Open Access Article
Open Access ArticleSubterranean marvels: microbial communities in caves and underground mines and their promise for natural product discovery
Paris S.
Salazar-Hamm
 a,
Frances E.
Homan
b,
Shyleigh A.
Good
b,
Jennifer J. M.
Hathaway
a,
Ashley E.
Clements
b,
Evelyn G.
Haugh
b and
Lindsay K.
Caesar
a,
Frances E.
Homan
b,
Shyleigh A.
Good
b,
Jennifer J. M.
Hathaway
a,
Ashley E.
Clements
b,
Evelyn G.
Haugh
b and
Lindsay K.
Caesar
 *b
*b
aDepartment of Biology, University of New Mexico, Albuquerque, NM, USA
bDepartment of Chemistry and Biochemistry, James Madison University, Harrisonburg, VA, USA. E-mail: caesarlk@jmu.edu
First published on 14th February 2025
Abstract
Covering: 2014 to 2024
Since the dawn of human history, caves have played an intimate role in our existence. From our earliest ancestors seeking shelter from the elements to more recent generations harnessing cave substances for medicinal purposes, caves have served as essential resources and havens. The last 40 years of geomicrobiology research has replaced the outdated perception of subterranean environments as lifeless and unchanging with the realization that vibrant microbial communities have adapted to thrive in extreme conditions over millions of years. The ability of subterranean microbial communities to withstand nutrient deprivation and darkness creates a unique reservoir of untapped biosynthetic potential. These communities offer exciting prospects for medicine (e.g., antimicrobial and antitumor therapies) and biotechnology (e.g., redox chemical properties and biomineralization). This article highlights the significance of caves and mines as reservoirs of microbial diversity, the potential impact of their bioactive compounds on the fields of healthcare and biotechnology, and the significant challenges that must be overcome to access and harness the biotechnological potential of subterranean microbial communities. Additionally, it emphasizes the conservation efforts needed to protect these delicate ecosystems, ensuring the preservation of both ancient traditions and tomorrow's medicines.
1. Introduction
Subterranean environments have long held a significant place in human history. Caves have both served as shelters for our earliest ancestors and repositories of traditional knowledge. As early as 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 years ago, humans began excavating their own subterranean environments, creating man-made mines that have provided essential resources that have shaped societies throughout history.1 This is unsurprising given that around 15% of the ice-free surface of the Earth is karstified and close to 17% of the world's population lives in karst environments.2 Despite the extensive utilization of caves and mines by humans, many subterranean environments remain undisturbed by anthropogenic activities and represent pristine environments with remarkable biodiversity. Beneath their seemingly unchanging façade, subterranean environments host intricate microbial communities that have evolved to thrive in extreme oligotrophic conditions over millions of years. The adaptations of these cave-dwelling microorganisms to survive under extreme darkness and nutrient scarcity are made possible, at least in part, through the development of specialized metabolic pathways encoding natural product molecules with diverse biological activities.
000 years ago, humans began excavating their own subterranean environments, creating man-made mines that have provided essential resources that have shaped societies throughout history.1 This is unsurprising given that around 15% of the ice-free surface of the Earth is karstified and close to 17% of the world's population lives in karst environments.2 Despite the extensive utilization of caves and mines by humans, many subterranean environments remain undisturbed by anthropogenic activities and represent pristine environments with remarkable biodiversity. Beneath their seemingly unchanging façade, subterranean environments host intricate microbial communities that have evolved to thrive in extreme oligotrophic conditions over millions of years. The adaptations of these cave-dwelling microorganisms to survive under extreme darkness and nutrient scarcity are made possible, at least in part, through the development of specialized metabolic pathways encoding natural product molecules with diverse biological activities.
In recent years, a number of notable review papers have been published on the topic of microbial communities in caves and their biotechnological applications.3–7 These reviews provide important background information on the medicinal and biotechnological properties of prokaryotic communities, primarily actinomycetes, in caves, and we encourage the interested reader to explore these manuscripts for additional commentary on the subject. Our review concentrates on the underground communities in terrestrial settings (i.e., caves and mines). While previous reviews have primarily focused on bacteria, this review also covers studies of fungal communities, including bioactive cave-dwelling fungi and the devastating pathogen Pseudogymnoascus destructans. We highlight the challenges associated with accessing the untapped bio- and chemodiversity of underground systems, with particular emphasis on cultivation techniques for maximizing microbial diversity as well as strategies to “turn on” cryptic biosynthetic gene clusters in laboratory settings. Finally, we emphasize the importance of cave conservation and environmental stewardship, highlighting the unfortunate negative impacts of mining and tourism and how to sustainably access fragile communities for the discovery of bioactive metabolites.
2. Subterranean ecosystems
Subterranean ecosystems are comprised of distinct ecological zones ranging from the illuminated entrance to the pitch-black deep interior that have profound effects on both natural ecological communities and human culture. This section provides a detailed examination of the structure of subterranean ecosystems, specifically terrestrial caves, as well as the ways in which ancient peoples engaged with these unique environments. By investigating the roles of caves in shaping human practices and beliefs, we uncover the deep cultural connections that have persisted throughout human history.2.1 The anatomy of caves
Cave systems consist of a network of subterranean, interconnected, oligotrophic environments that are characterized by high humidity, low consistent temperatures, and limited organic nutrients.8 These environments can form under a variety of geological settings including volcanic, limestone, granite, gypsum, mud, marble, glacial, and boulderous, with most formations occurring by water erosion.8 The three most well-studied cave types are epigenic limestone caves (formed by water-driven dissolution of limestone), hypogenic sulfuric acid caves (formed from the bottom up by sulfuric acid), and lava tubes (formed from the cooling, evacuation, and crusting of the lava drainage channel).9 Because caves are largely isolated from surface energy input, they are considered nutrient-limited environments. The level of nutrients and organic matter is largely influenced by mineralogical composition, tourism, animal activity, runoff, and drip water.10Caves habitats can be divided into four distinct zones: the entrance zone, the twilight zone, the transition zone, and the deep interior (Fig. 1). The entrance zone is where the surface and subterranean environments intersect. This is followed by the twilight zone which is characterized by variable temperatures and low levels of light. While plants and other autotrophic organisms struggle to grow in the twilight zone, photosynthetic organisms such as lichens and algae have the capacity to grow here as well as other heterotrophic and mixotrophic organisms. The transition zone of the cave experiences complete darkness, but still has some temperature variability. The deep interior of caves is characterized by complete darkness, relatively constant temperature, high humidity, and fixed CO2 pressure regardless of surface conditions. This section of the cave is of particular interest because of the highly specialized microbes capable of surviving in the extreme nutrient-limited environments using energy from the surrounding rocks, infiltrating water, and air.5,11,12 While the microbial communities of the entrance and twilight zones are highly mediated by outside forces, such as the movement of water, wind, soils, and animals, a high barrier of dispersion has largely preserved the transition and deep interior zones, resulting in unique microbial and metabolic diversity.
 | ||
| Fig. 1 Schematic representation of cave zones. Created in BioRender. Caesar, L. (2025) https://BioRender.com/q65d669. | ||
2.2 Human history in caves: folk medicines and cultural significance
The association between humans and caves is so profound that our earliest human ancestors are commonly referred to as ‘cavemen’. Indeed, there is considerable anthropologic evidence that prehistoric people, including now-extinct relatives of modern humans (e.g., Neanderthals and Cro-Magnon), found physical protection in caves.13–16 The first evidence of early Neanderthals was found in a cave in Germany,14 and additional evidence of Neanderthal-like skulls found inside Spy Cave in Belgium have provided key insights into when Neanderthal populations disappeared from Eurasia over 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 years ago.16 Although caves were utilized extensively as shelters, they were appreciated for more than mere physical protection. Some viewed caves as spiritual portals, uncontaminated from the outside world. Human remains suggest caves served as burial sites for some Indigenous communities.17,18 Numerous artifacts have also been recovered from caves, for example, ceramic vessels coated in copal incense residue from the caves of Naj Tunic,19 indicating that Indigenous peoples including the Mayans conducted ritual events in them.17,18 One of the most widely performed ritual events in caves was the rite of passage, which was required for Indigenous males to enter adulthood.9,10 Now, show caves host more than 150 million visitors worldwide;20 thus they are considered places with great geoheritage significance.21
000 years ago.16 Although caves were utilized extensively as shelters, they were appreciated for more than mere physical protection. Some viewed caves as spiritual portals, uncontaminated from the outside world. Human remains suggest caves served as burial sites for some Indigenous communities.17,18 Numerous artifacts have also been recovered from caves, for example, ceramic vessels coated in copal incense residue from the caves of Naj Tunic,19 indicating that Indigenous peoples including the Mayans conducted ritual events in them.17,18 One of the most widely performed ritual events in caves was the rite of passage, which was required for Indigenous males to enter adulthood.9,10 Now, show caves host more than 150 million visitors worldwide;20 thus they are considered places with great geoheritage significance.21
Cave walls have long held stories of ancient civilizations including depictions of animals, handprints, and geometric patterns,9 and many revere prehistoric caves as the world's first museums.15 Gypsum, a soft sulfate mineral within caves, was used by some Indigenous groups for paint, as evidenced by Mammoth and Salt caves in central Kentucky (USA).18 One of the most famous caves for paleolithic art is Lascaux Cave in Montignac, France, containing over 600 paintings dating between 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–18
000–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 years ago.15 The majority of paintings in Lascaux depict bison, aurochs, and horses. Interestingly, anthropomorphic images in Western European paleolithic cave art are exceedingly rare and simplistic, usually located in the deepest galleries among dense concentrations of drawings. In fact, the only known anthropomorphic image in Lascaux, a simple stick figure with a bird-like head, is hidden at the bottom of a hard-to-reach well.22
000 years ago.15 The majority of paintings in Lascaux depict bison, aurochs, and horses. Interestingly, anthropomorphic images in Western European paleolithic cave art are exceedingly rare and simplistic, usually located in the deepest galleries among dense concentrations of drawings. In fact, the only known anthropomorphic image in Lascaux, a simple stick figure with a bird-like head, is hidden at the bottom of a hard-to-reach well.22
In addition to their spiritual and artistic uses, caves have a long-intertwined history with medicine. For example, the presence of indigenous artifacts and preserved human fecal matter near deposits of selenite and mirabilite suggest these minerals may have been consumed for their laxative effects.17,18 Salt caves have been used to treat respiratory illnesses due to their high humidity and presence of anti-inflammatory ions (e.g., Ca2+, Mg2+, and I−), a practice known as speleotherapy.23 This efficacy of speleotherapy is in part supported by a small study of 22 participants who stayed at “Wieliczka” Salt Mine Resort where the health of all participants significantly improved after three weeks.23 However, the most famous and widely utilized medicinal cave substance is moonmilk. Moonmilk is a viscous white substance primarily composed of CaCO3 that forms small pools in caves when hydrated. Etruscans and Romans used moonmilk as an emollient to induce lactation, while followers of Christianity revered moonmilk as blessings from angels (first century to present day).24 European peasants applied moonmilk on their livestock between the 11th and 15th centuries, realizing that moonmilk healed wounds at an exponentially faster rate than letting the wound resolve on its own. Because of these healing powers, they believed that moonmilk was created by supernatural entities such as gnomes.25 When Conrad Gesner published a document about the healing properties of moonmilk in 1555, moonmilk became more prominent in the pharmaceutical industry.11 Pharmacies prescribed and sold moonmilk to treat heartburn until the early 19th century.24 While the diverse microbial communities inhabiting moonmilk are influenced by microclimatic conditions including temperature and CO2 availability,26 the healing properties of moonmilk likely stem from the bioactive compounds produced by symbiotic Streptomyces species which are involved in the biomineralization of calcite in these unique formations.5 Moonmilk-associated Streptomyces strains have exhibited antibacterial and antifungal properties, although only a few bioactive compounds have been identified (discussed in Sections 4.1 and 5.1).5,27
3. Microbial diversity in subterranean environments
It is with the utmost difficulty that we attempt to describe microbial diversity in subterranean systems. The definition of subterranean environments is quite broad with differences in geology, climate, and organic inputs that can have immense influences on microbial diversity. Although the diminished light and nutrients are constraints, they have also created unique conditions for convergent evolution to subterranean environments. Microbial species richness and diversity generally decreases with increasing distance from the aboveground entrance, likely tied to available biomass;28 however, there have been instances reported of high biodiversity and richness even compared to regional surface environments.29 Microbes are essential members of these communities playing important roles in primary production, decomposition, and biogeochemical processing. Here, we discuss the biotic and abiotic factors that shape subterranean microbial communities. We focus on bacterial and fungal diversity because of their importance in natural product production; nonetheless, we also acknowledge that archaea, cyanobacteria, and microalgae are present in subterranean systems with notable roles.30–32 For example, archaea contribute greatly to, and possibly modulate, nitrogen cycling in caves.33 However, the role of these groups in natural product production is scarce at best.3.1 Bacterial diversity
Microbial research of subterranean environments has largely focused on bacteria. This is both because of their wide-ranging metabolic capabilities allowing them to thrive in nutrient-limited environments but also their roles in sustaining these ecosystems. For example, a study of karst caves in southwest China leveraged over 200 bacterial genomes and eight metagenomic datasets, identifying genes involved in carbon, nitrogen, and sulfur metabolism essential for biogeochemical cycling.34 Chemolithotrophs contribute to mineral weathering and formation of secondary structures in caves by breaking down inorganic compounds in the rocks and walls for energy. The chemical composition of the cave bedrock modulates the diversity and metabolic responses of these cave bacteria.35 For example, ferromanganese deposits deep in Lechuguilla Cave (New Mexico, USA) were in part produced by manganese and iron-oxidizing bacteria.36,37 Microbial biofilms are recognized to aid in cave pearl formation,38 and bacteria-induced carbonate precipitation is important for formation of soda straw and popcorn speleothems.39Early cultivation from the 1900s revealed that caves contained undescribed bacteria different from aboveground soil communities.40,41 As with other ecosystems, the development of DNA-based sequencing has allowed for culture-independent studies revealing that the cave microbiome is more complex than a few species of highly specialized bacteria.42,43 In fact, recent studies demonstrate that cave communities are still largely undescribed. Research from Hawaiian lava caves (USA) and fumaroles found that ∼70% of the taxa found could not be classified at lower taxonomic levels.44 Similar results have been found in karst caves where up to 19% of the sequences recovered belonged to unclassified phyla.45 The percentage of sequences belonging to unknown taxa was >90% in the Sukinda chromite mine in India.46
Caves across the world are inhabited by members of the phyla Pseudomonadota, Actinomycetota, and Acidobacteriota.42,47,48 However, the abundance of these groups shifts based on geological history of caves, as illustrated in a recent review of bacterial metabarcoding (16S rRNA) studies of limestone caves, sulfuric acid speleogenetic caves, and volcanic caves.9 While limited in its scope (105 samples from 22 caves), it reveals the distinct differences in bacterial composition and structure of higher taxonomic groups in caves based on rock type.9 Other variables, such as sample type (i.e., air, water, rock, and sediment), native minerals (e.g., carbon, nitrogen, and copper), seasonality, and bat activity, also influence bacterial communities.35,48–51 While the weight of these factors as key determinants of microbial composition and function is not simple, salinity and pH have been highlighted across other ecosystems to particularly affect bacterial constituents.52,53
Bacterial residents of mines similarly are inhabited by diverse members of Pseudomonadota, Actinomycetota, Bacillota, Bacteroidota, which are largely driven by contamination of heavy metals and pH.54 Effluents draining from abandoned mines, known as acid mine drainage, are environmental pollutants due to their high acidity and presence of dissolved metals (e.g., iron, manganese, copper, nickel, and zinc). Chen et al.55 documented a shift in bacterial community composition correlating to pH of an acid mine drainage, specifically by members within Pseudomonadota and Nitrospirota. Bioprospecting efforts have targeted heavy metal tolerant bacteria in mines across Europe,54 and this may be an avenue worth continued investigation for bioremediation potential.
Taxa within Actinomycetota are estimated to produce over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bioactive compounds of which 70–80% are produced by members of the genus Streptomyces.56 Actinomycetota are renowned for their cellular and metabolic versatility, allowing for specialization and emerging speciation in subterranean environments.57–63 Volcanic caves in particular have been praised for their high diversity of Actinomycetota and their bioactive metabolites (discussed in Sections 5.1 and 5.2).64–66 While many species are harmless to humans, a few actinomycetes from caves can opportunistically cause infections including Nocardiopsis dassonvillei, which can cause a subcutaneous skin infection (actinomycetoma), and Inquilinus limosus, which has been associated with cystic fibrosis.67
000 bioactive compounds of which 70–80% are produced by members of the genus Streptomyces.56 Actinomycetota are renowned for their cellular and metabolic versatility, allowing for specialization and emerging speciation in subterranean environments.57–63 Volcanic caves in particular have been praised for their high diversity of Actinomycetota and their bioactive metabolites (discussed in Sections 5.1 and 5.2).64–66 While many species are harmless to humans, a few actinomycetes from caves can opportunistically cause infections including Nocardiopsis dassonvillei, which can cause a subcutaneous skin infection (actinomycetoma), and Inquilinus limosus, which has been associated with cystic fibrosis.67
Pseudomonadota, among the most dominant taxa in caves, vary in abundance and distribution within and between cave systems. For example, Gammaproteobacteria, mainly represented by the genus Pseudomonas, have been shown to be particularly abundant in tourist caves.68 Conversely, the genera Sphingomonas, Lysobacter and Polaromonas were more prevalent in pristine caves.68 Species of Pseudomonas isolated from caves have been documented for their antibacterial and antifungal activities65,69–74 (discussed in Sections 5.1 and 5.2). Rare members within this phylum are human pathogens, including Aurantimonas altamirensis, which was first described from Altamira Cave in Spain75 and was subsequently connected to nosocomial infections of cystic fibrosis patients.76
Acidobacteriota is a highly diverse phylum found across multiple ecosystems;77 however, they are poorly understood due to the difficulty of isolating them in culture.78 Acidobacteriota appear to be prominent in hydrogen sulfide rich environments, including the springs of Lower Kane Cave (Wyoming, USA)79 and cave wall biofilms of Frasassi cave system (Italy).80 This group requires further investigation for their metabolic and functional capabilities, but members may be of biotechnological interest. For example, members of the genus Blastocatella, found in moonmilk of both carbonate81 and lava caves,82 have been shown to contribute to ammonium removal in wastewater.83
3.2 Fungal diversity
Fungi that can persist in subterranean environments generally function as parasites or decomposers and play important roles in biogeochemical cycling of carbon, nitrogen, phosphorus, iron, manganese, and sulfur. Fungi can also participate in speleogenesis including the formation of needle fiber calcite.84–86 Endolithic species penetrate the rock substratum, which can cause biological weathering of rock surfaces, but also can contribute to stabilization and preservation over time.87 Due to limited organic matter in most subterranean environments, fungal richness tends to be lower than bacterial richness.49,88 Despite this, there have been over 1000 species of fungi reported from karst caves alone,89 and this is likely an underestimate of cave-associated fungi given the exclusion of glacier, lava, and sea caves. Fungal surveys of caves consistently report dominance of members within the phylum of Ascomycota followed by members of Basidiomycota and Mucoromycota.89,90 Fungi (especially within the phylum Ascomycota) thrive in anthropogenically impacted caves with additional human-mediated inputs of organic matter (discussed in Section 3.3.2).91 Like humans, bats also serve as vectors transporting environmental fungi into caves;92 however, transient spores may also be introduced by drip water or air currents. Natural sources of subterranean organic materials preferred by fungi include bat guano93–95 and cave insects or arachnids.96–100 Bat and arthropod populations likely have some influence on cave fungal community composition, but it fluctuates between caves and cave structures.49,101 Significant variability between fungal communities has been observed between caves of one system, locations within a cave, and sample type.28,102,103Subterranean mines offer similar conditions to caves, sometimes with the additional factor of heavy metals. Coal mines have been frequently used for timber storage in the USA and Europe, resulting in the dominance of wood decaying fungi, mainly within the phylum Basidiomycota.104 The Soudan Mine in Tower, Minnesota (USA) has both high iron-ore concentrations as well as an abundance of wood that remained from mining activities, supporting several phylogenetically distinct fungal species,105 some of which have been recently explored for bioactive potential (discussed in Section 5.2).106,107 Fungi tolerant of heavy metals and metalloid compounds isolated from subterranean mines may be useful for bioremediation. Armillaria rhizomorphs observed in Soudan Mine105 as well as Champion Mine, a copper rich mine in Michigan (USA),108 are suspected to play a role in absorption of metal ions and protection.109Trichoderma harzianum isolated from Libiola Mine (Italy) demonstrated the highest efficiency of the native fungal population for silver bioaccumulation.110Trichoderma virens and several members of Penicillium (P. griseopurpureum, P. janthinellum, P. canescens, and P. soppii) cultured from the soils of Pestarena gold mine (Italy) were tolerant to arsenic levels of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1.111
000 mg L−1.111
Despite a growing body of research, subterranean environments remain one of the largely underexplored areas on this planet. Notorious pathogens such as Histoplasma capsulatum, causing human histoplasmosis, and Pseudogymnoascus destructans, causing white-nose syndrome (WNS) (discussed in Section 3.3.3), have created a somewhat cynical view of cave-associated fungi. However, caves and mines offer copious opportunities for discovery of novel fungal diversity112–117 and fungal-derived natural products with both medicinal and biotechnological applications (discussed in Section 5).
