2020, 196, 117017 CrossRef.
- E. M.-L. Janssen, M. R. Jones, E. Pinto, F. Dörr, M. A. Torres, F. Rios Jacinavicius, H. Mazur-Marzec, K. Szubert, R. Konkel, L. Tartaglione, C. Dell'Aversano, A. Miglione, P. McCarron, D. G. Beach, C. O. Miles, D. P. Fewer, K. Sivonen, J. Jokela, M. Wahlsten, T. H. J. Niedermeyer, F. Schanbacher, P. Leão, M. Preto, P. M. D'Agostino, M. Baunach, E. Dittmann, M. Miguel-Gordo, R. Reher and S. Sieber, Zenodo, 2024 Search PubMed.
- D. W. Udwary, D. T. Doering, B. Foster, T. Smirnova, S. A. Kautsar and N. J. Mouncey, Nucleic Acids Res., 2024, gkae1060, DOI:10.1093/nar/gkae1060.
- M. M. Zdouc, K. Blin, N. L. L. Louwen, J. Navarro, C. Loureiro, C. D. Bader, C. B. Bailey, L. Barra, T. J. Booth, K. A. J. Bozhüyük, J. D. D. Cediel-Becerra, Z. Charlop-Powers, M. G. Chevrette, Y. H. Chooi, P. M. D’Agostino, T. de Rond, E. Del Pup, K. R. Duncan, W. Gu, N. Hanif, E. J. N. Helfrich, M. Jenner, Y. Katsuyama, A. Korenskaia, D. Krug, V. Libis, G. A. Lund, S. Mantri, K. D. Morgan, C. Owen, C.-S. Phan, B. Philmus, Z. L. Reitz, S. L. Robinson, K. S. Singh, R. Teufel, Y. Tong, F. Tugizimana, D. Ulanova, J. M. Winter, C. Aguilar, D. Y. Akiyama, S. A. A. Al-Salihi, M. Alanjary, F. Alberti, G. Aleti, S. A. Alharthi, M. Y. A. Rojo, A. A. Arishi, H. E. Augustijn, N. E. Avalon, J. A. Avelar-Rivas, K. K. Axt, H. B. Barbieri, J. C. J. Barbosa, L. G. Barboza Segato, S. E. Barrett, M. Baunach, C. Beemelmanns, D. Beqaj, T. Berger, J. Bernaldo-Agüero, S. M. Bettenbühl, V. A. Bielinski, F. Biermann, R. M. Borges, R. Borriss, M. Breitenbach, K. M. Bretscher, M. W. Brigham, L. Buedenbender, B. W. Bulcock, C. Cano-Prieto, J. Capela, V. J. Carrion, R. S. Carter, R. Castelo-Branco, G. Castro-Falcón, F. O. Chagas, E. Charria-Girón, A. A. Chaudhri, V. Chaudhry, H. Choi, Y. Choi, R. Choupannejad, J. Chromy, M. S. C. Donahey, J. Collemare, J. A. Connolly, K. E. Creamer, M. Crüsemann, A. A. Cruz, A. Cumsille, J.-F. Dallery, L. C. Damas-Ramos, T. Damiani, M. de Kruijff, B. D. Martín, G. D. Sala, J. Dillen, D. T. Doering, S. R. Dommaraju, S. Durusu, S. Egbert, M. Ellerhorst, B. Faussurier, A. Fetter, M. Feuermann, D. P. Fewer, J. Foldi, A. Frediansyah, E. A. Garza, A. Gavriilidou, A. Gentile, J. Gerke, H. Gerstmans, J. P. Gomez-Escribano, L. A. González-Salazar, N. E. Grayson, C. Greco, J. E. G. Gomez, S. Guerra, S. G. Flores, A. Gurevich, K. Gutiérrez-García, L. Hart, K. Haslinger, B. He, T. Hebra, J. L. Hemmann, H. Hindra, L. Höing, D. C. Holland, J. E. Holme, T. Horch, P. Hrab, J. Hu, T.-H. Huynh, J.-Y. Hwang, R. Iacovelli, D. Iftime, M. Iorio, S. Jayachandran, E. Jeong, J. Jing, J. J. Jung, Y. Kakumu, E. Kalkreuter, K. B. Kang, S. Kang, W. Kim, G. J. Kim, H. Kim, H. U. Kim, M. Klapper, R. A. Koetsier, C. Kollten, Á. T. Kovács, Y. Kriukova, N. Kubach, A. M. Kunjapur, A. K. Kushnareva, A. Kust, J. Lamber, M. Larralde, N. J. Larsen, A. P. Launay, N.-T.-H. Le, S. Lebeer, B. T. Lee, K. Lee, K. L. Lev, S.-M. Li, Y.-X. Li, C. Licona-Cassani, A. Lien, J. Liu, J. A. V. Lopez, N. V. Machushynets, M. I. Macias, T. Mahmud, M. Maleckis, A. M. Martinez-Martinez, Y. Mast, M. F. Maximo, C. M. McBride, R. M. McLellan, K. M. Bhatt, C. Melkonian, A. Merrild, M. Metsä-Ketelä, D. A. Mitchell, A. V. Müller, G.-S. Nguyen, H. T. Nguyen, T. H. J. Niedermeyer, J. H. O’Hare, A. Ossowicki, B. O. Ostash, H. Otani, L. Padva, S. Paliyal, X. Pan, M. Panghal, D. S. Parade, J. Park, J. Parra, M. P. Rubio, H. T. Pham, S. J. Pidot, J. Piel, B. Pourmohsenin, M. Rakhmanov, S. Ramesh, M. H. Rasmussen, A. Rego, R. Reher, A. J. Rice, A. Rigolet, A. Romero-Otero, L. R. Rosas-Becerra, P. Y. Rosiles, A. Rutz, B. Ryu, L.-A. Sahadeo, M. Saldanha, L. Salvi, E. Sánchez-Carvajal, C. Santos-Medellin, N. Sbaraini, S. M. Schoellhorn, C. Schumm, L. Sehnal, N. Selem, A. D. Shah, T. K. Shishido, S. Sieber, V. Silviani, G. Singh, H. Singh, N. Sokolova, E. C. Sonnenschein, M. Sosio, S. T. Sowa, K. Steffen, E. Stegmann, A. B. Streiff, A. Strüder, F. Surup, T. Svenningsen, D. Sweeney, J. Szenei, A. Tagirdzhanov, B. Tan, M. J. Tarnowski, B. R. Terlouw, T. Rey, N. U. Thome, L. R. Torres Ortega, T. Tørring, M. Trindade, A. W. Truman, M. Tvilum, D. W. Udwary, C. Ulbricht, L. Vader, G. P. van Wezel, M. Walmsley, R. Warnasinghe, H. G. Weddeling, A. N. M. Weir, K. Williams, S. E. Williams, T. E. Witte, S. M. W. Rocca, K. Yamada, D. Yang, D. Yang, J. Yu, Z. Zhou, N. Ziemert, L. Zimmer, A. Zimmermann, C. Zimmermann, J. J. J. van der Hooft, R. G. Linington, T. Weber and M. H. Medema, Nucleic Acids Res., 2024, 53, D678–D690 CrossRef.
- N. B. Cech, M. H. Medema and J. Clardy, Nat. Prod. Rep., 2021, 38, 1947–1953 RSC.
- G. Hjorleifsson Eldjarn, A. Ramsey, J. J. J. van der Hooft, K. R. Duncan, S. Soldatou, J. Rousu, R. Daly, J. Wandy and S. Rogers, PLoS Comput. Biol., 2021, 17, e1008920 CrossRef CAS PubMed.
- C. S. McCaughey, J. A. van Santen, J. J. J. van der Hooft, M. H. Medema and R. G. Linington, Nat. Chem. Biol., 2021, 18, 295–304 CrossRef.
- T. F. Leão, M. Wang, R. da Silva, A. Gurevich, A. Bauermeister, P. W. P. Gomes, A. Brejnrod, E. Glukhov, A. T. Aron, J. J. R. Louwen, H. W. Kim, R. Reher, M. F. Fiore, J. J. J. van der Hooft, L. Gerwick, W. H. Gerwick, N. Bandeira and P. C. Dorrestein, PNAS Nexus, 2022, 1, pgac257 CrossRef PubMed.
- T. C. E. Liebergesell, E. G. Murdock and A. W. Puri, Anal. Chem., 2024, 96, 16330–16337 CrossRef CAS.
- D. S. May, C. M. Crnkovic, A. Krunic, T. A. Wilson, J. R. Fuchs and J. E. Orjala, ACS Chem. Biol., 2020, 1015, 758–765 CrossRef.
- P. Cimermancic, M. H. Medema, J. Claesen, K. Kurita, L. C. Wieland Brown, K. Mavrommatis, A. Pati, P. A. Godfrey, M. Koehrsen, J. Clardy, B. W. Birren, E. Takano, A. Sali, R. G. Linington and M. A. Fischbach, Cell, 2014, 158, 412–421 CrossRef CAS PubMed.
- E. Kalkreuter, G. Pan, A. J. Cepeda and B. Shen, Trends Pharmacol. Sci., 2020, 41, 13–26 CrossRef CAS PubMed.
- T. F. Smith and M. S. Waterman, J. Mol. Biol., 1981, 147, 195–197 CrossRef CAS.
- Hindra, T. Huang, D. Yang, J. D. Rudolf, P. Xie, G. Xie, Q. Teng, J. R. Lohman, X. Zhu, Y. Huang, L.-X. Zhao, Y. Jiang, Y. Duan and B. Shen, J. Nat. Prod., 2014, 77, 2296–2303 CrossRef CAS.
- H. Gross, V. O. Stockwell, M. D. Henkels, B. Nowak-Thompson, J. E. Loper and W. H. Gerwick, Chem. Biol., 2007, 14, 53–63 CrossRef CAS.
- R. Xin, Y. Zhang, K. Zhang, Y. Yang, Y. Ma and Z. Niu, Sci. Total Environ., 2024, 909, 168516 CrossRef CAS PubMed.
- M. D. Mungan, M. Alanjary, K. Blin, T. Weber, M. H. Medema and N. Ziemert, Nucleic Acids Res., 2020, 48, W546–W552 CrossRef CAS.
- M. L. Beck, S. Song, I. E. Shuster, A. Miharia and A. S. Walker, J. Ind. Microbiol. Biotechnol., 2023, 50, kuad024 CrossRef CAS PubMed.
- O. Riedling, A. S. Walker and A. Rokas, Microbiol. Spectrum, 2024, 12, e0340023 CrossRef.
- A. S. Walker and J. Clardy, J. Chem. Inf. Model., 2021, 61, 2560–2571 CrossRef CAS PubMed.
- M. Gahl, H. W. Kim, E. Glukhov, W. H. Gerwick and G. W. Cottrell, J. Nat. Prod., 2024, 87, 567–575 CrossRef CAS.
- D. Meijer, M. A. Beniddir, C. W. Coley, Y. M. Mejri, M. Öztürk, J. J. J. van der Hooft, M. H. Medema and A. Skiredj, Nat. Prod. Rep., 2024, 42, 654–662 RSC.
- R. Reher, H. W. Kim, C. Zhang, H. H. Mao, M. Wang, L. F. Nothias, A. M. Caraballo-Rodriguez, E. Glukhov, B. Teke, T. Leao, K. L. Alexander, B. M. Duggan, E. L. Van Everbroeck, P. C. Dorrestein, G. W. Cottrell and W. H. Gerwick, J. Am. Chem. Soc., 2020, 142, 4114–4120 CrossRef CAS PubMed.
- H. W. Kim, C. Zhang, G. W. Cottrell and W. H. Gerwick, Magn. Reson. Chem., 2022, 60, 1070–1075 CrossRef CAS PubMed.
- H. W. Kim, C. Zhang, R. Reher, M. Wang, K. L. Alexander, L. F. Nothias, Y. K. Han, H. Shin, K. Y. Lee, K. H. Lee, M. J. Kim, P. C. Dorrestein, W. H. Gerwick and G. W. Cottrell, J. Cheminf., 2023, 15, 71 Search PubMed.
- F. Huber, L. Ridder, S. Verhoeven, J. H. Spaaks, F. Diblen, S. Rogers and J. J. J. van der Hooft, PLoS Comput. Biol., 2021, 17, e1008724 CrossRef CAS PubMed.
- F. Huber, S. van der Burg, J. J. J. van der Hooft and L. Ridder, J. Cheminf., 2021, 13, 84 Search PubMed.
- M. A. Hoffmann, L. F. Nothias, M. Ludwig, M. Fleischauer, E. C. Gentry, M. Witting, P. C. Dorrestein, K. Dührkop and S. Böcker, Nat. Biotechnol., 2022, 40, 411–421 CrossRef CAS.
- J. Wandy and R. Daly, BMC Bioinf., 2021, 22, 603 CrossRef.
- A. M. Eren, E. Kiefl, A. Shaiber, I. Veseli, S. E. Miller, M. S. Schechter, I. Fink, J. N. Pan, M. Yousef, E. C. Fogarty, F. Trigodet, A. R. Watson, C. Esen Ö, R. M. Moore, Q. Clayssen, M. D. Lee, V. Kivenson, E. D. Graham, B. D. Merrill, A. Karkman, D. Blankenberg, J. M. Eppley, A. Sjödin, J. J. Scott, X. Vázquez-Campos, L. J. McKay, E. A. McDaniel, S. L. R. Stevens, R. E. Anderson, J. Fuessel, A. Fernandez-Guerra, L. Maignien, T. O. Delmont and A. D. Willis, Nat. Microbiol., 2021, 6, 3–6 CrossRef CAS.
- S. Smiles, Self-help: With illustrations of character and conduct, John Murray, Albemarle Street, London, 1859 Search PubMed.
- A. E. Kadjo and A. S. Eustáquio, J. Ind. Microbiol. Biotechnol., 2023, 50, kuad044 CrossRef CAS.
- J. Wan, N. Ma and H. Yuan, Eng. Microbiol., 2023, 3, 100085 CrossRef CAS PubMed.
- B. A. Neilan, L. A. Pearson, J. Muenchhoff, M. C. Moffitt and E. Dittmann, Environ. Microbiol., 2013, 15, 1239–1253 CrossRef CAS.
- A. Guljamow, M. Kreische, K. Ishida, A. Liaimer, B. Altermark, L. Bähr, C. Hertweck, R. Ehwald and E. Dittmann, Appl. Environ. Microbiol., 2017, 83, e01510–e01517 CrossRef.
- Q. Gao and F. Garcia-Pichel, Nat. Rev. Microbiol., 2011, 9, 791–802 CrossRef CAS.
- E. Esquenazi, A. C. Jones, T. Byrum, P. C. Dorrestein and W. H. Gerwick, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5226–5231 CrossRef CAS.
- T. Galica, N. Borbone, J. Mareš, A. Kust, A. Caso, G. Esposito, K. Saurav, J. Hájek, K. Řeháková, P. Urajová, V. Costantino and P. Hrouzek, Appl. Environ. Microbiol., 2021, 87, e0312820 CrossRef.
- S. Breinlinger, T. J. Phillips, B. N. Haram, J. Mareš, J. A. Martínez Yerena, P. Hrouzek, R. Sobotka, W. M. Henderson, P. Schmieder, S. M. Williams, J. D. Lauderdale, H. D. Wilde, W. Gerrin, A. Kust, J. W. Washington, C. Wagner, B. Geier, M. Liebeke, H. Enke, T. H. J. Niedermeyer and S. B. Wilde, Science, 2021, 371, eaax9050 CrossRef CAS PubMed.
- J. C. Meeks and J. Elhai, Microbiol. Mol. Biol. Rev., 2002, 66, 94–121 CrossRef CAS PubMed.
- D. Dehm, J. Krumbholz, M. Baunach, V. Wiebach, K. Hinrichs, A. Guljamow, T. Tabuchi, H. Jenke-Kodama, R. D. Süssmuth and E. Dittmann, ACS Chem. Biol., 2019, 14, 1271–1279 CrossRef CAS PubMed.
- J. Krumbholz, K. Ishida, M. Baunach, J. E. Teikari, M. M. Rose, S. Sasso, C. Hertweck and E. Dittmann, Angew. Chem., Int. Ed., 2022, 61, e202204545 CrossRef CAS PubMed.
- T. Soule, V. Stout, W. D. Swingley, J. C. Meeks and F. Garcia-Pichel, J. Bacteriol., 2007, 189, 4465–4472 CrossRef CAS.
- A. Liaimer, E. J. N. Helfrich, K. Hinrichs, A. Guljamow, K. Ishida, C. Hertweck and E. Dittmann, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 1862–1867 CrossRef CAS.
- J. Elhai, A. Vepritskiy, A. M. Muro-Pastor, E. Flores and C. P. Wolk, J. Bacteriol., 1997, 179, 1998–2005 CrossRef CAS.
- B. W. Jester, H. Zhao, M. Gewe, T. Adame, L. Perruzza, D. T. Bolick, J. Agosti, N. Khuong, R. Kuestner, C. Gamble, K. Cruickshank, J. Ferrara, R. Lim, T. Paddock, C. Brady, S. Ertel, M. Zhang, A. Pollock, J. Lee, J. Xiong, M. Tasch, T. Saveria, D. Doughty, J. Marshall, D. Carrieri, L. Goetsch, J. Dang, N. Sanjaya, D. Fletcher, A. Martinez, B. Kadis, K. Sigmar, E. Afreen, T. Nguyen, A. Randolph, A. Taber, A. Krzeszowski, B. Robinett, D. B. Volkin, F. Grassi, R. Guerrant, R. Takeuchi, B. Finrow, C. Behnke and J. Roberts, Nat. Biotechnol., 2022, 40, 956–964 CrossRef CAS.
- A. Taton, C. Erikson, Y. Yang, B. E. Rubin, S. A. Rifkin, J. W. Golden and S. S. Golden, Nat. Commun., 2020, 11, 1688 CrossRef CAS.
- B. E. Rubin, S. Diamond, B. F. Cress, A. Crits-Christoph, Y. C. Lou, A. L. Borges, H. Shivram, C. He, M. Xu, Z. Zhou, S. J. Smith, R. Rovinsky, D. C. J. Smock, K. Tang, T. K. Owens, N. Krishnappa, R. Sachdeva, R. Barrangou, A. M. Deutschbauer, J. F. Banfield and J. A. Doudna, Nat. Microbiol., 2022, 7, 34–47 CrossRef CAS PubMed.
- P. D'Agostino, C. J. Seel, T. Gulder and T. Gulder, Nat. Chem. Biol., 2022, 18, 652–658 CrossRef.
- P. M. D'Agostino, R. R. de Castro, V. Uka, K. Saurav, T. M. Milzarek, A. Sester, M. P. Schneider and T. A. M. Gulder, bioRxiv, 2025, preprint, 2025.2002.2026.640319, DOI:10.1101/2025.02.26.640319.
- P. M. D'Agostino and T. A. M. Gulder, ACS Synth. Biol., 2018, 7, 1702–1708 CrossRef.
- C. Greunke, E. R. Duell, P. M. D'Agostino, A. Glöckle, K. Lamm and T. A. M. Gulder, Metab. Eng., 2018, 47, 334–345 CrossRef CAS.
- S. E. Ongley, X. Bian, Y. Zhang, R. Chau, W. H. Gerwick, R. Müller and B. A. Neilan, ACS Chem. Biol., 2013, 8, 1888–1893 CrossRef CAS.
- M. Katoch, R. Mazmouz, R. Chau, L. A. Pearson, R. Pickford and B. A. Neilan, Appl. Environ. Microbiol., 2016, 82, 6167–6173 CrossRef CAS.
- X. Ouyang, P. M. D'Agostino, M. Wahlsten, E. Delbaje, J. Jokela, P. Permi, G. Gaiani, A. Poso, P. Bartos, T. A. M. Gulder, H. Koistinen and D. P. Fewer, Org. Biomol. Chem., 2023, 21, 4893–4908 RSC.
- T. Liu, R. Mazmouz, S. E. Ongley, R. Chau, R. Pickford, J. N. Woodhouse and B. A. Neilan, ACS Chem. Biol., 2017, 12, 2021–2029 CrossRef CAS PubMed.
- A. C. Jones, S. Ottilie, A. S. Eustáquio, D. J. Edwards, L. Gerwick, B. S. Moore and W. H. Gerwick, FEBS J., 2012, 279, 1243–1251 CrossRef CAS.
- S.-H. Park, K. Lee, J. W. Jang and J.-S. Hahn, ACS Synth. Biol., 2019, 8, 346–357 CrossRef CAS PubMed.
- T. T. Selão, J. Exp. Bot., 2022, 73, 3057–3071 CrossRef.
- X. Liu, K. Tang and J. Hu, Microorganisms, 2024, 12, 1375 CrossRef CAS.
- J. Roulet, A. Taton, J. W. Golden, A. Arabolaza, M. D. Burkart and H. Gramajo, Metab. Eng., 2018, 49, 94–104 CrossRef CAS PubMed.
- A. Taton, A. Ecker, B. Diaz, N. A. Moss, B. Anderson, R. Reher, T. F. Leão, R. Simkovsky, P. C. Dorrestein, L. Gerwick, W. H. Gerwick and J. W. Golden, ACS Synth. Biol., 2020, 9, 3364–3376 CrossRef CAS.
- A. Taton, S. Rohrer, B. Diaz, R. Reher, A. M. Caraballo Rodriguez, M. L. Pierce, P. C. Dorrestein, L. Gerwick, W. H. Gerwick and J. W. Golden, ACS Chem. Biol., 2022, 17, 1910–1923 CrossRef CAS PubMed.
- G. Yang, M. A. Cozad, D. A. Holland, Y. Zhang, H. Luesch and Y. Ding, ACS Synth. Biol., 2018, 7, 664–671 CrossRef CAS PubMed.
- J. Yu, M. Liberton, P. F. Cliften, R. D. Head, J. M. Jacobs, R. D. Smith, D. W. Koppenaal, J. J. Brand and H. B. Pakrasi, Sci. Rep., 2015, 5, 8132 CrossRef CAS.
- D. Jaiswal, A. Sengupta, S. Sohoni, S. Sengupta, A. G. Phadnavis, H. B. Pakrasi and P. P. Wangikar, Sci. Rep., 2018, 8, 16632 CrossRef PubMed.
- A. Włodarczyk, T. T. Selão, B. Norling and P. J. Nixon, Commun. Biol., 2020, 3, 215 CrossRef.
- C. J. Knoot, Y. Khatri, R. M. Hohlman, D. H. Sherman and H. B. Pakrasi, ACS Synth. Biol., 2019, 8, 1941–1951 CrossRef CAS.
- A. J. Victoria, T. T. Selão, J. Moreno-Cabezuelo, L. A. Mills, G. A. R. Gale, D. J. Lea-Smith and A. J. McCormick, Plant Physiol., 2024, 196, 1674–1690 CrossRef CAS.
- J. A. van Santen, G. Jacob, A. L. Singh, V. Aniebok, M. J. Balunas, D. Bunsko, F. C. Neto, L. Castaño-Espriu, C. Chang, T. N. Clark, J. L. Cleary Little, D. A. Delgadillo, P. C. Dorrestein, K. R. Duncan, J. M. Egan, M. M. Galey, F. P. J. Haeckl, A. Hua, A. H. Hughes, D. Iskakova, A. Khadilkar, J.-H. Lee, S. Lee, N. LeGrow, D. Y. Liu, J. M. Macho, C. S. McCaughey, M. H. Medema, R. P. Neupane, T. J. O’Donnell, J. S. Paula, L. M. Sanchez, A. F. Shaikh, S. Soldatou, B. R. Terlouw, T. A. Tran, M. Valentine, J. J. J. van der Hooft, D. A. Vo, M. Wang, D. Wilson, K. E. Zink and R. G. Linington, ACS Cent. Sci., 2019, 5, 1824–1833 CrossRef CAS.
- A. Sengupta, D. Liu and H. B. Pakrasi, Methods Enzymol., 2022, 676, 403–432 CAS.
- J. Ungerer and H. B. Pakrasi, Sci. Rep., 2016, 6, 39681–39689 CrossRef CAS PubMed.
- K. Shabestary, J. Anfelt, E. Ljungqvist, M. Jahn, L. Yao and E. P. Hudson, ACS Synth. Biol., 2018, 7, 1669–1675 CrossRef CAS PubMed.
- M. Dann, E. M. Ortiz, M. Thomas, A. Guljamow, M. Lehmann, H. Schaefer and D. Leister, Nat. Plants, 2021, 7, 681–695 CrossRef CAS PubMed.
- H. Sun, G. Luan, Y. Ma, W. Lou, R. Chen, D. Feng, S. Zhang, J. Sun and X. Lu, Nat. Commun., 2023, 14, 1238 CrossRef CAS.
- K. Kukil, E. Englund, N. Crang, E. P. Hudson and P. Lindberg, Metab. Eng., 2023, 79, 27–37 CrossRef CAS.
- S. Arai, K. Hayashihara, Y. Kanamoto, K. Shimizu, Y. Hirokawa, T. Hanai, A. Murakami and H. Honda, Biotechnol. Bioeng., 2017, 114, 1771–1778 CrossRef CAS PubMed.
- P. M. D'Agostino, B. Al-Sinawi, R. Mazmouz, J. Muenchhoff, B. A. Neilan and M. C. Moffitt, BMC Microbiol., 2020, 20, 35 CrossRef.
- S. C. do Amaral, L. P. Xavier, V. Vasconcelos and A. V. Santos, Mar. Drugs, 2023, 21, 359 CrossRef CAS.
- M. Mutalipassi, G. Riccio, V. Mazzella, C. Galasso, E. Somma, A. Chiarore, D. de Pascale and V. Zupo, Mar. Drugs, 2021, 19, 227 CrossRef CAS.
- E. Briand, M. Bormans, M. Gugger, P. C. Dorrestein and W. H. Gerwick, Environ. Microbiol., 2016, 18, 384–400 CrossRef CAS.
- S. Diamond, D. Jun, B. E. Rubin and S. S. Golden, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E1916–E1925 CrossRef CAS PubMed.
- J. S. Markson, J. R. Piechura, A. M. Puszynska and E. K. O'Shea, Cell, 2013, 155, 1396–1408 CrossRef CAS PubMed.
- R. Arbel-Goren, V. Buonfiglio, F. Di Patti, S. Camargo, A. Zhitnitsky, A. Valladares, E. Flores, A. Herrero, D. Fanelli and J. Stavans, eLife, 2021, 10, e64348 CrossRef CAS.
- A. Bandyopadhyay, A. Sengupta, T. Elvitigala and H. B. Pakrasi, Nat. Commun., 2024, 15, 3712 CrossRef CAS PubMed.
- C. Köbler, N. M. Schmelling, A. Wiegard, A. Pawlowski, G. K. Pattanayak, P. Spät, N. M. Scheurer, K. N. Sebastian, F. P. Stirba, L. C. Berwanger, P. Kolkhof, B. Maček, M. J. Rust, I. M. Axmann and A. Wilde, Nat. Commun., 2024, 15, 7674 CrossRef.
- E. J. Davenport, M. J. Neudeck, P. G. Matson, G. S. Bullerjahn, T. W. Davis, S. W. Wilhelm, M. K. Denney, L. E. Krausfeldt, J. M. A. Stough, K. A. Meyer, G. J. Dick, T. H. Johengen, E. Lindquist, S. G. Tringe and R. M. L. McKay, Front. Microbiol., 2019, 10, 2081 CrossRef PubMed.
- Y. Xu, M. L. Jabbur, T. Mori, J. D. Young and C. H. Johnson, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2318690121 CrossRef CAS.
- B. Wang, C. Eckert, P. C. Maness and J. Yu, ACS Synth. Biol., 2018, 7, 276–286 CrossRef CAS PubMed.
- A. Cullen, L. A. Pearson, R. Mazmouz, T. Liu, A. H. Soeriyadi, S. E. Ongley and B. A. Neilan, Nat. Prod. Rep., 2019, 36, 1117–1136 RSC.
- P. Videau, K. N. Wells, A. J. Singh, W. H. Gerwick and B. Philmus, ACS Synth. Biol., 2016, 5, 978–988 CrossRef CAS PubMed.
- P. Videau, K. N. Wells, A. J. Singh, J. Eiting, P. J. Proteau and B. Philmus, ACS Synth. Biol., 2020, 9, 63–75 CrossRef CAS PubMed.
- K. N. Wells, P. Videau, D. Nelson, J. E. Eiting and B. Philmus, FEMS Microbiol. Lett., 2018, 365, fny164 CrossRef CAS PubMed.
- M. A. Skiba, M. M. Bivins, J. R. Schultz, S. M. Bernard, W. D. Fiers, Q. Dan, S. Kulkarni, P. Wipf, W. H. Gerwick, D. H. Sherman, C. C. Aldrich and J. L. Smith, ACS Chem. Biol., 2018, 13, 3221–3228 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |