Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Antarctic bacterial natural products: from genomic insights to drug discovery†
William
Medeiros
a,
Stanislava
Kralova
 bc,
Valéria
Oliveira
a,
Nadine
Ziemert
bc,
Valéria
Oliveira
a,
Nadine
Ziemert
 *de and
Ludek
Sehnal
*de and
Ludek
Sehnal
 *def
*def
aMicrobial Resources Division, Research Center for Chemistry, Biology, and Agriculture (CPQBA), Universidade Estadual de Campinas (UNICAMP), Paulínia, São Paulo, Brazil
bDivision of Microbial Ecology, Centre for Microbiology and Environmental Systems Science, University of Vienna, Vienna, Austria
cDepartment of Chemistry and Biochemistry, Faculty of AgriSciences, Mendel University in Brno, Brno, Czech Republic
dInterfaculty Institute of Microbiology and Infection Medicine Tübingen, Institute for Bioinformatics and Medical Informatics (IBMI), University of Tübingen, Tübingen, Germany. E-mail: nadine.ziemert@uni-tuebingen.de
eGerman Center for Infection Research (DZIF), Partner Site Tübingen, Tübingen, Germany
fMasaryk University, Faculty of Science, RECETOX, Kamenice 753/5, 625 00 Brno, Czech Republic. E-mail: ludek.sehnal@recetox.muni.cz
First published on 25th February 2025
Abstract
Covering: up to the end of 2024
Microbial life dominates the extreme continent Antarctica, playing a pivotal role in ecosystem functioning and serving as a reservoir of specialized metabolites known as natural products (NPs). NPs not only contribute to microbial adaptation to harsh conditions but also modulate microbial community structure. Long-term isolation and environmental pressures have shaped the genomes of Antarctic bacteria, suggesting that they also encode unique NPs. Since NPs are also an important source of drugs, we argue that investigating Antarctic bacterial NPs is essential not only for understanding their ecological role and evolution, but also for discovering new chemical structures, biosynthetic mechanisms, and potential new drugs. Yet, despite advances in omics technologies and increased scientific activities in Antarctica, relatively few new bacterial NPs have been discovered. The lack of systematic research activities focused on the exploration of Antarctic bacteria and their NPs constitutes a big problem considering the climate change issue, to which ecosystems in polar regions are the most sensitive areas on the Earth. Here, we highlight the currently available data on Antarctic bacteria, their biosynthetic potential, and the successful NP discoveries, while addressing the challenges in NP research and advocating for systematic, collaborative efforts aligned with the Antarctic Treaty System and the Antarctic Conservation Biogeographic Regions.
1. Introduction
Antarctica represents a primarily pristine and extreme environment that is biologically and climatically isolated from the rest of the world by the Antarctic Convergence. This natural boundary, where cold Antarctic waters meet and sink beneath warmer sub-Antarctic waters, creates a unique ecological barrier, maintaining distinct ecosystems and fostering the evolution of specialized life forms. Despite its isolation, Antarctica plays an essential role in global climate regulation and ocean ecosystem functioning.Microbes are the most dominant forms of life on this continent, where they play a significant role in biogeochemical cycles.1,2 Among the tools microbes use to interact with their environment, specialized metabolites – also known as natural products (NPs) – are particularly important. Produced notably by bacteria, these compounds help them adapt to extreme conditions and play a crucial role in shaping the composition and interactions of microbial communities, both within and between kingdoms.3,4 From the human perspective, investigating bacterial NPs in an environment as unique and extreme as Antarctica is crucial not only for the discovery of novel drugs but also for understanding their role in Antarctic ecosystems. These studies offer insights into the logic, structural diversity, biochemistry, and evolution of NP biosynthetic pathways under extreme conditions.
The increased scientific activities in Antarctica during the last decades and the broader availability of powerful omics technologies provided the first insights into microbial community composition, microbial diversity, and NP-encoding biosynthetic genes.5–7 Using both culture-dependent and -independent methods, a small set of studies showed high bacterial diversity, widespread endemism, and, importantly, a tremendous and unique diversity of NP biosynthetic genes as compared to sequences stored in public databases.5,7–10 Despite the indicated biosynthetic potential, relatively few bacterial NPs from Antarctic ecosystems have been discovered which is alarming in the light of the current speed of climate change (see ESI data†).
Although climate change influences all regions around the world, the polar regions are the most sensitive ones, since even moderate alterations in the environmental conditions have a significant impact on the landscape structure together with the associated ecosystems, many of which will completely vanish in the near future (e.g. cryoconites on melting glaciers).11,12 Antarctica is the most isolated and unexplored continent on the planet13 and the threat of perturbations due to climate change can undermine attempts to understand the unique evolutionary history of life on this continent, including diversity and evolution of NPs, a key to understanding microbial adaptations and ecological dynamics in this extreme environment.
This article will: (i) contrast data on different Antarctic ecosystems and the number of studies concerning bacterial NPs, while addressing the divergent evolution of Antarctic microbes and its implications for NP uniqueness; (ii) explore the biosynthetic potential for novel NP discovery revealed by (meta)genome mining; (iii) highlight successful NP discoveries using diverse methodological approaches; and (iv) emphasize the importance of systematic, collaborative research on bacterial NPs in Antarctic microbial communities aligned with the Antarctic Treaty System (ATS) and the Antarctic Conservation Biogeographic Regions (ACBRs).
2. Antarctic ecosystems and natural products research
Antarctica's extremely cold environment, limited nutrient availability, and short vegetative seasons make it an inhospitable region for most macroscopic organisms. As a result, microbes are the most dominant forms of life in terrestrial Antarctica, where they play a critical role in ecosystem functioning and biogeochemical cycles. They are also recognized as a promising source of NPs with significant potential for human applications.14 Over the last two decades, studies of microbiome composition and diversity across various Antarctic ecosystems5,15–17 have revealed highly diverse microbial communities with widespread endemism.8,18,19 However, our understanding of the evolution of these microbes and their NP biosynthetic pathways across different Antarctic ecosystems remains limited. Furthermore, climate change is driving significant shifts in microbial community structure, likely altering their metabolic repertoire and associated functions.202.1 Microbial life across Antarctic ecosystems
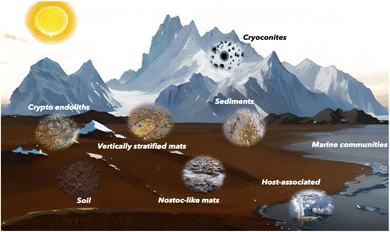
Antarctica is often described as a polar desert due to its extreme aridity and harsh conditions.21 Despite this characterization, the continent hosts a surprising diversity of ecosystems, particularly at the interface between glaciated and deglaciated regions. Deglaciated parts of Antarctica give rise to freshwater habitats such as lakes, melting ponds, streams, and seepages/wetlands.22 They also allow access to soils and sediments. In addition, glacial and snow-covered environments harbor specialized habitats unique to polar regions, including cryoconites, which are microbe-rich granules found on ice surfaces. Together, these environments constitute Antarctic terrestrial ecosystems.23 However, marine ecosystems are equally significant, hosting a critical part of Antarctic microbial diversity and ecological functioning.24–26 For a detailed description of Antarctic terrestrial and marine habitats, we direct interested readers to comprehensive reviews on these topics.22,27–29Environmental conditions shape the structure of microbial communities so that a wide range of community structures occur across Antarctic ecosystems (Fig. 1). Hotspots of Antarctic primary production include vertically stratified microbial mats in lakes, where stratification is driven by physicochemical conditions. These mats are characterized by upper layers dominated by cyanobacteria.30 Cyanobacteria are even more dominant in microbial mats typical of wetlands and seepages, where filaments of cyanobacteria from the genus Nostoc form the basis of the community. Nevertheless, the members of the phyla Pseudomonadota and Bacteriodota are equally abundant in many Antarctic ecosystems, including lakes or streams. Among these, Pseudomonadota represents the most diverse phylum within the microbial mats. Additionally, Bacillota, Actinomyceota, Chloroflexota, Verrucomicrobiota, and Deinococcota frequently occur in the Antarctic microbial mats with their composition strongly dependent on the mat type.22 Notably, a recent study showed that older lakes host significantly more diverse bacterial communities than young lakes formed by deglaciation.31 Despite this microbial diversity and the presence of well-recognized prolific producers of NPs, the biosynthetic potential of these microbial mats remains unexplored.
Cyanobacteria also dominate streams where simple biofilms can grow to vertically stratified microbial mats.32 In less microbially colonized ecosystems, such as rocks (lithic environments), cyanobacteria or lichen-dominated biofilms typically occur.33 A unique microbial community structure is found in cryoconites, where mineral granules create a distinctive ecological niche for glacial microbes.34 In soils, microbial community composition is largely driven by soil types.35 Marine ecosystems, by contrast, exhibit highly diverse microbial communities, varying spatially from coastal areas to the open ocean, and vertically from sunlit surface waters to the deep ocean floor.36 This remarkable diversity of microbial ecosystems strongly suggests the diversification of NPs and the presence of unique NP profiles across distinct environments.
2.2 Microbial evolution in Antarctica as a ground for investigation of NPs evolution
The evolution of microbial life in Antarctica has been strongly influenced by long-term glaciation and isolation from the rest of the world. Although various hypotheses exist regarding the global dispersion of microorganisms (e.g., the global ubiquity hypothesis), a recent study by Tytgat et al. (2023)37 provided new insights into the evolution of microorganisms in polar lakes. Their research revealed that only 1% of microbial taxonomic diversity, measured in operational taxonomic units (OTUs) and amplicon sequence variants (ASVs), detected in Antarctic lakes overlaps with that of other polar regions. This finding highlights a deep phylogenetic divergence of polar microbiota, with many clades restricted to specific biogeographical regions. Moreover, the same study showed that the net diversification rates of regionally restricted taxa significantly differ between polar regions. Notably, diversification rates are higher in unsaturated niches, allowing for the accumulation of new genotypes. Antarctica is likely an unsaturated niche,37 providing unique opportunities for microbial evolution, including the diversification of biosynthetic gene clusters (BGCs) encoding bacterial NPs. All these findings suggest long-term evolutionary divergence of bacteria in Antarctic lakes because of low interhemispheric dispersal and isolated diversification.Since NPs constitute grounds of molecular co-evolution and ecological relationships among different forms of life,38 it can be strongly anticipated that the long-term evolutionary divergence of polar microbiota has been reflected in the long-term evolution of pathways encoding NP biosynthesis associated with the accumulation of new genotypes in unsaturated Antarctic niches.
However, to our knowledge, no information is currently available on the biosynthetic potential of bacteria in Antarctic lakes or other freshwater ecosystems, nor on the evolution of NP biosynthetic pathways within these environments. To date, studies on bacterial NPs and their respective biosynthetic pathways in Antarctica have been limited to soils and sediments, using either culture-independent7,9,10 or culture-dependent39,40 approaches. Despite this, these studies provide valuable insights into the diversity and divergence of BGCs harbored by Antarctic bacteria.
2.3 Sequencing data availability across Antarctic ecosystems
One of the main objectives of this chapter is not only to show the types of diverse ecosystems that shape NP biosynthesis in Antarctic microbes but also to highlight what data are currently (un)available, what is their current use towards NP research, and what type of data from which type of ecosystem should be systematically produced.Next-generation sequencing has revolutionized bioprospecting strategies, enabling rapid access to microbial diversity and biosynthetic potential across ecosystems.41–43 Antarctic ecosystems represent no exception: an overview of the currently available shotgun metagenome sequencing data in four selected nucleotide sequence repositories is shown in Table 1. While samples from soils and sediments are available across all investigated databases, marine samples are present in ¾ of the databases. However, freshwater, glacial/snow, rock, and host-associated ecosystems remain strongly under-represented. Only 7 (0.25% of all) Antarctic soils or sediment samples were studied to identify the biosynthetic potential of microbes in these samples.9,10
| Environment | SRA | JGI | MGnify | MG rast | Ecosystemsa |
|---|---|---|---|---|---|
| a A1 – permafrost, A2 – upper soil layer, A3 – lake sediments, A4 – polygons; B1 – lake, B2 – melting pond, B3 – stream, B4 – wet land (seepages), B5 – wet rocks; C1 – plankton, C2 – marine sediments; C3 – water column; D1 – animals; D2 – plants, D3 – lichens, D4 – human; E1 – cryoconites, E2 – glacial lakes, E3 – snow surface; F1 – endolithic communities, F2 – airborne bacteria. | |||||
| Soils and sediments | 236 | 358 | 14 | 33 | A1, A3 |
| Freshwater ecosystems | 95 | 46 | — | — | B1, B2, B4 |
| Marine ecosystems | 208 | 134 | — | 8 | C1, C2, C3 |
| Host-associated | 49 | — | — | — | D4 |
| Glacial/snow ecosystems | 21 | 2 | — | — | E1 |
| Terrestrial and rocks | 239 | 102 | — | — | F1 |
| Others | 817 | 499 | — | 3 | A2, A4, B3, B5, D2–4, E2, F2 |
| TOTAL | 1665 | 1141 | — | — | — |
Another issue regarding the available data is the sequencing platform used for shotgun metagenome sequencing. Long-read sequencing data provide better resolution and completeness of BGCs and are therefore more suitable for BGC detection and characterization.48,49 However, long-read data from PacBio and Oxford Nanopore Technologies (ONT) constitute only 0.2% and 1.2%, respectively, of the available shotgun metagenome sequencing data stored in the Sequence Read Archive (SRA), while 95% of the Antarctic whole metagenome sequencing data have been generated using the Illumina sequencing platform. On one hand, short-read Illumina data can be problematic for the detection of BGCs encoding the production of polyketides and non-ribosomal peptides due to the highly repetitive nature of the biosynthetic genes.50 This may represent a bottleneck in the accurate mining of these Antarctic datasets.51,52 On the other hand, Illumina data are so far more accurate than long-read data, and thus, also useful, especially in follow-up experimental work. However, long-read sequencing is rapidly advancing53 and is expected to complement short-read sequencing, particularly enhancing metagenome studies in the future.
3. Biosynthetic potential in Antarctic bacteria
Exploring Antarctic bacterial genomes has uncovered genetic signatures indicative of evolutionary divergence, reflecting the long-term isolation and environmental pressures in Antarctica. This divergence is reflected in the unique enzymatic and biosynthetic potential of these bacterial communities.54,55 Antarctic bacterial metabolites exhibit distinct specialized adaptations to freezing temperatures, high UV radiation, and persistent nutrient limitation. These adaptations include cold-active enzymes, regulatory elements sensitive to environmental stressors, and structural modifications that enhance metabolite stability in extreme conditions.56–59 Comparative genomic studies have revealed that Antarctic-derived BGCs often contain unusual domain architectures, novel gene combinations, or regulatory features that distinguish them from their non-Antarctic counterparts, even when homologous BGCs are found in other environments.39,60 While certain biosynthetic pathways may be globally distributed, the selective pressures in Antarctica have likely contributed to the functional divergence of these clusters, leading to the production of specialized metabolites with potentially unique properties.39While the unique biosynthetic potential of Antarctic bacteria is evident, Fig. 2 highlights the uneven distribution of NP research efforts across the continent. Notably, studies are mostly concentrated in the Antarctic Peninsula, underscoring its importance as a primary site for such explorations. However, this focus also highlights that many other areas within Antarctica remain underexplored, suggesting significant opportunities for further investigation in less-studied regions and niches of this unique environment. Currently, available studies about the biosynthetic potential of Antarctic bacteria indicate a high diversity of terpene, non-ribosomal peptide, and polyketide BGCs, which are predicted to encode compounds with antitumor, antifungal, antibacterial, and biosurfactant properties.9,10,39,61 Unsurprisingly, the most striking BGC diversity and novelty are observed in understudied microbial phyla, particularly Acidobacteriota, Verrucomicrobiota, Gemmatimonadota, and rare Actinomycetota.7,9,62
Metagenomic analysis provides the most comprehensive approach for assessing the genetic NP potential in Antarctica. However, the exploration of bacterial biosynthetic potential within metagenomes is hindered by the limited availability of long-read sequencing data (see also Chapter 2). To date, only five studies have offered initial insights into the biosynthetic potential of Antarctic soil bacteria.7,9,10,62,63 These include two comprehensive metagenome analyses and three amplicon sequencing projects, focusing primarily on desert and maritime soils, as well as sediments. Despite their contributions, these studies have explored only a narrow range of Antarctic environments, leaving much of the continent's biosynthetic potential unexplored. A shotgun metagenomic analysis of Antarctic maritime soils by Waschulin et al. (2022)9 found that well-known NPs producers – Actinomycetota, Pseudomonadota, and Bacteroidota – encoded roughly 60% of all identified BGCs. Most BGCs were harbored in Actinomycetota, especially Streptomyces and Pseudonocardia. Less studied actinobacterial classes, Acidimicrobiia and Thermoleophilia, hosted the most unique BGCs, underlining the overlooked NP potential in these lineages. Additionally, the challenging-to-cultivate phyla Acidobacteriota and Verrucomicrobiota made up another 20% of the BGCs, with Acidobacteriota being particularly prevalent in Antarctic soils, making them promising subjects for further bioprospecting.64 Nevertheless, the study of Acidobacteriota is challenging worldwide due to their slow-growing nature and low genetic tractability. The isolation of the Acidobacteriota members and their successful genetic engineering to establish stable, fast-growing strains would be substantial achievements which would allow heterologous expression of BGCs from both other isolated strains and metagenomes worldwide.
The most abundant types of BGCs in Antarctic environments include terpenes, non-ribosomal peptide synthetases (NRPSs), polyketide synthases (PKSs) and bacteriocins, which encompass ribosomally synthesized and post-translationally modified peptides (RiPPs) and RiPP-like families.9,39 Supporting this, Medeiros et al. (2024)10 reported a high abundance of terpene BGCs in biofilms on Deception Island in Antarctica, influenced by both Antarctic and volcanic conditions.10,65 Their findings confirmed the presence of key BGC classes, including PKSs, NRPSs, terpenes, and RiPPs, all with significant drug discovery potential. These observations align with the results from the recently developed bioinformatics tool, BGC Atlas,66 which analyzed nearly two million BGCs across more than 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 metagenomes and identified a high abundance of terpenes in terrestrial samples.
000 metagenomes and identified a high abundance of terpenes in terrestrial samples.
Furthermore, Medeiros et al. (2024)10 highlighted the unique character of Antarctic BGCs, as evidenced by their limited overlap with entries in the MIBiG database,67 indicating untapped biosynthetic diversity. Notably, this study also revealed spatiotemporal variations in BGC distribution within the biofilm community, emphasizing the dynamic nature of biosynthetic activity.
Amplicon sequencing targeting NRPS and PKS domains identified Actinomycetota and Pseudomonadota as primary producers in Antarctic soils, with Cyanobacteria emerging as a notable source of NRPS.7,62,63 Additionally, these studies found evidence that a high percentage of adenylation (A) domains of NRPS and ketosynthase (KS) domain of PKS in Antarctic ecosystems are unassigned to known A/KS domains in reference databases and potentially endemic, suggesting untapped biosynthetic potential.
While the biosynthetic potential of terrestrial Antarctic microbiota has been explored to some extent, our understanding of marine counterparts remains limited (see also Chapter 2). Despite comprehensive studies on the structure and function of Antarctic marine microbial communities,68–72 investigations into their biosynthetic capabilities remain scarce. A global ocean microbiome survey, which included data from the Southern Ocean, highlighted the prevalence of NRPS and PKS BGCs.42 However, dedicated studies on Antarctic marine NP-synthesizing microbes are few. Notable exceptions include research on palmerolide biosynthesis in Verrucomicrobiota associated with the Antarctic ascidian Synoicum adareanum,73 and on the complexity of carotenoid biosynthesis in the Southern Ocean.74
Cultivation-based studies have complemented metagenomics by uncovering biotechnologically relevant NPs (comprehensively reviewed in 75–79). For example, Marisediminicola antarctica ZS314T, a likely endemic species isolated from Antarctica, exhibits diverse biosynthetic potential, including C50 carotenoids, oligosaccharides, salinixanthin, alkylresorcinol derivatives, and NRPS-encoded compounds.80 Similarly, Antarctic bacteria such as Planococcus and Rhodococcus strains produce carotenoids, surfactants, and siderophores, all of which hold significant biotechnological value.81 The unique evolutionary pressures of the Antarctic environment have shaped microbial biosynthetic capabilities, leading to novel BGCs. This is exemplified by an Antarctic Sphingomonas strain, which harbors divergent orthologous clusters related to pollutant degradation.60
Promising microorganisms from well-known antibiotic-producing phyla, such as Pseudomonadota, Actinomycetota, Cyanobacteriota, Bacillota, and Bacteroidota have been isolated from Antarctica.76 For instance, Streptomyces strain So13.3 produces potent antibiotics against Gram-positive bacteria, harboring 42 BGCs, many of which encode novel NRPS gene clusters.82 Similarly, Sphingomonas alpina So64.6b contains six distinct BGCs, including three with potential antibiotic activity.60 In addition, Antarctic isolates like Flavobacterium sp. Ant342 and Janthinobacterium sp. Ant5-2 produce the antimycobacterial pigments flexirubin and violacein, respectively.83 Aside from antimicrobial NPs, bacteria from Antarctic soils and plant rhizospheres exhibited diverse metabolic potentials, with some strains showing a high number of predicted genes for amino acid, carbohydrate, and xenobiotics metabolism, and anticancer/anticancerogenic compounds.61,84
4. NP – discoveries from Antarctica
In addition to genomic studies aimed at understanding the biosynthetic potential of bacterial NPs in Antarctica, phenotypic screening approaches have been widely employed to isolate bioactive compounds. These methods, which involve growth inhibition assays and other bioactivity tests, have successfully identified numerous bioactive bacterial strains from diverse Antarctic ecosystems.76,79,85–88 While many studies report the detection of bioactivities, only a small number have progressed to the purification and characterization of specific compounds. Based on our literature review, less than 40 structurally distinct bacterial NPs have been identified to date (end of the year 2024; see ESI data†). Nevertheless, these efforts have resulted in the discovery of several novel bacterial secondary metabolites,89–93 with a selection highlighted in Fig. 3.Contemporary studies have predominantly focused on quantifying and exploring the novelty of BGCs in bacterial strains.60,94–96 However, only a small number of isolate-centered NP studies incorporate chemical evaluations, which limits our understanding of the actual NPs produced by these microbes.80,84,97–99
Liao et al. (2019)80 showcase the power of integrating genomics and chemical analysis. Despite its relatively small genome (3.35 Mb), the Antarctic isolate M. antarctica ZS314T encoded five BGCs, including novel terpene, NRPS, PKS III, and oligosaccharide clusters. Detailed analysis of a terpene BGC suggested the production of a C50 carotenoid glucoside, a pigment known for its UV protection and antioxidant properties. This was supported by the extraction of a compound with UV-visible spectra and a molecular mass consistent with glycosylated carotenoids. While these findings highlight the potential of M. antarctica ZS314T to produce unique secondary metabolites, further purification and structural elucidation are needed to fully characterize the compound and its bioactivity.
The value of multifaceted approaches in Antarctic NP discovery was highlighted by Vitale et al. (2023).98 Genomic analysis of the Antarctic bacterium Lacinutrix shetlandiensis WUR7, which harbors only three biosynthetic gene clusters (BGCs), uncovered an unusual plant-like decarboxylase. This enzyme catalyzes the decarboxylation of L-tryptophan, removing its carboxyl group to produce tryptamine. Tryptamine serves as a precursor for various indole-based alkaloids. When the researchers supplemented the bacterial culture with L-tryptophan, they observed a significant increase in the production of indole alkaloids, including a novel compound named 8,9-dihydrocoscinamide B with antimicrobial activity against Staphylococcus aureus and MRSA (methicillin-resistant S. aureus) strains.
Chemical analysis also proves to be a powerful tool for NP discovery, as demonstrated by a study on Nocardiopsis sp. LX-1, an Antarctic krill isolate.100 This research, solely reliant on chemical analysis using molecular networking, successfully identified a novel antifungal compound named nocarpyrroline A, alongside 11 other structurally diverse secondary metabolites exhibiting antimicrobial activity. Nocarpyrroline A itself exhibited antifungal activity against Fusarium fujikuroi and antibacterial activity against Aeromonas hydrophila.
A pioneering study on the Antarctic bacterium Pseudomonas sp. ANT_H4 demonstrated the successful integration of genomic analysis with a fosmid expression system.101 This approach led to the identification of pyomelanin, a UV-protective pigment with remarkable multifunctional properties. Pyomelanin not only exhibited antioxidant activity and sun protection capabilities but also promoted plant growth, as demonstrated by its priming effects on Calendula officinalis hairy roots in vitro. These findings highlight the unique biosynthetic adaptations of Pseudomonas sp. ANT_H4 to extreme Antarctic conditions, with significant implications for biotechnological applications such as agricultural enhancers and skincare formulations.
Similarly, Chen et al. (2024)102 employed a combined strategy of genomics, chemical analysis, and heterologous expression to uncover weddellamycin, a metabolite produced by the Antarctic Streptomyces sp. DSS69. Weddellamycin exhibited potent anti-cancer activity, targeting cancer cell lines with significant efficacy, as well as broad-spectrum antimicrobial properties. The study also provided crucial insights into the biosynthetic pathway of weddellamycin, identifying not only the genes responsible for its synthesis but also key regulatory elements that influence its production. Remarkably, the pathway's structure suggests adaptations specific to the extreme Antarctic environment, including possible responses to cold stress and low-nutrient availability. These adaptations may enhance the efficiency and versatility of weddellamycin production, offering unique advantages for pharmaceutical applications. In addition to characterizing the compound, Chen et al. (2024)102 explored the heterologous expression of weddellamycin BGC, which successfully replicated its production in a model host. This approach paves the way for scalable production of weddellamycin and its derivatives, highlighting the potential for Antarctic microbes to serve as a sustainable source of bioactive compounds.
4.1 Challenges in Antarctic NP research
Modern NP discovery employs a powerful toolbox that integrates genomics, metagenomics, cheminformatics, metabolomics, and artificial intelligence to identify novel NPs from diverse sources. Antarctica represents still a pristine environment, although strongly influenced by climate change. The glacier ablation and global deglaciation speed increased during the last decade, which is significantly changing Antarctica including many unique ecosystems, some of which are irreversibly vanishing (e.g., cryoconites on melted glaciers).103 Despite that, Antarctica provides many underexplored ecosystems like freshwater, glacial, and rock environments that hold untapped potential. It also points out the urge for systematic biobanking of Antarctic samples and data. Limited sequencing data from these areas, coupled with the dominance of short-read sequencing technologies, further restricts the discovery and the resolution of complex BGCs. Incorporating long-read sequencing technologies could certainly enhance the detection and characterization of these clusters. Other innovative technologies that improve the field of NPs are expected, and systematic biobanking can conserve the valuable Antarctic samples and information for future re-analysis, from ecosystems that will not exist in that time.Moreover, while culture-independent methods have provided insights into the biosynthetic potential of Antarctic microbes, advanced culture-dependent approaches are also crucial. Integrating genomic, chemical, and synthetic biology approaches is necessary to unlock the full potential of Antarctic bacterial NPs. Successful studies combining these methodologies highlight the effectiveness of this integrated approach.
Although enzyme-centric explorations of Antarctic microbial sources have taken precedence in recent years,104–108 remarkably few studies have employed cutting-edge techniques to activate dormant or metagenome-sourced BGCs for NP production in Antarctic microbial life forms. Despite their rich biosynthetic potential, Antarctic bacterial BGCs remain largely unexplored using advanced methods, although promising examples exist, such as the above-mentioned studies utilizing heterologous expression to discover pyomelanin and weddellamycin.101,102
5. Future directions of NP research in Antarctica
A delicate balance between scientific exploration, and ethical, and environmental considerations is necessary for future exploration of the untapped NP diversity harbored by Antarctica's pristine environments. Antarctica is governed by the Antarctic Treaty System (ATS), a unique international agreement that prioritizes peace, scientific collaboration, and environmental protection. This system ensures that research activities uphold the highest ethical and sustainable standards. While the ATS promotes responsible research in Antarctica, the ACBRs109,110 provide a framework for targeted conservation efforts. This system divides Antarctica and surrounding islands into distinct biogeographic regions, allowing for more targeted and effective conservation strategies based on unique species assemblages, environmental conditions, and potential threats.5.1 Bioprospecting: balancing innovation with responsibility
The growing interest in NP research in Antarctica overlaps with the concept of “bioprospecting” – a search for valuable biological resources. While this presents exciting scientific opportunities, it also raises complex legal questions.111 One challenge lies in reconciling the ATS's principle of sharing scientific findings (ATCM Resolution 7, 2005) with intellectual property rights.112 While Resolution 5 on Biological Prospecting acknowledges the potential of bioprospecting to benefit humanity, it also emphasizes the importance of adhering to Article III (1) of the Antarctic Treaty. This article mandates the open and unrestricted sharing of scientific discoveries made in Antarctica – a cornerstone of collaborative research in the region. This can complicate the transition from basic research to commercialization, raising questions about ownership of potential benefits. Should they benefit the global community, scientists, research institutions, or commercial partners? Additionally, ensuring bioprospecting activities in compliance with Antarctica's nature reserve status is essential. As of 2021 (‘Antarctic Bioprospecting: SCAR Survey of Member Countries’), the ATS lacks comprehensive regulations and oversight mechanisms for bioprospecting activities, including the transition from research to commercialization. Addressing this gap requires collaboration among ATS member countries. The goal is to develop legislation that promotes scientific progress, maintains Antarctica's pristine environment, and ensures fair and ethical benefit-sharing on a global scale.5.2 Proposal for systematic collaborative research on Antarctic bacterial NPs
Our understanding and utilization of Antarctic bacterial NPs is currently limited, both in terms of potential applications and their ecological significance in extreme environments. While few studies have highlighted the evolutionary divergence of Antarctic bacteria and their BGCs or led to the identification of compounds with diverse bioactivities, there is a need to shift towards a more comprehensive approach that focuses on exploring novel NPs with bioactivities and understanding their evolution and role in extreme conditions, including the whole spectrum of Antarctic ecosystems. To bridge this gap and ensure sustainable and protective research on Antarctica's unique environment, a systematic and collaborative research effort is essential (Fig. 4). Key actions may include: | ||
| Fig. 4 A framework for sustainable and collaborative research on Antarctic microbial resources. Created in BioRender https://BioRender.com/d17z768. | ||
• Active biobanking initiatives: biobanking plays a critical role in the protection and availability of Antarctic microbial resources. By systematically collecting, preserving, and cataloging bacterial strains from various Antarctic ecosystems, biobanks ensure that these invaluable resources are safeguarded for future research and potential biotechnological applications. While pioneering efforts like the Australian Collection of Antarctic Microorganisms (ACAM)113 or the Culture Collection of Fungi from Extreme Environments (CCFEE, Italy)114 exist, the current landscape of Antarctic microbial biobanking remains fragmented. Given the significant research activity in Antarctica (75 research stations across 25 countries), a comprehensive assessment of existing biobanking efforts is necessary to identify potential gaps and avoid redundant sampling.
• Open sharing of Antarctic microbial strains: researchers should freely share isolated Antarctic microbial strains. To facilitate knowledge exchange and resource optimization, a centralized Antarctic platform, modeled after the Global Catalogue of Microorganisms (GCM),115 is essential. This platform would serve as a repository for strain information and accessibility details, facilitating resource sharing and collaboration within the Antarctic research community, minimizing redundant sampling, and promoting sustainable practices aligned with the ATS.
• Fostering transparency and collaboration through information sharing: transparent data sharing is crucial for maximizing research efficiency and Antarctic microbial discoveries. A comprehensive database modeled after the Global Biodiversity Information Facility (GBIF),116 could gather information on available bacterial strains, their DNA or protein sequences, metagenomic and transcriptomic profiles, and bioactivity results. Notably, Antarctica currently has the fewest entries on GBIF itself (https://www.GBIF.org, accessed 3 August 2024), highlighting the need for improved data sharing within the Antarctic research community.
• Joint studies using larger datasets: collaborative efforts should focus on conducting extensive joint studies utilizing larger datasets and diverse sources. This approach can provide a more comprehensive understanding of microbial diversity and biosynthetic potential than individual research endeavors. It also promotes efficient use of resources, significantly reducing the environmental footprint of Antarctic research.117 Additionally, research networks often provide additional funding opportunities for participating teams, further supporting collaborative initiatives and enhancing the overall impact of the research (e.g. European COST Actions or Biodiversa).
• Improved coordination of field studies and sampling expeditions: Antarctic logistics and operations are fossil fuel intensive.117,118 By optimizing these activities, researchers can minimize their environmental footprint while maximizing data collection and scientific output. Coordinated efforts, like the Antarctic Circumnavigation Expedition (ACE) organized by the Swiss Polar Institute, would help cover a wide range of ecological niches and seasonal variations and capture a broader diversity of microbial life and their metabolites. Careful planning and collaboration can reduce redundant efforts and minimize negative environmental impact of these expeditions.
• Cross-disciplinary collaboration: bringing together teams with diverse scientific backgrounds can enrich the research process. Collaboration between microbiologists, bioinformaticians, chemists, geologists and ecologists can provide a holistic approach to studying Antarctic microbial communities and their environmental drivers (such as during the ACE expeditions). This diversity in expertise ensures that research considers both ecological impacts and potential applications, fostering a balanced approach to sustainability and innovation.
• Regular workshops and conferences: regular workshops and conferences dedicated to Antarctic microbial research are crucial for fostering collaboration, knowledge exchange, and the dissemination of latest findings. While the International Conference on Polar and Alpine Microbiology (PAM) and the SCAR (Scientific Committee on Antarctic Research) Open Science Conference are valuable platforms, additional dedicated events are needed, such as those focused on NPs or other biotechnologically valuable outcomes. These gatherings should prioritize sustainable research practices, data sharing, and resource optimization to ensure scientific progress aligns with environmental protection.
To ensure the success of this proposal/initiative, we advocate for the involvement of both governmental bodies and international organizations. Governments can provide crucial funding, logistical support, and policy alignment with the ATS, while international organizations such as the Scientific Committee on Antarctic Research (SCAR), the International Council for Science (ICSU), the European Polar Board (EPB) or the United Nations Environment Programme (UNEP) can facilitate collaboration, data sharing, and sustainability frameworks. These actions will not only advance NP research but also significantly benefit the broader Antarctic microbiology community, as well as contribute to the preservation of Antarctica and its microbial resources.
6. Conclusion
The NP research in Antarctica holds great potential due to the unique environmental conditions that foster the evolution of distinct microbial communities. These microorganisms produce specialized metabolites, which are crucial for their survival and have significant biotechnological and medicinal potential. Despite advances in omics technologies, the discovery of new NPs from Antarctic bacteria remains limited, highlighting several pressing issues.The evolutionary divergence of Antarctic bacteria suggests the presence of unique BGCs encoding novel bioactive compounds. However, a comprehensive understanding of BGC diversity across Antarctic ecosystems is lacking, limiting the realization of their full biosynthetic potential. Moreover, Antarctic NP research must adhere to the ethical and legal frameworks established by the ATS. Sustainable, collaborative research efforts and transparent data sharing are essential to ensure responsible exploration and utilization of these microbial resources. By addressing these challenges through systematic and collaborative research, we can advance our understanding on NPs research on three major levels: (i) deciphering NPs ecological and evolutionary roles, (ii) uncovering their evolutionary history, and (iii) discovering novel NPs and unlocking their potential for transformative applications in medicine and biotechnology.
7. Data availability
No primary research results, software or code have been included, and no new data were generated or analyzed as part of this review.8. Author contributions
W. M., S. K. and L. S. were responsible for manuscript writing and preparation of graphical content. All authors were responsible for manuscript editing.9. Conflicts of interest
The authors declare no conflict of interest.10. Acknowledgements
This work was supported by the German Centre for Infection Research (DZIF) [grant number TTU09.704] to N. Z., by the Cluster of Excellence EXC2124 Controlling Microbes to Fight Infection (CMFI) – project ID 390838134, and by the Czech Antarctic Research Programme (CARP). WM has been supported by the Sao Paulo Research Foundation – FAPESP (grant 2020/11534-0). SK has been funded by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 101020356 and by the standard grant project GACR 25-17343S. LS has been funded by the European Union – Horizon Europe under the Marie Skłodowska-Curie Action (project NAfrAM – grant agreement No. 10106428). We also thank Libera Lo Presti for her invaluable comments on the manuscript. We acknowledge the use of OpenArt AI (https://openart.ai/) for Fig. 1 and the BioRender for Fig. 4 and Graphical Abstract.11. References
- D. A. Cowan, T. P. Makhalanyane, P. G. Dennis and D. W. Hopkins, Front. Microbiol., 2014, 5, 154 CrossRef PubMed.
- P. Han, X. Tang, H. Koch, X. Dong, L. Hou, D. Wang, Q. Zhao, Z. Li, M. Liu, S. Lücker and G. Shi, Nat. Commun., 2024, 15, 3143 CrossRef CAS PubMed.
- R. Mackelprang, P. Vaishampayan and K. Fisher, mSystems, 2022, 7, e01419–e01421 CrossRef PubMed.
- L. Weisskopf, S. Schulz and P. Garbeva, Nat. Rev. Microbiol., 2021, 19, 391–404 CrossRef CAS PubMed.
- I. S. Pessi, Y. Lara, B. Durieu, P. D. C. Maalouf, E. Verleyen and A. Wilmotte, FEMS Microbiol. Ecol., 2018, 94, fiy042 CrossRef PubMed.
- E. E. Jackson, I. Hawes and A. D. Jungblut, Polar Biol., 2021, 44, 823–836 CrossRef.
- A. Rego, A. G. G. Sousa, J. P. Santos, F. Pascoal, J. Canário, P. N. Leão and C. Magalhães, Microorganisms, 2020, 8, 279 CrossRef PubMed.
- W. Vyverman, E. Verleyen, A. Wilmotte, D. A. Hodgson, A. Willems, K. Peeters, B. Van de Vijver, A. De Wever, F. Leliaert and K. Sabbe, Polar Sci., 2010, 4, 103–113 CrossRef.
- V. Waschulin, C. Borsetto, R. James, K. K. Newsham, S. Donadio, C. Corre and E. Wellington, ISME J., 2022, 16, 101–111 CrossRef CAS PubMed.
- W. Medeiros, K. Hidalgo, T. Leão, L. M. de Carvalho, N. Ziemert and V. Oliveira, Microbiol. Spectr., 2024, 12, e0024424 CrossRef PubMed.
- B. Worm and H. K. Lotze, in Climate Change, ed. T. M. Letcher, Elsevier, 3rd edn, 2021, pp. 445–464 Search PubMed.
- A. G. Fountain, J. L. Campbell, E. A. G. Schuur, S. E. Stammerjohn, M. W. Williams and H. W. Ducklow, BioScience, 2012, 62, 405–415 CrossRef.
- R. B. Aronson, S. Thatje, J. B. McClintock and K. A. Hughes, Ann. N. Y. Acad. Sci., 2011, 1223, 82–107 CrossRef PubMed.
- A. G. Atanasov, S. B. Zotchev, V. M. Dirsch and C. T. Supuran, Nat. Rev. Drug Discov., 2021, 20, 200–216 CrossRef CAS PubMed.
- D. Obbels, E. Verleyen, M.-J. Mano, Z. Namsaraev, M. Sweetlove, B. Tytgat, R. Fernandez-Carazo, A. De Wever, S. D’hondt, D. Ertz, J. Elster, K. Sabbe, A. Willems, A. Wilmotte and W. Vyverman, FEMS Microbiol. Ecol., 2016, 92, fiw041 CrossRef PubMed.
- C.-W. Chong, D. A. Pearce and P. Convey, Front. Microbiol., 2015, 6, 1058 Search PubMed.
- B. Tytgat, E. Verleyen, M. Sweetlove, S. D’hondt, P. Clercx, E. Van Ranst, K. Peeters, S. Roberts, Z. Namsaraev, A. Wilmotte, W. Vyverman and A. Willems, FEMS Microbiol. Ecol., 2016, 92, fiw126 CrossRef PubMed.
- J. P. Kociolek, K. Kopalová, S. E. Hamsher, T. J. Kohler, B. Van de Vijver, P. Convey and D. M. McKnight, Polar Biol., 2017, 40, 1185–1196 CrossRef.
- E. Verleyen, B. Van de Vijver, B. Tytgat, E. Pinseel, D. A. Hodgson, K. Kopalová, S. L. Chown, E. Van Ranst, S. Imura, S. Kudoh, W. Van Nieuwenhuyze, A. Consortium, K. Sabbe and W. Vyverman, Ecography, 2021, 44, 548–560 CrossRef.
- H. J. Griffiths, V. J. Cummings, A. Van de Putte, R. J. Whittle and C. L. Waller, Nat. Rev. Earth Environ., 2024, 5, 645–664 CrossRef.
- J. van Mieghem and P. van Oye, Biogeography and Ecology in Antarctica, Springer Science & Business Media, 2013 Search PubMed.
- V. V. Doytchinov and S. G. Dimov, Life, 2022, 12, 916 CrossRef CAS PubMed.
- L. Sehnal, Czech Polar Rep., 2015, 5, 200–209 CrossRef.
- K. C. Lee, T. Caruso, S. D. J. Archer, L. N. Gillman, M. C. Y. Lau, S. C. Cary, C. K. Lee and S. B. Pointing, Front. Microbiol., 2018, 9, 2619 CrossRef PubMed.
- A. Lo Giudice, G. Caruso, C. Rizzo, M. Papale and M. Azzaro, J. Environ. Sustain., 2019, 2, 297–310 CAS.
- N. C. van Gestel, H. W. Ducklow and E. Bååth, Glob. Change Biol., 2020, 26, 2280–2291 CrossRef PubMed.
- D. Wilkins, S. Yau, T. J. Williams, M. A. Allen, M. V. Brown, M. Z. DeMaere, F. M. Lauro and R. Cavicchioli, FEMS Microbiol. Rev., 2013, 37, 303–335 CrossRef CAS PubMed.
- T. P. Makhalanyane, M. W. Van Goethem and D. A. Cowan, Curr. Opin. Biotechnol., 2016, 38, 159–166 CrossRef CAS PubMed.
- R. Cavicchioli, Nat. Rev. Microbiol., 2015, 13, 691–706 CrossRef CAS PubMed.
- J. Seckbach and A. Oren, Microbial Mats: Modern and Ancient Microorganisms in Stratified Systems, Springer Science & Business Media, 2010 Search PubMed.
- J. Kollár, K. Kopalová, J. Kavan, K. Vrbická, D. Nývlt, L. Nedbalová, M. Stibal and T. J. Kohler, FEMS Microbiol. Ecol., 2023, 99, fiad087 CrossRef PubMed.
- D. J. Van Horn, C. R. Wolf, D. R. Colman, X. Jiang, T. J. Kohler, D. M. McKnight, L. F. Stanish, T. Yazzie and C. D. Takacs-Vesbach, FEMS Microbiol. Ecol., 2016, 92, fiw148 CrossRef PubMed.
- A. Mezzasoma, C. Coleine, C. Sannino and L. Selbmann, Microb. Ecol., 2022, 83, 328–339 CrossRef PubMed.
- P. Rozwalak, P. Podkowa, J. Buda, P. Niedzielski, S. Kawecki, R. Ambrosini, R. S. Azzoni, G. Baccolo, J. L. Ceballos, J. Cook, B. Di Mauro, G. F. Ficetola, A. Franzetti, D. Ignatiuk, P. Klimaszyk, E. Łokas, M. Ono, I. Parnikoza, M. Pietryka, F. Pittino, E. Poniecka, D. L. Porazinska, D. Richter, S. K. Schmidt, P. Sommers, J. Souza-Kasprzyk, M. Stibal, W. Szczuciński, J. Uetake, Ł. Wejnerowski, J. C. Yde, N. Takeuchi and K. Zawierucha, Sci. Total Environ., 2022, 807, 150874 CrossRef CAS PubMed.
- S. S. Mantri, T. Negri, H. Sales-Ortells, A. Angelov, S. Peter, H. Neidhardt, Y. Oelmann and N. Ziemert, mSystems, 2021, 6, e0101821 CrossRef PubMed.
- J. A. Cram, C.-E. T. Chow, R. Sachdeva, D. M. Needham, A. E. Parada, J. A. Steele and J. A. Fuhrman, ISME J., 2015, 9, 563–580 CrossRef PubMed.
- B. Tytgat, E. Verleyen, M. Sweetlove, K. Van den Berge, E. Pinseel, D. A. Hodgson, S. L. Chown, K. Sabbe, A. Wilmotte, A. Willems, The Polar Lake Sampling Consortium and W. Vyverman, Sci. Adv., 2023, 9, eade7130 CrossRef CAS PubMed.
- P. S. Suresh, S. Kumari, D. Sahal and U. Sharma, Eur. J. Med. Chem., 2023, 260, 115748 CrossRef CAS PubMed.
- N. Benaud, R. J. Edwards, T. G. Amos, P. M. D'Agostino, C. Gutiérrez-Chávez, K. Montgomery, I. Nicetic and B. C. Ferrari, Environ. Microbiol., 2020, 23, 3646–3664 CrossRef PubMed.
- S. Soldatou, G. H. Eldjárn, A. Ramsay, J. J. J. van der Hooft, A. H. Hughes, S. Rogers and K. R. Duncan, Mar. Drugs, 2021, 19, 103 CrossRef PubMed.
- J. Chen, Y. Jia, Y. Sun, K. Liu, C. Zhou, C. Liu, D. Li, G. Liu, C. Zhang, T. Yang, L. Huang, Y. Zhuang, D. Wang, D. Xu, Q. Zhong, Y. Guo, A. Li, I. Seim, L. Jiang, L. Wang, S. M. Y. Lee, Y. Liu, D. Wang, G. Zhang, S. Liu, X. Wei, Z. Yue, S. Zheng, X. Shen, S. Wang, C. Qi, J. Chen, C. Ye, F. Zhao, J. Wang, J. Fan, B. Li, J. Sun, X. Jia, Z. Xia, H. Zhang, J. Liu, Y. Zheng, X. Liu, J. Wang, H. Yang, K. Kristiansen, X. Xu, T. Mock, S. Li, W. Zhang and G. Fan, Nature, 2024, 633, 371–379 CrossRef CAS PubMed.
- L. Paoli, H.-J. Ruscheweyh, C. C. Forneris, F. Hubrich, S. Kautsar, A. Bhushan, A. Lotti, Q. Clayssen, G. Salazar, A. Milanese, C. I. Carlström, C. Papadopoulou, D. Gehrig, M. Karasikov, H. Mustafa, M. Larralde, L. M. Carroll, P. Sánchez, A. A. Zayed, D. R. Cronin, S. G. Acinas, P. Bork, C. Bowler, T. O. Delmont, J. M. Gasol, A. D. Gossert, A. Kahles, M. B. Sullivan, P. Wincker, G. Zeller, S. L. Robinson, J. Piel and S. Sunagawa, Nature, 2022, 607, 111–118 CrossRef CAS PubMed.
- A. Gavriilidou, S. A. Kautsar, N. Zaburannyi, D. Krug, R. Müller, M. H. Medema and N. Ziemert, Nat. Microbiol., 2022, 7, 726–735 CrossRef CAS PubMed.
- E. W. Sayers, E. E. Bolton, J. R. Brister, K. Canese, J. Chan, D. C. Comeau, R. Connor, K. Funk, C. Kelly, S. Kim, T. Madej, A. Marchler-Bauer, C. Lanczycki, S. Lathrop, Z. Lu, F. Thibaud-Nissen, T. Murphy, L. Phan, Y. Skripchenko, T. Tse, J. Wang, R. Williams, B. W. Trawick, K. D. Pruitt and S. T. Sherry, Nucleic Acids Res., 2022, 50, D20–D26 CrossRef CAS PubMed.
- K. Palaniappan, I.-M. A. Chen, K. Chu, A. Ratner, R. Seshadri, N. C. Kyrpides, N. N. Ivanova and N. J. Mouncey, Nucleic Acids Res., 2020, 48, D422–D430 CAS.
- L. Richardson, B. Allen, G. Baldi, M. Beracochea, M. L. Bileschi, T. Burdett, J. Burgin, J. Caballero-Pérez, G. Cochrane, L. J. Colwell, T. Curtis, A. Escobar-Zepeda, T. A. Gurbich, V. Kale, A. Korobeynikov, S. Raj, A. B. Rogers, E. Sakharova, S. Sanchez, D. J. Wilkinson and R. D. Finn, Nucleic Acids Res., 2023, 51, D753–D759 CrossRef CAS PubMed.
- F. Meyer, D. Paarmann, M. D'Souza, R. Olson, E. Glass, M. Kubal, T. Paczian, A. Rodriguez, R. Stevens, A. Wilke, J. Wilkening and R. Edwards, BMC Bioinf., 2008, 9, 386 CrossRef CAS PubMed.
- M. W. Van Goethem, A. R. Osborn, B. P. Bowen, P. F. Andeer, T. L. Swenson, A. Clum, R. Riley, G. He, M. Koriabine, L. Sandor, M. Yan, C. G. Daum, Y. Yoshinaga, T. P. Makhalanyane, F. Garcia-Pichel, A. Visel, L. A. Pennacchio, R. C. O'Malley and T. R. Northen, Commun. Biol., 2021, 4, 1–10 CrossRef PubMed.
- R. Sánchez-Navarro, M. Nuhamunada, O. S. Mohite, K. Wasmund, M. Albertsen, L. Gram, P. H. Nielsen, T. Weber and C. M. Singleton, mSystems, 2022, 7, e0063222 CrossRef PubMed.
- N. Ziemert, S. Podell, K. Penn, J. H. Badger, E. Allen and P. R. Jensen, PLoS One, 2012, 7, e34064 CrossRef CAS PubMed.
- J. P. Gomez-Escribano, J. F. Castro, V. Razmilic, G. Chandra, B. Andrews, J. A. Asenjo and M. J. Bibb, BMC Genom., 2015, 16, 485 CrossRef PubMed.
- R. H. Baltz, J. Ind. Microbiol. Biotechnol., 2021, 48, kuab044 CrossRef CAS PubMed.
- M. Sereika, R. H. Kirkegaard, S. M. Karst, T. Y. Michaelsen, E. A. Sørensen, R. D. Wollenberg and M. Albertsen, Nat. Methods, 2022, 19, 823–826 CrossRef CAS PubMed.
- J. R. Ottoni, T. Rodrigues e Silva, V. Maia de Oliveira and M. R. Zambrano Passarini, Biocatal. Agric. Biotechnol., 2020, 23, 101452 CrossRef.
- D. Albanese, C. Coleine, O. Rota-Stabelli, S. Onofri, S. G. Tringe, J. E. Stajich, L. Selbmann and C. Donati, Microbiome, 2021, 9, 63 CrossRef CAS PubMed.
- T. Collins and G. Feller, Essays Biochem., 2023, 67, 701–713 CrossRef CAS PubMed.
- Z.-B. Zhang, Y.-L. Xia, G.-H. Dong, Y.-X. Fu and S.-Q. Liu, Int. J. Mol. Sci., 2021, 22, 1781 CrossRef CAS PubMed.
- K. Snopková, D. Čejková, K. Dufková, I. Sedláček and D. Šmajs, Arch. Microbiol., 2020, 202, 447–454 CrossRef PubMed.
- K. S. Siddiqui and R. Cavicchioli, Annu. Rev. Biochem., 2006, 75, 403–433 CrossRef CAS PubMed.
- K. Núñez-Montero, D. Rojas-Villalta and L. Barrientos, Front. Microbiol., 2022, 13, 1007225 CrossRef PubMed.
- L. J. Silva, E. J. Crevelin, D. T. Souza, G. V. Lacerda-Júnior, V. M. de Oliveira, A. L. T. G. Ruiz, L. H. Rosa, L. A. B. Moraes and I. S. Melo, Sci. Rep., 2020, 10, 13870 CrossRef CAS PubMed.
- C. Borsetto, G. C. A. Amos, U. N. da Rocha, A. L. Mitchell, R. D. Finn, R. F. Laidi, C. Vallin, D. A. Pearce, K. K. Newsham and E. M. H. Wellington, Microbiome, 2019, 7, 78 CrossRef PubMed.
- N. Benaud, E. Zhang, J. van Dorst, M. V. Brown, J. A. Kalaitzis, B. A. Neilan and B. C. Ferrari, FEMS Microbiol. Ecol., 2019, 95, fiz031 CrossRef CAS PubMed.
- G. Varliero, P. H. Lebre, B. Adams, S. L. Chown, P. Convey, P. G. Dennis, D. Fan, B. Ferrari, B. Frey, I. D. Hogg, D. W. Hopkins, W. Kong, T. Makhalanyane, G. Matcher, K. K. Newsham, M. I. Stevens, K. V. Weigh and D. A. Cowan, Microbiome, 2024, 12, 9 CrossRef PubMed.
- V. B. Centurion, G. V. Lacerda-Júnior, A. W. F. Duarte, T. R. Silva, L. J. Silva, L. H. Rosa and V. M. Oliveira, Sci. Total Environ., 2021, 758, 143671 CrossRef CAS PubMed.
- C. Bağcı, M. Nuhamunada, H. Goyat, C. Ladanyi, L. Sehnal, K. Blin, S. A. Kautsar, A. Tagirdzhanov, A. Gurevich, S. Mantri, C. von Mering, D. Udwary, M. H. Medema, T. Weber and N. Ziemert, Nucleic Acids Res., 2024, gkae953 Search PubMed.
- S. A. Kautsar, K. Blin, S. Shaw, J. C. Navarro-Muñoz, B. R. Terlouw, J. J. J. van der Hooft, J. A. van Santen, V. Tracanna, H. G. Suarez Duran, V. Pascal Andreu, N. Selem-Mojica, M. Alanjary, S. L. Robinson, G. Lund, S. C. Epstein, A. C. Sisto, L. K. Charkoudian, J. Collemare, R. G. Linington, T. Weber and M. H. Medema, Nucleic Acids Res., 2020, 48, D454–D458 Search PubMed.
- M. Moreno-Pino, A. Cristi, J. F. Gillooly and N. Trefault, Sci. Rep., 2020, 10, 645 CrossRef CAS PubMed.
- S. Cao, W. Zhang, W. Ding, M. Wang, S. Fan, B. Yang, A. Mcminn, M. Wang, B. Xie, Q.-L. Qin, X.-L. Chen, J. He and Y.-Z. Zhang, Microbiome, 2020, 8, 47 CrossRef CAS PubMed.
- A. I. Garber, J. R. Zehnpfennig, C. S. Sheik, M. W. Henson, G. A. Ramírez, A. R. Mahon, K. M. Halanych and D. R. Learman, mSphere, 2021, 6, e0077021 CrossRef PubMed.
- T. Alarcón-Schumacher, S. Guajardo-Leiva, M. Martinez-Garcia and B. Díez, mSystems, 2021, 6, e0039621 CrossRef PubMed.
- A. Dutta, E. Connors, R. Trinh, N. Erazo, S. Dasarathy, H. W. Ducklow, D. K. Steinberg, O. M. Schofield and J. S. Bowman, Front. Microbiol., 2023, 14, 1168507 CrossRef PubMed.
- A. E. Murray, C.-C. Lo, H. E. Daligault, N. E. Avalon, R. W. Read, K. W. Davenport, M. L. Higham, Y. Kunde, A. E. K. Dichosa, B. J. Baker and P. S. G. Chain, mSphere, 2021, 6, e0075921 CrossRef PubMed.
- W. Y. Cho and P. C. Lee, Microorganisms, 2024, 12, 390 CrossRef CAS PubMed.
- G. A. Quinn and P. J. Dyson, npj Antimicrob. Resist., 2024, 2, 1–9 CrossRef PubMed.
- K. Núñez-Montero and L. Barrientos, Antibiotics, 2018, 7, 90 CrossRef PubMed.
- M. B. Silva, A. O. Feitosa, I. G. O. Lima, J. R. S. Bispo, A. C. M. Santos, M. S. A. Moreira, P. E. A. S. Câmara, L. H. Rosa, V. M. Oliveira, A. W. F. Duarte and A. C. Queiroz, An. Acad. Bras. Cienc., 2022, 94, e20210840 CrossRef PubMed.
- J.-T. Liu, X.-L. Lu, X.-Y. Liu, Y. Gao, B. Hu, B.-H. Jiao and H. Zheng, Mini-Rev. Med. Chem., 2013, 13, 617–626 CrossRef CAS PubMed.
- V. C. Tripathi, S. Satish, S. Horam, S. Raj, A. lal, J. Arockiaraj, M. Pasupuleti and D. K. Dikshit, Polar Sci., 2018, 18, 147–166 CrossRef.
- L. Liao, S. Su, B. Zhao, C. Fan, J. Zhang, H. Li and B. Chen, Mar. Drugs, 2019, 17, 388 CrossRef CAS PubMed.
- M. Styczynski, A. Rogowska, K. Gieczewska, M. Garstka, A. Szakiel and L. Dziewit, Molecules, 2020, 25, 4357 CrossRef CAS PubMed.
- K. Núñez-Montero, C. Lamilla, M. Abanto, F. Maruyama, M. A. Jorquera, A. Santos, J. Martinez-Urtaza and L. Barrientos, Sci. Rep., 2019, 9, 7488 CrossRef PubMed.
- N. Mojib, R. Philpott, J. P. Huang, M. Niederweis and A. K. Bej, Antonie van Leeuwenhoek, 2010, 98, 531–540 CrossRef CAS PubMed.
- A. C. da Silva, C. T. C. da C. Rachid, H. E. de Jesus, A. S. Rosado and R. S. Peixoto, Polar Biol., 2017, 40, 1393–1407 CrossRef.
- Y. Tian, Y.-L. Li and F.-C. Zhao, Mar. Drugs, 2017, 15, 28 CrossRef PubMed.
- T. R. Silva, A. W. F. Duarte, M. R. Z. Passarini, A. L. T. G. Ruiz, C. H. Franco, C. B. Moraes, I. S. de Melo, R. A. Rodrigues, F. Fantinatti-Garboggini and V. M. Oliveira, Polar Biol., 2018, 41, 1505–1519 CrossRef.
- T. R. Silva, R. S. N. Tavares, R. Canela-Garayoa, J. Eras, M. V. N. Rodrigues, I. A. Neri-Numa, G. M. Pastore, L. H. Rosa, J. A. A. Schultz, H. M. Debonsi, L. R. G. Cordeiro and V. M. Oliveira, Mar. Biotechnol., 2019, 21, 416–429 CrossRef CAS PubMed.
- T. R. E. Silva, L. C. F. Silva, A. C. de Queiroz, M. S. Alexandre Moreira, C. A. de Carvalho Fraga, G. C. A. de Menezes, L. H. Rosa, J. Bicas, V. M. de Oliveira and A. W. F. Duarte, Crit. Rev. Biotechnol., 2021, 41, 809–826 CrossRef PubMed.
- C. Bruntner, T. Binder, W. Pathom-aree, M. Goodfellow, A. T. Bull, O. Potterat, C. Puder, S. Hörer, A. Schmid, W. Bolek, K. Wagner, G. Mihm and H.-P. Fiedler, J. Antibiot., 2005, 58, 346–349 CrossRef CAS PubMed.
- P. Tedesco, I. Maida, F. Palma Esposito, E. Tortorella, K. Subko, C. C. Ezeofor, Y. Zhang, J. Tabudravu, M. Jaspars, R. Fani and D. de Pascale, Mar. Drugs, 2016, 14, 83 CrossRef PubMed.
- H. Zhang, K. Saurav, Z. Yu, A. Mándi, T. Kurtán, J. Li, X. Tian, Q. Zhang, W. Zhang and C. Zhang, J. Nat. Prod., 2016, 79, 1610–1618 CrossRef CAS PubMed.
- C. Buonocore, P. Tedesco, G. A. Vitale, F. Palma Esposito, R. Giugliano, M. C. Monti, M. V. D'Auria and D. de Pascale, Mar. Drugs, 2020, 18, 269 CrossRef CAS PubMed.
- Y. Xiao, F. Yan, Y. Cui, J. Du, G. Hu, W. Zhai, R. Liu, Z. Zhang, J. Fang, L. Chen and X. Yu, Front. Microbiol., 2023, 13, 1085063 CrossRef PubMed.
- Y. Lim, I. Kang and J.-C. Cho, Sci. Rep., 2020, 10, 1–12 CrossRef PubMed.
- J. Schultz, M. T. D. Parise, D. Parise, L. G. Medeiros, T. J. Sousa, R. B. Kato, A. P. T. Uetanabaro, F. Araújo, R. T. J. Ramos, S. de Castro Soares, B. Brenig, V. A. de Carvalho Azevedo, A. Góes-Neto and A. S. Rosado, Microorganisms, 2022, 10, 1673 CrossRef CAS PubMed.
- Y. Wang, L. Ma, J. He, Z. Liu, S. Weng, L. Wang, J. He and C. Guo, BMC Microbiol., 2021, 21, 288 CrossRef CAS PubMed.
- S. Habib, S. A. Ahmad, W. L. Wan Johari, M. Y. Abd Shukor, S. A. Alias, J. Smykla, N. H. Saruni, N. S. Abdul Razak and N. A. Yasid, Int. J. Mol. Sci., 2020, 21, 6138 CrossRef CAS PubMed.
- G. A. Vitale, G. G. January, E. Oppong-Danquah, G. Della Sala, F. Palma Esposito, D. Tasdemir and D. de Pascale, PNAS Nexus, 2023, 2, pgad221 CrossRef PubMed.
- J. M. C. Shaffer, L.-A. Giddings, R. M. Samples and J. A. Mikucki, Front. Microbiol., 2023, 14, 1156033 CrossRef PubMed.
- T. Shi, Y.-J. Li, Z.-M. Wang, Y.-F. Wang, B. Wang and D.-Y. Shi, Mar. Drugs, 2023, 21, 127 CrossRef CAS PubMed.
- M. Styczynski, A. Rogowska, C. Nyabayo, P. Decewicz, F. Romaniuk, C. Pączkowski, A. Szakiel, R. Suessmuth and L. Dziewit, Microb. Cell Fact., 2022, 21, 261 CrossRef CAS PubMed.
- L. Chen, K. Liu, J. Hong, Z. Cui, W. He, Y. Wang, Z. Deng and M. Tao, Mar. Drugs, 2024, 22, 189 CrossRef CAS PubMed.
- M. C. Dryak and E. M. Enderlin, J. Glaciol., 2020, 66, 457–470 CrossRef.
- Y. Wang, H. Han, B. Cui, Y. Hou, Y. Wang and Q. Wang, Bioengineered, 2017, 8, 742–749 CrossRef CAS PubMed.
- Y. Hou, C. Qiao, Y. Wang, Y. Wang, X. Ren, Q. Wei and Q. Wang, Mar. Drugs, 2019, 17, 147 CrossRef CAS PubMed.
- A. C. Sanchez, M. C. Ravanal, B. A. Andrews and J. A. Asenjo, Protein Expr. Purif., 2019, 155, 78–85 CrossRef CAS PubMed.
- F. Karakaş and A. Arslanoğlu, Sci. Rep., 2020, 10, 22063 CrossRef PubMed.
- E. Rzoska-Smith, R. Stelzer, M. Monterio, S. C. Cary and A. Williamson, Front. Microbiol., 2023, 14, 1156817 CrossRef PubMed.
- A. Terauds, S. L. Chown, F. Morgan, H. J. Peat, D. J. Watts, H. Keys, P. Convey and D. M. Bergstrom, Divers. Distrib., 2012, 18, 726–741 CrossRef.
- A. Terauds and J. R. Lee, Divers. Distrib., 2016, 22, 836–840 CrossRef.
- R. Puig-Marcó, Mar. Genomics, 2014, 17, 73–78 CrossRef PubMed.
- M. W. Tvedt, Polar Rec., 2011, 47, 46–55 CrossRef.
- J. Bowman, The Australian Collection of Antarctic Microorganisms, Ver. 1, Australian Antarctic Division, https://data.aad.gov.au/metadata/ACAM, 2012, accessed 07.01.2024 Search PubMed.
- L. Selbmann, S. Onofri, L. Zucconi, D. Isola, M. Rottigni, C. Ghiglione, P. Piazza, M. C. Alvaro and S. Schiaparelli, MycoKeys, 2015, 10, 57–71 CrossRef.
- L. Wu, Q. Sun, H. Sugawara, S. Yang, Y. Zhou, K. McCluskey, A. Vasilenko, K.-I. Suzuki, M. Ohkuma, Y. Lee, V. Robert, S. Ingsriswang, F. Guissart, D. Philippe and J. Ma, BMC Genom., 2013, 14, 933 CrossRef PubMed.
- J. L. Edwards, BioScience, 2004, 54, 485–486 CrossRef.
- P. Elshout, J. Chappellaz, T. Gibéryen, C. Hansen, J. Jania, K. Jones-Williams, J. Nolan, B. Reverdy, E. Topp-Jørgensen, A. Yılmaz and R. Badhe, Synthesis Report on the Environmental Impacts of Polar Research and Logistics in the Polar Regions, Zenodo, 2023, DOI:10.5281/zenodo.79072342023.
- R. R. Cordero, E. Sepúlveda, S. Feron, A. Damiani, F. Fernandoy, S. Neshyba, P. M. Rowe, V. Asencio, J. Carrasco, J. A. Alfonso, P. Llanillo, P. Wachter, G. Seckmeyer, M. Stepanova, J. M. Carrera, J. Jorquera, C. Wang, A. Malhotra, J. Dana, A. L. Khan and G. Casassa, Nat. Commun., 2022, 13, 984 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4np00045e |
| This journal is © The Royal Society of Chemistry 2025 |