3.3 Factors influencing microbial diversity in subterranean environments
Subterranean ecosystems present a diverse range of conditions that significantly influence the microbial life inhabiting them. Here, we discuss the impacts of harsh conditions on the development of specialized niches that support unique microbial communities adapted to thrive in extreme environments, as well as the impact of mining and tourism industries on microbial diversity through the introduction of non-native species and alteration of otherwise stable environmental conditions. We address the impact of WNS on bat populations and its broader implications for cave ecosystems. Understanding these factors is crucial to understand the complexity and fragility of subterranean microbial ecosystems and to develop strategies to protect and study these habitats.While our review primarily focuses on terrestrial environments, it is worth mentioning that anchialine ecosystems, consisting of both microeukaryotic and prokaryotic communities, represent reservoirs of new biodiversity anticipated to host unique biometabolic activity.129,130 Such ecosystems are unique in their anoxic conditions, which vary based on ocean depth, and their distinct salinity, temperature, and pH zones that influence species distribution.131
Finally, the unique conditions of caves may offer insight into microbial life beyond planet Earth. Subsurface environments on Mars hold particular promise for astrobiology given their protection from extreme winds and ultraviolet, cosmic, and solar ionizing radiation.132 Identifying Martian caves through remote sensing is challenging due to poor visibility and unknown structural integrity. Analogous formations on Earth, including lava tubes and basaltic caves, may provide insights into volcanic terrain on Mars, allowing for development of exploration strategies to investigate Martian subsurface environments.82,133,134
In some delicate cases, subterranean artifacts have been disrupted or destroyed by even brief human contact. For example, 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 years-old cave paintings in Lascaux Cave (Montignac, France) were contaminated with algae, bacteria, and fungi contributed by human visitation that introduced humidity, warmth, and light.84 Aerosolized bacterial and fungal spores, in part attributed to human activities, have also contributed to biodeterioration of paleolithic paintings in the Cave of Altamira (Cantabria, Spain).148 Castañar Cave (Cáceres, Spain), notable for its spectacular mineral morphologies including aragonite and calcite speleothems, has suffered from two human-mediated fungal outbreaks.149,150 The different nature of organic carbon introduced into Castañar Cave in 2008 (human vomit) and 2021 (environmental debris transported by construction workers) provoked unique disturbances and ecological changes to delicate cave fungal communities.151 Within caves, deep interior zones may be the most fragile because total organic carbon concentrations are less than 2 mg L−1,152 making even cursory human interactions risky. While education and exploration can be worthwhile ventures, efforts should be taken to minimize human impacts and preserve the natural conditions of cave systems.153
000 years-old cave paintings in Lascaux Cave (Montignac, France) were contaminated with algae, bacteria, and fungi contributed by human visitation that introduced humidity, warmth, and light.84 Aerosolized bacterial and fungal spores, in part attributed to human activities, have also contributed to biodeterioration of paleolithic paintings in the Cave of Altamira (Cantabria, Spain).148 Castañar Cave (Cáceres, Spain), notable for its spectacular mineral morphologies including aragonite and calcite speleothems, has suffered from two human-mediated fungal outbreaks.149,150 The different nature of organic carbon introduced into Castañar Cave in 2008 (human vomit) and 2021 (environmental debris transported by construction workers) provoked unique disturbances and ecological changes to delicate cave fungal communities.151 Within caves, deep interior zones may be the most fragile because total organic carbon concentrations are less than 2 mg L−1,152 making even cursory human interactions risky. While education and exploration can be worthwhile ventures, efforts should be taken to minimize human impacts and preserve the natural conditions of cave systems.153
4. Natural products discovered from subterranean environments
In the last decade, exploration of natural products derived from subterranean environments has significantly advanced our understanding of subterranean microbiota. In this section, we review the diverse natural products discovered from cave- and mine-dwelling bacteria and fungi between 2014–2024 and examine the methodologies utilized in their discovery. While most compounds have been discovered using traditional bioactivity- or taxonomy-directed approaches, the potential of omics-guided strategies is high and remains underutilized.4.1 Examples of natural products
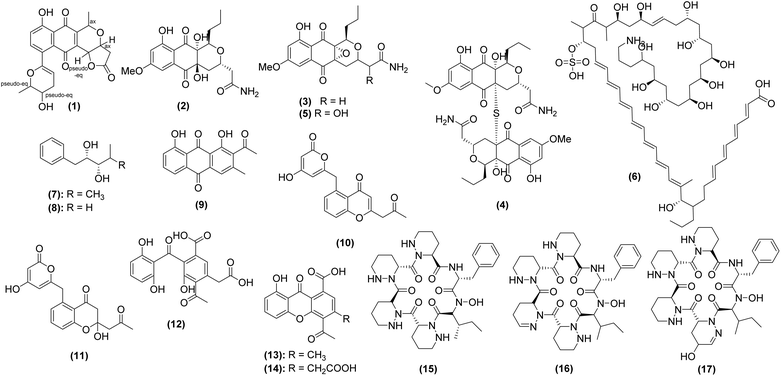
Prior to 2014, only eight compounds were discovered and characterized from caves, and these are covered in previous reviews.4–6 Searches in PubMed, Google Scholar, Scopus, and ScienceDirect resulted in 17 articles that describe the isolation and/or characterization of 90 polyketides, peptides, terpenoids, and hybrid molecules (Tables 1 and 2). Of the compounds, 50 (56%) are derived from bacteria (Fig. 2 and 3) and 40 (44%) were of fungal origin (Fig. 4 and 5). Thirty-seven (41%) demonstrated one or more bioactivities (discussed in Section 5). Thirty-four (38%) compounds were novel, having only been discovered from subterranean environments (Fig. 2 and 4).| No. | Compound name | Strain name | Cave of origin | Analytical technique | Ref. |
|---|---|---|---|---|---|
| a Compounds were identified by matching accurate masses to natural products databases. Without MS/MS fragmentation patterns or NMR spectra, these compounds should be considered putative. | |||||
| Novel compounds | |||||
| 1 | Xiakemycin A | Streptomyces sp. CC8-201 | Chongqing City, China | NMR | 171 |
| 2 | Hypogeamicin A | Nonomuraea specus | Hardin's cave system, Ashland City, Tennessee, USA | 172 and 173 | |
| 3 | Hypogeamicin B | ||||
| 4 | Hypogeamicin C | ||||
| 5 | Hypogeamicin D | ||||
| 6 | Funisamine | Streptosporangium sp. KDCAGE35 | Various cave systems, Tennessee, USA | 172 | |
| 7 | (2S, 3S, 4S)-4-methyl-1-phenylhexane-2,3-diol | Streptomyces sp. CB09001 | Karstic cave in Xiangxi, China | 174 | |
| 8 | (2S, 3S)-4-methyl-1-phenylpentane-2,3-diol | ||||
| 9 | Huanglongmycin A | 175 | |||
| 10 | Huanglongmycin B | ||||
| 11 | Huanglongmycin C | ||||
| 12 | Huanglongmycin D | 176 | |||
| 13 | Huanglongmycin E | ||||
| 14 | Huanglongmycin F | ||||
| 15 | Lunaemycin A | Streptomyces lunaelactis MM109 | Grotte des Collemboles, Comblain-au-Pont, Belgium | 177 | |
| 16 | Lunaemycin B1 | ||||
| 17 | Lunaemycin D | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Known compounds | |||||
| 18 | Diazepinomicin | Streptomyces sp. | Iron Curtain Cave, Canada | LC-MS/MS | 178 |
| 19 | 14-Deoxychaxalactin B | Streptomyces sp. IB 2014/I/78-8 | Bolshaya Oreshnaya Cave, Siberia | LC-MSa | 179 |
| 20 | Cyclodysidin D | ||||
| 21 | Stylissazole B | ||||
| 22 | Gyrophoric acid | ||||
| 23 | Okicenone | Micromonospora sp. BBHARD22 | Various cave systems, Tennessee, USA | NMR | 172 |
| 24 | Aloesaponarin II | ||||
| 25 | Actinomycin C2 | Streptomyces sp. BCCAGE06 | |||
| 26 | Propeptin 1 | Microbispora sp. BCCAGE54 | |||
| 27 | Propeptin 2 | ||||
| 28 | Tetarimycin B | ||||
| 29 | Xenocyloin B | Streptomyces sp. CB09001 | Karstic cave in Xiangxi, China | 174 | |
| 30 | Xenocyloin C | ||||
| 31 | Xenocyloin D | ||||
| 32 | Lumichrome | ||||
| 33 | Thymidine | ||||
| 34 | Hexadecanamide | Paenibacillus sp. 23TSA30-6 | Krubera-Voronja Cave, Georgia | GC-MS | 180 |
| 35 | Octadecanamide | Paenibacillus spp. 23TSA30-6 and 28ISP30-2 | |||
| 36 | (Z)-Octadec-9-enamide | ||||
| 37 | Cyclic dipeptide cyclo(Pro–Phe) | ||||
| 38 | (1-Methyl-2,2-diphenylcyclopropyl) sulfanylbenzene | Paenibacillus sp. 28ISP30-2 | |||
| 39 | Diisooctyl phthalate | Streptomyces sp. GLD25 | Gueldaman Cave, Akbou-Algeria | 181 | |
| 40 | 6-Hydroxy-heptanoic acid | ||||
| 41 | Hexadecanoic acid | ||||
| 42 | Benzeneacetic acid | ||||
| 43 | 3-(3,5-di-tert-Butyl-4-hydroxyphenyl)propionic acid | ||||
| 44 | Cycloheximide | Streptomyces sp. MM99 | Grotte des Collemboles, Comblain-au-Pont, Belgium | LC-MS/MS | 177 |
| 45 | Dehydrocycloheximide | ||||
| 46 | Ferroverdin A | Streptomyces lunaelactis MM109 | |||
| 47 | Phenazine-1-carboxylic acid | Pseudomonas yamanorum GZD14026 | Bats swabbed in Ge-zi Cave and Temple Cave, China | GC-MS | 182 |
| 48 | Octanoic acid | ||||
| 49 | Isoprenol | ||||
| 50 | 3-tert-Butyl-4-hydroxyanisole | ||||
| No. | Compound name | Strain name | Cave of origin | Ref. |
|---|---|---|---|---|
| Novel compounds | ||||
| 51 | Sulfurasperine A | Aspergillus fumigatus GZWMJZ-152 | Fangjing mountain, Guizhou province, China | 184 |
| 52 | Sulfurasperine B | |||
| 53 | Sulfurasperine C | |||
| 54 | Sulfurasperine D | |||
| 55 | 4-Methoxy-7-methylbenzo[d]thiazole-5,6-diol | |||
| 56 | 2-Hydroxymethyl-4-methoxy-7-methylbenzo[d]thiazole-5,6-diol | |||
| 57 | Pseudoanguillosporoin C | Cadophora sp. 10-5-2 M | Soudan underground iron mine, Minnesota, USA | 106 |
| 58 | Soudanone A | |||
| 59 | Soudanone B | |||
| 60 | Soudanone C | |||
| 61 | Soudanone D | |||
| 62 | Soudanone E | |||
| 63 | Soudanone F | |||
| 64 | Soudanone G | |||
| 65 | Oidiolactone G | Oidiodendron truncatum | 107 | |
| 66 | Epi-oidiolactone G | |||
| 67 | Oidiolactone H | |||
| 68 | Oidiolactone I | |||
| 69 | Oidiolactone J | |||
| 70 | Oidiolactone K | |||
| 71 | Oidiolactone L | |||
| 72 | 5-Chloroparietin | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Known compounds | ||||
| 73 | Sulochrin | Aspergillus fumigatus GZWMJZ-152 | Fangjing mountain, Guizhou province, China | 184 |
| 74 | Monomethylsulochrin | |||
| 75 | 3-Hydroxy-5-methoxy-2-methylbenzoquinone | |||
| 76 | Pseudoangillosporin A | Cadophora sp. 10-5-2 M | Soudan underground iron mine, Minnesota, USA | 106 |
| 77 | Nectriapyrone | |||
| 78 | Isosclerone | |||
| 79 | 3,8-Dihydroxy-3-hydroxymethyl-6-methoxy-4,5-dimethylisochroman-1-one | |||
| 80 | 7-Hydroxy-3-(1-hydroxyethyl)-5-methoxy-3,4-dimethylisobenzofuran-1(3H)-one | |||
| 81 | 3-Acetyl-7-hydroxy-5-methoxy-3,4-dimethylisobenzofuran-1(3H)-one | |||
| 82 | PR 1388 | Oidiodendron truncatum | 107 | |
| 83 | Oidiolactone C | |||
| 84 | Oidiolactone D | |||
| 85 | Oidiolactone E | |||
| 86 | Oidiodendronic acid | |||
| 87 | LL-Z1271α | |||
| 88 | LL-Z1271β | |||
| 89 | Physcion | |||
| 90 | Emodin | |||
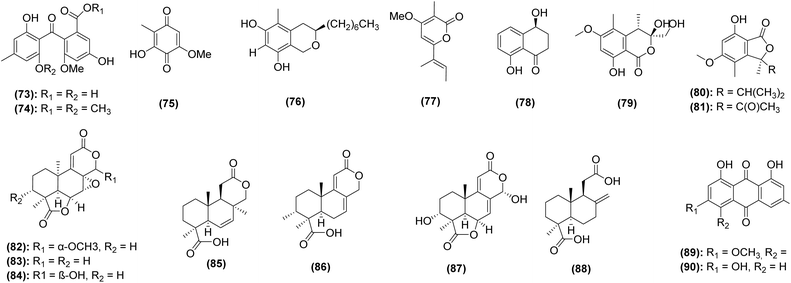
Streptomyces are the most prominent source of natural products discovered from caves in the last decade. Thirty-one (34%) of the compounds were from Streptomyces, which also represented 62% of the bacterial-derived compounds. Of the 19 strains whose natural products have been investigated since 2014, 16 are bacterial and only 3 are fungal, illustrating that fungi are underrepresented in natural products studies of subterranean environments. Further, of the 553 natural products derived from fungi published in 2023, none were reported from caves.183 The importance of exploring this under-researched niche is emphasized by the fact that the three subterranean fungal strains in this review yielded 40 total natural products (44% of total compounds), of which more than half were novel. Interestingly, a strain of Aspergillus fumigatus isolated from cave soil collected near Fanjing Mountain, China, produced six new compounds,184 suggesting that even well-studied species,185 when adapted to cave environments, may possess novel biosynthetic potential.
4.2 Approaches to natural products discovery
Natural products discovery strategies typically fall into one of two categories: traditional and omics-guided approaches. Traditional approaches have been used for nearly 100 years and include prioritization of natural products by bioactivity-guided fractionation and/or taxonomic novelty of their producing organism.186 Omics-guided approaches, while still incorporating some of the steps utilized in traditional approaches, take advantage of genomics, metabolomics, proteomics, and/or transcriptomics datasets to guide natural products discovery efforts.183Although researchers have increasingly turned to omics-guided discovery in the last decade,187 the majority of studies involving natural products discovery from subterranean ecosystems utilized traditional approaches. Indeed, of the 17 total studies conducted between 2014 and 2024, 12 (71%) use traditional approaches.69,106,107,171,173–176,179,181,184,188,189 These studies account for the identification of 72 total compounds (80%), including 30 novel compounds (81% of total novel compounds). Only three studies utilize omics-guided approaches (18%),172,177,180 while two additional studies utilize a combination of traditional and non-traditional approaches (12%).178,182 Hybrid approaches have led to the identification of only three compounds in the last decade (3%, none of them novel), and omics-guided strategies have led to the identification of 15 compounds (17%, four of them novel, accounting for 11% of all novel compounds discovered in this timeframe).
While these numbers may cause one to question the advantages of using non-traditional approaches to natural products discovery, it is worth noting that over 30% of novel bacterial compounds were discovered using omics-guided strategies (from just three studies total), particularly those involving genome mining to evaluate biosynthetic potential177,180 and/or mass spectrometry-based comparative metabolomics to identify target metabolites.172,177,180 Thus far, no studies involving subterranean fungi have utilized hybrid or omics-guided strategies, and incorporating these new approaches could accelerate discovery of novel fungal natural products.
Regardless of approach, researchers must utilize analytical tools including gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS), and/or nuclear magnetic resonance (NMR) to identify natural product molecules. While the majority of studies covered in this review utilized robust identification and dereplication methods, including full structure elucidation using NMR and matching fragmentation spectra of experimental data to those of authentic standards using GC-MS or LC-MS/MS, some studies only utilized accurate masses obtained by LC-MS to those found in natural products databases.179,188 In one such case, authors identified dichloranthrabenzoxocinones as putative bioactive constituents from subterranean Streptomyces spp. by matching experimentally determined accurate masses to those in the Dictionary of Natural Products.188 However, when inspecting the mass spectrometry data, it is clear that the associated ions do not contain the isotopic distribution patterns characteristic of chlorine-containing molecules and that the molecules were misidentified. This case study emphasizes the limitations of simple database matching for dereplication of natural products to avoid incorrect annotation of identified natural products.
5. Biotechnological and medicinal potential of subterranean microorganisms
Extracts from cave microorganisms, and in some cases, purified compounds from them, have been evaluated for numerous biological activities. In this section, we describe antibacterial, antifungal, cytotoxic, antioxidant, anti-inflammatory, and other biological activities of cave microorganisms with potential use in human society. Although most studies evaluating biotechnological potential of cave microbiota do not investigate the chemical constituents responsible for their activity, when possible, known active constituents and structure–activity relationships are described. A summary of in vitro and in vivo bioactivity studies on cave microorganisms can be found in Tables 3–6.5.1 Antibacterial properties
Antibacterial resistance is a global health and economic issue. In 2019, antibacterial resistance was directly attributed to 1.27 million deaths worldwide.190 Drug-resistant strains of Escherichia coli and Staphylococcus aureus are among the World Health Organization's major concerns, contributing to more than 900![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 infections and 230
000 infections and 230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths each year.191 Microbial communities in caves have demonstrated in vitro antibacterial activity against such bacterial pathogens. Since 2014, bacterial isolates from caves have been investigated for bioactivity against methicillin-resistant S. aureus (MRSA) (25 papers) and E. coli (20 papers), among others (Table 3). A compilation of studies investigating the antibacterial potential of subterranean microorganisms since 2014 are provided in Table 3. It is worth noting that most of these studies evaluate antimicrobial activities of strains or strain extracts using cross-streak or disk diffusion assays, and the identity and strength of individual active constituents remains unknown.
000 deaths each year.191 Microbial communities in caves have demonstrated in vitro antibacterial activity against such bacterial pathogens. Since 2014, bacterial isolates from caves have been investigated for bioactivity against methicillin-resistant S. aureus (MRSA) (25 papers) and E. coli (20 papers), among others (Table 3). A compilation of studies investigating the antibacterial potential of subterranean microorganisms since 2014 are provided in Table 3. It is worth noting that most of these studies evaluate antimicrobial activities of strains or strain extracts using cross-streak or disk diffusion assays, and the identity and strength of individual active constituents remains unknown.
| Bioactive strain(s) | Pathogens tested | Bioactive agent(s) | Cave of origin | Ref. |
|---|---|---|---|---|
a Authors identified active constituents as dichloranthrabenzoxocinones using accurate masses and database matching. However, the isotope patterns of the detected ions did not contain the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 isotope pattern characteristic of chlorine-containing molecules, and as such, were likely misidentified.
b Putative bioactive compounds identified by GC-MS analysis of bioactive extracts.
c Putative bioactive compounds identified by presence of biosynthetic gene clusters in the microbial genomes.
d Putative bioactive compounds identified by LC-MS based molecular networking. 1 isotope pattern characteristic of chlorine-containing molecules, and as such, were likely misidentified.
b Putative bioactive compounds identified by GC-MS analysis of bioactive extracts.
c Putative bioactive compounds identified by presence of biosynthetic gene clusters in the microbial genomes.
d Putative bioactive compounds identified by LC-MS based molecular networking.
|
||||
| Aspergillus fumigatus, Trichoderma yunnanense | S. aureus, P. aeruginosa | Not determined | Sthreepura Cave – Kuruwita, Sri Lanka | 193 |
| Streptomyces sp. CC8-201 | S. aureus | Compound 1 | Karst cave in Chongqing City, China | 171 |
| Six strains of Bacillus spp., Rhodococcus sp. P209 | S. aureus | Not determined | Rogers Belmont Cave, Warren County, Virginia, USA | 194 |
| Brevibacterium frigoritolerans, Bacillus thuringiensis, B. weihenstephanensis, B. cereus, Bacillus sp., Pseudomonas sp., Saccharopolyspora erythraea | S. epidermidis, B. subtilis, S. aureus, E. coli | Not determined | Kadıini Cave, Antalya, Turkey | 70 |
| Four Streptomyces spp. and Erwinia sp. | M. luteus, M. smegmatis, ESBL-producing E. coli, S.aureus, A. baumanni | Not determined | Helmcken Falls Cave, Wells Gray Provincial Park, British Columbia | 195 |
| Fictibacillus nanhaiensis, Bacillus humi, B. eiseniae, Pseudomonas mosselii | S. typhi, S. aureus | Not determined | Hindu Kush, India | 71 |
| Nine Streptomyces spp. | B. subtilis, S. carnosus, E. coli, P. putida | Not determined | Badzheyskaya and Okhotnichya caves in Siberia | 196 |
| Toxopsis calypsus, Phormium melanochroun | S. aureus, E. faecalis, E. coli, P. aeruginosa | Not determined | Francthi Cave in Peloponnese, Greece | 197 |
| Streptomyces spp. M4_24 and M5_8 | S. aureus, S. enterica, Enterococcus sp., E. coli, B. subtilis, B. megaterium, B. cereus, P. aeruginosa | Not determineda | Szczelina Chochołowska cave, Tatra mountains, Poland | 188 |
| 11 strains belonging to nine genera (Microbacterium, Arthrobacter, Candidimonas, Dietzia, Pseudarthrobacter, Caulobacter, Delfia, Pseudomonas, Bacillus) | S. aureus, E. coli, E. cloacae, Pseudomonas sp., E. falcium | Not determined | Scarisoara Ice Cave, Romania | 72 |
| Streptomyces sp. GLD22 | E. coli, P. aeruginosa, B. subtilis, B. cereus, S. aureus | 2-tert-Butyl-4,6-bis(3,5-di-tert-butyl-4-hydroxybenzyl)phenol, dibutyl phthalate, Cyclo(leucyloprolyl)b | Gueldaman cave, Algeria | 181 |
| Streptomyces sp. CB09001 | S. aureus, E. coli, K. pneumoniae, P. aeruginosa | Compounds 9–11 | Xiangxi, China | 175 |
| Paenibacillus spp. 23TSA30-6 and 28ISP30-2 w | E. coli, M. luteus, B. thuringiensis, Pseudomonas sp. | Fusaricidins, polymyxins, and tridecaptinsc | Krubera-Voronja Cave, Western Caucasus | 180 |
| Actinomycetota strains GSF102, and GSF201 | B. subtilis, K. pneumoniae | Not determined | Parque Nacional dos Campos Ferruginosos National Park, southeastern Amazon | 198 |
| Five strains belonging to three genera (Pseudomonas, Flavobacterium, Rhodococcus) | E. coli, S. aureus | Not determined | Raspberry rising Cave located in the Columbia mountain range, British Columbia, Canada | 73 |
| Streptomyces sp. GLD25 | P. aeruginosa, E. coli, K. pneumoniae, B. subtilis, B. cereus, S. aureus | Compounds 39–43 | Algeria | 181 |
| Bacillus spp. 1350R2-TSA30-6 and 1410WF1-TSA30-2 | B. cereus, E. faecalis, L. monocytogenes, S. aureus, Rhodococcus sp | Diisobutyl phthalate and pyrrolopyrazinesb | Krubera-Voronja Cave | 199 |
| Paenibacillus polymyxa AC30 and Paenibacillus peoriae AC32 | S. aureus, Salmonella sp., Klebsiella sp., E. coli, P. aeruginosa., Acinetobacter sp. | Not determined | Mossy cave in Summan region, Saudi Arabia | 200 |
| 21 strains belonging to 11 genera (Streptomyces, Psychrobacillus, Lysinbacillus, Cupriavidus, Micromonospora, Fontibacillus, Nonomuraea, Kocuria, Pseudonocardia, Mesorhizobium, Bacillus) | S. aureus, E. faecalis, B. cereus, K. pneumoniae | Not determined | Oceania, Fiji | 201 |
| Five Streptomyces spp. | S. aureus, M. luteus, B. subtilis, E. coli, L. monocytogenes | Not determined | Chaabe Cave, Algeria | 202 |
| 38 strains belonging to ten genera (Agrobacterium, Aerococcus, Bacillus, Kocuria, Lysobacter, Micrococcus, Pseudomonas, Rhodococcus, Sphingomonas, Streptomyces) | E. coli, S. enterica, B. cereus, K. pneumoniae, B. subtilis, S. aureus, L. monocytogenes, S. pseudointermedius | Not determined | Slovenian karst caves | 74 |
| 65 Streptomyces spp., five Bacillus spp., Pseudomonas sp., Nocardia sp., and Erwinia sp. | M. luteus, S. aureus, M. smegmatis, E. coli, A. baumannii, P. aeruginosa, K. pneumoniae | Not determined | Helmcken Falls Cave, Wells Gray Provincial Park, British Columbia | 65 |
| 38 Strains belonging to six families (Streptomycetaceae, Nocardiaceae, Micrococcaceae, Microbacteriaceae, Micromonosporaceae, Pseudonocardiaceae) | S. aureus, B. subtilis, M. luteeaus, K. pneumoniae, E. coli, C. freundii, P. aeruginosa | Not determined | Grotte des Collemboles, Belgium | 192 |
| Streptomyces lunaelactis | K, rhizophila, B. subtilis, S. aureus | Compounds 15–17 | 177 | |
| 28 Streptomyces spp. and three unidentified strains | K. pneumoniae, E. coli, C. freudii, P. aeruginosa, S. aureus, B. subtilis, M. luteus | Not determined | 182 | |
| Micrococcus sp. | S. aureus and S. epidermidis | Azaserine, adefovir, dipivoxil, valclavam and leucomycin A7/A4b | Parsık Cave, Turkey | 203 |
| Two Crossiella spp. | B. cereus, A. baumannii, S. aureus, E. coli, P aeruginosa | Not determined | Six caves in Spain, one of which is Altamira Cave | 47 |
| Two Streptomyces spp. | E. coli, P. aeruginosa, and B. subtilis | Diketopiperazinesd | Iron Curtain Cave, Chilliwack, Canada | 178 |
| 12 Streptomyces spp. and two Arthrobacter spp. | S. typhimurium, E. coli, P. aeruginosa, Proteus sp., L. monocytogenes, L. innocua, S. aureus | Not determined | Two Canadian caves and 12 Portuguese volcanic cave | 204 |
| 16 Actinobacteria from six genera (Streptomyces, Nocardioides, Agromyces, Oerskovia, Micromonospora, and Actinoplanes) | S. aureus, E. coli, B. cinerea | Not determined | Shuanghe Cave, China | 205 |
| 23 Actinobacteria from five genera (Streptomyces, Kocuria, Micromonospora, Saccharomonospora, and Streptosporangium) | M. luteus, E. coli, B. subtilis, S. aureus | Not determined | Hampoeil Cave, Iran | 206 |
| 136 Bacterial isolates, including members of Streptomyces, Micrococcus, Actinobacteria, Actinomycetales, Virgibacillus, and Kocuria genera | S. aureus, P. aeruginosa, E. coli, M. luteus, B. subtilis | Not determined | Pukzing Cave, India | 207 |
Four studies investigated bacterial isolates collected from moonmilk for antibacterial activity.177,182,188,192Streptomyces spp. (M4_24 and M5_8) were collected from the Szczelina Chochołowska Cave in the Tatra Mountains, Poland and evaluated using the cross-streak method.188 Both strains exhibit strong antibacterial activities against Salmonella enterica (inhibition zone: M4_24 = 11.5 mm; M5_8 = 8.0 mm), and M5_8 additionally inhibited E. coli (inhibition zone = 8.5 mm). Bacterial isolates from La Grotte des Collemboles, Belgium were also evaluated using the cross-streak method against a variety of Gram-positive and Gram-negative bacterial pathogens.182,192 The majority of these strains (67.5%) were identified as Streptomyces spp. with varied bioactive potential. While many strains showed greater than 10 mm zones of inhibition against Bacillus subtilis (58%) and Micrococcus luteus (61%) under at least one growth condition, only 13% of tested strains inhibited growth of S. aureus with more than a 10 mm zone of inhibition, with a maximum inhibition zone of 30 mm compared to the 45 mm maximum for both other Gram-positive organisms. Although a good portion of the tested strains showed activity against Klebsiella pneumoniae (45%, maximum inhibition zone of 44 mm), activity against the other Gram-negative organisms was limited, with only 15%, 16%, and 9% of bacterial isolates showing activity (zone of inhibition ≥ 10 mm) against E. coli, Citrobacter freundii, and Pseudomonas aeruginosa, respectively.182,192
The majority of studies investigating the antibacterial properties of cave microbiota do not explore the chemistry behind these bioactivities; however, a small subset of studies have identified the bioactive molecules responsible for the observed activities. For example, researchers studying Streptomyces lunaelactis isolated from moonmilk in La Grotte des Collemboles (Belgium) utilized genomic data from multiple S. lunaelactis strains to identify biosynthetic gene cluster (BGC) sequences that were not conserved across the species. They overlaid this data with LC-MS/MS fragmentation data to identify a suite of antibacterial molecules called lunaemycins (compounds 15–17) which were associated with this gene cluster. In silico analysis of the lunaemycin BGC along with LC-MS/MS, 1H, and 13C NMR data enabled structural elucidation of this novel group of molecules.177 Agar diffusion assays showed that lunaemycins A and B1 exhibited stronger antibacterial activity against Gram-positive bacteria than lunaemycin D. Lunaemycin A was further studied by in vitro experiments to determine MIC values; this compound exhibited the greatest activity against B. subtilis, E. faecalis, and Staphyloccoccus. spp. (MIC = 0.12 μg mL−1 for all strains). Additional bioactive compounds produced by Streptomyces include xiakemycin A (compound 1) and huanglongmycins A–C (compounds 9–11). Compound 1 was found to exhibit antibacterial activity against Gram-positive bacteria such as Enterococcus faecalis (MIC = 16 μg mL−1),171 while compounds 9–11 exhibited only weak antibacterial activity, with MICs against Staphylococcus spp., E. coli, K. pneumonia, and P. aeruginosa ≥64 μg mL−1.175
5.2 Antifungal properties
It is estimated that 6.5 million fungal infections occur annually, directly leading to around 2.5 million deaths.208 Despite this, there are only three main classes of antifungal drugs available: azoles, echinocandins, and polyenes.209,210 A multitude of bacterial and fungal strains isolated from caves, underground mines, or bats have been found to possess in vitro antifungal activity against fungal pathogens. Since 2014, 22 papers have investigated the antifungal capacity of subterranean microorganisms, 11 of which focus on members of genus Streptomyces (Table 4). Thirty other genera have been investigated, only of four of which were fungal. Over half of the studies investigated in vitro antifungal activity against Candida albicans, a yeast that naturally occurs in the human microbiome but whose overgrowth can cause candidiasis. One additional study reported antifungal activity against C. glabrata,188 which causes 28% of Candida bloodstream infections, second only to C. albicans (39%).211 These studies report moderate to weak antifungal activities from the tested organisms/purified compounds, with zones of inhibition ranging from 5–22 mm and MICs ranging from 12–40 μg mL−1. Given the lack of antifungal medications effective against Candida spp., the discovery of new antifungals should be a critical priority.212 In addition to anti-Candida activities, crude extracts from cave microbes have been tested against nine genera of opportunistic fungal pathogens.47,106,107,182,196In vitro zones of inhibition ranged from 3 mm to >20 mm and MICs from 12.5–50 μg mL−1 (Table 4). It is worth noting that studies in which bioactive constituents were not identified utilized co-culture assays (e.g., cross-streak or agar plug) and not chemical extracts, so the potency of individual chemical constituents cannot be estimated.| Bioactive strain(s) | Pathogens tested | Bioactive agent(s) | Cave of origin | Ref. |
|---|---|---|---|---|
| Streptomyces spp. | Rasamsonia argillacea, Penicillium chrysogenum, Aspergillus fumigatus, Trichophyton mentagrophytes, Candida albicans and C. albicans ‘R’ | Compound 44 | Grotte des Collemboles, Comblain-au-Pont, Belgium | 182 |
| Cadophora sp. 10-5-2 M | Cryptococcus neoformans and Candida albicans | Compounds 57, 58, 77, and 78 | Soudan underground iron mine, Tower, Minnesota, USA | 106 |
| Oidiodendron truncatum | C. neoformans, C. albicans, and Pseudogymnoascus destructans | Compounds 68, 69, 82, and 83 | 107 | |
| Pseudogymnoascus spp. | P. destructans | Not determined | 170 | |
| Pseudomonas yamanorum, P. brenneri, and P. fragi | P. destructans | Compounds 47–50 | Bats swabbed in Ge-zi Cave in Jilin, China and Temple Cave in Liaoning, China | 69 |
| 36 Bacterial strains from 5 genera (Luteipulveratus, Streptomyces Nocardiopsis, Rhodococcus, and Streptosporangium) | P. destructans | Not determined | Bats swabbed in New Mexico, USA (Carlsbad Caverns National Park, El Malpais Conservation Area, Fort Stanton-Snowy River Cave National Conservation Area, and Bureau of Land Management caves 45 and 55) and Arizona, USA (Grand Canyon-Parashant National Monument) | 160 |
| Streptomyces sp. MM56 | Candida spp. | Not determined | Szczelina Chochołowska Cave, Tatra Mountains, Poland | 188 |
| Streptomyces spp. | Candida spp. | Not determined | Chaabe Cave, Algeria | 202 |
| Actinobacteria isolate TB64, actinomycetes isolate TA62, and Bacilli isolates TB48, SB1, and SC3 | C. albicans | Not determined | Parsık Cave, Turkey | 221 |
| Streptomyces spp. | Saccharomyces cerevisiae, C. albicans | Not determined | Badzheyskaya Cave, Krasnoyarsk Krai, Siberia, Russia | 196 |
| Pseudomonas fluorescens | P. destructans | Not determined | Virginia, USA | 165 and 166 |
| Crosiella spp. ON669108 and ON669109 | Aspergillus versicolor, Penicillium chrysogenum, Cladosporium cladosporioides, Ochroconis lascauxensis | Not determined | Altamira Cave, Spain | 47 |
| Brevibacillus borstelensis and Pseudomonas mosselii | C. albicans | Not determined | Koat Maqbari and Smasse-Rawo Caves, Hindu Kush Mountain Range, Pakistan | 71 |
| 42 Bacterial strains from 11 genera (Lactococcus, Bacillus, Paenibacillus, Curtobacterium, Rhodococcus, Streptomyces, Psychrobacter, Achromobacter, Erwinia, Serratia, and Pseudomonas) | P. destructans | Not determined | Bats swabbed in various locations across Western Canada | 162 |
| 18 bacterial strains from 16 genera (Arthrobacter, Lysobacter, Bacillus, Agromyces, Rhodococcus, Rhizobium,Achromobacter, Aminobacter, Sphingomonas,Pseudomonas, Luteibacter,Streptomyces,Microbacterium,Nocardia, Corynebacterium,Enterococcus) | P. destructans | Not determined | Bats swabbed in Eastern and central Tennessee, USA | 161 |
| Pseudomonas spp. | P. destructans | Not determined | Bats swabbed in New York and Virginia, USA | 159 |
| Two isolates of Cutaneotrichosporon moniliiforme | P. destructans | Not determined | Bats swabbed in Arkansas, West Virginia, Iowa, Pennsylvania, Wisconsin, Alabama, Kentucky, New York, Missouri, and Oklahoma, USA | 216 |
| Streptomyces sp. GLD25 | C. albicans and F. oxysporum | Compounds 39–43 | Gueldaman Cave, Akbou-Algeria | 181 |
| Actinomycete strain PM100 | C. albicans | Not determined | Helmcken Falls Cave, Wells Gray Provincial Park, British Columbia, Canada | 195 |
| 65 Strains of Streptomyces spp. and five strains of Bacillus spp., Pseudomonas spp., Nocardia spp., and Erwinia spp. | C. albicans | Not determined | 65 | |
| 3 Streptomyces strains | P. anomala | Not determined | Hampoeil cave, Iran | 206 |
| 106 Bacterial strains including members of Streptomyces, Micrococcus, Actinobacteria, Actinomycetales, Virgibacillus, and Kocuria genera | C. albicans | Not determined | Pukzing Cave, India | 207 |
To date, only three studies have identified bioactive compounds responsible for the observed antifungal activities. Two additional studies identified putative bioactive constituents from antifungal bacterial strains using GC-MS181 or LC-MS/MS,182 but individual constituents were not purified or tested individually. This includes cycloheximide, a known inhibitor of eukaryotic protein synthesis, and its precursor (compounds 44–45), which were identified as major constituents from Streptomyces spp. isolated from a cave moonmilk deposit in Grotte des Collemboles in Belgium182 as well as compounds 39–43, identified in extracts of cave-derived Streptomyces from Gueldaman Cave in Akbou-Algeria.181 The three studies that have definitively identified antifungal constituents discovered weak to moderate antifungal activity, at best. For example, the fungus Cadophora sp. 10-5-2 M collected from the Soudan Mine (Minnesota, USA) yielded 14 secondary metabolites (compounds 57–64 and 76–81), four of which exhibited weak antifungal activity. Only isosclerone (compound 78) inhibited the growth of both C. albicans (MIC = 40 μg mL−1) and Cryptococcus neoformans (MIC = 30 μg mL−1), while pseudoanguillosporin C (compound 57), soudanone A (compound 58), and nectriapyrone (compound 77) only inhibited the growth of C. neoformans with MICs from 20–40 μg mL−1.106 A concerted effort has been undertaken to investigate the bat microbiome for antifungal activity against P. destructans, the cause of WNS (discussed in Section 3.3.3). P. destructans has the ability to cause skin lesions on bats,213 weakening regulatory processes including thermoregulation, gas exchange, and water balance,214,215 and decreasing their likelihood of surviving hibernation. Several authors have identified candidate bacteria69,159–162 and fungi216 with antagonism against P. destructans in vitro. Follow-up studies, though few, have shown particular promise of bat-derived strains of the bacterium Pseudomonas fluorescens, which has successfully been used as a treatment in situ.165,166 Although the bioactive compounds from bat-derived strains of P. fluorescens have not yet been identified, other authors have identified promising secondary metabolites in other strains of the bacterium.217 Another species of Pseudomonas, P. yamanorum, isolated from bats in China, was found to produce four compounds that inhibited P. destructans (compounds 47–50).69 The main inhibitory compound, phenazine-1-carboxylic acid (compound 47), was determined to have a MIC of 50.12 μg mL−1 and an IC50 of 32.08 μg mL−1. Compounds 48–50, all volatile organic compounds, demonstrated inhibition of P. destructans at concentrations of 10 ppm (compound 48) and 100 ppm (compounds 49–50). Though they demonstrate only moderate antifungal abilities, the production of these compounds supports the role of the bat microbiome in protection from WNS. Several researchers have leveraged standard genome mining approaches to explore the secondary biosynthetic potential of bat-associated Streptomyces;218,219 however, they have yet to confirm which natural products were directly correlated to the inhibition of P. destructans. A few fungi have also shown bioactivity against P. destructans. For example, a preliminary screening of bat-associated yeasts yielded two strains of Cutaneotrichosporon moniliiforme that inhibited P. destructans under certain conditions.216 Non-pathogenic Pseudogymnoascus spp., also isolated from bat hibernacula, have been shown to inhibit the growth of P. destructans.170 Notably, pH, salinity, temperature, and nitrogen source appear to have an effect on antifungal activity, and additional chemical analyses are required to identify the associated products.220
Beyond the bat microbiome, the fungus Oidiodendron truncatum, isolated from wood in the Soudan Mine (Minnesota, USA), demonstrated antifungal activity against multiple zoonotic fungal pathogens including P. destructans. Fourteen secondary metabolites produced by O. truncatum were identified (compounds 65–72 and 82–90), the strongest being PR 1388 (compound 82) with antifungal activity against P. destructans (MIC = 7.5 μg mL−1), C. albicans (MIC = 20 μg mL−1), and C. neoformans (MIC = 17.5 μg mL−1). Compound 82 was determined to be non-cytotoxic toward primary fibroblast cell cultures from bat species Myotis septentrionalis (IC50 = 75.6 μM) and Myotis grisescens (IC50 = 102.7 μM) as well as humans (IC50 > 100 μM).107 Although this in vitro screening for cytotoxicity may not accurately reflect potential irritation or toxicity toward bat skin, the strong anti-P. destructans and non-cytotoxic activity of O. truncatum indicates its promise as a treatment for WNS.
5.3 Cytotoxic and antiproliferative properties
Cancer is a leading cause of death worldwide, accounting for nearly 10 million deaths in 2020.222 Both bacterial extracts from caves and purified natural products from them have been shown to possess chemopreventive properties in vitro against a variety of cancer cell types, including colon,173 breast,171,188 lung,171,175,223 and melanoma cells (Table 5).224 For example, the cytotoxic effects of two cave-derived Bacillus subtilis strains were evaluated against murine melanoma cells (B16F10). Organic extracts produced during the stationary phase of growth showed highest cytotoxicity, with an IC50 value of 83.99 μg mL−1 against B16F10 cells and no impact on the normal cell lines evaluated, indicating a high degree of selectivity.224 In another study, a Nonomuraea strain was isolated from cave soil in Pha Tup Cave Forest Park in Thailand and tested against human small cell lung cancer (NCI-H187), human oral cavity cancer (KB), and human breast cancer (MCF7) cell lines and found to have IC50 values of 3.48 μg mL−1, 16.11 μg mL−1, and >50 μg mL−1 respectively.223 Although the extracts in these studies were only weakly or moderately active, it is possible that further efforts to purify cytotoxic agents would result in concentration of activity.| Bioactive strain(s) | Cell lines evaluated | Bioactive agent(s) | Cave of origin | Ref. |
|---|---|---|---|---|
a Authors identified active constituents as dichloranthrabenzoxocinones using accurate masses and database matching. However, the isotope patterns of the detected ions did not contain the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 isotope pattern characteristic of chlorine-containing molecules, and as such, were likely misidentified. 1 isotope pattern characteristic of chlorine-containing molecules, and as such, were likely misidentified.
|
||||
| Nonomuraea specus | TCT-1 cells | Compound 2 | Hardin's Cave, Tennessee | 173 |
| Two isolates of Streptomyces sp. MM56 | T47D cells | Not determineda | Szczelina Chocholowka Cave, Poland | 188 |
| Bacillus subtilis | B16F10 cells | Not determined | Pedra da Chaoeria Cave, Brazil | 224 |
| Streptomyces sp. CB09001 | A549, SKOVV3, Hela, Caco-20 cells | Compound 9 | Karstic cave in Xiangxi, China | 175 |
| Nonomuraea sp. PT708 | NCI-H187 cells | Not determined | Pha Tup Cave Forest Park, Thailand | 223 |
| Streptomyces sp. CC8-201 | A549, MCF-7, HepG-2, HeLa, HCT-116, SH-SY57, PC-3 cells | Compound 1 | Karst cave in Chongquing City, China | 171 |
Several researchers have studied the inhibitory effects of purified compounds from cave microorganisms as well, with IC50 values of individual constituents in the micromolar or high nanomolar ranges. Hypogeamicins A–D (compounds 2–5) were purified from the cave-derived actinomycete Nonomuraea specus and subjected to a suite of biological assays. Interestingly, compound 2, the only dimeric hypogeamicin, was the only compound to possess cytotoxic activity against colon cancer cells (IC50 = 6.4–12.8 μM), indicating that dimerization is essential for chemopreventive activity.173 Huanglongmycins (compounds 9–13) from Streptomyces sp. CB09001 were evaluated against non-small cell lung cancer (A549), epithelial cancer (SKOVV3), and epithelial colorectal adenocarcinoma (Caco-20) cells and demonstrated moderate cytotoxicity against A549 (IC50 = 13.8 μM) and weak activities against all other cell lines tested (IC50 = 40–45 μM).175 The most potent cytotoxic natural product yet discovered from caves is the pyranapthoquinone xiakemycin A (compound 1), which was found to have in vitro activity against A549, MCF7, hepatoma (HepG-2), cervical cancer (HeLa), colon carcinoma (HCT-116), neuroblastoma (SH-SY57), and human prostate cancer (PC-3) cells with IC50 values ranging from 0.43–2.77 μM.171 Several in vitro activities have been conducted on the cytotoxic effects of cave-derived natural products; however, the efficacy of these compounds in in vivo systems has yet to be determined. As such, no conclusive evidence yet exists to confirm the use of cave-derived natural products as anticancer agents, and more robust animal studies followed by clinical trials are essential to support the utilization of these constituents for cancer treatment.
5.4 Other bioactivities and potential applications
Oxidative damage to cellular components may cause downstream complications leading to cardiovascular diseases, carcinogenesis, and neurodegeneration.181 In addition to their antimicrobial and chemopreventive activities, compounds isolated from caves have been shown to possess antioxidant and anti-inflammatory properties (Table 6). Modes of action include radical scavenging activity,181,184 attenuation of oxidative stress,184 and inhibition of inducible nitric oxide synthase (iNOS) proteins.174 Multiple assays exist to evaluate radical scavenging through evaluation of hydrogen transfer and/or electron transfer ability, including oxygen radical absorbance capacity (ORAC), 2,2-diphenyl-1-picrylhydrazyl (DHHP), and ferric reducing antioxidant power (FRAP) assays. Chemical investigation of fermented A. fumigatus GZWMJZ-152 revealed a suite of anti-inflammatory compounds (compounds 52–56 and 73–75). Compounds 55, 56, 73, and 75 showed radical-scavenging activity in the DPPH assay with IC50 values ranging from 3–17 μM, and compounds 52, 54, and 73–74 showed oxygen radical absorbance capacity values in the low μM range. Compounds 52–53 also protected PC12 cells against H2O2 oxidative damage.184 In another study, a crude extract from Streptomyces sp. GLD25 illustrated weak antioxidant activities in both DHHP and FRAP assays. While individual constituents were not isolated, authors were able to identify several putatively antioxidant metabolites (compounds 40–43) via GC-MS.181 Finally, Jiang et al.174 investigated compounds 29–33 for anti-inflammatory effects via iNOS inhibition and found that compound 29 showed potent iNOS inhibition, while compounds 31 and 33 showed only moderate iNOS inhibitory effects.| Bioactive strain(s) | Bioactivity tested | Bioactive agent(s) | Cave of origin | Ref. |
|---|---|---|---|---|
| a Although individual compounds were not identified, FT-IR analysis revealed the presence of carbonyl and carboxyl functional groups, indicating that organic compounds including carboxylic acids, amides, and ketones, were available for copper ion capture. | ||||
| Aspergillus fumigatus GZWMJZ-152 | Antioxidant capacity (DPPH assay, ORAC assay, and cell viability assay in PC12 cells) | Compounds 52–56 and 73–75 | Cave near Fanjing Mountain of Guizhou province, China | 184 |
| Streptomyces sp. GLD25 | Antioxidant capacity (DPPH assay, FRAP assay) | Compounds 40–43 | Gueldaman Cave GLD1, Akbou-Algeria | 181 |
| Streptomyces sp. CB09001 | Anti-inflammatory activity (iNOS inhibition, COX-2 protein expression) | Compounds 29, 31, and 33 | Karstic cave in Xiangxi, China | 174 |
| 10 Microbial strains (eight gram-positive bacteria, one gram-negative bacterium, and one yeast fungus) | Enzymatic activity (proteolytic, cellulolytic, amylolytic, nitrogen fixation, and phosphate solubilization activities) | Not determined | Cave GEM-1462 in Parque Nacional dos Campos Ferruginosos National Park, Brazil | 198 |
| 49 Bacterial strains from 9 genera (Paenibacillus, Staphylococcus, Streptococcus, Salimicrobium, Lysinibacillus, Aeromonas, Proteus, and Clostridium) | Enzymatic activity (proteolytic, cellulolytic, amylolytic, and lipolytic activities) | Not determined | Gumki Cave, Garhwhal Himalaya, India | 225 |
| 28 Strains of Actinomycetota belonging to 13 genera (Streptomyces, Agromyces, Nocardioides, Propionicimonas, Microbacterium, Arthrobacter, Nocardia, Pseudoarthrobacter, Micrococcus, Rhodococcus, Kocuria, Oerskovia, Microterricola) | Enzymatic activity (amylase, gelatinase, cellulase, DNase, urease, and casein hydrolysing activities) | Not determined | Parsık Cave, Turkey | 203 |
| 61 Strains of Actinobactera belonging to 11 genera (Micromonospora, Kocuria, Streptomyces, Micrococcus, Promicromonospora, Rhodococcus, Actinomadura, Nonomuraea, Nocardia, Cornebacterium, Streptosporangium) | Enzymatic activity (amylase, protease, esterase, lipase, DNase), and resistance to heavy metals (Zn, Cu, Cd, Ni, Pb) | Not determined | Hampoeil cave, Iran | 206 |
| 15 Bacterial isolates belonging to three genera (Serratia, Dickeya, Nissabacter) | Biocontrol activity against phytopathogens, plant growth promoting activity | Not determined | Seven caves from the iron Quadrangle, Minas Gerais, Brazil | 229 |
| Four bacterial isolates | Biocontrol activity against phytopathogens, plant growth promoting activity | Not determined | Lime Cave of Andaman and Nicobar Islands, India | 230 |
| Rhodococcus erythropolis | Copper biosorption capacity | Not determineda | Sossego mine, Brazil | 227 |
| Trichoderma harzanium | Nickel accumulation capacity | Not determined | Libiola mine, Italy | 228 |
| Silver accumulation capacity | Not determined | 110 | ||
| Seven strains belonging to three genera (Chaetomium, Penicillium, Trichoderma) | Arsenic volatilization capacity | Not determined | Pastarena gold mine complex, Italy | 111 |
Cave microorganisms are known to produce various enzymes with potential uses in environmental bioremediation as well as in the detergent, cosmetic, and textile industries.203,225 For example, ten microbial strains isolated from different zones (entrance/twilight, transition, and deep interior) of the GEM-1462 cave in the southeastern Amazon exhibited proteolytic activity, along with varying degrees of cellulolytic, amylolytic, phosphate solubilization, and starch/casein degradation activities. Strains isolated from the deep interior zone produced the highest enzymatic indices (particularly proteolytic activities), followed by those from the transition zone and twilight/entrance zones.198 The enzymatic activities of 49 isolates from Gumki Cave, India belonging to Paenibacillus, Staphylococcus, Streptococcus, Salimicrobia, Lysinibacillus, Aeromonas, Proteus, and Clostridium genera also showed high promise for enzymatic production. Of the 90% of isolates with some enzyme production, 75% were lipase producers, 47% were amylase producers, 24% produced protease, and 12% produced cellulase.225 In Parsık Cave (Turkey), 28 Actinomycetota strains showed amylase, gelatinase, casein hydrolase, cellulase, DNase, and/or urease activities, with Streptomyces exfoliatus showing the greatest enzymatic potential.203
Mining activities produce vast quantities of toxic metal wastes, including copper, nickel, and arsenic, which significantly contaminate our soils and waterways and pose serious risks to the environment. The effective detoxification and removal of metal contaminants from polluted environments has increasingly moved towards bioremediation by specialized microorganisms as a sustainable solution to mitigate the negative environmental impacts of mining.226 Given the presence of toxic pollutants, mines house organisms that have adapted unique enzymatic activities to function in harsh conditions and break down toxic pollutants, priming them for utilization in bioremediation. For instance, Rhodococcus erythropolis, isolated from the Sossego Mine in Brazil, demonstrates impressive copper biosorption capabilities, reaching up to 101.90 mg of copper absorption per gram of biomass. Physical adsorption and ion exchange mechanisms by this bacterium are responsible for its notable ability to capture Cu2+ ions, and highlight its potential for use in environmental treatment of metal residues from waterways.227 Fungi isolated from mines have also showed promise for use in bioremediation.110,111,228Trichoderma harzanium, isolated from sulfide-rich waste rock dumps from the Libiola Mine in Italy, showed remarkable Ni2+ tolerance, capable of hyperaccumulating up to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg of nickel per kg of biomass.228 In a later study, this same strain possessed significant silver accumulation capabilities, with an uptake capacity of 46.36% taken at an initial concentration of 330 mg L−1.110 Additional fungal isolates from the decommissioned Pastarena gold mine complex located in the Anzasca Valley, Italy, belonging to Penicillium, Trichoderma, and Chaetomium genera, showed promise to effectively manage arsenic contamination, primarily through volatilization.111 While these results are promising, the chemical mechanisms behind these activities remain poorly understood, and further investigations are warranted.
000 mg of nickel per kg of biomass.228 In a later study, this same strain possessed significant silver accumulation capabilities, with an uptake capacity of 46.36% taken at an initial concentration of 330 mg L−1.110 Additional fungal isolates from the decommissioned Pastarena gold mine complex located in the Anzasca Valley, Italy, belonging to Penicillium, Trichoderma, and Chaetomium genera, showed promise to effectively manage arsenic contamination, primarily through volatilization.111 While these results are promising, the chemical mechanisms behind these activities remain poorly understood, and further investigations are warranted.
One particularly promising yet understudied area of investigation is the agricultural uses of cave microorganisms. Farda et al.3 published an excellent review outlining the unique adaptations of cave microorganisms that make them amenable to use in soil environments. The use of plant growth promoting (PGPR) bacteria is a promising method to enhance crop productivity and manage plant diseases. PGPR bacteria promote plant growth through mechanisms including phosphorus solubilization, hormone production, and phytopathogen antagonism.3,230 Given that caves are rich in carbonates, phosphates, sulfates, and potassium-rich sediments, they are a promising source of mineral-solubilizing microbes with PGPR activities.3 Despite this potential, only two studies have investigated cave isolates as bioinoculants and plant growth promoters.229,230 In one study, 563 cave isolates from ferruginous caves in Brazil were tested against Xanthomonas citri subsp. citri (citrus canker) and later evaluated for bioactivity against fusariosis (Fusarium oxysporum) and bean anthracnose (Colletotrichum lindemuthianum). Twenty strains inhibited F. oxysporum, 15 of which also inhibited C. lindemuthianum. These strains were also evaluated for their ability to solubilize inorganic phosphates, fix nitrogen, and produce siderophores and hydrolytic enzymes. All strains fixed nitrogen, produced proteases and siderophores, and showed motility and biofilm forming abilities, and all but one solubilized phosphates. These bacteria, primarily from the genera Serratia, Nissabacter, and Dickeya could be important candidates for future investigations into sustainable agriculture.229 A similar study of four strains isolated from Lime Cave on the Island of Baratang (Andaman and Nicobar Islands, India) showed that one strain had antagonistic effects against Sclerotium rolfsii, Pythium aphanidermatum, and Rhizoctonia solani, three strains produced indole acetic acid and solubilized phosphate, two strains had protease activity, and one strain produced siderophores.230
6. Accessing the untapped potential of subterranean microbial chemistry
Molecular approaches employing metabarcoding or metagenomics are unparalleled in their ability to provide a bird's-eye perspective of microbial communities in an environment. While these culture-independent approaches can provide insight into the biosynthetic potential of microbial communities, cultivation is required to discern the functional metabolic activity under the growth conditions studied.192,231 In this section, we review the culture-dependent and culture-independent approaches to access potential of cave microbial chemistry and some of the lingering challenges of these techniques.6.1 Culture-dependent approaches
Culture-dependent approaches play a crucial role in studying microbial diversity and functionality. Cultures enable diverse downstream analyses including bioactivity assays, metabolomics analysis, and whole genome sequencing (WGS). Unfortunately, uncultivability is a major challenge in microbiology, as many species exist in the environment in a viable but non-cultivable state.192 This so-called “great plate count anomaly” highlights the discrepancy between the total number of microbial cells in an environmental specimen and the isolable population of that sample. It has been estimated that only 0.1–1% of species can be cultured under common laboratory conditions, and for cave microorganisms, this percentage is at an even lower 0.02%.192,232 To overcome this challenge, innovations in cultivation techniques are being developed to increase the recovery yield of microorganisms in a laboratory setting.The pre-treatment of microbial samples requires careful thought by researchers to maximize cell counts, diversity, and/or novelty depending on the project goals. Physical pre-treatments of samples include air drying, moist heat, dry heat, and microwave irradiation.66,231 Moist heating (50 °C for 5–6 minutes) can stimulate or inhibit different Actinomycetota, typically favoring slow-growing bacteria at the expense of dominant fast-growers.231 For researchers aiming to isolate rare Actinomycetota, air drying is a useful method given that dry spores have low respiration rates and can survive for longer periods of time. Fang et al.234 found that drying samples at 40 °C for two days yielded the highest cultivability compared to those dried at higher temperatures or at room temperature. Their team also noted a positive correlation between the cultivability of spore-forming Actinomycetota and pre-treatment temperatures.234 Interestingly, numerous researchers have documented a paradoxical effect of inoculum dilution on final plate counts, in which the median viable cell counts obtained in 1000-fold dilutions were an order of magnitude higher than those obtained with only a 10- or 100-fold dilution, potentially due to a negative impact of overcrowding or antibiosis on cell viability.192,231
Cultivation media used to isolate cave microbiota range from routine media including soil extract agar (SEA), malt-yeast extract agar (MYA; ISP2), glycerol-asparagine agar (GAA; ISP5), or tryptic soy agar (TSA) to selective media like Actinomycete isolation agar (AI), Hickey–Tresner medium (HT), pyruvate agar, and Reasoner's 2 agar (R2A).66,73,231,232,234,235 While many novel organisms have been isolated from these sources, media preparations that reduce organic carbon levels to those more accurately mimicking the low concentrations in caves have found great success in targeting oligotrophic organisms.232 Inconveniently but perhaps unsurprisingly given the immense diversity of cave environments themselves, there is no consistent “winner-takes-all” medium for maximizing microbial cultivability. While some studies have found low-nutrient TWA as the best medium for isolating oligotrophs from caves,231,232 others have found that full-strength R2A yielded higher numbers of bacteria than diluted versions of the same medium or other minimal media.71 Selective media like HV agar or even pyruvate agar showed maximum cultivability and bacterial diversity in some cases.232,234 Bender et al.232 found that the most nutrient-rich media, including soil agar and ISP2, were particularly poor in culturing isolates from cave environments. While this could be explained by osmotic stress of cave-associated bacteria in the presence of high levels of nutrients, it could also be explained by the fact that standard preparation procedures of nutrient rich media, particularly those containing added phosphates, can result in the formation of toxic reactive oxygen species during autoclaving that can impact cell growth. Adam et al.192 found that nutrient-rich ISP5 media performed quite well for isolating hard-to-culture and rare Actinomycetota from moonmilk when components of the media were autoclaved separately but not when they were prepared using standard procedures.
Supplementation of isolation media with chemical modifiers has also been shown to influence microbial cultivability. In a study evaluating the impact of pH and calcium salts on isolation of cave Actinomycetota, Fang et al.234 found that the highest number of colony-forming units were obtained at a neutral pH as opposed to alkaline or nearly neutral pH, suggesting that neutral conditions facilitate easier maintenance of cytoplasmic pH within cells. In the same study, both the type and concentration of calcium salts significantly affected isolation efforts, with CaCO3 yielding more colony forming units than CaCl2 or Ca(CH3COO)2. Higher CFUs were observed at 0.1% or 0.01% than at 1% w/v or in the absence of salts. Calcium ions are crucial for spore-forming microorganisms, with CaCO3 stimulating the most growth of rare heterotrophic bacteria.234 Supplementation of culture media with low concentrations of antibiotics has shown promise for the selection of slow-growing microbial species. Bender et al.232 found that although antibiotic treatment with chloramphenicol and nalidixic acid reduced overall colony counts and species diversity, it increased the proportion of slow-growing oligotrophs that may represent rare species, emphasizing the differential selection pressures exerted by nutrient composition and antibiotic presence.
Finally, incubation temperature and time significantly impact the cultivability of cave microorganisms, with optimal conditions varying both by species and location. While temperatures between 28–30 °C generally yield more isolates, temperatures around 5 °C can improve isolation of rare psychrophiles.66,236 Optimal temperatures are also media specific. For example, recovery of bacteria from caves in the Hindu Kush mountain range in Pakistan was highest at 37 °C when samples were plated on full-strength R2A, but when plated on half-strength R2A, 17 °C incubation temperatures resulted in higher colony counts.71 Notably, extending incubation time from two to four (or even ten) weeks consistently increases colony counts and diversity across cave systems, allowing for the isolation of rare and slow-growing species,192,232 so a combination of varying temperatures and prolonged incubation time is recommended for researchers aiming to maximize diversity of cave isolates.
Cultivation, WGS, and LC-MS can be effective strategies for prioritizing bacterial strains for natural product discovery, but for fungal genomes that are larger with more repetitive elements and often poor annotation, this strategy requires considerable time and financial resources. Initial investigations of alternative low-cost methodologies suggest ketoacyl synthase alpha subunit (KSα) gene homology may be used as a proxy for a strain's total biosynthetic capacity.238 PKS II systems, the simplest type of PKS pathways, contain only a single representative of each domain: ketosynthase alpha (KSα), ketosynthase beta (KSβ), and the acyl carrier protein, and the presence of one of these domains can be representative of an entire PKS II gene cluster. Researchers isolated 467 bacterial isolates from the bat skin microbiome and found that between 34–60% contained KSα sequences, depending on bat species. Among these, 21% of KSα sequences had less than 85% homology to known sequences, suggesting that the associated BGCs may encode novel polyketide products. WGS of a 16-strain subset of these bacterial isolates revealed that lower KSα homology correlated with higher overall BGC novelty. These findings (although notably discovered using a small sample size) suggest that KSα gene homology may predict a strain's biosynthetic capacity, allowing for quicker strain prioritization through a simple and cost-effective PCR screening.238 However, this approach has its biases because KSα is not distributed evenly across all bacterial diversity, thus selecting for known natural product producers.
A bottleneck to accessing this untapped biosynthetic potential is the well-documented observation that most microbial BGCs are transcriptionally inactive under laboratory conditions, likely because biosynthesis is energetically expensive and organisms grown in controlled monocultures lack the environmental cues required to induce metabolite formation.172,237 Numerous strategies have been taken to activate silent gene clusters under laboratory conditions, including “brothological” methods involving varying cultivation parameters such as medium composition, pH, and temperature to more effectively mimic the organisms' natural environment (or challenge the organism in unique ways), assessing changes to secondary metabolism under different phases of microbial growth, or by adding defined chemical or biological stressors to cultivation media to model environmental stimuli.172 Such additives include histone deacetylase inhibitors,240 subinhibitory concentrations of microbially-derived antibiotics,241 heavy metals,242 and co-cultures with other microorganisms.243
Although studies evaluating the impact of culture conditions on biosynthetic gene expression in cave microorganisms are limited, they emphasize that growth conditions have significant impacts on both BGC transcription180,237 and bioactivity.182,195 For instance, Lebedeva et al.180 conducted transcription analysis of two cave-derived Paenibacillus strains and found that certain genes were transcribed at significantly higher rates during the transition phase, while others peaked during the stationary phase. Higher overall transcription was also noted in half-strength medium compared to full-strength medium, although results varied significantly from one BGC to another. In a related study, transcriptional analysis of 91 additional strains from the same location similarly demonstrated that for a subset of BGCs, growth phase and nutrient levels impacted transcription, with some genes showing higher expression in the stationary phase and others in the exponential phase.237 Biological activity, presumably resulting from the change in expression of secondary metabolites, can also be impacted by cultivation conditions. For example, Streptomyces spp. isolated from moonmilk deposits possessed strong antimicrobial activities against a suite of microorganisms.182 Researchers compared BGC profiles to observed bioactivities and found that there was no correlation between the global antimicrobial activity of a strain and the number of NRPS and PKS genes in the genome. In some strains, antimicrobial activity was elicited by culturing them in minimal medium supplemented with GlcNAc, a known elicitor of antibiotics under nutrient-poor conditions. Other isolates did not have antimicrobial phenotype under any tested condition but still possessed numerous BGCs, indicating that the lack of bioactivity was due to inappropriate culture conditions rather than a lack of biosynthetic potential.182 Perhaps unsurprisingly given their location in lightless conditions, exposure to UV light can also change the behavior of cave microorganisms. Rule and Cheeptham195 tested 176 actinomycetes against microbial pathogens with and without UV light exposure and found that 70% of the strains had antimicrobial activity under at least one growth condition. Approximately 20% were active under both conditions, 17% were active only with UV light exposure, and 33% were active only under no light. Notably, Streptomyces spp. exhibited the most significant change in antibacterial activity in UV light versus darkness. These isolates lost activity against Acinetobacter baumannii, Mycobacterium smegmatis, multi-drug resistant (MDR) S. aureus, extended-spectrum beta-lactamases-containing (ESBL) E. coli, M. luteus, and C. albicans when placed in UV light.
Somewhat surprisingly, only a single study has evaluated changes in expressed secondary metabolites using untargeted metabolomics.172 In this study, 20 phylogenetically diverse Actinomycetota from caves in Tennessee (USA) were exposed to subinhibitory concentrations of antibiotics (rifampin and streptomycin), rare earth metals (lanthanum or scandium), or co-cultured with mycolic acid-containing bacteria Tsukamurella pulmonis or Rhodococcus sp. BBSNAI13 and evaluated for changes to their secondary metabolite profiles. Comparative metabolomics using LC-MS analysis revealed significant changes in secondary metabolism, with over 30% of detected features increasing at least tenfold under at least one treatment (Fig. 6A). Among these upregulated features, several known natural products were identified, compounds 2–5 and 23–28, along with a novel aminopolyol polyketide, funisamine (compound 6) (Fig. 6B). Notably, the specific stimuli that triggered upregulation were both strain- and metabolite-specific, highlighting the somewhat unpredictable microbial responses to environmental conditions.172
 | ||
| Fig. 6 Impacts of environmental stimuli (antibiotics, rare earth metals, and co-culture) on secondary metabolism of subterranean microorganisms. (A) Percent of total detected features with 10-fold or higher increase in abundance in stimuli vs. control conditions. (B) Fold-changes of identified natural products across stimuli conditions of subinhibitory concentrations of rifampicin (Rif) and streptomycin (Str), rare earth metal exposure of lanthanum (La) and scandium (Sc), and co-culture with T. pulmonis (Tp) or R. wratis (Rw). Adapted with permission from Covington et al. 2018.172 | ||
6.2 Culture-independent approaches
Modern sequencing approaches circumvent the necessity of culturing by providing a comprehensive view of the genetic and/or metabolic diversity of the community members contained within a complex sample. Depending on the specific aims, investigators may leverage targeted or untargeted approaches involving short- or long-read sequencing for in silico prioritization and characterization of natural products reviewed below.In natural product discovery, amplicon sequencing can be leveraged for large-scale screening of a targeted gene region. Signature genes and conserved protein domains of enzymes responsible for the biosynthesis of diverse families of bioactive compounds have been identified as putative targets.246 Researchers have employed degenerate PCR primers that target the ketosynthase (KS) and adenylation (AD) domains of the PKS and NRPS pathways, respectively.247–249 PKS and NRPS pathways are known for prolific production of bioactive compounds.250 For example, they are among the most common BGC classes of Streptomyces isolated from bats in caves from New Mexico and Arizona (USA).251 Rego et al.252 provides a proof-of-concept that these targets (KS and AD domains) are anticipated to divulge metabolites biosynthesized by cryptic genes or by uncultured microorganisms that have adapted to unique niches. One comparative study screened the AD domain to explore diversity and richness of NRPS biosynthesis in cave-derived sediments from a lava tube and a limestone cave in Canada. They discovered that the sequence clusters could be distinguished based on if they were limestone or volcanic cave origin.204 This approach can therefore both identify ecological variables that influence biosynthetic capacity and identify metabolites that are biosynthesized by uncultured microorganisms within the constraints of known BGC classes.
7. Challenges and future outlook
Subterranean microbial communities represent a largely untapped reservoir of natural products with diverse activities useful in biotechnology, including antimicrobial, anticancer, and bioremediative properties. In the last ten years, there have been 59 studies exploring the biotechnological potential of thousands of strains of subterranean bacteria and fungi, more than 750 of which have shown at least one bioactivity. Despite these significant findings, only 30% of these studies have identified the natural products produced by these microorganisms (and these studies investigate only 19 microorganisms, <3% of the bioactive strains investigated since 2014), highlighting a considerable gap in our knowledge. Fungi are particularly underrepresented in the subterranean research landscape, as only three studies focusing on natural products discovery from subterranean microorganisms have involved fungi. Strikingly, the three strains investigated in these studies were responsible for the production of more than half of the novel compounds discovered from these environments in the last decade, highlighting the tremendous unrealized potential of subterranean fungi.It is clear based on existing studies that microbial diversity and biosynthetic potential of cave microorganisms is high. However, accessing this untapped biosynthetic potential presents significant challenges. Bacterial uncultivability remains one of the key problems in modern-day microbiology, and a large majority of “known” microorganisms have been identified only through genome-based approaches with no culturable representatives.192,255 This challenge is compounded by the fact that each microorganism has their own optimal nutrient and physical growth requirements, and that for most novel species, these requirements are unknown.11 Factors such as sample collection and processing methods, media composition, and storage temperature and time all have significant impacts on the cultivation of microorganisms, and tailored approaches to maximize microbial recovery are required. Even when microorganisms are successfully cultured, the production of secondary metabolites is inconsistent as the majority of BGCs remain transcriptionally inactive under laboratory conditions. Strategies to activate these silent BGCs are many but require significant experimentation and optimization. It is worth noting that while many natural products chemists have turned to heterologous expression as a valuable tool for accessing metabolite products of cryptic BGCs,187 such technologies have not yet been exploited in subterranean microorganisms.
In recent years, cultivation-independent techniques have enhanced our understanding of microbial diversity and evolution, allowing for genome-based identification of novel bacterial groups and assessment of their biotechnological potential without the limitations of culturing and single organism isolation.256,257 However, despite ongoing improvement of standard methods, all platforms inevitably miss mutations and contain sequencing artifacts.258 Many computational tools are limited to algorithms that search for conserved enzyme motifs; however, improved strategies of data training or incorporating phylogenomics could offer discovery of novel natural products.259 Additionally, metagenomic sequence data can provide insights into traits involving primary metabolism, substrate utilization, and oxygen requirements, allowing researchers to design and optimize specialized media tailored to specific metabolic needs.260 Such strategies in studies involving subterranean microorganisms remains underutilized, but could help to unlock the potential of previously unculturable microorganisms from these unique environments. The larger issue now, beyond identification, is prioritization of laborious experimental procedures to characterize the compounds with the greatest biomedical or biotechnological potential. Studies that combine culture-dependent and culture-independent methodologies, such as those utilized by Suárez-Moo et al.261 who leveraged culturing, metagenomic sequencing, and genome mining of microbial communities in a karst coastal sinkhole in Yucatán, Mexico, may provide the greatest opportunities to elucidate unexplored microbial genomes and metabolic functions.
Of course, the study of natural products in subterranean ecosystems is only possible if the delicate communities contained within these environments are protected, and it is essential that researchers adopt a conservation-oriented mindset when exploring caves and mines. Alien species pose significant threats to subterranean habitats, threatening biodiversity and access to the untapped biotechnological potential contained within these environments.262 Mining, in particular, introduces substantial environmental hazards, including acid mine drainage and toxic element contamination, which can impact neighboring ecosystems and persist long after mining activities cease.263 Interestingly, the same mines that produce such environmental hazards contain microbes with specialized metabolic pathways privileged for bioremediative activities 124 including the detoxification of pollutants such as arsenic, vanadium, and cyanide.264,265 Specific conservation methods, such as limiting visitor numbers in show caves, enforcing hygiene protocols to prevent the introduction of non-native species, and establishing guidelines for rehabilitation of mining sites, are essential to safeguard indigenous subterranean communities.153,266,267
8. Data availability
No primary research results, software, or code have been included and no new data were generated in this review.9. Conflicts of interest
There are no conflicts to declare.10. Acknowledgements
This review was supported by the National Science Foundation (award #2334070) to L. Caesar and P. Salazar-Hamm. The authors would like to thank Dr Diana Northup and Dr Ángel Garcia for their valuable insights. We would also like to acknowledge the use of ChatGPT as an organizational tool in the development of this review paper, with all AI-generated content being thoroughly reviewed and edited by human authors.11. References
- C. E. Gregory, A Concise History of Mining, CRC Press, London, 2021 Search PubMed.
- N. Goldscheider, Z. Chen, A. S. Auler, M. Bakalowicz, S. Broda, D. Drew, J. Hartmann, G. Jiang, N. Moosdorf, Z. Stevanovic and G. Veni, Hydrogeol. J., 2020, 28, 1661–1677 CrossRef CAS.
- B. Farda, R. Djebaili, I. Vaccarelli, M. Del Gallo and M. Pellegrini, Microorganisms, 2022, 10, 453 CrossRef CAS PubMed.
- S. Zada, W. Sajjad, M. Rafiq, S. Ali, Z. Hu, H. Wang and R. Cai, Microb. Ecol., 2022, 84, 676–687 CrossRef CAS PubMed.
- K. Kosznik-Kwaśnicka, P. Golec, W. Jaroszewicz, D. Lubomska and L. Piechowicz, Microorganisms, 2022, 10, 222 CrossRef PubMed.
- P. Gatinho, C. Salvador, A. M. Silva and A. T. Caldeira, Sustainability, 2023, 15, 7471 CrossRef CAS.
- N. N. Kato, G. S. Arini, R. R. Silva, M. E. Bichuette, J. A. P. Bitencourt and N. P. Lopes, J. Braz. Chem. Soc., 2024, 35, e CAS.
- H. A. Barton, M. R. Taylor and N. R. Pace, Geomicrobiol. J., 2004, 21, 11–20 CrossRef CAS.
- P. Turrini, A. Chebbi, F. P. Riggio and P. Visca, Front. Microbiol., 1370520, DOI:10.3389/fmicb.2024.1370520.
- E. L. Marques, G. S. Silva, J. C. Dias, E. Gross, M. S. Costa and R. P. Rezende, Microorganisms, 2019, 7, 33 CrossRef CAS PubMed.
- S. Ghosh, N. Kuisiene and N. Cheeptham, Biochem. Pharmacol., 2017, 134, 18–34 CrossRef CAS PubMed.
- T. L. Poulson and W. B. White, Science, 1969, 165, 971–981 CrossRef CAS PubMed.
- J. R. Pellini, M. A. Salerno and A. Zarankin, Coming to Senses: Topics in Sensory Archaeology, Cambridge Scholars Publishing, 2015 Search PubMed.
- M. Hofreiter, S. Münzel, N. J. Conard, J. Pollack, M. Slatkin, G. Weiss and S. Pääbo, Curr. Biol., 2007, 17, R122–R123 CrossRef CAS PubMed.
- H. P. Blum, Int. Forum Psychoanal., 2011, 20, 196–204 CrossRef.
- T. Devièse, G. Abrams, M. Hajdinjak, S. Pirson, I. De Groote, K. Di Modica, M. Toussaint, V. Fischer, D. Comeskey, L. Spindler, M. Meyer, P. Semal and T. Higham, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2022466118 CrossRef PubMed.
- L. E. Sponsel, Chang. World Relig. Map Sacred Places Identities Pract. Polit., 2015, pp. 503–522 Search PubMed.
- G. M. Crothers, Am. Antiq., 2012, 77, 524–541 CrossRef.
- J. E. Brady and P. A. Peterson, Re-envisioning Ancient Maya Ritual Assemblages, in Religion, Archaeology and the Material World, ed. L. Fogelin, 2008, pp. 78–96 Search PubMed.
- A. Cigna and E. Burri, Int. J. Speleol., 2000, 29, 1–27 Search PubMed.
- Á. Garcia and A. Shank, Geol. Soc. Spec. Publ., 2024, 543, 37–50 CrossRef.
- J.-M. Geneste, Archaeol. Ethnol. Anthropol. Eurasia, 2017, 45, 29–40 CrossRef.
- S. Mętel, M. Kostrzon, J. Adamiak, H. Gattner, D. Kościelecka, A. Sosulska, E. Szczygieł and J. Golec, Ther. Adv. Respir. Dis., 2020, 14, 1753466620926952 CrossRef PubMed.
- A. C. Pizzorusso, Carbonates Evaporites, 2021, 36, 75 CrossRef.
- J. Roth, Nat. Notes Crater Lake, 1995, vol. 6 Search PubMed.
- M. Theodorescu, R. Bucur, P.-A. Bulzu, L. Faur, E. A. Levei, I. C. Mirea, O. Cadar, R. L. Ferreira, M. Souza-Silva and O. T. Moldovan, Microb. Ecol., 2023, 86, 2847–2857 CrossRef PubMed.
- G. A. Quinn, A. M. Banat, A. M. Abdelhameed and I. M. Banat, J. Med. Microbiol., 2020, 69, 1040–1048 CrossRef CAS PubMed.
- M. J. Vaughan, W. Nelson, C. Soderlund, R. M. Maier and B. M. Pryor, Microb. Ecol., 2015, 70, 175–187 CrossRef CAS PubMed.
- O. S. Hershey, J. Kallmeyer, A. Wallace, M. D. Barton and H. A. Barton, Front. Microbiol., 02823, DOI:10.3389/fmicb.2018.02823.
- X. Cheng, X. Xiang, Y. Yun, W. Wang, H. Wang and P. L. E. Bodelier, Front. Microbiol., 1068595, DOI:10.3389/fmicb.2023.1068595.
- C.-M. Joanna, M. Andrzej, C.-M. Joanna and M. Andrzej, in Cyanobacteria, IntechOpen, 2018 Search PubMed.
- J. Mulec, Acta Carsologica, 2008, 37 DOI:10.3986/ac.v37i1.167.
- R. Zhao, H. Wang, H. Yang, Y. Yun and H. A. Barton, Geomicrobiol. J., 2017, 34, 511–523 CrossRef CAS.
- H.-Z. Zhu, Z.-F. Zhang, N. Zhou, C.-Y. Jiang, B.-J. Wang, L. Cai, H.-M. Wang and S.-J. Liu, Appl. Environ. Microbiol., 2021, 87, e024400 Search PubMed.
- Microbial Interactions Drive Distinct Taxonomic and Potential Metabolic Responses to Habitats in Karst Cave Ecosystem, Microbiology Spectrum, https://journals.asm.org/doi/10.1128/spectrum.01152-21, accessed October 2, 2024.
- D. E. Northup, S. M. Barns, L. E. Yu, M. N. Spilde, R. T. Schelble, K. E. Dano, L. J. Crossey, C. A. Connolly, P. J. Boston, D. O. Natvig and C. N. Dahm, Environ. Microbiol., 2003, 5, 1071–1086 CrossRef PubMed.
- M. N. Spilde, D. E. Northup, P. J. Boston, R. T. Schelble, K. E. Dano, L. J. Crossey and C. N. Dahm, Geomicrobiol. J., 2005, 22, 99–116 CrossRef CAS.
- L. A. Melim and M. N. Spilde, J. Sediment. Res., 2018, 88, 344–364 CrossRef CAS.
- K. Koning, R. McFarlane, J. T. Gosse, S. Lawrence, L. Carr, D. Horne, N. Van Wagoner, C. N. Boddy and N. Cheeptham, Front. Microbiol., 2022, 13, 933388, DOI:10.3389/fmicb.2022.933388.
- C. Fliermans and E. Schmidt, Int. J. Speleol., 1977, 9, 1, DOI:10.5038/1827-806X.9.1.1.
- M. Mason-Williams, Int. J. Speleol., 1967, 2, 389–395 CrossRef.
- K. H. Lavoie, A. S. Winter, K. J. H. Read, E. M. Hughes, M. N. Spilde and D. E. Northup, PLoS One, 2017, 12, e0169339 CrossRef PubMed.
- O. S. Hershey and H. A. Barton, in Cave Ecology, ed. O. T. Moldovan, Ľ. Kováč and S. Halse, Springer International Publishing, Cham, 2018, pp. 69–90 Search PubMed.
- R. D. Prescott, T. Zamkovaya, S. P. Donachie, D. E. Northup, J. J. Medley, N. Monsalve, J. H. Saw, A. W. Decho, P. S. G. Chain and P. J. Boston, Front. Microbiol., 934708, DOI:10.3389/fmicb.2022.934708.
- R. Addesso, J. L. Gonzalez-Pimentel, I. M. D'Angeli, J. De Waele, C. Saiz-Jimenez, V. Jurado, A. Z. Miller, B. Cubero, G. Vigliotta and D. Baldantoni, Microb. Ecol., 2021, 81, 884–896 CrossRef CAS PubMed.
- S. K. Pradhan, N. R. Singh, U. Kumar, S. R. Mishra, R. C. Perumal, J. Benny and H. Thatoi, Ecol. Genet. Genomics, 2020, 15, 100054 Search PubMed.
- J. L. Gonzalez-Pimentel, I. Dominguez-Moñino, V. Jurado, L. Laiz, A. T. Caldeira and C. Saiz-Jimenez, Microorganisms, 2022, 10, 1575 CrossRef CAS PubMed.
- H.-Z. Zhu, Z.-F. Zhang, N. Zhou, C.-Y. Jiang, B.-J. Wang, L. Cai and S.-J. Liu, Front. Microbiol., 01726, DOI:10.3389/fmicb.2019.01726.
- J. J. Hathaway, P. S. Salazar-Hamm, N. A. Caimi, D. O. Natvig, D. C. Buecher and D. E. Northup, Geomicrobiol. J., 2024, 41, 82–97 CrossRef CAS.
- I. M. D'Angeli, D. I. Serrazanetti, C. Montanari, L. Vannini, F. Gardini and J. De Waele, Sci. Total Environ., 2017, 598, 538–552 CrossRef PubMed.
- J. J. M. Hathaway, M. G. Garcia, M. M. Balasch, M. N. Spilde, F. D. Stone, M. D. L. N. E. Dapkevicius, I. R. Amorim, R. Gabriel, P. A. V. Borges and D. E. Northup, Geomicrobiol. J., 2014, 31, 205–220 CrossRef PubMed.
- G. Zhang, J. Bai, C. C. Tebbe, Q. Zhao, J. Jia, W. Wang, X. Wang and L. Yu, Environ. Microbiol., 2021, 23, 1020–1037 CrossRef CAS PubMed.
- K. Zhang, Y. Shi, X. Cui, P. Yue, K. Li, X. Liu, B. M. Tripathi and H. Chu, mSystems, 2019, 4, 10–1128, DOI:10.1128/msystems.00225-18.
- A. R. Sprocati, C. Alisi, F. Tasso, A. Fiore, P. Marconi, F. Langella, G. Haferburg, A. Nicoara, A. Neagoe and E. Kothe, Environ. Sci. Pollut. Res., 2014, 21, 6824–6835 CrossRef CAS PubMed.
- L. Chen, J. Li, Y. Chen, L. Huang, Z. Hua, M. Hu and W. Shu, Environ. Microbiol., 2013, 15, 2431–2444 CrossRef CAS PubMed.
- A. S. Abdel-Razek, M. E. El-Naggar, A. Allam, O. M. Morsy and S. I. Othman, Processes, 2020, 8, 470 CrossRef CAS.
- M.-X. Han, B.-Z. Fang, Y. Tian, W.-Q. Zhang, J.-Y. Jiao, L. Liu, Z.-T. Zhang, M. Xiao, D.-Q. Wei and W.-J. Li, Int. J. Syst. Evol. Microbiol., 2017, 67, 633–639 CrossRef CAS PubMed.
- Q.-Q. Li, M.-X. Han, B.-Z. Fang, J.-Y. Jiao, L. Liu, Z.-W. Yang, W.-Q. Zhang, D.-Q. Wei and W.-J. Li, Int. J. Syst. Evol. Microbiol., 2017, 67, 2998–3003 CrossRef CAS PubMed.
- B.-Z. Fang, M.-X. Han, L. Liu, Z.-T. Zhang, W.-L. Liu, J.-T. Shen, Y. Wang, W.-Q. Zhang, D.-Q. Wei and W.-J. Li, Int. J. Syst. Evol. Microbiol., 2017, 67, 2357–2362 CrossRef CAS PubMed.
- L.-Y. Zhang, H. Ming, X.-L. Meng, B.-Z. Fang, J.-Y. Jiao, N. Salam, X.-T. Zhang, W.-J. Li and G.-X. Nie, Antonie van Leeuwenhoek, 2019, 112, 179–186 CrossRef CAS PubMed.
- L. Tuo, L. Guo, S.-W. Liu, J.-M. Liu, Y.-Q. Zhang, Z.-K. Jiang, X.-F. Liu, L. Chen, J. Zu and C.-H. Sun, Int. J. Syst. Evol. Microbiol., 2015, 65, 3305–3312 CrossRef CAS PubMed.
- Z. Fang, X. Zhao, Q. Wu, S. Li, Q. Liu, L. Tan and Q. Weng, Int. J. Syst. Evol. Microbiol., 2022, 72, 005445 CAS.
- K. Lipun, W. F. A. Teo, P. Suksaard, W. Pathom-Aree and K. Duangmal, Int. J. Syst. Evol. Microbiol., 2020, 70, 5296–5303 CrossRef CAS PubMed.
- C. Riquelme, J. J. Marshall Hathaway, M. de L. N. Enes Dapkevicius, A. Z. Miller, A. Kooser, D. E. Northup, V. Jurado, O. Fernandez, C. Saiz-Jimenez and N. Cheeptham, Front. Microbiol., 01342, DOI:10.3389/fmicb.2015.01342.
- N. Cheeptham, T. Sadoway, D. Rule, K. Watson, P. Moote, L. Soliman, N. Azad, K. Donkor and D. Horne, Int. J. Speleol., 2013, 42, 35–47 CrossRef.
- P. Rangseekaew and W. Pathom-Aree, Front. Microbiol., 2019, 10, 387 CrossRef PubMed.
- V. Jurado, L. Laiz, V. Rodriguez-Nava, P. Boiron, B. Hermosin, S. Sanchez-Moral and C. Saiz-Jimenez, Int. J. Speleol., 2010, 39, 15–24 CrossRef.
- F. Biagioli, C. Coleine, E. Piano, G. Nicolosi, A. Poli, V. Prigione, A. Zanellati, C. Varese, M. Isaia and L. Selbmann, Sci. Rep., 2023, 13, 689 CrossRef CAS PubMed.
- Z. Li, A. Li, J. R. Hoyt, W. Dai, H. Leng, Y. Li, W. Li, S. Liu, L. Jin and K. Sun, et al. , Microb. Biotechnol., 2022, 15, 469–481 CrossRef CAS PubMed.
- N. Dogruoz-Güngor, B. Candıroglu and G. Altug, J. Cave Karst Stud., 2020, 82, 106–115 CrossRef.
- M. Yasir, Braz. J. Microbiol., 2018, 49, 248–257 CrossRef CAS PubMed.
- V. I. Paun, P. Lavin, M. C. Chifiriuc and C. Purcarea, Sci. Rep., 2021, 11, 514 CrossRef CAS PubMed.
- S. Ghosh, G. Kam, M. Nijjer, C. Stenner and N. Cheeptham, Int. J. Speleol., 2020, 49, 6 Search PubMed.
- J. Ambrožič Avguštin, P. Petrič and L. Pašić, Int. J. Speleol., 2019, 48, 9, DOI:10.5038/1827-806X.48.3.2272.
- V. Jurado, J. M. Gonzalez, L. Laiz and C. Saiz-Jimenez, Int. J. Syst. Evol. Microbiol., 2006, 56, 2583–2585 CrossRef CAS PubMed.
- R. E. Mendes, G. A. Denys, T. R. Fritsche and R. N. Jones, J. Clin. Microbiol., 2009, 47, 514–515 CrossRef PubMed.
- A. M. Kielak, C. C. Barreto, G. A. Kowalchuk, J. A. van Veen and E. E. Kuramae, Front. Microbiol., 00744, DOI:10.3389/fmicb.2016.00744.
- S. Kalam, A. Basu, I. Ahmad, R. Z. Sayyed, H. A. El-Enshasy, D. J. Dailin and N. L. Suriani, Front. Microbiol., 580024, DOI:10.3389/fmicb.2020.580024.
- D. B. Meisinger, J. Zimmermann, W. Ludwig, K.-H. Schleifer, G. Wanner, M. Schmid, P. C. Bennett, A. S. Engel and N. M. Lee, Environ. Microbiol., 2007, 9, 1523–1534 CrossRef CAS PubMed.
- D. S. Jones, H. L. Albrecht, K. S. Dawson, I. Schaperdoth, K. H. Freeman, Y. Pi, A. Pearson and J. L. Macalady, ISME J., 2012, 6, 158–170 CrossRef CAS PubMed.
- S. Park, Y.-J. Cho, D. Jung, K. Jo, E.-J. Lee and J.-S. Lee, Front. Microbiol., 00613, DOI:10.3389/fmicb.2020.00613.
- J. J. Medley, J. J. M. Hathaway, M. N. Spilde and D. E. Northup, Appl. Sci., 2024, 14, 6500 CrossRef CAS.
- E. Ouyang, Y. Liu, J. Ouyang and X. Wang, Environ. Technol., 2019, 40, 329–341 CrossRef CAS PubMed.
- F. Bastian, V. Jurado, A. Nováková, C. Alabouvette and C. Saiz-Jimenez, Microbiology, 2010, 156, 644–652 CrossRef CAS PubMed.
- A. S. Engel, L. A. Stern and P. C. Bennett, Geology, 2004, 32, 369–372 CrossRef CAS.
- S. Bindschedler, L. Millière, G. Cailleau, D. Job and E. P. Verrecchia, Geomicrobiol. J., 2012, 29, 301–313 CrossRef CAS.
- M. Hoppert, C. Flies, W. Pohl, B. Günzl and J. Schneider, Environ. Geol., 2004, 46, 421–428 CrossRef CAS.
- L. Epure, I. N. Meleg, C.-M. Munteanu, R. D. Roban and O. T. Moldovan, Geomicrobiol. J., 2014, 31, 116–127 CrossRef.
- K. Vanderwolf, D. Malloch, D. McAlpine and G. Forbes, Int. J. Speleol., 2013, 42, 77–96 CrossRef.
- D. E. Northup and K. H. Lavoie, in Microbial Life of Cave Systems, ed. A. Summers Engel, De Gruyter, 2015, pp. 161–192 Search PubMed.
- S. K. Carmichael, B. T. Zorn, C. M. Santelli, L. A. Roble, M. J. Carmichael and S. L. Bräuer, Environ. Microbiol. Rep., 2015, 7, 592–605 CrossRef CAS PubMed.
- P. J. Kearns, A. S. Winter, D. C. Woodhams and D. E. Northup, Microb. Ecol., 2023, 86, 1565–1574 CrossRef PubMed.
- R. Ogorek, M. Dylag, B. Kozak, Z. Visnovska, D. Tancinova and A. Lejman, J. Cave Karst Stud., 2016, 78, 41–49 CrossRef.
- A. O. B. Cunha, J. D. P. Bezerra, T. G. L. Oliveira, E. Barbier, E. Bernard, A. R. Machado and C. M. Souza-Motta, PLoS One, 2020, 15, e0243494 CrossRef CAS PubMed.
- I. G. Wasti, F. A. A. Khan, H. Bernard, N. H. Hassan, T. Fayle and J. S. Sathiya Seelan, Mycology, 2021, 12, 188–202 CrossRef CAS PubMed.
- J. A. Yoder, J. B. Benoit, B. S. Christensen, T. J. Croxall and H. H. Hobbs III, J. Cave Karst Stud., 2009, 71, 116–120 Search PubMed.
- J. B. Benoit, J. A. Yoder, L. W. Zettler and H. H. Hobbs III, Ann. Entomol. Soc. Am., 2004, 97, 989–993 CrossRef.
- K. M. Jensen, L. Rodrigues, T. Pape, A. Garm, S. Santamaria and A. S. P. S. Reboleira, J. Invertebr. Pathol., 2019, 166, 107206 CrossRef PubMed.
- A. Nováková, A. Kubátová, F. Sklenář and V. Hubka, Czech Mycol., 2018, 70, 101–121 CrossRef.
- J. L. V. R. Carvalho, J. M. S. Lima, E. Barbier, E. Bernard, J. D. P. Bezerra and C. M. Souza-Motta, Braz. J. Microbiol., 2022, 53, 2077–2091 CrossRef PubMed.
- V. Jurado, T. Martin-Pozas, A. Fernandez-Cortes, J. M. Calaforra, S. Sanchez-Moral and C. Saiz-Jimenez, Microb. Ecol., 2024, 87, 80 CrossRef PubMed.
- Z.-F. Zhang and L. Cai, J. Biogeogr., 2019, 46, 1504–1518 CrossRef.
- B. Man, H. Wang, Y. Yun, X. Xiang, R. Wang, Y. Duan and X. Cheng, Front. Microbiol., 01400, DOI:10.3389/fmicb.2018.01400.
- W. E. Eslyn and F. F. Lombard, For. Prod. J. Search PubMed.
- B. W. Held, C. E. Salomon and R. A. Blanchette, PLoS One, 2020, 15, e0234208 CrossRef CAS PubMed.
- Y. Rusman, B. W. Held, R. A. Blanchette, S. Wittlin and C. E. Salomon, J. Nat. Prod., 2015, 78, 1456–1460 CrossRef CAS PubMed.
- Y. Rusman, M. B. Wilson, J. M. Williams, B. W. Held, R. A. Blanchette, B. N. Anderson, C. R. Lupfer and C. E. Salomon, J. Nat. Prod., 2020, 83, 344–353 CrossRef CAS PubMed.
- F. H. Erbisch and N. Harry, Mycologia, 1979, 71, 652–655 CrossRef.
- D. M. Rizzo, R. A. Blanchette and M. A. Palmer, Can. J. Bot., 1992, 70, 1515–1520 CrossRef CAS.
- G. Cecchi, P. Marescotti, S. Di Piazza and M. Zotti, J. Environ. Sci. Health, Part B, 2017, 52, 191–195 CrossRef CAS PubMed.
- S. Crognale, A. D'Annibale, L. Pesciaroli, S. R. Stazi and M. Petruccioli, Front. Microbiol., 02202, DOI:10.3389/fmicb.2017.02202.
- Z. F. Zhang, F. Liu, X. Zhou, X. Z. Liu, S. J. Liu and L. Cai, Pers.: Mol. Phylogeny Evol. Fungi., 2017, 39, 1–31 CAS.
- A. F. Leão, T. O. Condé, Y. L. G. Dutra, A. W. C. Rosado, P. H. Grazziotti, S. de Carvalho Neves, L. M. S. Fraga, M. C. M. Kasuya and O. L. Pereira, Braz. J. Microbiol., 2024, 55, 1569–1585 CrossRef PubMed.
- M. L. S. Pereira, J. L. V. R. Carvalho, J. M. S. Lima, E. Barbier, E. Bernard, J. D. P. Bezerra and C. M. Souza-Motta, Mycol. Prog., 2022, 21, 345–357, DOI:10.1007/s11557-021-01760-2.
- V. C. S. Alves, R. A. Lira, J. M. S. Lima, R. N. Barbosa, D. M. Bento, E. Barbier, E. Bernard, C. M. Souza-Motta and J. D. P. Bezerra, Fungal Syst Evol., 2022, 10, 139–167 CrossRef CAS PubMed.
- L. Martinelli, P. Zalar, N. Gunde-Cimerman, A. Azua-Bustos, K. Sterflinger and G. Piñar, Extremophiles, 2017, 21, 755–773 CrossRef CAS PubMed.
- C. M. Visagie, N. Yilmaz, K. Vanderwolf, J. B. Renaud, M. W. Sumarah, J. Houbraken, R. Assebgui, K. A. Seifert and D. Malloch, Fungal Syst Evol., 2020, 5, 1–15 CrossRef CAS PubMed.
- A. Espino del Castillo, H. Beraldi-Campesi, P. Amador-Lemus, H. I. Beltrán and S. L. Borgne, Int. J. Speleol., 2018, 47, 10, DOI:10.5038/1827-806X.47.2.2161.
- F. Ruiz-Blas, V. Muñoz-Hisado, E. Garcia-Lopez, A. Moreno, M. Bartolomé, M. Leunda, E. Martinez-Alonso, A. Alcázar and C. Cid, Front. Microbiol., 1110091, DOI:10.3389/fmicb.2023.1110091.
- T. Brad, C. Itcus, M.-D. Pascu, A. Persoiu, A. Hillebrand-Voiculescu, L. Iancu and C. Purcarea, Sci. Rep., 2018, 8, 10096 CrossRef PubMed.
- A. Hillebrand-Voiculescu, C. Itcus, I. Ardelean, D. Pascu, A. Persoiu, A. Rusu, T. Brad, E. Popa, B. P. Onac and C. Purcarea, Acta Carsologica, 2014, 43 DOI:10.3986/ac.v43i2-3.604.
- B. M. Tebo, R. E. Davis, R. P. Anitori, L. B. Connell, P. Schiffman and H. Staudigel, Front. Microbiol., 2015, 6, 179, DOI:10.3389/fmicb.2015.00179.
- L. Connell and H. Staudigel, Biology, 2013, 2, 798–809 CrossRef PubMed.
- L. Newsome and C. Falagán, GeoHealth, 2021, 5, e2020GH000380 CrossRef PubMed.
- K. Burow, A. Grawunder, M. Harpke, S. Pietschmann, R. Ehrhardt, L. Wagner, K. Voigt, D. Merten, G. Büchel and E. Kothe, FEMS Microbiol. Lett., 2019, 366, fnz167 CrossRef CAS PubMed.
- J. Shen, A. C. Smith, M. J. Barnett, A. Morgan and P. M. Wynn, J. Geophys. Res. G: Biogeosciences, 2022, 127, e2022JG006866 CrossRef.
- N. Dopffel, B. A. An-Stepec, P. Bombach, M. Wagner and E. Passaris, Int. J. Hydrogen Energy, 2024, 58, 1478–1485 CrossRef CAS.
- R. A. Daly, M. A. Borton, M. J. Wilkins, D. W. Hoyt, D. J. Kountz, R. A. Wolfe, S. A. Welch, D. N. Marcus, R. V. Trexler, J. D. MacRae, J. A. Krzycki, D. R. Cole, P. J. Mouser and K. C. Wrighton, Nat. Microbiol., 2016, 1, 1–9 Search PubMed.
- M. Ivarsson, S. Bengtson, H. Drake and W. Francis, in Advances in Applied Microbiology, ed. S. Sariaslani and G. M. Gadd, Academic Press, 2018, vol. 102, pp. 83–116 Search PubMed.
- C. Escudero, M. Oggerin and R. Amils, Int. Microbiol., 2018, 21, 3–14 CrossRef PubMed.
- K. Kajan, N. Cukrov, N. Cukrov, R. Bishop-Pierce and S. Orlić, Microb. Ecol., 2022, 83, 257–270 CrossRef CAS PubMed.
- L. Sam, A. Bhardwaj, S. Singh, F. J. Martin-Torres, M.-P. Zorzano and J. A. Ramírez Luque, Remote Sens., 2020, 12, 1970 CrossRef.
- F. Sauro, R. Pozzobon, M. Massironi, P. De Berardinis, T. Santagata and J. De Waele, Earth-Sci. Rev., 2020, 209, 103288 CrossRef.
- R. J. Léveillé and S. Datta, Planet. Space Sci., 2010, 58, 592–598 CrossRef.
- S. Šebela, G. Baker and B. Luke, Geoheritage, 2019, 11, 1163–1175 CrossRef.
- N. Nikolic, N. Zarubica, B. Gavrilovic, D. Predojevic, I. Trbojevic, G. Subakov Simic and S. Popovic, J. Cave Karst Stud., 2020, 82, 69–81 CrossRef.
- J. Burgoyne, R. Crepeau, J. Jensen, H. Smith, G. Baker and S. D. Leavitt, Microorganisms, 2021, 9, 1188 CrossRef CAS PubMed.
- Z. Havlena, T. L. Kieft, G. Veni, R. D. Horrocks and D. S. Jones, Appl. Environ. Microbiol., 2021, 87, e026955 CrossRef PubMed.
- E. L. S. Marques, J. C. T. Dias, G. S. Silva, C. P. Pirovani and R. P. Rezende, Genet. Mol. Res., 2016, 15 DOI:10.4238/gmr.15038611.
- S. Leuko, K. Koskinen, L. Sanna, I. M. D'Angeli, J. De Waele, P. Marcia, C. Moissl-Eichinger and P. Rettberg, PloS One, 2017, 12, e0180700 CrossRef PubMed.
- Z. Bontemps, L. Alonso, T. Pommier, M. Hugoni and Y. Moënne-Loccoz, Sci. Total Environ., 2022, 816, 151492 CrossRef CAS PubMed.
- D. W. Griffin, M. A. Gray, M. B. Lyles and D. E. Northup, Geomicrobiol. J., 2014, 31, 175–185 CrossRef.
- K. H. Lavoie and D. E. Northup, in Proceedings of the 17th National Cave and Karst Management Symposium, The NCKMS Steering Committee Albany, NY, USA, 2006, pp. 40–47 Search PubMed.
- L. Alonso, T. Pommier, B. Kaufmann, A. Dubost, D. Chapulliot, J. Doré, C. J. Douady and Y. Moënne-Loccoz, Mol. Ecol., 2019, 28, 3383–3394 CrossRef PubMed.
- J. Shapiro and A. Pringle, Am. Midl. Nat., 2010, 163, 76–86 CrossRef.
- J. Mulec, J. Nat. Conserv., 2014, 22, 132–141 CrossRef.
- A. Fernandez-Cortes, S. Cuezva, S. Sanchez-Moral, J. C. Cañaveras, E. Porca, V. Jurado, P. M. Martin-Sanchez and C. Saiz-Jimenez, Environ. Sci. Pollut. Res., 2011, 18, 1037–1045 CrossRef CAS PubMed.
- C. Saiz-Jimenez, S. Cuezva, V. Jurado, A. Fernandez-Cortes, E. Porca, D. Benavente, J. C. Cañaveras and S. Sanchez-Moral, Science, 2011, 334, 42–43 CrossRef CAS PubMed.
- V. Jurado, E. Porca, S. Cuezva, A. Fernandez-Cortes, S. Sanchez-Moral and C. Saiz-Jimenez, Sci. Total Environ., 2010, 408, 3632–3638 CrossRef CAS PubMed.
- T. Martin-Pozas, A. Nováková, V. Jurado, A. Fernandez-Cortes, S. Cuezva, C. Saiz-Jimenez and S. Sanchez-Moral, Front. Microbiol., 2022, 13, 869661, DOI:10.3389/fmicb.2022.869661.
- T. Martin-Pozas, A. Nováková, V. Jurado, S. Cuezva, A. Fernandez-Cortes, C. Saiz-Jimenez and S. Sanchez-Moral, Microb. Ecol., 2024, 87, 53 CrossRef PubMed.
- H. A. Barton, in Microbial Life of Cave Systems, ed. A. Summers Engel, De Gruyter, 2015, pp. 79–104 Search PubMed.
- N. A. Rachıd and N. D. Güngör, Int. J. Life Sci. Biotechnol., 2021, 4, 311–323 CrossRef.
- D. S. Blehert, A. C. Hicks, M. Behr, C. U. Meteyer, B. M. Berlowski-Zier, E. L. Buckles, J. T. Coleman, S. R. Darling, A. Gargas and R. Niver, et al. , Science, 2009, 323, 227 CrossRef CAS PubMed.
- C. V. Avena, L. W. Parfrey, J. W. Leff, H. M. Archer, W. F. Frick, K. E. Langwig, A. M. Kilpatrick, K. E. Powers, J. T. Foster and V. J. McKenzie, Front. Microbiol., 2016, 7, 1753 Search PubMed.
- A. S. Winter, J. J. Hathaway, J. C. Kimble, D. C. Buecher, E. W. Valdez, A. Porras-Alfaro, J. M. Young, K. J. Read and D. E. Northup, PeerJ, 2017, 5, e3944 CrossRef PubMed.
- H. L. Lutz, E. W. Jackson, P. W. Webala, W. S. Babyesiza, J. C. Kerbis Peterhans, T. C. Demos, B. D. Patterson and J. A. Gilbert, Msystems, 2019, 4, 10–1128 Search PubMed.
- H. G. Shapiro, A. S. Willcox, M. L. Verant and E. V. Willcox, Wildl. Soc. Bull., 2021, 45, 422–429 CrossRef.
- J. R. Hoyt, T. L. Cheng, K. E. Langwig, M. M. Hee, W. F. Frick and A. M. Kilpatrick, PLoS One, 2015, 10, e0121329 CrossRef PubMed.
- P. S. Hamm, N. A. Caimi, D. E. Northup, E. W. Valdez, D. C. Buecher, C. A. Dunlap, D. P. Labeda, S. Lueschow and A. Porras-Alfaro, Appl. Environ. Microbiol., 2017, 83, e030577 CrossRef PubMed.
- M. Grisnik, O. Bowers, A. J. Moore, B. F. Jones, J. R. Campbell and D. M. Walker, FEMS Microbiol. Ecol., 2020, 96, fiz193 CrossRef CAS PubMed.
- A. Forsythe, N. Fontaine, J. Bissonnette, B. Hayashi, C. Insuk, S. Ghosh, G. Kam, A. Wong, C. Lausen and J. Xu, et al. , Sci. Rep., 2022, 12, 9895 CrossRef CAS PubMed.
- Y. Lu, H. Ren, Z. Li, H. Leng, A. Li, W. Dai, L. Huang, J. Feng and K. Sun, Appl. Environ. Microbiol., 2024, 90, e006933 Search PubMed.
- V. Lemieux-Labonté, A. Simard, C. K. Willis and F.-J. Lapointe, Microbiome, 2017, 5, 1–14 CrossRef PubMed.
- T. L. Cheng, H. Mayberry, L. P. McGuire, J. R. Hoyt, K. E. Langwig, H. Nguyen, K. L. Parise, J. T. Foster, C. K. Willis and A. M. Kilpatrick, et al. , J. Appl. Ecol., 2017, 54, 701–708 CrossRef.
- J. R. Hoyt, K. E. Langwig, J. P. White, H. M. Kaarakka, J. A. Redell, K. L. Parise, W. F. Frick, J. T. Foster and A. M. Kilpatrick, Sci. Rep., 2019, 9, 9158 CrossRef PubMed.
- Y.-S. Kim, S.-Y. Lee, C.-U. Chung, J.-S. Park, Y.-J. Kim and J.-K. Oem, Diversity, 2023, 15, 198 CrossRef CAS.
- L. J. A. N. Johnson, A. N. Miller, R. A. McCleery, R. McClanahan, J. A. Kath, S. Lueschow and A. Porras-Alfaro, Appl. Environ. Microbiol., 2013, 79, 5465–5471 CrossRef CAS PubMed.
- P. Becker, C. van den Eynde, F. Baert, E. D'hooge, R. De Pauw, A.-C. Normand, R. Piarroux and D. Stubbe, Mycologia, 2023, 115, 484–498 CrossRef CAS PubMed.
- M. B. Wilson, B. W. Held, A. H. Freiborg, R. A. Blanchette and C. E. Salomon, PloS One, 2017, 12, e0178968 CrossRef PubMed.
- Z. Jiang, L. Guo, C. Chen, S. Liu, L. Zhang, S. Dai, Q. He, X. You, X. Hu and L. Tuo, et al. , J. Antibiot., 2015, 68, 771–774 CrossRef CAS PubMed.
- B. C. Covington, J. M. Spraggins, A. E. Ynigez-Gutierrez, Z. B. Hylton and B. O. Bachmann, Appl. Environ. Microbiol., 2018, 84, e011255 CrossRef PubMed.
- D. K. Derewacz, C. R. McNees, G. Scalmani, C. L. Covington, G. Shanmugam, L. J. Marnett, P. L. Polavarapu and B. O. Bachmann, J. Nat. Prod., 2014, 77, 1759–1763 CrossRef CAS PubMed.
- L. Jiang, H. Pu, X. Qin, J. Liu, Z. Wen, Y. Huang, J. Xiang, Y. Xiang, J. Ju and Y. Duan, et al. , Nat. Prod. Res., 2021, 35, 144–151 CrossRef CAS PubMed.
- L. Jiang, H. Pu, J. Xiang, M. Su, X. Yan, D. Yang, X. Zhu, B. Shen, Y. Duan and Y. Huang, Front. Chem., 2018, 6, 254 CrossRef PubMed.
- L. Jiang, J. Xiang, S. Zhu, D. Tang, B. Gong, H. Pu, Y. Duan and Y. Huang, Nat. Prod. Res., 2022, 36, 1725–1733 CrossRef CAS PubMed.
- L. Martinet, A. Naômé, L. C. D. Rezende, D. Tellatin, B. Pignon, J.-D. Docquier, F. Sannio, D. Baiwir, G. Mazzucchelli, M. Frédérich and S. Rigali, Int. J. Mol. Sci., 2023, 24, 1114 CrossRef CAS PubMed.
- J. T. Gosse, S. Ghosh, A. Sproule, D. Overy, N. Cheeptham and C. N. Boddy, Front. Microbiol., 01020, DOI:10.3389/fmicb.2019.01020.
- D. V. Axenov-Gibanov, I. V. Voytsekhovskaya, B. T. Tokovenko, E. S. Protasov, S. V. Gamaiunov, Y. V. Rebets, A. N. Luzhetskyy and M. A. Timofeyev, PLoS One, 2016, 11, e0149216 CrossRef PubMed.
- J. Lebedeva, G. Jukneviciute, R. Čepaitė, V. Vickackaite, R. Pranckutė and N. Kuisiene, Front. Microbiol., 612483, DOI:10.3389/fmicb.2020.612483.
- F. Z. Djebbah, N. A. Al-Dhabi, M. V. Arasu, L. Belyagoubi, F. Kherbouche, D. E. Abdelouahid and B. Ravindran, J. King Saud Univ. Sci., 2022, 34, 101719 CrossRef.
- M. Maciejewska, D. Adam, L. Martinet, A. Naômé, M. Całusińska, P. Delfosse, M. Carnol, H. A. Barton, M.-P. Hayette, N. Smargiasso, E. De Pauw, M. Hanikenne, D. Baurain and S. Rigali, Front. Microbiol., 2016, 7, 1455 CrossRef PubMed.
- Y. Shi, M. Ji, J. Dong, D. Shi, Y. Wang, L. Liu, S. Feng and L. Liu, Mycology, 2024, 15, 283–321 CrossRef CAS PubMed.
- Y. Xu, W. Liu, D. Wu, W. He, M. Zuo, D. Wang, P. Fu, L. Wang and W. Zhu, J. Nat. Prod., 2022, 85, 433–440 CrossRef CAS PubMed.
- H.-W. Seo, N. S. Wassano, M. S. Amir Rawa, G. R. Nickles, A. Damasio and N. P. Keller, J. Fungi, 2024, 10, 266 CrossRef CAS PubMed.
- P. A. Jose, A. Maharshi and B. Jha, Microbiol. Res., 2021, 246, 126708 CrossRef CAS PubMed.
- L. K. Caesar, R. Montaser, N. P. Keller and N. L. Kelleher, Nat. Prod. Rep., 2021, 38, 2041–2065 RSC.
- W. Jaroszewicz, P. Bielańska, D. Lubomska, K. Kosznik-Kwaśnicka, P. Golec, Ł. Grabowski, E. Wieczerzak, W. Dróżdż, L. Gaffke and K. Pierzynowska, et al. , Antibiotics, 2021, 10, 1212 CrossRef CAS PubMed.
- M. Maciejewska, I. S. Pessi, A. Arguelles-Arias, P. Noirfalise, G. Luis, M. Ongena, H. Barton, M. Carnol and S. Rigali, Antonie van Leeuwenhoek, 2015, 107, 519–531 CrossRef CAS PubMed.
- C. J. L. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. Robles Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. Kashef Hamadani, E. A. P. Kumaran, B. McManigal, S. Achalapong, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F.-X. Babin, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, J. A. Berkley, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, R. Clotaire Donatien, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. De Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. Duong Bich, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, C. Garcia, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, Y. Hsia, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, A. W. J. Jenney, M. Khorana, S. Khusuwan, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, K. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, A. Manoharan, F. Marks, J. May, M. Mayxay, N. Mturi, T. Munera-Huertas, P. Musicha, L. A. Musila, M. M. Mussi-Pinhata, R. N. Naidu, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, T. J. Ochoa, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, P. Ounchanum, G. D. Pak, J. L. Paredes, A. Y. Peleg, C. Perrone, T. Phe, K. Phommasone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, S. Rattanavong, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, V. D. Rosenthal, K. E. Rudd, N. Russell, H. S. Sader, W. Saengchan, J. Schnall, J. A. G. Scott, S. Seekaew, M. Sharland, M. Shivamallappa, J. Sifuentes-Osornio, A. J. Simpson, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Tigoi, C. Turner, P. Turner, H. R. Van Doorn, S. Velaphi, A. Vongpradith, M. Vongsouvath, H. Vu, T. Walsh, J. L. Walson, S. Waner, T. Wangrangsimakul, P. Wannapinij, T. Wozniak, T. E. M. W. Young Sharma, K. C. Yu, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- N. Daneman, D. Fridman, J. Johnstone, B. J. Langford, S. M. Lee, D. M. MacFadden, K. Mponponsuo, S. N. Patel, K. L. Schwartz and K. A. Brown, eClinicalMedicine, 2022, 56, 101781 CrossRef PubMed.
- D. Adam, M. Maciejewska, A. Naômé, L. Martinet, W. Coppieters, L. Karim, D. Baurain and S. Rigali, Antibiotics, 2018, 7, 28 CrossRef PubMed.
- E. I. Silva, P. Jayasingha, S. Senanayake, A. Dandeniya and D. H. Munasinghe, Int. J. Speleol., 2021, 50, 4, DOI:10.5038/1827-806X.50.1.2343.
- J. T. Farmer, A. V. Shimkevitch, P. S. Reilly, K. D. Mlynek, K. S. Jensen, M. T. Callahan, K. L. Bushaw-Newton and J. B. Kaplan, J. Appl. Microbiol., 2014, 117, 1663–1673 CrossRef CAS PubMed.
- D. Rule and N. Cheeptham, Int. J. Speleol., 2013, 42, 147–153 CrossRef.
- I. V. Voytsekhovskaya, D. V. Axenov-Gribanov, S. A. Murzina, S. N. Pekkoeva, E. S. Protasov, S. V. Gamaiunov and M. A. Timofeyev, PeerJ, 2018, 6, e5832 CrossRef PubMed.
- V. Lamprinou, K. Tryfinopoulou, E. Velonakis, A. Vatopoulos, S. Antonopoulou, E. Fragopoulou, A. Pantazidou and A. Economou-Amilli, Int. J. Speleol., 2015, 44, 231–238 CrossRef.
- B. Lima, R. Scherer, U. Albino, F. Siqueira, J. Bitencourt and S. C. Santos, Sci. Plena, 2023, 19, 046201 CAS.
- A. Klusaite, V. Vickackaite, B. Vaitkeviciene, R. Karnickaite, D. Bukelskis, I. Kieraite-Aleksandrova and N. Kuisiene, Int. J. Speleol., 2016, 45, 275–287 CrossRef.
- S. D. Jastaniah, R. H. Amasha and A. H. Alanbari, IOSR Int. J. Pharm. Biol. Sci., 2019, 14, 26–33 Search PubMed.
- A. Pipite, P. J. Lockhart, P. A. McLenachan, K. Christi, D. Kumar, S. Prasad and R. Subramani, Front. Microbiol., 1012867, DOI:10.3389/fmicb.2022.1012867.
- L. Belyagoubi, N. Belyagoubi-Benhammou, V. Jurado, J. Dupont, S. Lacoste, F. Djebbah, F. Ounadjela, S. Benaissa, S. Habi, A. Abdelouahid and C. Saiz-Jimenez, Int. J. Speleol., 2018, 47, 189–199 CrossRef.
- N. A. Rachid, N. D. Güngör and J. Matthey, Technol. Rev., 2023, 67, 159–170 CAS.
- C. Riquelme, M. de L. Enes Dapkevicius, A. Z. Miller, Z. Charlop-Powers, S. Brady, C. Mason and N. Cheeptham, Appl. Microbiol. Biotechnol., 2017, 101, 843–857 CrossRef CAS PubMed.
- Y. Long, J. Jiang, X. Hu, J. Zhou, J. Hu and S. Zhou, World J. Microbiol. Biotechnol., 2019, 35, 153 CrossRef PubMed.
- J. Hamedi, M. Kafshnouchi and M. Ranjbaran, Saudi J. Biol. Sci., 2019, 26, 1587–1595 CrossRef CAS PubMed.
- M. Iquebal, A. Passari, J. Jagannadham, F. Ahmad, V. Leo, G. Singh, S. J, A. Rai, D. Kumar and B. Singh, Unveiling of Unique Microbiome Resource Having High Antimicrobial Peptide Activity Endowed With Agriculture and Industrial Applications From Pukzing Cave, 2021 Search PubMed.
- D. W. Denning, Lancet Infect. Dis., 2024, 24, e428–e438 CrossRef PubMed.
- J. Berman and D. J. Krysan, Nat. Rev. Microbiol., 2020, 18, 319–331 CrossRef CAS PubMed.
- A. Arastehfar, T. Gabaldón, R. Garcia-Rubio, J. D. Jenks, M. Hoenigl, H. J. F. Salzer, M. Ilkit, C. Lass-Flörl and D. S. Perlin, Antibiotics, 2020, 9, 877 CrossRef CAS PubMed.
- M. T. Toda, S. R. Williams, E. L. Berkow, M. M. Farley, L. H. Harrison, L. Bonner, K. M. Marceaux, R. Hollick, A. Y. Zhang, W. Schaffner, S. R. Lockhart, B. R. Jackson and S. Vallabhaneni, MMWR Surveill. Summ., 2019, vol. 68, pp. 1–15 Search PubMed.
- S. Bhattacharya, S. Sae-Tia and B. C. Fries, Antibiotics, 2020, 9, 312 CrossRef CAS PubMed.
- C. U. Meteyer, E. L. Buckles, D. S. Blehert, A. C. Hicks, D. E. Green, V. Shearn-Bochsler, N. J. Thomas, A. Gargas and M. J. Behr, J. Vet. Diagn. Invest., 2009, 21, 411–414 CrossRef PubMed.
- M. L. Verant, C. U. Meteyer, J. R. Speakman, P. M. Cryan, J. M. Lorch and D. S. Blehert, BMC Physiol., 2014, 14, 10 CrossRef PubMed.
- D. M. Reeder, C. L. Frank, G. G. Turner, C. U. Meteyer, A. Kurta, E. R. Britzke, M. E. Vodzak, S. R. Darling, C. W. Stihler, A. C. Hicks, R. Jacob, L. E. Grieneisen, S. A. Brownlee, L. K. Muller and D. S. Blehert, PLoS One, 2012, 7, e38920 CrossRef CAS PubMed.
- K. J. Vanderwolf, L. J. Campbell, T. L. Goldberg, D. S. Blehert and J. M. Lorch, ISME J., 2021, 15, 909–920 CrossRef CAS PubMed.
- M. G. Bangera and L. S. Thomashow, J. Bacteriol., 1999, 181, 3155–3163 CrossRef CAS PubMed.
- P. S. Hamm, N. A. Caimi, D. E. Northup, E. W. Valdez, D. C. Buecher, C. A. Dunlap, D. P. Labeda and A. Porras-Alfaro, Antonie van Leeuwenhoek, 2019, 112, 1297–1305 CrossRef CAS PubMed.
- P. S. Hamm, C. A. Dunlap, M. W. Mullowney, N. A. Caimi, N. L. Kelleher, R. J. Thomson, A. Porras-Alfaro and D. E. Northup, Antonie van Leeuwenhoek, 2020, 113, 2213–2221 CrossRef CAS PubMed.
- C. Gasparetti, P. Buzzini, M. R. Cramarossa, B. Turchetti, U. M. Pagnoni and L. Forti, Enzyme Microb. Technol., 2006, 39, 1341–1346 CrossRef CAS.
- B. Çandiroğlu, N. D. Güngör and J. Matthey, Technol. Rev., 2020, 64, 466–479 Search PubMed.
- J. Ferlay, M. Colombet, I. Soerjomataram, D. M. Parkin, M. Piñeros, A. Znaor and F. Bray, Int. J. Cancer, 2021, 149, 778–789 CrossRef CAS PubMed.
- N. Nakaew, R. Sungthong, J. Ortega-Calvo and S. Lumyong, in Microbes in applied research: Current advances and challenges, World Scientific, 2012, pp. 474–480 Search PubMed.
- R. S. dos Santos, U. B. Albino, K. S. Paludo, S. Y. S. Silva, M. N. Oliveira, D. de Alexandria Santos, A. M. do Rosário Marinho, P. B. Marinho, T. da Costa Lima and G. de F. N. dos Santos, et al. , Sci. Plena, 2021, 17 Search PubMed.
- R. Rautela, S. Rawat, R. Rawat, P. Verma and A. Bhatt, Environ. Conserv. J., 2017, 18, 115–122 CrossRef CAS.
- A. S. Ayangbenro, O. S. Olanrewaju and O. O. Babalola, Front. Microbiol., 01986, DOI:10.3389/fmicb.2018.01986.
- M. dos, P. G. Baltazar, L. H. Gracioso, I. R. Avanzi, B. Karolski, J. A. S. Tenório, C. A. O. do Nascimento and E. A. Perpetuo, J. Mater. Res. Technol., 2019, 8, 475–483 CrossRef.
- G. Cecchi, E. Roccotiello, S. Di Piazza, A. Riggi, M. G. Mariotti and M. Zotti, J. Environ. Sci. Health, Part B, 2017, 52, 166–170 CrossRef CAS PubMed.
- C. G. Lemes, I. F. Cordeiro, C. H. De Paula, A. K. Silva, F. F. Do Carmo, L. H. Kamino, F. M. Carvalho, J. C. Caicedo, J. A. Ferro and L. M. Moreira, Sustainability, 2021, 13, 9354 CrossRef CAS.
- G. Venkadesaperumal, N. Amaresan and K. Kumar, Braz. J. Microbiol., 2014, 45, 1271–1281 CrossRef CAS PubMed.
- B. Herzog Velikonja, R. Tkavc and L. Pašić, et al. , Int. J. Speleol., 2014, 43, 5 Search PubMed.
- K. E. Bender, K. Glover, A. Archey and H. A. Barton, Int. J. Speleol., 49, 3 Search PubMed.
- D. B. Raudabaugh, N. A. Rivera, G. C. Anchor, E. Bach, A. N. Miller and N. E. Mateus-Pinilla, Diversity, 2021, 13, 188 CrossRef CAS.
- B.-Z. Fang, N. Salam, M.-X. Han, J.-Y. Jiao, J. Cheng, D.-Q. Wei, M. Xiao and W.-J. Li, Front. Microbiol., 2017, 8, 1535 CrossRef PubMed.
- T. Sadoway, D. Rule, K. Watson, P. Moote, L. C. Soliman, N. Azad, K. Donkor and D. Horne, et al. , Int. J. Speleol., 2013, 42, 5 Search PubMed.
- L. Laiz, M. Gonzalez-Delvalle, B. Hermosin, A. Ortiz-Martinez and C. Saiz-Jimenez, Geomicrobiol. J., 2003, 20, 479–489 CrossRef CAS.
- D. Bukelskis, D. Dabkeviciene, L. Lukoseviciute, A. Bucelis, I. Kriaučiūnas, J. Lebedeva and N. Kuisiene, Front. Microbiol., 02149, DOI:10.3389/fmicb.2019.02149.
- P. S. Salazar-Hamm, J. J. M. Hathaway, A. S. Winter, N. A. Caimi, D. C. Buecher, E. W. Valdez and D. E. Northup, FEMS Microbes, 2022, 3, xtac012 CrossRef PubMed.
- H. Jeong, S.-K. Choi, C.-M. Ryu and S.-H. Park, Front. Microbiol., 2019, 10, 467 CrossRef PubMed.
- M. T. Henke, A. A. Soukup, A. W. Goering, R. A. McClure, R. J. Thomson, N. P. Keller and N. L. Kelleher, ACS Chem. Biol., 2016, 11, 2117–2123 CrossRef CAS PubMed.
- F. Xu, B. Nazari, K. Moon, L. B. Bushin and M. R. Seyedsayamdost, J. Am. Chem. Soc., 2017, 139, 9203–9212 CrossRef CAS PubMed.
- F. M. Locatelli, K.-S. Goo and D. Ulanova, Metallomics, 2016, 8, 469–480 CrossRef CAS PubMed.
- G. Pishchany, E. Mevers, S. Ndousse-Fetter, D. J. Horvath, C. R. Paludo, E. A. Silva-Junior, S. Koren, E. P. Skaar, J. Clardy and R. Kolter, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10124–10129 CrossRef CAS PubMed.
- V. Taleski, I. Dimkić, B. Boev, I. Boev, S. Živković and S. Stanković, FEMS Microbiol. Ecol., 2020, 96, fiaa155 CrossRef CAS PubMed.
- B. Thompson, D. Richardson, R. Vangundy, A. B. Cahoon and J. Cave, Karst Stud., 2019, 244–253 CrossRef.
- N. Ziemert, M. Alanjary and T. Weber, Nat. Prod. Rep., 2016, 33, 988–1005 RSC.
- A. Ayuso-Sacido and O. Genilloud, Microb. Ecol., 2005, 49, 10–24 CrossRef CAS PubMed.
- W. Liu, J. Ahlert, Q. Gao, E. Wendt-Pienkowski, B. Shen and J. S. Thorson, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 11959–11963 CrossRef CAS PubMed.
- M. Metsä-Ketelä, V. Salo, L. Halo, A. Hautala, J. Hakala, P. Mäntsälä and K. Ylihonko, FEMS Microbiol. Lett., 1999, 180, 1–6 CrossRef.
- M. L. Timmermans, Y. P. Paudel and A. C. Ross, Mar. Drugs, 2017, 15, 235 CrossRef PubMed.
- M. Montoya-Giraldo, K. R. Piper, O. O. Ikhimiukor, C. J. Park, N. A. Caimi, D. C. Buecher, E. W. Valdez, D. E. Northup and C. P. Andam, Microb. Genomics, 2024, 10, 001238 CAS.
- A. Rego, A. G. G. Sousa, J. P. Santos, F. Pascoal, J. Canário, P. N. Leão and C. Magalhães, Microorganisms, 2020, 8, 279 CrossRef PubMed.
- A. Wiseschart, W. Mhuantong, S. Tangphatsornruang, D. Chantasingh and K. Pootanakit, BMC Microbiol., 2019, 19, 144 CrossRef PubMed.
- B. Samanta, S. Sharma and R. Budhwar, Curr. Microbiol., 2023, 80, 317 CrossRef CAS PubMed.
- L. A. Hug, B. J. Baker, K. Anantharaman, C. T. Brown, A. J. Probst, C. J. Castelle, C. N. Butterfield, A. W. Hernsdorf, Y. Amano, K. Ise, Y. Suzuki, N. Dudek, D. A. Relman, K. M. Finstad, R. Amundson, B. C. Thomas and J. F. Banfield, Nat. Microbiol., 2016, 1, 1–6 Search PubMed.
- K. L. Cross, J. H. Campbell, M. Balachandran, A. G. Campbell, C. J. Cooper, A. Griffen, M. Heaton, S. Joshi, D. Klingeman, E. Leys, Z. Yang, J. M. Parks and M. Podar, Nat. Biotechnol., 2019, 37, 1314–1321 CrossRef CAS PubMed.
- J. Schultz, F. Modolon, R. S. Peixoto and A. S. Rosado, Front. Microbiol., 2023, 14, 1167718 CrossRef PubMed.
- S. Sandmann, A. O. de Graaf, B. A. van der Reijden, J. H. Jansen and M. Dugas, PLoS One, 2017, 12, e0171983 CrossRef PubMed.
- P. N. Tran, M.-R. Yen, C.-Y. Chiang, H.-C. Lin and P.-Y. Chen, Appl. Microbiol. Biotechnol., 2019, 103, 3277–3287 CrossRef CAS PubMed.
- S. Liu, C. D. Moon, N. Zheng, S. Huws, S. Zhao and J. Wang, Microbiome, 2022, 10, 76 CrossRef CAS PubMed.
- P. Suárez-Moo and A. Prieto-Davó, MicrobiologyOpen, 2024, 13, e1407 CrossRef PubMed.
- G. Nicolosi, S. Mammola, L. Verbrugge and M. Isaia, Biol. Rev. Cambridge Philos. Soc., 2023, 98, 849–867 CrossRef PubMed.
- M. C. Laker, Mining, 2023, 3, 205–220 CrossRef.
- L. Hao, B. Zhang, C. Feng, Z. Zhang, Z. Lei and K. Shimizu, Chemosphere, 2021, 263, 128246 CrossRef CAS PubMed.
- L. C. Razanamahandry, H. Karoui, H. Andrianisa and H. Yacouba, Afr. J. Environ. Sci. Technol., 2017, 11, 272–291 CrossRef CAS.
- B. Candiroglu and N. Dogruoz Gungor, Eur. J. Biol., 2017, 76, 36–42 CrossRef.
- J. R. Hoyt, A. M. Kilpatrick and K. E. Langwig, Nat. Rev. Microbiol., 2021, 19, 196–210 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |