Open Access Article
Open Access ArticleHuman milk as a complex natural product
Julie A.
Talbert
and
Steven D.
Townsend
 *
*
Department of Chemistry, Vanderbilt University, Nashville, Tennessee 37240, USA. E-mail: steven.d.townsend@vanderbilt.edu
First published on 20th January 2025
Abstract
Covering: up to the end of 2024
Breastfeeding is one of the most effective ways to promote child health. However, characterizing the chemistry that fortifies the benefits of breastfeeding remains a grand challenge. Current efforts in the community are focused on characterizing the roles of the different carbohydrates, proteins, and fats in milk. The goal of this review is to highlight and describe current knowledge about the major classes of macromolecules in human milk and their potential role in infant health and wellness.
1 Introduction
Traditionally, natural products have played a central role in the intellectual and experimental growth of organic chemistry. Undeniably, a disproportionate amount of clinical therapeutics are natural products or are natural product derived. On occasion, a natural product can possess the requisite pharmacological characteristics to render it a clinically beneficial medication. More often, however, are examples where a natural product serves as “the lead”, providing a structural platform, which the organic chemist can build upon to derive new chemical matter and a valuable therapeutic.Historically, the most important sources for natural products have been plants, aquatic invertebrates, and soil-bound microorganisms. While these studies have produced a bounty, it is likely that many valuable natural compounds have escaped discovery. Moreover, we have limited what we define as a natural product. It is in this vein that we draw the reader's attention to human milk. At the molecular level, human milk is a complex mixture where the composition reflects the secretory activity of the mother's mammary gland. Blood group also plays a major role in milk composition, particularly in the context of oligosaccharide profile, vida infra. Previously, human milk has been considered as a source of new therapeutics. In this writing, we make the case for human milk, in its homogeneous form, as a natural product.
During early life, milk fulfills all nutritional requirements for the developing infant. Given its dynamic nature and ability to meet the needs of the child in real time, the World Health Organization and United Nations Children's Fund recommend exclusive breastfeeding for at least 6 months after birth and to continue for up to 2 years of age or longer.1 Recently, infant food products have been developed that share greater homology to human milk, with many being supplemented with small amounts of naturally occurring compounds such as human milk oligosaccharides (HMOs).2 While these products feature molecules native to human milk, a homologous, synthetic alternative to human milk does not exist. Consequently, it is imperative to critically evaluate the natural products that make human milk the gold standard for infant nutrition.
2 The origin of human milk
In 1758, Linnaeus named animals with the ability to produce milk Mammalia. The evolution of the mammary gland, the glandular organ that generates milk, is difficult to establish due to a lack of evidence supporting the origin of soft tissue organs from fossils. However, it is well established that lactation appears to be a reproductive feature that predates the evolution of mammals. A persuasive theory for the evolution of the mammary gland and lactation was elegantly curated by Oftedal.3 The molecular components of human milk arise from a variety of sources – some are produced and secreted by the epithelium of the mammary gland. Others are produced by cells found in the milk. Lastly, select molecules are taken from maternal blood and transported across the mammary epithelium into the milk.3 Stages of lactation
Colostrum is the initial type of milk produced by a mother. While this “first milk” can vary in color, it is often deep yellow or orange due to its high concentration of β-carotene. Colostrum, produced in low volume during the immediate postnatal period, is rich in immune-enhancing and developmental components such as immunoglobulins (e.g. secretory IgA), glycoproteins (e.g. lactoferrin), leukocytes, and epidermal growth factor.4–6 Interestingly, colostrum has a lower concentration of fat and carbohydrate, compared to mature milk, which hints to its primary role as being protective and developmental rather than nutritional. As tight junction closure commences in the epithelium of the mammary gland, the concentration of lactose increases (triggered by a decline in the sodium to potassium ratio) and the production of transitional milk begins. The timing of this secretory activation process varies among women, but typically occurs over the first week postpartum. While intermediate milk has molecular homology with colostrum, it also represents a period of increased milk production to support the nutritional and developmental needs of the infant. After 14 days postpartum, milk is considered mature. Contrasting to the dynamic nature of milk observed early in life, the molecular components remain relatively stable once milk reaches maturity.4 Human milk composition
The composition of human breast milk is well-established (Table 1). Milk is ca. 87% water. The major macromolecules can be organized into three categories: carbohydrates (7%), fat (4%), and protein (1%). Counter to infant formula, the nutritional and protective molecules in breast milk are dynamic and vary depending on maternal diet and health, mammary gland physiology, and the needs of the child.| Component | Concentration (g L−1) | % (w/v) of breast milk |
|---|---|---|
| Water | — | 87% |
| Carbohydrates | 70 | 7% |
| Fat | 35–40 | 4% |
| Protein | 11 | 1% |
| Growth factors, hormones, cytokines, etc. | 0.9 | <1% |
4.1 Carbohydrates
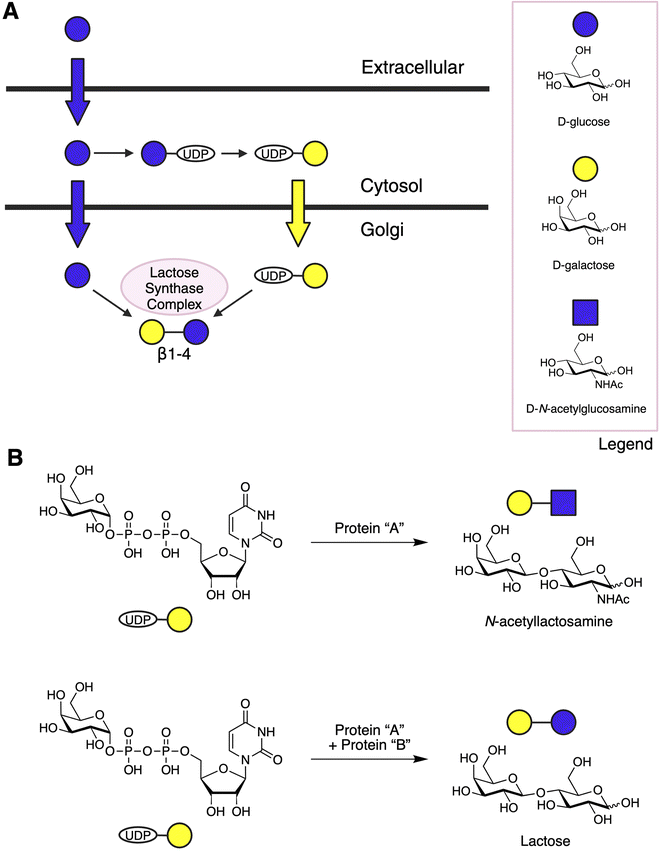
The lactose synthase complex consists of two components: the “A” protein and the “B” protein. The “A” protein is a constitutively expressed β1-4 galactosyltransferase (β4GalT1) that typically transfers UDP-galactose to terminal N-acetylglucosamine (GlcNAc) during glycoconjugate biosynthesis. However, in the presence of ⍺-lactalbumin (described in detail below), also known as protein “B”, which is expressed in response to lactation hormones, β4GalT1 shifts its acceptor specificity from GlcNAc to glucose, yielding lactose (Fig. 1B).8
For infants to benefit from lactose ingestion, the disaccharide must be hydrolyzed by lactase into its monosaccharide components, D-glucose and D-galactose. Lactase is produced in sufficient quantities to digest about 60–70 g of lactose daily in the small intestine. However, not all lactose from breast milk is absorbed by infants, a process known as physiological lactose malabsorption. This malabsorbed lactose is fermented in the colon to produce short chain fatty acids (SCFAs), hydrogen, carbon dioxide, methane, and lactic acid.9,10 Commensal bacteria in the infant gut, particularly Bifidobacteria and Lactobacilli, preferentially salvage this leftover lactose, promoting the healthy development of the infant's microbiota.11
Lactose intolerance (LI) is a common gastrointestinal condition where the body does not have enough lactase to break down all consumed lactose. Roughly 70% of the world's population suffers from LI due to lactase non-persistence (LNP), a gradual decline in lactase expression after weaning.9 Individuals who tolerate lactose beyond early childhood were most likely selected after the introduction of dairy farming and the growing consumption of cow's milk over 5000 years ago.12 Fortunately, lactose intolerance is rare for children under five years of age; therefore, LI is an uncommon concern for breastfeeding mothers.
By the late 19th century, it was established that breastfed infants exhibited significantly greater resistance to disease compared to those fed bovine milk.16 As the common structural feature of all HMOs is their lactose core, human mammary glands evolved mechanisms to diversify lactose biosynthetically via addition of lacto-N-biose (Galβ1-3GlcNAc-) or N-acetyllactosamine (Galβ1-4GlcNAc-) (Fig. 2B). Elongation via lacto-N-biose appears to terminate further chain growth, while elongation via N-acetyllactosamine permits continued chain extension. Lactose or the elongated oligosaccharides can be further diversified by addition of an L-fucose or N-acetylneuraminic acid residue (Figs. 2C–E). However, fucosylation is limited to ⍺1-2, ⍺1-3, or ⍺1-4 linkages, while sialylation occurs in ⍺2-3 or ⍺2-6 linkages. Although the biosynthesis of lactose in mammary glands is known, the specificity of how lactose is extended to form HMOs remains poorly understood.
Across 89 publications, it was found that lacto-N-tetraose (LNT) and lacto-N-neotetraose (LNnT) are the most abundant neutral core HMOs (Fig. 2C), with percent abundances of 6.61% and 3.35%, respectively.17 2′-FL and lacto-N-fucopentaose I (LNFPI) are the most abundant fucosylated HMOs (Fig. 2D) at 20.50% and 7.50% abundance, respectively. Finally, 6′-sialyllactose (6′-SL) and 3′-sialyllactose (3′-SL) are the most abundant sialylated HMOs with abundances of 3.63% and 1.71%, respectively. Putative compositions listed of the HMO fraction were based on g L−1.
The oligosaccharide composition of breast milk is influenced by the mother's secretor status and blood group characteristics. Mothers with an active Se locus, which encodes for the ⍺1-2-fucosyltransferase FUT2, are referred to as secretors. In contrast, nonsecretors lack FUT2, resulting in milk with limited levels of ⍺1-2-fucosylated HMOs. However, Se-mothers can produce low amounts of ⍺1-2-fucosylated HMOs, likely due to the activity of other FUT genes.18 Individuals with an active Le locus, which encodes for the ⍺1-4-fucosyltransferase FUT3, are classified as Le positive. These two loci create four distinct breast milk groups: Se+Le+, Se+Le−, Se−Le+, and Se−Le−, with typical frequency in the global population being 70%, 9%, 20%, and 1%, respectively.17
HMOs modulate the infant gut in various ways. First, commensal bacteria harbor the enzymes necessary to break down HMOs into their monosaccharide components to use as energy. This fermentation process generates short-chain fatty acids (SCFAs), which acidify the environment, promoting the growth of beneficial bacteria.19 Further, SCFAs are associated with activation of the immune and inflammatory response.20 HMOs also function as antiadhesive molecules through two mechanisms: by binding directly to pathogens or by binding to the intestinal epithelial cells, inducing conformational changes to their receptors.21–23
Our team and others have extensively reported on the antimicrobial activity of HMOs outside the infant gut against Group B Streptococcus (GBS).24–26 Further, we have shown that HMOs disrupt biofilms formed by GBS, methicillin-resistant Staphylococcus aureus, and Acinetobacter baumannii.27,28 HMOs have also been shown to interfere with the attachment of Haemophilus influenzae and Streptococcus pneumoniae to respiratory epithelium.29 With extensive knowledge of the antimicrobial and antiadhesive properties of HMOs, future research will address characterizing their mechanism of action.
4.2 Fat
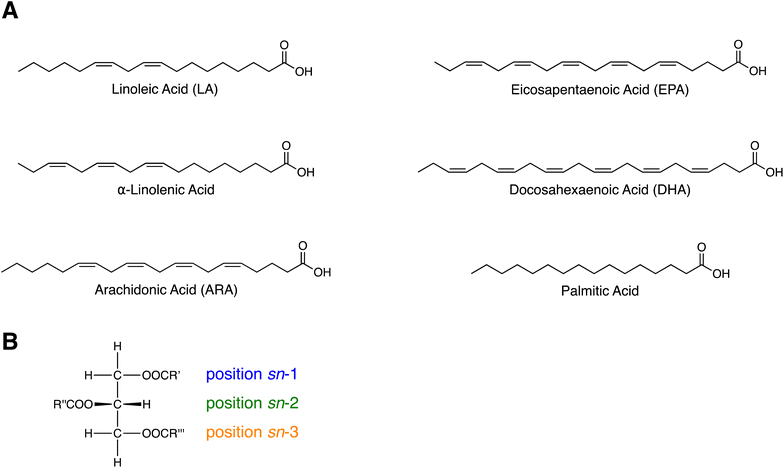
Fat is the 2nd largest component of human milk and plays a critical role in infant nutrition. Mechanistically, fats are critical to inflammatory responses, immune function, and cellular growth. Additionally, 50% of the child's energy supply originates from fat.30 Mature milk contains ca. 40 g L−1 and there's triple the fat present in hindmilk than in foremilk.31 The fats present in breast milk are readily digestible and absorbed due to the presence of bile salt-stimulated lipases that complement pancreatic lipases.32 The major fats present in human milk are the fatty acid triglycerides, and two essential fatty acids linoleic acid (LA) and α-linolenic acid (Fig. 3).33 LA and α-linolenic acid are precursors of arachidonic acid (ARA) and eicosapentaenoic acid (EPA), respectively. EPA can eventually be converted to docosahexaenoic acid (DHA) (Fig. 3).Fats are important for inflammatory responses, immune function, and growth of the infant. In triglycerides, the sn-2 position is often occupied by palmitic acid (Fig. 3). The specific positioning of this fatty acid is critical as changing the position to sn-1 or sn-3 leads to impaired absorption of calcium and fat, negatively affecting bone accretion.34 Further, palmitic acid at the sn-2 position is beneficial for intestinal and immunological health outcomes of the infant. In a mouse model for spontaneous enterocolitis, sn-2 palmitic acid supplementation resulted in decreased intestinal injury and inflammation.35
In the early postnatal period of preterm infants, there is a delicate balance of DHA, ARA, and LA levels. Indeed, decreased DHA levels are associated with an increased chance of chronic lung disease and decreased ARA levels are associated with an increased risk of late-onset sepsis.36 Further, an increased LA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DHA ratio is associated with an increased risk of chronic lung disease and late-onset sepsis. Löfqvist found that low levels of ARA are also associated with an increased risk of retinopathy of prematurity, an eye disease occurring when abnormal blood vessels grow in the retina.37 Various studies have investigated correlations between these polyunsaturated fatty acids and intestinal injury, inflammatory markers, and necrotizing enterocolitis (NEC), as recently reviewed by Martin.38 Across these studies, it was found that in vitro and in vivo studies consistently demonstrate the anti-inflammatory actions of these fatty acids, yet more research is needed to draw conclusions about benefits of fatty acid-supplemented feedings against NEC.
DHA ratio is associated with an increased risk of chronic lung disease and late-onset sepsis. Löfqvist found that low levels of ARA are also associated with an increased risk of retinopathy of prematurity, an eye disease occurring when abnormal blood vessels grow in the retina.37 Various studies have investigated correlations between these polyunsaturated fatty acids and intestinal injury, inflammatory markers, and necrotizing enterocolitis (NEC), as recently reviewed by Martin.38 Across these studies, it was found that in vitro and in vivo studies consistently demonstrate the anti-inflammatory actions of these fatty acids, yet more research is needed to draw conclusions about benefits of fatty acid-supplemented feedings against NEC.
Other complex lipids are delivered to the infant through milk fat globule membranes (MFGMs) and exosomes. MFGM is a triple membrane structure that encases milk fat and contains phospholipids, sphingolipids, cholesterol, and various proteins (Fig. 4A).39 The delivery of MFGMs and exosomes also brings individual components, including sphingolipids, such as sphingomyelin and gangliosides (Fig. 4B), which modulate neonatal intestinal development, gut microbiota establishment, and inflammation.40–43 Indeed, Blesso and his group observed that adult mice fed a diet containing 0.25% (wt/wt) milk sphingomyelin exhibited a lower abundance of gram-negative bacteria and a higher abundance of Bifidobacteria in their feces compared to those fed a high fat diet.44 Similar results were found in preterm infants fed a ganglioside-supplemented diet compared to infants fed a control milk formula in which fecal contents of the ganglioside-supplemented group had lower amounts of Escherichia coli and higher amounts of Bifidobacteria.45 This microbiota mediation could explain the decreased proinflammatory signaling in various models investigating ganglioside supplementation as E. coli is known to produce lipopolysaccharide (LPS), which triggers immune cells to release proinflammatory cytokines.46–48
Fat content in milk is closely related to maternal diet and weight gain during pregnancy. Interestingly, consumption of breads, snacks, fast foods, and margarines by lactating mothers can cause an increase in trans fatty acids in breast milk.49 Trans fatty acid concentrations have adverse effects on infant growth and development and are inversely related to LA and α-linolenic acid.50 ARA also correlates with ARA-rich food intake from lactating mothers, and EPA and DHA are also closely related.51,52 Consequently, vegetarians have very low levels of DHA in their milk because of the lack of fish or other foods in their diet.53 Therefore, it is recommended to take up to 300 mg of DHA per day to maintain enough DHA in breast milk.54
4.3 Protein
Human milk contains a diverse portfolio of proteins with various functions that contribute to the short- and long-term beneficial health outcomes of breastfeeding. The protein content of milk at birth is ca. 20 g L−1 and decreases to ca. 11 g L−1 at six months postpartum. Human milk proteins can be classified into three categories: whey, casein, and mucin, however, mucin is only present in the milk fat globule membrane (MFGM). Human milk is whey dominant, yet the whey![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) casein ratio fluctuates during stages of lactation (Fig. 5). Certain proteins facilitate infant development by providing amino acids, stimulating intestinal growth and maturation, governing the composition of the microbiome, and enhancing learning. Other proteins are present to improve the bioavailability of vitamins and minerals. Lastly, there are proteins that provide defense against pathogens.
casein ratio fluctuates during stages of lactation (Fig. 5). Certain proteins facilitate infant development by providing amino acids, stimulating intestinal growth and maturation, governing the composition of the microbiome, and enhancing learning. Other proteins are present to improve the bioavailability of vitamins and minerals. Lastly, there are proteins that provide defense against pathogens.
| Protein | Stage of lactation (days postpartum) | |||
|---|---|---|---|---|
| Colostrum ∼ (0–5) | Early ∼ (6–15) | Transitional ∼ (16–30) | Mature ∼ (30–360) | |
| α-Lactalbumin | 4.56 | 4.3 | 3.52 | 2.85 |
| Lactoferrin | 6.15 | 3.65 | 2.46 | 1.76 |
| Osteopontin | 0.18 | 1.49 | — | 0.138 |
| Secretory IgA | 5.45 | 1.5 | 1.10 | 0.138 |
| Lysozyme | 0.32 | 0.30 | 0.28 | 0.38 |
| Totals | 20.6 | 15.7 | 14.8 | 11.1 |
 | ||
| Fig. 6 PDB structures of α-lactalbumin,109 lactoferrin,110 osteopontin,111 secretory IgA,112 and lysozyme.113 Helices shown in blue, β-sheets shown in orange, loops shown in pink. | ||
4.3.1.1 α-Lactalbumin. α-Lactalbumin is a small, acidic, Ca2+ binding protein that constitutes about 25–35% of the total protein of human breast milk.58–61 Concentrations of α-lactalbumin range from 2-3 g L−1 in breast milk. Structurally, α-lactalbumin is a single-chain polypeptide of 123 amino acids, with high levels of tryptophan, lysine, and cysteine, providing a well-balanced supply of essential amino acids for the growing infant.58 Clinical studies have demonstrated that increasing the proportion of α-lactalbumin to other breast milk proteins results in plasma levels of essential amino acids comparable to those found in breastfed infants.62 Further, Hernell's group found in a double-blind randomized control trial that infants fed an α-lactalbumin-enriched formula exhibited growth patterns similar to breastfed infants.63 Similar findings were reported in a study comparing age-appropriate growth, head circumference, and plasma essential amino acids concentrations, where infants receiving α-lactalbumin-enriched formula showed growth outcomes more comparable to breastfed infants than those fed standard formula.64
The metal binding-capabilities of α-lactalbumin facilitate enhanced mineral absorption in infants, which is crucial for proper growth and development. In its unbound state, α-lactalbumin exists in the molten globule-like state, but the binding of metals such as Ca2+, Mg2+, Mn2+, Na+, and K+, stabilizes its structure.59,65 α-Lactalbumin is hypothesized to bind various metals due to its structural flexibility in its molten globule-like state and presence of coordinating amino acid residues.
α-Lactalbumin plays a pivotal role in breast milk as it serves as the “B” component of the two-protein lactase synthase complex in the golgi of mammary cells (Fig. 1). During milk production, α-lactalbumin is transported from the inner surface of the golgi to mammary secretory vesicles and finally to the alveolar lumen. In the lactose synthase complex, α-lactalbumin interacts with β1-4-galactosyltransferase (β4GalT1), the “A” component, which typically transfers galactose to terminal GlcNAc residues. Yet, in the presence of α-lactalbumin, β4GalT1's substrate specificity shifts almost exclusively to glucose, enabling the synthesis of lactose.66 Once α-lactalbumin binds β4GalT1, a large conformational change occurs in the sugar-binding region of the enzyme. This reorganization causes a difference in the hydrophobic pocket where GlcNAc usually binds, limiting such an interaction. Further, α-lactalbumin directly stabilizes glucose via binding its O1 hydroxyl group, increasing binding efficiency. α-Lactalbumin's interaction with β4GalT1 lowers the Km for glucose by 1000-fold, significantly enhancing the production of lactose over other disaccharides.67
4.3.1.2 Lactoferrin. Lactoferrin (LF) is an 80 kDa glycoprotein that exists in two primary isoforms: an apo-form (iron-free) and a holo-form (iron-bound). The concentration of LF in milk is highest in colostrum at ca. 6 g L−1 and drops to ca. 3 g L−1 after a month of lactation.56,68 Numerous studies have highlighted the antimicrobial and immunomodulatory activates of LF. LF exhibits antimicrobial effects against viral, fungal, parasitic, and bacterial pathogens.69,70 This activity often extends from its iron-binding abilities where the chelation of this essential nutrient starves invading microorganisms; a process termed “nutritional immunity”.71–73 Beyond its iron-chelating role, LF also exerts direct antimicrobial effects by binding to bacterial cell wells causing destabilization.74–76
A randomized control trial in humans showed that bovine LF supplementation, either alone or in combination with Lactobacillus rhamnosus GG decreased late onset sepsis very low birth weight (VLBW) neonates.77 A subsequent study further demonstrated that bovine LF prevented invasive fungal infections in VLBW infants.78 High levels of fecal LF in neonates were positively correlated with the establishment of the gut microbiota, illustrating LF's key role in beneficial organism habitation.79 Additionally, a hydration drink containing 1.0 g L−1 recombinant human LF and 0.2 g L−1 lysozyme reduced the duration of diarrhea in 5- to 35- month-olds hospitalized for diarrhea in Peru.80
LF is also critical for maintaining immune homeostasis, serving as a bridge between innate and adaptive immunity through modulation of antigen-presenting cells (APCs) like macrophages, dendritic cells, and B cells.81 In macrophages, LF enhances antigen presentation while reducing excessive inflammation. LF also aids dendritic cell maturation and migration, boosts B cell antibody production, and helps balance pro- and anti-inflammatory actions in T cells. In piglets, bovine LF intake modulated immune development by activating a balanced T-helper-1/2 cytokine response as reviewed by Donovan.82 Together, LF is critical for microbiome maturation.
4.3.1.3 Osteopontin. Osteopontin (OPN) is a multifunctional bioactive protein with a concentration of ca. 0.140 g L−1 in human milk,83 though levels vary depending on the stage of lactation and the mother's geographic location.84 This 314-amino acid protein is negatively charged, glycosylated, and highly phosphorylated.85 OPN is critical in the development of the infant's immune system by influencing the function of macrophages, dendritic cells, and T cells.86,87 Notably, OPN is an early regulator of T helper 1 cell-mediated immunity by activating IL-12 secretion and inhibiting IL-10 production.88 OPN also enhances host resistance to infection and facilitates phagocytosis by binding to bacterial pathogens, making them more recognizable by phagocytes.89–91
Donovan's group investigated the effects of bovine OPN in infant rhesus monkeys. In this study, the control group was fed a standard milk-based formula, while the test groups received either a formula containing 0.125 g L−1 bovine milk OPN or were nursed by their mothers.92 When comparing intestinal transcriptomes, they found that the formula supplemented with OPN resulted in only 217 differently expressed genes (DEGs) compared to the nursed monkeys', whereas the standard formula led to 1017 DEGs. This suggests that adding OPN to formula shifted the gene expression closer to that of nursed monkeys. Another study by Sørensen explored the effects of OPN on intestinal Caco-2 cells.93 They found that bovine and human milk OPN activated the expression of 322 and 239 genes, respectively. Analyzing 131 genes that were similarly expressed in response to both human and bovine OPN revealed that biological processes related to the ubiquitin system, DNA binding, transcription, and translation were impacted by OPN. Collectively, these studies demonstrate OPN's ability to modulate gene expression in the infant gut.
4.3.1.4 Secretory immunoglobulin A. Secretory immunoglobulin A (SIgA) is the most abundant immunoglobulin in human milk, with its highest concentration found in colostrum at ca. 2.5 g L−1, decreasing to ca. 1 g L−1 between days 8–12, and stabilizing to ca. 0.7 g L−1 in mature milk.94 SIgA consists of two IgA monomers linked by a J chain, and during its translocation across the epithelium, it acquires the secretory component from the polymeric Ig receptor. There are two forms of SIgA: a T-cell dependent version that is monoclonal and forms high-affinity interactions, and a T-cell independent form, which is polyclonal and binds with lower affinity.95,96
SIgA plays a crucial role in shaping the microbiota by interacting with both pathogenic and commensal organisms. Against pathogens, SIgA facilitates the clumping of bacteria, which are then moved through and out of the intestine.97 SIgA also prevents the translocation of pathogens across the intestinal epithelium and promotes the colonization of symbiotic bacteria by supporting their biofilm formation.98,99 In this vein, several commensal organisms such as Bacteroides fragilis and members of Lachnospiraceae have evolved to increase SIgA binding and enhance their colonization. Other probiotics including Bifidobacterium and Enterobacteriaceae are abundant in IgA+ fractions obtained from feces, which explains their early establishment of the infant microbiota.100 How SIgA selectively induces clumping and removal of pathogens versus promoting colonization of commensals remains unclear.
By promoting the colonization of beneficial microorganisms in the maturing infant gut, SIgA also contributes to immune function. Evidence also exists demonstrating SIgA's immune dampening abilities, which promotes a more regulated immune response as the infant matures.101,102 As weaning occurs and the external source of SIgA decreases, the infant's body compensates by producing its own SIgA via active intestinal transport,103 underscoring the antibody's role in immune defense and microbiota development during early growth.
4.3.1.5 Lysozyme. Lysozyme is a 15 kDa protein responsible for lysing bacteria by hydrolyzing the β1-4 linkages between N-acetylglucosamine and N-acetylmuramic acid monosaccharides in peptidoglycan, a key component of bacterial cell walls.104 The concentration of lysozyme in human milk varies, ranging from ca. 0.37 g L−1 in colostrum, ca. 0.27 g L−1 in transitional milk, and ca. 0.24–0.89 g L−1 in mature milk.105 As lysozyme disrupts bacterial cell walls, leading to cell death, it is believed to selectively target pathogenic organisms. Several organisms have evolved mechanisms of resistance to lysozyme, such as peptidoglycan modifications. Yet, lysozyme has also been shown to activate the pro-inflammatory immune response, which is designed to target pathogenic organisms.106 Indeed, a study by Elizabeth Maga's group found that six-week-old pigs fed goat milk enriched with lysozyme had higher abundance of bacteria associated with positive gut health (Bifidobacteriaceae and Lactobacillaceae) and a lower abundance of bacteria associated with disease (Mycobacteriaceae, Streptococcaceae, and Campylobacterales).107 Further, they showed that lysozyme can modulate the inflammatory response in the gut. Pigs fed the human lysozyme-enriched goat milk had a significantly higher level of the anti-inflammatory cytokine TGF-β1 in a portion of their small intestine, without an increase in proinflammatory cytokines.108 Together, lysozyme's dual function of selectively eliminating pathogenic bacteria while modulating the immune system helps protect the infant from infections, supporting the development of a healthy microbiota.
| Protein | Stage of lactation (days postpartum) | ||
|---|---|---|---|
| Early ∼ (0–10) | Transitional ∼ (11–30) | Mature ∼ (30–360) | |
| β-casein | 1.29 | 1.46 | 1.03 |
| α-S1-casein | 0.34 | 0.33 | 0.33 |
| κ-casein | 0.86 | 0.80 | 0.55 |
| Totals | 2.49 | 2.59 | 1.92 |
Functionally, casein provides essential amino acids to the infant. Casein's ability to bind and transport divalent cations, such as zinc and calcium, suggests it may also aid in the absorption of these nutrients, although more research is needed to confirm this activity.117,118 With heavy glycosylation, κ-casein has been shown to bind pathogens, including Helicobacter pylori, thereby preventing attachment and infection.119 In 2023, Szeto and Zhao found that κ-casein was positively correlated with Clostridium butyricum, an intestinal symbiotic bacterium that has a strong butyric acid production capacity.120 This SCFA reduces the production of inflammatory cytokines by reducing the permeability of the intestinal epithelium, which is crucial for early intestinal immune system development.121 Casein is useful in breast milk as an antimicrobial and a provider of essential amino acids.
Mucins serve multiple functions, but are best known for protecting infants from infection, specifically via their glycosylated structures. Interestingly, most infants who are exclusively breastfed by human immunodeficiency virus (HIV)-positive mothers remain uninfected even after repeated exposure through breastmilk.123 Mucins have been implicated in this phenomenon with Saeland illustrating that MUC1 blocks infection by HIV-1.124 During mother-to-child transmission of HIV, the virus typically targets dendritic cells in mucosal areas, facilitating transmission to T-cells. However, O-linked glycans on MUC1 efficiently bind dendritic cell receptors, thereby blocking HIV-1 transmission. Additionally, mucins inhibit viral replication by binding to rotavirus and Norwalk virus.125,126
Mucins are also effective against bacterial colonization. MUC1 and MUC4 have been shown to block Salmonella enterica serovar typhimurium invasion of Caco-2 cells.127 MUC1 was also shown to provide a protective barrier against H. pylori colonization in a mouse model.128 Together, the mucins found in MFGM serve to protect the developing microbiota from unwanted habitation by viral and bacterial pathogens.
4.4 Hormones and growth factors
Human breast milk contains various biologically active factors such as hormones, peptide growth factors, and cytokines (Fig. 7). One such hormone, erythropoietin (epo), is responsible for increasing red blood cells, which may prevent premature anemia.129,130 Epo is also critical for tightening intestinal junctions in infants, protecting the child from any milk-borne pathogens or viruses.131 Further, epo helps maintain the integrity of the mammary epithelium, preventing HIV particles from seeping from the blood into milk.132 | ||
| Fig. 7 A representative schematic showing the distribution of hormones and growth factors found in breast milk and what they provide to the growing infant. | ||
Calcitonin is a growth-regulating hormone present in human milk. Serum levels of calcitonin are higher in pregnant and lactating women as compared to non-pregnant women and are not influenced by breastfeeding.133,134 Therefore, it is assumed that these increased levels are to protect the healthy maternal skeleton by opposing the action of 1,25-dihhydroxycholecalciferol (1,25[OH]2D3), a bone-resorbing hormone that exhibits increased levels during pregnancy as the mother needs excess calcium.133 In the context of infant health, calcitonin has been proposed to play a role in the maturation of enteric neurons.135
Human breast milk also contains adiponectin and leptin, which are adipose-derived hormones that regulate metabolism and body composition. Adiponectin promotes tissue insulin sensitivity, stimulates glucose uptake, and decreases energy expenditure.136,137 Leptin helps regulate appetite by informing the infant's brain of remaining energy sources.138,139 While some studies suggest a link between these hormones and a reduced risk of obesity, results have been inconsistent.140–143 These variations may stem from differences in study methodologies, breast milk composition, and other health-related factors.
Epidermal growth factor (EGF) plays a vital role in the maturation and healing processes of the intestinal mucosa. EGF levels in human breast milk are highest in the first days after birth at roughly 100 ng mL−1 and then gradually decrease during lactation.144,145 EGF is resilient against low pH and digestive enzymes, allowing it to freely reach the intestines where it stimulates DNA synthesis, cell division, protein synthesis, and the absorption of critical nutrients.146,147 EGF also heals damaged mucosa by correcting alterations in tight junction proteins induced by the proinflammatory cytokine TNF-⍺.148,149 Heparin-binding EGF (HB-EGF), a member of the EGF family, is primarily responsible for the protection of the intestinal epithelium from hypoxic necrosis and cytokine-induced apoptosis.150–154 HB-EGF provides these functions mainly by decreasing nitrogen and oxygen reactive species production.155
Vascular endothelial growth factor (VEGF) and hepatocyte growth factor (HGF) are two of the most abundant growth factors in breast milk, especially in colostrum.156,157 VEGF protects against angiogenesis and vasculogenesis, promotes growth, and protects the infant's gastrointestinal system.158–161 Similarly, HGF stimulates epithelial cell proliferation and also contributes to tissue regeneration, defense, and longevity.162–164
The insulin-like growth factor (IGF) family, specifically IGF-I and IGF-II, is found in its highest levels in the colostrum.165,166 This family enhances cell survival, stimulates the proliferation of intestinal stem cells, and promotes erythropoiesis.167–169 In rat models, IGF-I protects against necrotizing enterocolitis (NEC) and reduces the inflammatory response.170 One study by van Goudoever's group found that adding double the usual amount of IGF-I in colostrum improved gut barrier function by day 14.171 Finally, both IGF-I and -II were shown to promote cell migration of human umbilical cord vascular endothelial cells, which protects and develops the maturing intestinal epithelium.172,173
Proper nervous system development requires both brain-derived neurotrophic factor (BDNF) and glial cell line-derived neurotrophic factor (GDNF). BDNF plays a role in nervous system development and maintenance and has been implicated in the modulation of learning and memory.174 Additionally, BDNF regulates enteric neuronal activity, increasing gastrointestinal motility.175 GDNF is critical for neuronal survival in the enteric nervous system.176,177 Together, BDNF and GDNF stimulate neurodevelopment of the infant during breastfeeding.
5 Outlook
Ontology. Despite its importance in advancing infant health and wellness, there are major challenges preventing our complete understanding of human milk. Currently, we understand milk to be a personally curated collection of key nutrients, macromolecules, and microorganisms used by the infant. However, we understand little about the ontology of human milk, that is, the mechanistic interplay between the components of the milk and how they complement each other to correctly guide growth and development. In a related vein, we have a poor understanding of the feedback loop between mother and child that governs how and when components change. Key gaps in this area include characterizing variability in human milk composition based on environmental determinants of health and evaluating effects of maternal nutrient supplementation during lactation on human milk composition.Defining milk quality. Given the large number of complex molecules present in milk, the greatest key gap in analysis is that there is no single analytical technique capable of all aspects of milk analysis. Mass spectrometry is the most enabling tool to characterize the composition of human milk. New technologies, particularly those capable of separating molecules with near identical polarity, such as HMOs, would be a great advance toward defining global reference values for macromolecules, micronutrients, and microorganisms in human milk.
Microbial metabolites. Microorganisms and their metabolites are increasingly being used as therapeutics. Even though human milk has a robust microbiome, to date, characterization of microbial metabolites in human milk, and their impact on the infant's health is incomplete. This is a key gap as these metabolites may potentiate developmental, metabolic, or immune programming in the child. Breast milk microbiome-associated metabolites are an intriguing avenue for new therapies. It would be informative to characterize which metabolites produced by the various microbiota of the mother are transported to the mammary gland during lactation. Moreover, it is unclear whether levels of diet-derived maternal microbial metabolites in human milk can be modulated by dietary intervention or by supplementation. Taken together, maternal and microbial metabolites represent promising targets for ‘postbiotic’ intervention during nursing. Given the current state of analytical chemistry, it is possible to use multi-omic approaches, data science, and statistical analysis to characterize this facet of human physiology.
After water, human milk is the 2nd most important liquid in existence. Yet, many unknowns remain. In a way, milk is an example of precision nutrition – its components unambiguously determined and produced by the mother specifically for her infant. It is undefined, however, how this occurs. We hypothesize that the retrograde flux that occurs during nursing is a channel for bidirectional signaling or the exchange of metabolites between the infant's mouth and the mammary gland. We recognize, however, that the composition of mother's milk may also be the result of environmental and genetic factors. Taken together, human milk science provides exciting opportunities to improve maternal–child health using frontier areas such as artificial intelligence, synthetic biology, machine learning, and advanced mass spectrometry. The ultimate deliverable of addressing these key gaps is characterizing new maternal and child nutrition interventions that may inform the politics of health policies.
6 Data availability
As this is a review article, there is no new data associated with this manuscript.7 Conflicts of interest
There are no conflicts to declare.8 Acknowledgements
Financial support was provided the National Science Foundation (CHE-1847804) to S.D.T. The reviewers are acknowledged for their assistance in strengthening the manuscript.9 References
- World Health Organization, The Global Strategy for Infant and Young Child Feeding, Geneva, Switzerland, 2003 Search PubMed.
- B. Koletzko, S. Baker, G. Cleghorn, U. F. Neto, S. Gopalan, O. Hernell, Q. S. Hock, P. Jirapinyo, B. Lonnerdal, P. Pencharz, H. Pzyrembel, J. Ramirez-Mayans, R. Shamir, D. Turck, Y. Yamashiro and D. Zong-Yi, J. Pediatr. Gastroenterol. Nutr., 2005, 41, 584–599 CrossRef PubMed.
- O. T. Oftedal, J. Mammary Gland Biol. Neoplasia, 2002, 7, 225–252 CrossRef PubMed.
- C. Castellote, R. Casillas, C. Ramírez-Santana, F. J. Pérez-Cano, M. Castell, M. G. Moretones, M. C. López-Sabater and A. Franch, J. Nutr., 2011, 141, 1181–1187 CrossRef CAS PubMed.
- W. W. Pang and P. E. Hartmann, J. Mammary Gland Biol. Neoplasia, 2007, 12, 211–221 CrossRef.
- J. K. Kulski and P. E. Hartmann, Aust. J. Exp. Biol. Med. Sci., 1981, 59, 101–114 CrossRef CAS.
- K. Y. Wojcik, D. J. Rechtman, M. L. Lee, A. Montoya and E. T. Medo, J. Am. Diet. Assoc., 2009, 109, 137–140 CrossRef PubMed.
- B. Ramakrishnan, E. Boeggeman and P. K. Qasba, Biochem. Biophys. Res. Commun., 2002, 291, 1113–1118 CrossRef CAS.
- R. G. Heine, F. Alrefaee, P. Bachina, J. C. De Leon, L. Geng, S. Gong, J. A. Madrazo, J. Ngamphaiboon, C. Ong and J. M. Rogacion, WAO J., 2017, 10, 41 Search PubMed.
- D. L. Bissett and R. L. Anderson, J. Bacteriol., 1974, 117, 318–320 CrossRef CAS PubMed.
- R. Francavilla, M. Calasso, L. Calace, S. Siragusa, M. Ndagijimana, P. Vernocchi, L. Brunetti, G. Mancino, G. Tedeschi, E. Guerzoni, F. Indrio, L. Laghi, V. L. Miniello, M. Gobbetti and M. De Angelis, PAI, 2012, 23, 420–427 Search PubMed.
- F. J. Simoons, AJDD, 1970, 15, 695–710 CrossRef CAS PubMed.
- G. Coppa, P. Pierani, L. Zampini, I. Carloni, A. Carlucci and O. Gabrielli, Acta Paediatr., 1999, 88, 89–94 CrossRef CAS PubMed.
- O. Gabrielli, L. Zampini, T. Galeazzi, L. Padella, L. Santoro, C. Peila, F. Giuliani, E. Bertino, C. Fabris and G. V. Coppa, Pediatrics, 2011, 128, e1520–e1531 CrossRef PubMed.
- M. Polonovski and J. Montreuil, C. R. Hebd. Seances Acad. Sci., 1954, 238, 2263–2264 CAS.
- C. Kunz, Adv Nutr., 2012, 3, 430s–439s CrossRef CAS PubMed.
- B. Soyyılmaz, M. H. Mikš, C. H. Röhrig, M. Matwiejuk, A. Meszaros-Matwiejuk and L. K. Vigsnæs, Nutrients, 2021, 13, 2737 CrossRef.
- D. S. Newburg, G. M. Ruiz-Palacios and A. L. Morrow, Annu. Rev. Nutr., 2005, 25, 37–58 CrossRef CAS PubMed.
- D. J. Morrison and T. Preston, Gut Microbes, 2016, 7, 189–200 CrossRef.
- N. Zheng, Y. Gao, W. Zhu, D. Meng and W. A. Walker, PLoS One, 2020, 15, e0229283 CrossRef CAS.
- C. Walsh, J. A. Lane, D. van Sinderen and R. M. Hickey, J. Funct. Foods, 2020, 72, 104074 CrossRef CAS.
- K. Le Doare, B. Holder, A. Bassett and P. S. Pannaraj, Front. Immunol., 2018, 9 CrossRef CAS PubMed.
- V. Triantis, L. Bode and R. J. J. Van Neerven, Front. Pediatr., 2018, 6 CrossRef.
- N. J. Andreas, A. Al-Khalidi, M. Jaiteh, E. Clarke, M. J. Hyde, N. Modi, E. Holmes, B. Kampmann and K. Mehring Le Doare, Clin. Transl. Immunology, 2016, 5, e99 CrossRef.
- K. M. Craft and S. D. Townsend, Acc. Chem. Res., 2019, 52, 760–768 CrossRef CAS PubMed.
- D. L. Ackerman, R. S. Doster, J. H. Weitkamp, D. M. Aronoff, J. A. Gaddy and S. D. Townsend, ACS Infect. Dis., 2017, 3, 595–605 CrossRef CAS.
- S. K. Spicer, R. E. Moore, J. Lu, M. A. Guevara, D. R. Marshall, S. D. Manning, S. M. Damo, S. D. Townsend and J. A. Gaddy, ACS Infect. Dis., 2021, 7, 3254–3263 CrossRef CAS PubMed.
- D. L. Ackerman, K. M. Craft, R. S. Doster, J. H. Weitkamp, D. M. Aronoff, J. A. Gaddy and S. D. Townsend, ACS Infect. Dis., 2018, 4, 315–324 CrossRef CAS PubMed.
- B. Andersson, O. Porras, L. A. Hanson, T. Lagergard and C. Svanborg-Eden, J. Infect. Dis., 1986, 153, 232–237 CrossRef CAS.
- O. Ballard and A. L. Morrow, Pediatr. Clin. North Am., 2013, 60, 49–74 CrossRef PubMed.
- M. A. Underwood, Pediatr. Clin. North Am., 2013, 60, 189–207 CrossRef PubMed.
- E. M. Straarup, L. Lauritzen, J. Faerk, C. E. Høy and K. F. Michaelsen, JPGN, 2006, 42, 293–299 CAS.
- M. Guo, in Functional Foods, ed. M. Guo, Woodhead Publishing, 2nd edn, 2025, pp. 327–362, DOI:10.1016/B978-0-443-19100-8.00012-9.
- M. E. D. Q. Leite, J. Lasekan, G. Baggs, T. Ribeiro, J. Menezes-Filho, M. Pontes, J. Druzian, D. L. Barreto, C. O. De Souza, Â. Mattos and H. Costa-Ribeiro, BMC Pediatr., 2013, 13, 215 CrossRef PubMed.
- P. Lu, F. Bar-Yoseph, L. Levi, Y. Lifshitz, J. Witte-Bouma, A. C. J. M. De Bruijn, A. M. Korteland-Van Male, J. B. Van Goudoever and I. B. Renes, PLoS One, 2013, 8, e65878 CrossRef CAS.
- C. R. Martin, D. A. Dasilva, J. E. Cluette-Brown, C. Dimonda, A. Hamill, A. Q. Bhutta, E. Coronel, M. Wilschanski, A. J. Stephens, D. F. Driscoll, B. R. Bistrian, J. H. Ware, M. M. Zaman and S. D. Freedman, J. Pediatr., 2011, 159, 743–749 CrossRef CAS PubMed.
- C. A. Löfqvist, S. Najm, G. Hellgren, E. Engström, K. Sävman, A. K. Nilsson, M. X. Andersson, A.-L. Hård, L. E. H. Smith and A. Hellström, JAMA Ophthalmol., 2018, 136, 271 CrossRef PubMed.
- D. Ramiro-Cortijo, P. Singh, Y. Liu, E. Medina-Morales, W. Yakah, S. D. Freedman and C. R. Martin, Nutrients, 2020, 12 Search PubMed.
- N. Timby, M. Domellöf, B. Lönnerdal and O. Hernell, Adv Nutr., 2017, 8, 351–355 CrossRef CAS PubMed.
- C. Martin, M. Patel, S. Williams, H. Arora and B. Sims, Innate Immune, 2018, 24, 278–284 CrossRef CAS.
- X. Wang, X. Yan, L. Zhang, J. Cai, Y. Zhou, H. Liu, Y. Hu, W. Chen, S. Xu, P. Liu, T. Chen, J. Zhang, Y. Cao, Z. Yu and S. Han, Mol. Nutr. Food Res., 2019, 63, 1801247 CrossRef PubMed.
- R. Gao, R. Zhang, T. Qian, X. Peng, W. He, S. Zheng, Y. Cao, A. Pierro and C. Shen, Pediatr. Surg. Int., 2019, 35, 1363–1368 CrossRef PubMed.
- H. Miyake, C. Lee, S. Chusilp, M. Bhalla, B. Li, M. Pitino, S. Seo, D. L. O'Connor and A. Pierro, Pediatr. Surg. Int., 2020, 36, 155–163 CrossRef PubMed.
- G. H. Norris, C. Jiang, J. Ryan, C. M. Porter and C. N. Blesso, J. Nutr. Biochem., 2016, 30, 93–101 CrossRef CAS PubMed.
- R. Rueda, J. L. Sabatel, J. Maldonado, J. A. Molina-Font and A. Gil, J. Pediatr., 1998, 133, 90–94 CrossRef CAS PubMed.
- K. L. Schnabl, B. Larsen, J. E. Van Aerde, G. Lees, M. Evans, M. Belosevic, C. Field, A. B. Thomson and M. T. Clandinin, J. Pediatr. Gastroenterol. Nutr., 2009, 49, 382–392 CrossRef CAS PubMed.
- E. J. Park, M. Suh, B. Thomson, A. B. R. Thomson, K. S. Ramanujam and M. T. Clinin, Glycobiology, 2005, 15, 935–942 CrossRef CAS PubMed.
- E. J. Park, M. Suh, B. Thomson, D. W. Ma, K. Ramanujam, A. B. Thomson and M. T. Clandinin, Shock, 2007, 28, 112–117 CrossRef CAS PubMed.
- S. M. Innis and D. J. King, Am. J. Clin. Nutr., 1999, 70, 383–390 CrossRef CAS PubMed.
- T. Decsi, Acta Paediatr., 2003, 92, 1369–1371 CrossRef CAS PubMed.
- M. Del Prado, S. Villalpando, A. Elizondo, M. Rodríguez, H. Demmelmair and B. Koletzko, Am. J. Clin. Nutr., 2001, 74, 242–247 CrossRef CAS PubMed.
- A. R. Weseler, C. E. Dirix, M. J. Bruins and G. Hornstra, J. Nutr., 2008, 138, 2190–2197 CrossRef CAS PubMed.
- C. L. Jensen, M. Maude, R. E. Anderson and W. C. Heird, Am. J. Clin. Nutr., 2000, 71, 292s–299s CrossRef CAS PubMed.
- M. Fleith and M. T. Clandinin, Crit. Rev. Food Sci. Nutr., 2005, 45, 205–229 CrossRef CAS PubMed.
- F. Haschke, N. Haiden and S. K. Thakkar, Ann. Nutr. Metab., 2016, 69, 16–26 CrossRef PubMed.
- B. Lönnerdal, P. Erdmann, S. K. Thakkar, J. Sauser and F. Destaillats, J. Nutr. Biochem., 2017, 41, 1–11 CrossRef PubMed.
- T. Nagatomo, S. Ohga, H. Takada, A. Nomura, S. Hikino, M. Imura, K. Ohshima and T. Hara, CEI, 2004, 138, 47–53 CAS.
- B. Lönnerdal and E. L. Lien, Nutr. Rev., 2003, 61, 295–305 CrossRef.
- E. A. Permyakov, Biomolecules, 2020, 10, 1210 CrossRef CAS PubMed.
- B. Lönnerdal, E. Forsum, M. Gebre-Medhin and L. Hambraeus, Am. J. Clin. Nutr., 1976, 29, 1134–1141 CrossRef PubMed.
- P. Montagne, M. L. Cuillière, C. Molé, M. C. Béné and G. Faure, J. Pediatr. Gastroenterol. Nutr., 1999, 29, 75–80 CAS.
- E. L. Lien, Am. J. Clin. Nutr., 2003, 77, 1555s–1558s CrossRef CAS PubMed.
- O. Sandström, B. Lönnerdal, G. Graverholt and O. Hernell, Am. J. Clin. Nutr., 2008, 87, 921–928 CrossRef PubMed.
- J. Trabulsi, R. Capeding, J. Lebumfacil, K. Ramanujam, P. Feng, S. McSweeney, B. Harris and P. Derusso, Eur. J. Clin. Nutr., 2011, 65, 167–174 CrossRef CAS.
- Y. Hiraoka, T. Segawa, K. Kuwajima, S. Sugai and N. Murai, Biochem. Biophys. Res. Commun., 1980, 95, 1098–1104 CrossRef CAS PubMed.
- P. K. Qasba, S. Kumar and K. Brew, Crit. Rev. Biochem. Mol. Biol., 1997, 32, 255–306 CrossRef CAS PubMed.
- W. A. Klee and C. B. Klee, Biochem. Biophys. Res. Commun., 1970, 39, 833–841 CrossRef CAS PubMed.
- M. Albenzio, A. Santillo, I. Stolfi, P. Manzoni, A. Iliceto, M. Rinaldi and R. Magaldi, Am. J. Perinatol., 2016, 33, 1085–1089 CrossRef PubMed.
- M. Sienkiewicz, A. Jaśkiewicz, A. Tarasiuk and J. Fichna, Crit. Rev. Food Sci. Nutr., 2022, 62, 6016–6033 CrossRef CAS PubMed.
- L. Adlerova, A. Bartoskova and M. Faldyna, Veterinární medicína, 2008, 53, 457–468 CrossRef CAS.
- E. D. Weinberg, JAMA, 1975, 231, 39–41 CrossRef CAS PubMed.
- K. W. Becker and E. P. Skaar, FEMS Microbiol. Rev., 2014, 38, 1235–1249 CrossRef CAS PubMed.
- R. R. Arnold, M. F. Cole and J. R. McGhee, Science, 1977, 197, 263–265 CrossRef CAS PubMed.
- R. T. Ellison, T. J. Giehl and F. M. Laforce, Infect. Immun., 1988, 56, 2774–2781 CrossRef CAS PubMed.
- E. Elass-Rochard, A. Roseanu, D. Legrand, M. Trif, V. Salmon, C. Motas, J. Montreuil and G. Spik, Biochem. J., 1995, 312, 839–845 CrossRef CAS PubMed.
- C. C. Yen, C. Y. Lin, K. Y. Chong, T. C. Tsai, C. J. Shen, M. F. Lin, C. Y. Su, H. L. Chen and C. M. Chen, J. Infect. Dis., 2009, 199, 590–598 CrossRef CAS PubMed.
- P. Manzoni, JAMA, 2009, 302, 1421 CrossRef CAS PubMed.
- P. Manzoni, I. Stolfi, H. Messner, S. Cattani, N. Laforgia, M. G. Romeo, L. Bollani, M. Rinaldi, E. Gallo, M. Quercia, M. Maule, M. Mostert, L. Decembrino, R. Magaldi, F. Mosca, F. Vagnarelli, L. Memo, P. M. Betta, M. Stronati and D. Farina, Pediatrics, 2012, 129, 116–123 CrossRef PubMed.
- P. Mastromarino, D. Capobianco, G. Campagna, N. Laforgia, P. Drimaco, A. Dileone and M. E. Baldassarre, BioMetals, 2014, 27, 1077–1086 CrossRef CAS PubMed.
- N. Zavaleta, D. Figueroa, J. Rivera, J. Sánchez, S. Alfaro and B. Lönnerdal, JPGN, 2007, 44, 258–264 CAS.
- T. Siqueiros-Cendón, S. Arévalo-Gallegos, B. F. Iglesias-Figueroa, I. A. García-Montoya, J. Salazar-Martínez and Q. Rascón-Cruz, Acta Pharmacol. Sin., 2014, 35, 557–566 CrossRef PubMed.
- S. M. Donovan, J. Pediatr., 2016, 173, S16–S28 CrossRef CAS PubMed.
- L. Schack, A. Lange, J. Kelsen, J. Agnholt, B. Christensen, T. E. Petersen and E. S. Sørensen, JDS, 2009, 92, 5378–5385 CAS.
- S. Bruun, L. N. Jacobsen, X. Ze, S. Husby, H. M. Ueno, K. Nojiri, S. Kobayashi, J. Kwon, X. Liu, S. Yan, J. Yang, G. Zachariassen, L. Chen, W. Zhou, B. Christensen and E. S. Sørensen, JPGN, 2018, 67, 250–256 CAS.
- R. Jiang and B. Lönnerdal, Curr. Opin. Clin. Nutr. Metab. Care, 2016, 19, 214–219 CAS.
- E. Y. Lin, W. Xi, N. Aggarwal and M. L. Shinohara, Int. Immunol., 2023, 35, 171–180 CrossRef CAS PubMed.
- K. X. Wang and D. T. Denhardt, Cytokine Growth Factor Rev., 2008, 19, 333–345 CrossRef CAS PubMed.
- S. Ashkar, G. F. Weber, V. Panoutsakopoulou, M. E. Sanchirico, M. Jansson, S. Zawaideh, S. R. Rittling, D. T. Denhardt, M. J. Glimcher and H. Cantor, Science, 2000, 287, 860–864 CrossRef CAS PubMed.
- G. J. Nau, L. Liaw, G. L. Chupp, J. S. Berman, B. L. M. Hogan and R. A. Young, Infect. Immun., 1999, 67, 4223–4230 CrossRef CAS PubMed.
- E. E. Rollo, S. J. Hempson, A. Bansal, E. Tsao, I. Habib, S. R. Rittling, D. T. Denhardt, E. R. Mackow and R. D. Shaw, J. Virol., 2005, 79, 3509–3516 CrossRef CAS PubMed.
- L. Schack, R. Stapulionis, B. Christensen, E. Kofod-Olsen, U. B. Skov Sørensen, T. Vorup-Jensen, E. S. Sørensen and P. HöLlsberg, J. Immunol., 2009, 182, 6943–6950 CrossRef CAS PubMed.
- S. M. Donovan, M. H. Monaco, J. Drnevich, A. S. Kvistgaard, O. Hernell and B. Lönnerdal, J. Nutr., 2014, 144, 1910–1919 CrossRef CAS PubMed.
- B. Christensen, A. J. Buitenhuis, L. N. Jacobsen, M. S. Ostenfeld and E. S. Sørensen, Nutrients, 2023, 15, 1166 CrossRef CAS PubMed.
- S. Trend, T. Strunk, M. L. Lloyd, C. H. Kok, J. Metcalfe, D. T. Geddes, C. T. Lai, P. Richmond, D. A. Doherty, K. Simmer and A. Currie, Br. J. Nutr., 2016, 115, 1178–1193 CrossRef CAS PubMed.
- K. E. Huus, C. Petersen and B. B. Finlay, Nat. Rev. Immunol., 2021, 21, 514–525 CrossRef CAS PubMed.
- J. J. Bunker and A. Bendelac, Immunity, 2018, 49, 211–224 CrossRef CAS PubMed.
- D. Hoces, M. Arnoldini, M. Diard, C. Loverdo and E. Slack, Immunology, 2020, 159, 52–62 CrossRef CAS PubMed.
- R. Randal Bollinger, M. L. Everett, D. Palestrant, S. D. Love, S. S. Lin and W. Parker, Immunology, 2003, 109, 580–587 CrossRef PubMed.
- P. E. Orndorff, A. Devapali, S. Palestrant, A. Wyse, M. L. Everett, R. R. Bollinger and W. Parker, Infect. Immun., 2004, 72, 1929–1938 CrossRef CAS PubMed.
- A. Janzon, J. K. Goodrich, O. Koren, J. L. Waters and R. E. Ley, mSystems, 2019, 4, e00612–e00619 CrossRef PubMed.
- Z. Al Nabhani, S. Dulauroy, R. Marques, C. Cousu, S. Al Bounny, F. Déjardin, T. Sparwasser, M. Bérard, N. Cerf-Bensussan and G. Eberl, Immunity, 2019, 50, 1276–1288 CrossRef CAS PubMed.
- N. L. Harris, I. Spoerri, J. F. Schopfer, C. Nembrini, P. Merky, J. Massacand, J. F. Urban, A. Lamarre, K. Burki, B. Odermatt, R. M. Zinkernagel and A. J. Macpherson, J. Immunol., 2006, 177, 6256–6262 CrossRef CAS PubMed.
- E. W. Rogier, A. L. Frantz, M. E. C. Bruno, L. Wedlund, D. A. Cohen, A. J. Stromberg and C. S. Kaetzel, Proc. Natl. Acad. Sci. U.S.A., 2014, 111, 3074–3079 CrossRef CAS PubMed.
- D. M. Chipman and N. Sharon, Science, 1969, 165, 454–465 CrossRef CAS PubMed.
- P. Montagne, M. L. Cuillière, C. Molé, M. C. Béné and G. Faure, Adv. Exp. Med. Biol., 2001, 501, 241–247 CrossRef CAS PubMed.
- S. A. Ragland and A. K. Criss, PLoS Pathog., 2017, 13, e1006512 CrossRef PubMed.
- E. A. Maga, P. T. Desai, B. C. Weimer, N. Dao, D. Kültz and J. D. Murray, Appl. Environ. Microbiol., 2012, 78, 6153–6160 CrossRef CAS PubMed.
- C. A. Cooper, D. R. Brundige, W. A. Reh, E. A. Maga and J. D. Murray, Transgenic Res., 2011, 20, 1235–1243 CrossRef CAS PubMed.
- N. Chandra, K. Brew and K. R. Acharya, Biochemistry, 1998, 37, 4767–4772 CrossRef CAS PubMed.
- M. Haridas, B. F. Anderson and E. N. Baker, Acta Crystallogr., Sect. D:Biol. Crystallogr., 1995, 51, 629–646 CrossRef CAS PubMed.
- J. Du, S. Hou, C. Zhong, Z. Lai, H. Yang, J. Dai, D. Zhang, H. Wang, Y. Guo and J. Ding, J. Mol. Biol., 2008, 382, 835–842 CrossRef CAS PubMed.
- Y. Wang, G. Wang, Y. Li, Q. Zhu, H. Shen, N. Gao and J. Xiao, Cell Research, 2020, 30, 602–609 CrossRef CAS PubMed.
- K. H. Nam, Appl. Sci., 2022, 12, 4363 CrossRef CAS.
- S. M. Sood, P. J. Herbert and C. W. Slatter, JDS, 1997, 80, 628–633 CAS.
- K. A. Dingess, I. Gazi, H. W. P. Van Den Toorn, M. Mank, B. Stahl, K. R. Reiding and A. J. R. Heck, Int. J. Mol. Sci., 2021, 22, 8140 CrossRef CAS PubMed.
- Y. Liao, D. Weber, W. Xu, B. P. Durbin-Johnson, B. S. Phinney and B. Lönnerdal, J. Proteome Res., 2017, 16, 4113–4121 CrossRef CAS PubMed.
- M. Hansen, B. Sandström and B. Lönnerdal, Pediatr. Res., 1996, 40, 547–552 CrossRef CAS PubMed.
- B. Teucher, G. Majsak-Newman, J. R. Dainty, D. McDonagh, R. J. FitzGerald and S. J. Fairweather-Tait, Am. J. Clin. Nutr., 2006, 84, 162–166 CrossRef CAS PubMed.
- M. Strömqvist, P. Falk, S. Bergström, L. Hansson, B. Lönnerdal, S. Normark and O. Hernell, J. Pediatr. Gastroenterol. Nutr., 1995, 21, 288–296 Search PubMed.
- M. Xi, D. Liang, Y. Yan, S. Duan, H. Leng, H. Yang, X. Shi, X. Na, Y. Yang, C. Yang, I. M.-Y. Szeto and A. Zhao, Front. Microbiol., 2023, 14 Search PubMed.
- M. K. Stoeva, J. Garcia-So, N. Justice, J. Myers, S. Tyagi, M. Nemchek, P. J. McMurdie, O. Kolterman and J. Eid, Gut Microbes, 2021, 13, 1907272 CrossRef PubMed.
- J. A. Peterson, M. Hamosh, C. D. Scallan, R. L. Ceriani, T. R. Henderson, N. R. Mehta, M. Armand and P. Hamosh, Pediatr. Res., 1998, 44, 499–506 CrossRef CAS PubMed.
- H. M. Coovadia, N. C. Rollins, R. M. Bland, K. Little, A. Coutsoudis, M. L. Bennish and M.-L. Newell, Lancet, 2007, 369, 1107–1116 CrossRef PubMed.
- E. Saeland, M. A. de Jong, A. A. Nabatov, H. Kalay, T. B. Geijtenbeek and Y. van Kooyk, Mol. Immunol., 2009, 46, 2309–2316 CrossRef CAS PubMed.
- R. H. Yolken, J. A. Peterson, S. L. Vonderfecht, E. T. Fouts, K. Midthun and D. S. Newburg, JCI, 1992, 90, 1984–1991 CrossRef CAS PubMed.
- N. Ruvoën-Clouet, E. Mas, S. Marionneau, P. Guillon, D. Lombardo and L. P. Jacques, Biochem. J., 2006, 393, 627–634 CrossRef PubMed.
- B. Liu, Z. Yu, C. Chen, D. E. Kling and D. S. Newburg, J. Nutr., 2012, 142, 1504–1509 CrossRef CAS PubMed.
- M. A. McGuckin, A. L. Every, C. D. Skene, S. K. Linden, Y. T. Chionh, A. Swierczak, J. McAuley, S. Harbour, M. Kaparakis, R. Ferrero and P. Sutton, Gastroenterology, 2007, 133, 1210–1218 CrossRef CAS PubMed.
- J. C. Kett, PIR, 2012, 33, 186–187 Search PubMed.
- V. Soubasi, G. Kremenopoulos, E. Diamanti, C. Tsantali, K. Sarafidis and D. Tsakiris, J. Pediatr., 1995, 127, 291–297 CrossRef CAS PubMed.
- S.-R. Shiou, Y. Yu, S. Chen, M. J. Ciancio, E. O. Petrof, J. Sun and E. C. Claud, J. Biol. Chem., 2011, 286, 12123–12132 CrossRef CAS PubMed.
- J. E. Arsenault, A. L. Webb, I. N. Koulinska, S. Aboud, W. W. Fawzi and E. Villamor, J. Infect. Dis., 2010, 202, 370–373 CrossRef PubMed.
- J. Stevenson, C. Hillyard, I. Macintyre, H. Cooper and M. Whitehead, Lancet, 1979, 314, 769–770 CrossRef PubMed.
- J. Struck, P. De Almeida, A. Bergmann and N. G. Morgenthaler, Horm. Metab. Res., 2002, 34, 460–465 CrossRef CAS PubMed.
- P. J. Wookey, K. Turner and J. B. Furness, Cell Tissue Res., 2012, 347, 311–317 CrossRef CAS PubMed.
- G. Çatlı, N. Olgaç Dündar and B. N. Dündar, J. Clin. Res. Pediatr. Endocrinol., 2014, 6, 192–201 CrossRef PubMed.
- F. Savino, E. Petrucci and G. Nanni, Acta Paediatr., 2008, 97, 701–705 CrossRef CAS PubMed.
- M. G. Ross and M. Desai, Ann. Nutr. Metab., 2014, 64, 36–44 CrossRef CAS PubMed.
- J. M. Friedman, AJCN, 2009, 89, 973S–979S CAS.
- J. G. Woo, M. L. Guerrero, M. Altaye, G. M. Ruiz-Palacios, L. J. Martin, A. Dubert-Ferrandon, D. S. Newburg and A. L. Morrow, Breastfeed Med., 2009, 4, 101–109 CrossRef PubMed.
- M. Weyermann, H. Brenner and D. Rothenbacher, Epidemiology, 2007, 18, 722–729 CrossRef PubMed.
- D. Chan, S. Goruk, A. B. Becker, P. Subbarao, P. J. Mandhane, S. E. Turvey, D. Lefebvre, M. R. Sears, C. J. Field and M. B. Azad, IJO, 2018, 42, 36–43 CAS.
- F. K. Uysal, E. E. Onal, Y. Z. Aral, B. Adam, U. Dilmen and Y. Ardiçolu, Clin. Nutr., 2002, 21, 157–160 CrossRef CAS PubMed.
- J. R. Moran, M. E. Courtney, D. N. Orth, R. Vaughan, S. Coy, C. D. Mount, B. J. Sherrell and H. L. Greene, J. Pediatr., 1983, 103, 402–405 CrossRef CAS PubMed.
- B. Dvorak, C. C. Fituch, C. S. Williams, N. M. Hurst and R. J. Schanler, Pediatr. Res., 2003, 54, 15–19 CrossRef CAS PubMed.
- L. C. Read, F. M. Upton, G. L. Francis, J. C. Wallace, G. W. Dahlenberg and F. John Ballard, Pediatr. Res., 1984, 18, 133–139 CrossRef CAS PubMed.
- C. J. Chang and J. C. Chao, J. Pediatr. Gastroenterol. Nutr., 2002, 34, 394–401 CAS.
- L. Khailova, K. Dvorak, K. M. Arganbright, C. S. Williams, M. D. Halpern and B. Dvorak, Pediatr. Res., 2009, 66, 140–144 CrossRef CAS PubMed.
- A. S. Tarnawski, Front. Biosci., 1999, 4, d303–d309 CrossRef PubMed.
- J. Feng, O. N. El-Assal and G. E. Besner, J. Pediatr. Surg., 2006, 41, 742–747 CrossRef PubMed.
- A. Radulescu, H.-Y. Zhang, C.-L. Chen, Y. Chen, Y. Zhou, X. Yu, I. Otabor, J. K. Olson and G. E. Besner, J. Surg. Res., 2011, 171, 540–550 CrossRef CAS PubMed.
- N. Tanaka, M. Sasahara, M. Ohno, S. Higashiyama, Y. Hayase and M. Shimada, Brain Res., 1999, 827, 130–138 CrossRef CAS PubMed.
- S. B. Pillai, M. A. Turman and G. E. Besner, J. Pediatr. Surg., 1998, 33, 973–978 CrossRef CAS PubMed.
- M. P. Michalsky, A. Kuhn, V. Mehta and G. E. Besner, J. Pediatr. Surg., 2001, 36, 1130–1135 CrossRef CAS PubMed.
- M. A. Kuhn, G. Xia, V. B. Mehta, S. Glenn, M. P. Michalsky and G. E. Besner, Antioxid. Redox Signal, 2002, 4, 639–646 CrossRef CAS PubMed.
- T. Ozgurtas, I. Aydin, O. Turan, E. Koc, I. M. Hirfanoglu, C. H. Acikel, M. Akyol and M. K. Erbil, Cytokine, 2010, 50, 192–194 CrossRef CAS PubMed.
- A. Loui, E. Eilers, E. Strauss, A. Pohl-Schickinger, M. Obladen and P. Koehne, J. Hum. Lact., 2012, 28, 522–528 CrossRef PubMed.
- C. Hirai, H. Ichiba, M. Saito, H. Shintaku, T. Yamano and S. Kusuda, J. Pediatr. Gastroenterol. Nutr., 2002, 34, 524–528 Search PubMed.
- S. Indumathi, M. Dhanasekaran, J. S. Rajkumar and D. Sudarsanam, Cytotechnology, 2013, 65, 385–393 CrossRef CAS PubMed.
- J. D. Reynolds, Paediatr. Drugs, 2001, 3, 263–272 CrossRef CAS PubMed.
- A. DiBiasie, Neonatal Netw., 2006, 25, 393–403 CrossRef PubMed.
- K. Hoshimoto and T. Ohkura, Br. J. Biomed. Sci., 2000, 57, 215–217 CAS.
- H. Funakoshi and T. Nakamura, Clin. Chim. Acta, 2003, 327, 1–23 CrossRef CAS PubMed.
- H. Itoh, A. Itakura, O. Kurauchi, M. Okamura, H. Nakamura and S. Mizutani, Horm. Metab. Res., 2002, 34, 16–20 CrossRef CAS PubMed.
- S. R. Milsom, W. F. Blum and A. J. Gunn, Horm. Res., 2008, 69, 307–311 CAS.
- C. G. Prosser, J. Mammary Gland Biol. Neoplasia, 1996, 1, 297–306 CrossRef CAS PubMed.
- T. J. Povsic, T. A. Kohout and R. J. Lefkowitz, J. Biol. Chem., 2003, 278, 51334–51339 CrossRef CAS PubMed.
- L. Van Landeghem, M. A. Santoro, A. T. Mah, A. E. Krebs, J. J. Dehmer, K. K. McNaughton, M. A. Helmrath, S. T. Magness and P. K. Lund, Faseb. J., 2015, 29, 2828–2842 CrossRef CAS PubMed.
- P. J. Kling, K. M. Taing, B. Dvorak, S. S. Woodward and A. F. Philipps, Growth Factors, 2006, 24, 218–223 CrossRef CAS PubMed.
- F. Tian, G. R. Liu, N. Li and G. Yuan, Eur. Rev. Med. Pharmacol. Sci., 2017, 21, 4711–4719 CAS.
- W. E. Corpeleijn, I. Van Vliet, D. A. H. De Gast-Bakker, S. R. Van Der Schoor, M. S. Alles, M. Hoijer, D. Tibboel and J. B. Van Goudoever, JPGN, 2008, 46, 184–190 CAS.
- O. H. Lee, S. K. Bae, M. H. Bae, Y. M. Lee, E. J. Moon, H. J. Cha, Y. G. Kwon and K. W. Kim, Br. J. Cancer, 2000, 82, 385–391 CrossRef CAS PubMed.
- S. Shigematsu, K. Yamauchi, K. Nakajima, S. Iijima, T. Aizawa and K. Hashizume, Endocr. J., 1999, 46, S59–S62 CrossRef CAS PubMed.
- A. M. Ismail, G. M. Babers and M. A. El Rehany, Breastfeed Med., 2015, 10, 277–282 CrossRef PubMed.
- W. Boesmans, P. Gomes, J. Janssens, J. Tack and P. V. Berghe, Gut, 2008, 57, 314–322 CrossRef CAS PubMed.
- M. P. Sánchez, I. Silos-Santiago, J. Frisén, B. He, S. A. Lira and M. Barbacid, Nature, 1996, 382, 70–73 CrossRef PubMed.
- M. Fichter, M. Klotz, D. L. Hirschberg, B. Waldura, O. Schofer, S. Ehnert, L. K. Schwarz, C. V. Ginneken and K. H. Schäfer, Mol. Nutr. Food Res., 2011, 55, 1592–1596 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |