DOI:
10.1039/D4NH00654B
(Communication)
Nanoscale Horiz., 2025,
10, 1465-1477
pH-Triggered delivery of pirarubicin–gemcitabine duo using polymeric nanoparticles for synergistic breast cancer therapy†
Received
19th December 2024
, Accepted 22nd April 2025
First published on 21st May 2025
Abstract
Combination chemotherapy using nanocarriers presents a promising approach to overcome the restrictions associated with conventional chemotherapy, particularly by enhancing drug stability in the bloodstream, modulating pharmacokinetics to improve therapeutic efficacy and minimizing adverse side effects on the patient's health. In pursuit of an optical treatment approach for breast cancer, various chemotherapeutic drug combinations with advanced nanocarriers are being extensively explored. This study investigated the development of pirarubicin and gemcitabine co-loaded polymeric nanoparticles for synergistic activity against breast cancer cells. To enable sustained and site-specific delivery within the tumor microenvironment, both pirarubicin and gemcitabine were chemically conjugated to a polylactic acid-based block copolymer via a pH-responsive “Schiff's base” linkage. The synthesized polymer–drug conjugates were subsequently formulated into Pira–Gem co-loaded block copolymeric nanoparticles, demonstrating good stability and minimal toxicity towards non-cancerous cells. Pira–Gem co-loaded nanoparticles exhibited a significantly higher percentage of drug release under acidic pH conditions, (characteristic of tumor microenvironments) compared with physiological pH conditions. Furthermore, they showed superior cellular uptake on 2D adherent cancer cell lines relative to free drugs in in vitro studies. Both apoptotic analysis and cell proliferation inhibition studies revealed that the co-loaded nanoparticles exhibited a synergistic therapeutic effect across multiple breast cancer cell lines, surpassing the efficacy of Pira/Gem single drug-loaded nanoparticles and their free drug counterparts. These findings suggest that the Pira–Gem co-loaded nanoformulation holds considerable promise for breast cancer therapy and requires further exploration as a potential treatment strategy.
New concepts
The integration of nanotechnology and combination chemotherapy introduces a paradigm shift in breast cancer treatment by addressing the limitations of conventional approaches. This study pioneers the development of pirarubicin (Pira) and gemcitabine (Gem) co-loaded polymeric nanoparticles, leveraging the synergistic potential of dual-drug therapy for enhanced anti-cancer efficacy. Using a polylactic acid-based block copolymer with drugs that was chemically conjugated via pH-responsive Schiff's base linkages, the design ensured targeted and sustained drug release in the acidic tumor microenvironment. The smart nanoformulation exhibited remarkable stability, selective cytotoxicity toward cancer cells, and minimal off-target effects. In vitro evaluations confirmed superior drug release, efficient cellular uptake, and potent apoptosis induction compared with free drugs and single drug-loaded formulations. By orchestrating a finely tuned interplay between pharmacokinetics, cellular targeting, and controlled release, the Pira–Gem nanoparticles established a versatile and promising platform for precision oncology. This innovative approach redefines the potential of combination nanomedicine as a cornerstone for next-generation breast cancer therapies.
|
1. Introduction
Breast cancer has been a prevalent cause of death in women globally over the last several decades. As per the 2020 statistics, approximately 2.3 million cases of breast cancer were diagnosed, with 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resulting in death.1 In addition to surgery or radiation therapy, chemotherapy has been the primary treatment given to patients with solid breast tumors.2,3 Among various categories of chemotherapeutic agents, anthracyclines are the most prominently researched class of drugs for their anti-cancer properties.4 Doxorubicin (Dox), the most well-known anthracycline, is an FDA-approved drug for solid tumors and metastatic breast cancer treatment.5 However, its associated cardiac toxicity and other side effects prompt the ongoing search for less toxic and more efficient analogues of Dox. Interestingly, pirarubicin (THP-Dox) is a semi-synthetic analogue of doxorubicin, with its 4-O position substituted with a tetrahydropyranyl (THP) ring, which has emerged as a potent substitute. According to pre-clinical trials, the latter demonstrated reduced cardiac toxicity while maintaining similar anti-tumor efficacy as that of Dox. Moreover, it showed faster intracellular uptake and competent anti-cancer activity even in Dox-resistant tumor cell lines.6–8
000 resulting in death.1 In addition to surgery or radiation therapy, chemotherapy has been the primary treatment given to patients with solid breast tumors.2,3 Among various categories of chemotherapeutic agents, anthracyclines are the most prominently researched class of drugs for their anti-cancer properties.4 Doxorubicin (Dox), the most well-known anthracycline, is an FDA-approved drug for solid tumors and metastatic breast cancer treatment.5 However, its associated cardiac toxicity and other side effects prompt the ongoing search for less toxic and more efficient analogues of Dox. Interestingly, pirarubicin (THP-Dox) is a semi-synthetic analogue of doxorubicin, with its 4-O position substituted with a tetrahydropyranyl (THP) ring, which has emerged as a potent substitute. According to pre-clinical trials, the latter demonstrated reduced cardiac toxicity while maintaining similar anti-tumor efficacy as that of Dox. Moreover, it showed faster intracellular uptake and competent anti-cancer activity even in Dox-resistant tumor cell lines.6–8
Chemotherapy treatment is often administered as a combination of multiple anti-cancer drugs to attack the malignant cells through diverse mechanisms to simultaneously minimize the risk of multi-drug resistance (MDR) and maximize cancer cell death.9,10 An additional advantage of synergistic chemotherapy over other synergistic strategies for cancer therapy lies in its potential for personalized medicine and comparatively easier clinical transition.11–17 Gemcitabine (2′,2′-difluoro 2′-deoxy cytidine, dFdC), a pyrimidine-based antagonist belonging to the antimetabolite category of chemotherapeutics, has been approved as the first line of treatment for solid breast tumors. However, its therapeutic activity is limited due to factors like its extremely short half-life (∼32 min after infusion) in the bloodstream caused by enzymatic deamination, non-selective targeting of cancer cells, severe side effects and development of drug-resistance in cells after prolonged exposure.18,19 To enhance the effectiveness of gemcitabine, it is usually combined with other chemotherapy drugs to achieve a synergistic effect. However, in the case of combination therapy, the varying pharmacokinetics of different drugs often complicate the dose optimization at the tumor site and increase the chances of adverse effects.20–22 Nanotechnology-based drug delivery systems have shown tremendous potential in addressing these limitations. The leaky vasculature and poor lymphatic drainage system of cancer cells provide an opportunity for nano-vehicles to preferentially accumulate at the tumor site due to the EPR effect.23,24
Amidst a wide variety of nano-formulations, polymeric nanoparticles offer numerous advantages like convenient tailoring of polymers, adjustable molecular weight, decent homogeneity, and excellent stability. Furthermore, the concept of “smart” polymeric nanoparticles utilizes the dissimilarities between the tumor microenvironment and healthy cells to achieve stimuli-responsive or site-specific drug delivery. One such approach exploits the difference of pH between the physiological environment (pH 7.4) and acidic tumor microenvironment (pH 5.0), which has driven the development of pH-triggered nanoparticles that are liable to unload drugs selectively in an acidic milieu.25,26 Schiff's base, widely known for being more sensitive to acidic environments, undergoes hydrolysis in the presence of an acid to give the parent amine and aldehyde/ketone substrates.27 In this study, both pirarubicin and gemcitabine have been conjugated to an amphiphilic PLA–PEG block copolymer through a linker molecule, levulinic acid via Schiff's base linkage to obtain a pH-responsive nanoparticle system. The system is designed to achieve prolonged drug release, reduced off-target cytotoxicity and maximum therapeutic efficacy on breast cancer cells. Polylactic acid (PLA), a biodegradable polymer, has been selected because of the hydrophobicity it provides to build the core of the nanoparticles responsible for their long-lasting stability.28 Polyethylene glycol (PEG) has been attached to PLA to provide a hydrophilic element in the nanoparticle, which helps in immune evasion and improves its circulatory half-life.29
 |
| | Fig. 1 Schematic of the synthesis of the block copolymer mPEG-b-PLA, attachment of levulinic acid and subsequent conjugation of pirarubicin/gemcitabine to the block copolymer. | |
2. Materials and methods
2.1. Materials
Both pirarubicin (Pira) and gemcitabine (Gem) were procured from MedChem Express (New Jersey, USA). L-Lactide, methoxy polyethylene glycol (5 kDa), levulinic acid and Pluronic® F-127 were purchased from Sigma-Aldrich (USA). Dialysis tubing (Regenerated Cellulose, 29 mm, MWCO 3.5 kDa) was obtained from Spectra Por (USA). Amicon ultracentrifuge filters were bought from Millipore (Billerica, MA, USA). Dulbecco's modified Eagle medium (DMEM), fetal bovine serum (FBS) and penicillin–streptomycin antibiotic solution were purchased from Gibco (MA, USA). The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay kit was purchased from HiMedia Laboratories (India). Human breast cancer cell lines, including MCF-7, MDA-MB-468, MDA-MB-231 and SUM-149, as well as healthy human and mice cell lines HEK293 and NIH3T3 were donated by the Dana Farber Cancer Institute (DFCI), Boston, USA. All solvents used in the experiments were obtained from Merck (India).
2.2. Synthesis of the mPEG-b-PLA block copolymer
The synthesis of the mPEG-b-PLA block copolymer was carried out using a previously reported protocol.30 Briefly, it involved a bulk polymerization technique in which mPEG was used as an initiator to induce ring opening polymerization of L-lactide in the presence of stannous octoate as the catalyst. 1.1 g of L-lactide (7.64 mmoles) and 2.2 g of mPEG (0.44 mmoles) were fused in a round bottom flask at 90 °C, followed by the addition of 15 μL Sn(Oct)2 (0.005 w/w% of L-lactide). The reaction was then stirred continuously for 4 h at 160 °C under a nitrogen environment. The product was then dissolved in dichloromethane, and precipitated using a mixture of cold diethyl ether and methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). The obtained polymer was vacuum dried and stored at −20 °C till further use.
4). The obtained polymer was vacuum dried and stored at −20 °C till further use.
2.3. Attachment of levulinic acid to mPEG-b-PLA
Levulinic acid (4-oxopentanoic acid) acts as a linker between the polymer and the drug. Attachment of levulinic acid to the block copolymer, mPEG-b-PLA, was done using the Steglich esterification reaction protocol.31 Briefly, 31 mg of levulinic acid (2 mM) was dissolved in anhydrous DMF and its carboxyl group was activated using 110 mg DCC (4 mM) at 0 °C. After 20 min, 1 g of mPEG-b-PLA (1 mM) was added to the reaction mixture. After its complete dissolution, 98 mg of DMAP (6 mM) was mixed as the catalyst. The reaction continued for the next 24 hours at room temperature until dicyclohexyl urea (DCU) was separated out from the solution. After DCU removal using PTFE syringe filters, the polymer mPEG-b-PLA–LA was precipitated using a mixture of cold diethyl ether and methanol, followed by drying under vacuum.
2.4. Conjugation of Pira/Gem to mPEG-b-PLA–LA
For conjugation of Pira to mPEG-b-PLA–LA, 10 mg of pirarubicin (13.4 mmoles) and 216 mg of mPEG-b-PLA–LA (26.8 mmoles) were dissolved in anhydrous DMF in the presence of glacial acetic acid (30 μL), and stirred continuously for 24 hours under dark conditions. The final reaction product Pira-mPEG-b-PLA–LA conjugate was purified by using the dialysis method (Regenerated cellulose membrane, MCWO 3 kDa) against the DMF solvent to remove the unconjugated pirarubicin, where the sink media were replaced after every 24 hours for 3 days. Similarly, for the conjugation of Gem to mPEG-b-PLA–LA, 10 mg of Gem (38.5 μmoles) and 200 mg of mPEG-b-PLA–LA (19.3 μmoles) were dissolved in anhydrous DMF in the presence of glacial acetic acid (30 μL) and stirred continuously for 24 hours to maintain the hygroscopic conditions. The reaction product Gem–mPEG-b-PLA–LA conjugate was purified using the solvent extraction method. The conjugate was first extracted in DCM, and then precipitated using cold diethyl ether to obtain the final product.
2.5. Characterization of Pira/Gem–mPEG-b-PLA–LA conjugates
The formation of the block copolymer, mPEG-b-PLA was confirmed using 1H NMR spectroscopy (Bruker AC, 500 MHz) and gel permeation chromatography (GPCmax, Viscotek) equipped with refractive index (RI), right-angle light scattering (RALS) and low-angle light scattering (LALS) detectors. [GPC method; THF solvent as the mobile phase, 1 mL min−1 as flow rate, 10 μL as the injection volume, column temperature at 25 °C]. For 1H NMR analysis, chemical shifts are reported as δ (ppm) with tetramethyl silane (TMS) as the internal standard. Attachment of levulinic acid was verified by using 1H NMR and FTIR spectroscopy (PerkinElmer Spectrum 100, MA) via KBr pellet sampling. The NMR spectra were recorded in CDCl3 solvent at room temperature and were analyzed to calculate the number average molecular weight (Mn) and degree of polymerization of the polymer (DP).32 GPC data were used to obtain the weight average molecular weight (Mw) and poly dispersity index (PDI) of the polymers. The conjugation of Pira or Gem to mPEG-b-PLA–LA was further confirmed using 1H NMR spectroscopy.
2.6. Calculation of % drug conjugation efficacy
The % drug conjugation efficiency of Pira and Gem to mPEG-b-PLA–LA was calculated using an HPLC system (Agilent techs 1260 Infinity, USA) equipped with C-18 column, auto-sampler and both UV-visible and fluorescence detector via reversed phase chromatography. For Pira quantification, the HPLC method utilized 0.1% TFA in ACN as the mobile phase, 1 mL min−1 as the flow rate, 10 μL as the injection volume, 25 °C as the column temperature and fluorescence detection at λexcitation = 480 nm and λemission = 580 nm. A calibration curve was plotted using free Pira in the conc. range of 0.5 mg mL−1 to 0.0078 mg mL−1 mobile phase, following the same HPLC protocol. For Gem quantification, the HPLC method utilized ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (70
H2O (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) as the mobile phase, 1 mL min−1 as the flow rate, 10 μL as the injection volume, 25 °C as the column temperature and UV detection at λmax = 270 nm. Another calibration curve was plotted using free Gem in the conc. range of 300 μg mL−1 to 1 μg mL−1 mobile phase following the above-mentioned HPLC protocol. In both cases, the unconjugated drug content was quantified using HPLC and the % drug conjugation efficiency was calculated indirectly using the following equation:
30) as the mobile phase, 1 mL min−1 as the flow rate, 10 μL as the injection volume, 25 °C as the column temperature and UV detection at λmax = 270 nm. Another calibration curve was plotted using free Gem in the conc. range of 300 μg mL−1 to 1 μg mL−1 mobile phase following the above-mentioned HPLC protocol. In both cases, the unconjugated drug content was quantified using HPLC and the % drug conjugation efficiency was calculated indirectly using the following equation:| |  | (1) |
2.7. Preparation and characterization of nanoparticles using the Pira/Gem–mPEG-b-PLA–LA conjugate
Pira/Gem–mPEG-b-PLA–LA conjugate nanoparticles were prepared by employing the nanoprecipitation technique. For the nanoparticle preparation, 30 mg of polymer–drug conjugate was dissolved in 1 mL THF solvent and subsequently added dropwise into an aqueous medium containing 30 mg of Pluronic® F-127, an emulsifying agent, while stirring continuously at 600 rpm. The nanoparticles were stirred at room temperature until complete evaporation of THF was achieved. The resulting nanoparticles were then lyophilized using 15 mg glucose as the cryoprotectant. To determine the particle size and surface charge, the lyophilized nanoparticles were reconstituted in Milli-Q® water to a conc. of 10 mg mL−1. A 20 μL aliquot of this solution was further diluted to 1 mL, and analyzed using dynamic light scattering Instrument (DLS) on an Anton Paar Litesizer 500 instrument. Additionally, the nanoparticles size and morphology were examined using field emission scanning electron Microscopy (TESCAN MAGNA, USA) and transmission electron microscopy (JEOL-1400, USA), respectively. For TEM sample preparation, the diluted nanoparticles (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution of 10 mg mL−1 stock solution) were drop-casted on a carbon-coated copper TEM grid, followed by uranyl acetate staining. Additionally, Pira–Gem co-loaded nanoparticles were prepared using the Pira/Gem–mPEG-b-PLA–LA conjugate in three different drug ratios (Pira–Gem as 1
10 dilution of 10 mg mL−1 stock solution) were drop-casted on a carbon-coated copper TEM grid, followed by uranyl acetate staining. Additionally, Pira–Gem co-loaded nanoparticles were prepared using the Pira/Gem–mPEG-b-PLA–LA conjugate in three different drug ratios (Pira–Gem as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), following the afore-mentioned protocol.
3), following the afore-mentioned protocol.
2.8. Toxicity analysis of nanoparticles
The cytocompatibility of mPEG-b-PLA–LA NPs (w/o drug) was evaluated on human and murine healthy embryonic cells, i.e., HEK 293 and NIH 3T3, respectively. Both types of cells were cultured in DMEM complete media supplemented with 10% FBS and 1% antibiotic solution in a 5% CO2 humidified atmosphere at 37 °C. Cells were seeded with a density of 5000 cells per well in a flat bottom 96-well plate (Corning, USA). After cell adhesion, they were treated with a serially diluted conc. range of 5 mg mL−1 to 0.5 μg mL−1 of nanoparticles, followed by incubation for 72 hours in a CO2 incubator. Cell viability was then assessed using the MTT assay kit as per manufacturer's protocol, and the absorbance was recorded using an ELISA microplate reader (BioTek, Agilent Technologies) at λmax = 570 nm. For hemocompatibility analysis, 1 mL blood of Balb/c female mice was collected using retro-orbital bleeding and processed to separate the RBCs from the other blood components. A 200 μL volume of separated RCBs were washed thrice with 10 ml of PBS (pH 7.4). Thereafter, RBCs were incubated with an equal volume of varying concentrations of mPEG-b-PLA–LA nanoparticles (stock conc. 10 mg mL−1) at 37 °C for 1 hour with gentle shaking at 120 rpm. Following incubation, samples were centrifuged at 1500 rpm for 10 min, and the absorbance of the supernatant was recorded at λmax = 540 nm. The percentage hemolysis was calculated using 1% Triton X 100 as the positive control and PBS (pH 7.4) as the negative control. The experiment was performed in triplicate, and the data were plotted and analyzed using GraphPad Prism 9.0.
2.9.
In vitro drug release study of the Pira/Gem mPEG-b-PLA–LA nanoparticles
The in vitro drug release experiments of Pira/Gem loaded or Pira and Gem co-loaded (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mPEG-b-PLA–LA nanoparticles were performed under two different pH environments, i.e., neutral physiological pH (7.4) and tumor microenvironment pH (5.0). For the study, 30 mg of lyophilized Pira/Gem–mPEG-b-PLA–LA nanoparticles were reconstituted at a concentration of 10 mg mL−1 and equally distributed into 10 mL of each type of buffer. To simulate physiological conditions, the nanoparticles suspension was incubated under dark conditions at 37 °C with gentle stirring at 150 rpm. Thereafter, at pre-determined time intervals, the nanoparticles were filtered and washed using Amicon Centrifuge filters (MWCO 3 kDa, Millipore) at 4000 rpm to separate out the drug released per day. The residual nanoparticles were redispersed in fresh buffer every time, and the collected filtrates were lyophilized and drug content was quantified using the previously described HPLC detection method. The drug release experiments were conducted in triplicate to minimize manual errors. For Pira–Gem co-loaded (1
1) mPEG-b-PLA–LA nanoparticles were performed under two different pH environments, i.e., neutral physiological pH (7.4) and tumor microenvironment pH (5.0). For the study, 30 mg of lyophilized Pira/Gem–mPEG-b-PLA–LA nanoparticles were reconstituted at a concentration of 10 mg mL−1 and equally distributed into 10 mL of each type of buffer. To simulate physiological conditions, the nanoparticles suspension was incubated under dark conditions at 37 °C with gentle stirring at 150 rpm. Thereafter, at pre-determined time intervals, the nanoparticles were filtered and washed using Amicon Centrifuge filters (MWCO 3 kDa, Millipore) at 4000 rpm to separate out the drug released per day. The residual nanoparticles were redispersed in fresh buffer every time, and the collected filtrates were lyophilized and drug content was quantified using the previously described HPLC detection method. The drug release experiments were conducted in triplicate to minimize manual errors. For Pira–Gem co-loaded (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) nanoparticles, the filtrate collected at every time was divided in half and processed separately to quantify the individual drug release.
1) nanoparticles, the filtrate collected at every time was divided in half and processed separately to quantify the individual drug release.
2.10. Cellular uptake studies
To investigate the cellular internalization ability of the prepared nanoparticles as compared to the free drug, SUM-149 cells at a cell density of 5 × 105 cells per well were seeded in a 6-well culture plate (Nest Biotech, China) containing gelatin-coated coverslips inside each well. Cells were left in a CO2 incubator for 2 days to attain 60–70% confluency. Subsequently, cells were treated with 50 μM of either free Pira or Pira–mPEG-b-PLA–LA nanoparticles. Cellular uptake was analyzed both after 1 hour and 4 hours of incubation. Following the incubation period, the cells were washed 2–3 times with PBS solution to remove uninternalized drug or nanoparticles, after which 5 μL of DAPI was added to stain the nucleus of the cells. After 15 min, the cells were washed again 2–3 times with PBS to remove the excess dye. The coverslip was then mount inverted on a glass slide using a PBS: glycerol solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) for adhesion. The cells were observed under a confocal laser scanning microscope (Olympus FV1000) in bright field and fluorescent modes. Additionally, the Pira–mPEG-b-PLA–LA nanoparticles uptake was evaluated by FACS (BD Biosciences, LSRFortessaTM X-20) with the standard 10
1, v/v) for adhesion. The cells were observed under a confocal laser scanning microscope (Olympus FV1000) in bright field and fluorescent modes. Additionally, the Pira–mPEG-b-PLA–LA nanoparticles uptake was evaluated by FACS (BD Biosciences, LSRFortessaTM X-20) with the standard 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events recorded for each sample. Untreated cells were used as a negative control. Software FlowJo (Version 10.10.0) was used to plot and analyze the FACS data.
000 events recorded for each sample. Untreated cells were used as a negative control. Software FlowJo (Version 10.10.0) was used to plot and analyze the FACS data.
2.11. Cell migration inhibition assay
To assess the inhibitory effect of Pira–Gem co-loaded nanoparticles on cell migration, a scratch assay was conducted. MDA-MB-468 cells (5 × 105 cells per well) were seeded in a 6-well plate and incubated until reaching ∼60% confluency. A scratch was then created in each well using a pipette tip, and the initial wound area was imaged using a confocal microscope, serving as the pre-treatment samples. Following this, each well was treated with 10 μM of either Pira-loaded, Gem-loaded or Pira–Gem co-loaded nanoparticles, and the cells were incubated for an additional 24 hours. Post-treatment images were captured using the confocal microscope again. The extent of gap closure was analyzed using ImageJ software, allowing for quantification of the percentage gap closure in each treatment condition.| |  | (2) |
2.12. Apoptosis study
To evaluate the apoptotic potential of Pira–Gem co-loaded nanoparticles, the standard protocol of the annexin-V/PI assay kit (Invitrogen, Massachusetts, USA) was employed. SUM-149 cells were seeded in a 6-well culture plate and cultured until they reached approximately 50% confluency. The cells were then treated with a 10 μM concentration of either Pira/Gem-loaded nanoparticles, Pira–Gem co-loaded nanoparticles, or the free drugs. After 24 hours, the treated cells were washed thrice with cold PBS, trypsinized and centrifuged down at 1200 rpm. The resulting cell pellet was resuspended in annexin-binding buffer, followed by the addition of 100 μL of Alexa Fluor 488 conjugated annexin-V/PI working solution, prepared in annexin-binding buffer. The samples were incubated for 20 minutes, after which an equal volume of annexin-binding buffer was added to each sample. The prepared samples were subsequently analyzed using flow cytometry.
2.13. Cell proliferation inhibition assay
The cytotoxicity of the Pira/Gem–mPEG-b-PLA–LA nanoparticles, in comparison to their respective free drug formulations, was assessed on various breast cancer cell lines, including MCF-7, MDA-MB-468, MDA-MB-231, 4T1 and SUM-149. For these experiments, cells were seeded at a density of 5 × 103 cells per well in complete DMEM media in a flat bottom 96-well tissue culture plate and incubated in a CO2 environment for 24 hours. Subsequently, the cells were treated with serially diluted concentrations of Pira/Gem-loaded nanoparticles or free Pira/Gem, ranging from 100 μM to 0.001 μM, and further incubated for 72 hours. After the incubation period, the % viability of cells was assessed using the MTT assay protocol, and the absorbance was measured at λmax = 570 nm using an ELISA microplate reader. Additionally, the cytotoxic activity of Pira and Gem co-loaded mPEG-b-PLA–LA nanoparticles at different ratios (Pira–Gem-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) was compared with that of the individually drug-loaded Pira/Gem mPEG-b-PLA–LA nanoparticles to determine the most synergistic combination ratio. GraphPad Prism (Version 9.0) was used to plot the dose-dependent cytotoxicity curves and to calculate the IC50 values.
3) was compared with that of the individually drug-loaded Pira/Gem mPEG-b-PLA–LA nanoparticles to determine the most synergistic combination ratio. GraphPad Prism (Version 9.0) was used to plot the dose-dependent cytotoxicity curves and to calculate the IC50 values.
2.14. Combination index evaluation
The combination index values (CI value) of Pira–Gem co-loaded mPEG-b-PLA–LA nanoparticles at different ratios (Pira–Gem-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) were calculated and compared to those of free drug combinations across various breast cancer cell lines; MCF-7, MDA-MB-468 and SUM-149. A drug combination resulting in CI value <1, =1 or >1 indicates synergy, additive effect or antagonism, respectively. Fraction affected (Fa) vs. CI curves were generated using CompuSyn software.
3) were calculated and compared to those of free drug combinations across various breast cancer cell lines; MCF-7, MDA-MB-468 and SUM-149. A drug combination resulting in CI value <1, =1 or >1 indicates synergy, additive effect or antagonism, respectively. Fraction affected (Fa) vs. CI curves were generated using CompuSyn software.
2.15. Statistical analysis
All the experiments were conducted in triplicate to ensure data accuracy, and the data have been reported in mean ± SD form consistently throughout the study. Statistical analysis was performed using one-way ANOVA with Bonferroni's test for multiple comparisons. Significance levels were determined based on p values, with values <0.0001 (****), <0.0002 (***), <0.0021 (**), and <0.0332 (*) considered statistically significant, and 0.1234 (ns) was regarded as non-significant.
3. Results and discussion
3.1. Characterization of mPEG-b-PLA and LA conjugation
The amphiphilic block copolymer mPEG-b-PLA was successfully synthesized using the standard ring opening polymerization technique, with mPEG serving as the hydrophilic moiety and PLA as the hydrophobic counterpart (Fig. 1). The synthesis yield of mPEG-b-PLA was determined to be ∼82%. The 1H NMR spectra exhibited the characteristic peaks of the PLA, –CH and –CH3 protons at δ 5.2 (q) and δ 1.6 ppm (d), as well as PEG –OCH3 and –CH2 protons at δ 3.4 (s) and δ 3.6 (q) ppm, respectively (Fig. 2A). Additionally, Mn and DP were calculated from NMR spectra using a previously reported method (Table S1, ESI†). A comparison of the expected and actual feed ratios indicated minimum monomer loss, confirming efficient polymerization. According to the GPC analysis, Mn and Mw were determined to be ∼9200 and ∼9700 g mol−1, respectively, with a polydispersity index (PDI) value of 1.05. Subsequently, levulinic acid was attached to the –OH terminal end of mPEG-b-PLA, resulting in the formation of the mPEG-b-PLA–LA conjugate. This conjugation was further confirmed by 1H NMR, where new peaks corresponding to the –CH2 and –CH3 protons of levulinic acid appeared at {δ 2.7 and δ 2.8 ppm (dd)} and δ 2.21 ppm (s), respectively. Additionally, FTIR analysis further validated the conjugation of levulinic acid by revealing a new –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching peak at 1650 cm−1 (Fig. 2B). The –C
O stretching peak at 1650 cm−1 (Fig. 2B). The –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of mPEG-b-PLA–LA was then used to conjugate both pirarubicin or gemcitabine to obtain the Schiff's base polymer drug conjugates. The successful conjugation of both Pira and Gem individually to mPEG-b-PLA–LA was confirmed again using 1H NMR spectroscopy (Fig. 2C and D), where new benzylic proton peaks were observed between δ 7.0–δ 8.0 ppm, along with all the characteristic peaks of the block copolymer, validating the formation of the Pira/Gem–mPEG-b-PLA–LA conjugates.
O group of mPEG-b-PLA–LA was then used to conjugate both pirarubicin or gemcitabine to obtain the Schiff's base polymer drug conjugates. The successful conjugation of both Pira and Gem individually to mPEG-b-PLA–LA was confirmed again using 1H NMR spectroscopy (Fig. 2C and D), where new benzylic proton peaks were observed between δ 7.0–δ 8.0 ppm, along with all the characteristic peaks of the block copolymer, validating the formation of the Pira/Gem–mPEG-b-PLA–LA conjugates.
 |
| | Fig. 2 (A) 1H NMR spectra and (B) FT-IR spectra of the block copolymer mPEG-b-PLA and mPEG-b-PLA–LA. 1H NMR spectra of (C) Pira–Gem mPEG-b-PLA–LA and (D) Gem–mPEG-b-PLA–LA. | |
3.2. Drug conjugation efficiency
The conjugation efficiency of pirarubicin in the Pira–mPEG-b-PLA–LA conjugate was determined to be 38.2% with a drug loading of 0.18 mg per 10 mg of conjugate, corresponding to a drug loading percentage of 1.8%. In contrast, the conjugation efficiency of gemcitabine in the Gem–mPEG-b-PLA–LA conjugate was found to be 31.3%, with a drug loading of 0.16 mg per 10 mg of conjugate, equivalent to a drug loading percentage of 1.6%.
3.3. Preparation and characterization of nanoparticles
The polymer–drug conjugates Pira/Gem–mPEG-b-PLA–LA were utilized to prepare nanoparticles via the nanoprecipitation method. Pira–mPEG-b-PLA–LA nanoparticles exhibited an average size of 124.20 ± 3.64 nm with a PDI of 0.14 ± 0.02, while Gem–mPEG-b-PLA–LA nanoparticles displayed an average size of 180.50 ± 2.30 nm with a PDI of 0.08 ± 0.04. In both cases, the average size increased compared to the blank (w/o drug) mPEG-b-PLA–LA nanoparticles, which had an average size of 104.50 ± 4.21 nm and a PDI of 0.21 ± 0.05. A similar trend was observed in the surface charge density, as the zeta potential of the blank nanoparticles increased from −4.90 ± 0.25 mV to −12.22 ± 1.13 mV for the Pira nanoparticles and to −8.6 ± 1.40 mV for the Gem nanoparticles. The increase in the average particle size and surface charge density may be attributed to the preferential entrapment of the drug within the nanoparticle core, and the presence of negatively charged –OH groups following drug conjugation. In the case of Pira–Gem co-loaded nanoparticles, the particle size further increased to 196.98 ± 2.30 nm, while the zeta potential became slightly less negative, reaching −7.7 mV with an overall PDI of 0.272 ± 0.02. Furthermore, TEM (Fig. 3C) and FE-SEM (Fig. S1, ESI†) analysis confirmed the formation of uniformly sized, stable nanoparticles with core–shell structural characteristics. The particle size distribution (Fig. 3D), as determined by TEM, correlated well with the results obtained from DLS analysis. However, a slight discrepancy was noted between the sizes measured by TEM and DLS, which is likely due to the different principles underlying these techniques. TEM captures the dry state of the nanoparticles, offering direct visualization of individual particles, whereas DLS measures the hydrodynamic diameter in solution, which includes the hydration layer and any possible aggregation. Additionally, the polydispersity of the sample may further influence the DLS data, contributing to variation in the measured size (Table 1).
 |
| | Fig. 3 (A) Hydrodynamic size measurements, (B) zeta potential analysis, (C) transmission electron microscopy (TEM) images, and (D) particle size distribution with corresponding average size and standard deviation for blank, Pira-loaded, Gem-loaded, and Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) co-loaded mPEG-b-PLA–LA nanoparticles. 1) co-loaded mPEG-b-PLA–LA nanoparticles. | |
Table 1 DLS characterization data
| Nanoparticles |
Size (nm) |
Zeta potential (mV) |
PDI |
|
Blank Nps–mPEG-b-PLA–LA nanoparticles w/o any drug.
|
| Blank Npsa |
104.50 ± 4.21 |
−4.90 ± 0.25 |
0.21 ± 0.05 |
| Pira Nps |
124.20 ± 3.64 |
−12.22 ± 1.13 |
0.14 ± 0.02 |
| Gem Nps |
180.50 ± 2.30 |
−8.60 ± 1.40 |
0.08 ± 0.04 |
Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps 1) Nps |
196.98 ± 2.30 |
−7.70 ± 1.10 |
0.27 ± 0.02 |
3.4. Toxicity analysis of nanoparticles
The biocompatibility analysis is crucial for nanoparticles that are intended for biological treatment applications.33 The prepared mPEG-b-PLA–LA nanoparticles were evaluated for cytocompatibility using non-cancerous human cells HEK 293 and murine fibroblasts NIH 3T3 (Fig. 4A). The outcomes showed nearly 100% viability, even at the highest concentration of the nanoparticles tested (5 mg mL−1), indicating that the mPEG-b-PLA–LA nanoparticles exhibit excellent biocompatibility. For the hemocompatibility assessment, the percentage of hemolysis was measured after incubating red blood cells (RBCs) with mPEG-b-PLA–LA nanoparticles for 1 h. The findings revealed that even at the highest nanoparticles concentration used (8.4 mg mL−1), only ∼2.4% hemolysis was observed, with lower concentrations causing even less hemolysis (Fig. 4B). Importantly, the percentage change in hemolysis among different nanoparticle concentrations was found to be statistically non-significant, whereas all concentrations induced significantly lower hemolysis compared to the positive control, Triton X. Therefore, mPEG-b-PLA–LA nanoparticles were considered hemocompatible and suitable for use in treatment applications.
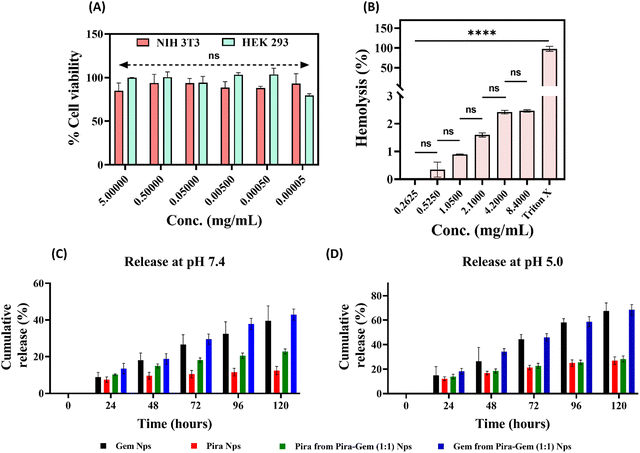 |
| | Fig. 4 (A) Cytocompatibility assessment of mPEG-b-PLA–LA nanoparticles on NIH 3T3 and HEK 293 cell lines using the MTT assay. (B) Hemocompatibility evaluation of mPEG-b-PLA–LA nanoparticles. (C) In vitro release profile of Pira and Gem from single-loaded and dual-loaded Pira/Gem–mPEG-b-PLA–LA nanoparticles at physiological pH (7.4). (D) Corresponding drug release profiles at acidic pH (5.0). Data are presented as mean ± SD (n = 3); p > 0.05 considered not significant (ns). | |
3.5.
In vitro release of nanoparticles
The Schiff's base linkage in polymer–drug conjugates has been employed to achieve pH-responsive drug release specifically in the tumor microenvironment. Pira–mPEG-b-PLA–LA nanoparticles showed ∼35% of cumulative Pira release in PBS buffer (pH 5.0), compared to only ∼12% at pH 7.4 (Fig. 5C). Similarly, Gem–mPEG-b-PLA–LA nanoparticles demonstrated a cumulative Gem release of ∼65% in PBS buffer (pH 5.0), while only ∼40% release was observed at pH 7.4 (Fig. 4D). Certainly, the release of both drugs was approximately doubled in acidic medium, confirming the pH-sensitive release mechanism. As expected, the cumulative release at pH 6.8 was higher than that observed at neutral pH and in serum conditions, but remained slightly lower than at pH 5.0. Moreover, the drug release in serum (10% FBS solution, pH 7.4) did not show any significant change, exhibiting only a marginal increase compared to PBS at pH 7.4 (Fig. S3, ESI†). Due to its hydrophilic nature, gemcitabine showed a higher release rate than the hydrophobic pirarubicin. In Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) co-loaded nanoparticles, the cumulative release of Pira was observed to be ∼21% and ∼28% at pH 7.4 and pH 5.0, respectively. In contrast, the co-loaded nanoparticles released ∼45% and ∼70% of Gem at pH 7.4 and pH 5.0, respectively (Fig. 4C and D). The contrasting physiochemical properties of the drugs, pirarubicin being hydrophobic and gemcitabine being hydrophilic, resulted in repulsive interactions when co-loaded, contributing to the enhanced release of both drugs.
1) co-loaded nanoparticles, the cumulative release of Pira was observed to be ∼21% and ∼28% at pH 7.4 and pH 5.0, respectively. In contrast, the co-loaded nanoparticles released ∼45% and ∼70% of Gem at pH 7.4 and pH 5.0, respectively (Fig. 4C and D). The contrasting physiochemical properties of the drugs, pirarubicin being hydrophobic and gemcitabine being hydrophilic, resulted in repulsive interactions when co-loaded, contributing to the enhanced release of both drugs.
 |
| | Fig. 5 (A) Confocal laser scanning microscopy (CLSM; Olympus) images showing the cellular uptake of Pira–mPEG-b-PLA–LA nanoparticles compared with free Pira in SUM-149 cells after incubation, captured at 60× magnification; scale bar: 50 μm. (B) Quantitative comparison of the mean fluorescence intensity (MFI) of free Pira and Pira-loaded nanoparticles in the nucleus and cytoplasm after 1 h and 4 h of incubation. Data are presented as mean ± SD; p > 0.05 considered not significant (ns). (C) Flow cytometric (FACS) analysis of cellular uptake to support CLSM findings. | |
3.6. Cellular uptake of nanoparticles
Effective therapeutic action requires drug-loaded nanoparticles to exhibit strong cellular internalization.34 In this study, the cellular uptake efficiency of Pira–mPEG-b-PLA–LA nanoparticles was compared with that of free Pira in triple negative breast cancer cells (SUM-149 cells) after 1 hour and 4 hours of treatment. Confocal microscopy images indicated that even after 1 h, Pira–mPEG-b-PLA–LA nanoparticles demonstrated significantly greater internalization as compared to free Pira. At the 1-hour mark, Pira–mPEG-b-PLA–LA nanoparticles were observed to be localized within the cellular cytoplasm and along the nuclear boundary (indicated by red fluorescence) with the nucleus stained using DAPI (blue fluorescence). In contrast, the free drug appeared dispersed around the cells, showing early signs of apoptosis. (Fig. 5A). After 4 hours of treatment, Pira–mPEG-b-PLA–LA nanoparticles were found to penetrate the nucleus, as evidenced by the overlap of red and blue fluorescence. In comparison, free Pira treated cells showed a higher population of cells undergoing apoptosis, accompanied by the loss of cell shape and morphology. Furthermore, FACS analysis revealed nearly ∼100% uptake of Pira–mPEG-b-PLA nanoparticles after 4 hours, confirming the superior uptake efficiency of the prepared nanoparticles (Fig. 5C).
3.7. Cell migration inhibition assay
The ability of a therapeutic drug to inhibit cell migration is a critical attribute for effective cancer treatment, as cell migration plays a central role in tumor progression and metastasis.35 The scratch assay determines the efficacy of a drug based on the percentage of gap closure, with lower values indicating higher inhibitory potential. In this study, Pira-loaded and Gem-loaded nanoparticles resulted in moderate migration inhibition, i.e., 29.25% and 48.25% gap closure, respectively, after 24 hours. In contrast, Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) co-loaded nanoparticles exhibited only 7.46% gap closure under identical conditions (Fig. 6A and B). The substantial reduction in cell migration highlights the synergistic therapeutic potential of the Pira–Gem combination. In control wells without any treatment, the scratch gap was completely closed within 24 hours, underscoring the aggressive migratory behavior of untreated cells. These findings provide compelling evidence of the superior migration inhibitory activity potential of Pira–Gem co-loaded nanoparticles compared to single-drug loaded formulations for combating metastatic breast cancer.
1) co-loaded nanoparticles exhibited only 7.46% gap closure under identical conditions (Fig. 6A and B). The substantial reduction in cell migration highlights the synergistic therapeutic potential of the Pira–Gem combination. In control wells without any treatment, the scratch gap was completely closed within 24 hours, underscoring the aggressive migratory behavior of untreated cells. These findings provide compelling evidence of the superior migration inhibitory activity potential of Pira–Gem co-loaded nanoparticles compared to single-drug loaded formulations for combating metastatic breast cancer.
 |
| | Fig. 6 (A) Representative images from the scratch (wound healing) assay of MDA-MB-468 breast cancer cells before and after treatment with Pira-loaded, Gem-loaded, or Pira–Gem co-loaded nanoparticles compared with the saline-treated control. Images were captured using a confocal microscope (20× magnification; scale bar: 200 μm). (B) Quantitative gap closure analysis expressed as the percentage wound closure performed using ImageJ software. Data are presented as mean ± SD (n = 3); p > 0.05 was considered not significant (ns). | |
3.8. Apoptosis study
The apoptotic potential of a drug is a critical determinant of its therapeutic efficacy, with higher percentages of cancer cell apoptosis correlating with an improved therapeutic index. The apoptosis analysis demonstrated that Pira–Gem co-loaded nanoparticles induced the highest level of cell death compared to Pira/Gem nanoparticles individually, as a significant proportion of cells were observed in the late-apoptotic and necrotic phases (Fig. 7A). In the case of Pira-loaded nanoparticles, an increased proportion of cells were in the early and late apoptotic phases relative to free Pira, where the majority of cells were in the necrotic or late-apoptotic phase. The effect is likely attributed to the sustained release profile of Pira from the nanoparticles. Similarly, Gem-loaded nanoparticles exhibited an approximately two-fold increase in early-apoptotic cell population compared to free Gem, potentially due to the enhanced stability of Gem in the nanoparticle formulation. The Pira–Gem combination demonstrated the highest level of SUM-149 cell death among all treatments, surpassing the effects of single-drug-loaded nanoparticles and free drugs, indicating a synergistic therapeutic effect.
 |
| | Fig. 7 (A) Flow cytometric analysis (FACS) of apoptosis in SUM-149 cells treated with Pira-loaded, Gem-loaded, and Pira–Gem co-loaded nanoparticles using annexin V–FITC/PI dual staining. Quadrants: Q1 (annexin V−/PI−, viable cell populations), Q2 (annexin V+/PI−, early apoptotic cell populations), Q3 (annexin V+/PI+, late apoptotic cell populations), and Q4 (annexin V−/PI+, necrotic cell populations). (B) Quantitative comparison of cell populations (%) in each apoptotic and necrotic phase across different treatments. Data are presented as mean ± SD (n = 3). Statistical analysis was performed using two-way ANOVA, followed by Tukey's multiple comparisons test. * and ** indicate significant differences in Q1 and Q3, while # and ## indicate significant differences in Q2 and Q4, respectively, compared with the free Pira-treated group. | |
3.9. Cytotoxic studies of nanoparticles
Pira–mPEG-b-PLA–LA nanoparticles exhibited slightly higher IC50 values compared to free pirarubicin across all the breast cancer cell lines tested (Fig. 8A). This is attributed to the slower drug release from the nanoparticles, resulting in reduced availability for therapeutic action compared to the free drug. In contrast, Gem–mPEG-b-PLA–LA nanoparticles demonstrated the increased stability and extended half-life of gemcitabine in the nanoformulation, as its conjugation to the polymer reduced its susceptibility to enzymatic degradation and renal clearance.36 Consequently, Gem–mPEG-b-PLA–LA nanoparticles displayed lower IC50 values across all breast cancer cell lines compared to the free gemcitabine. Furthermore, when single drug-loaded (Pira or Gem) mPEG-b-PLA–LA nanoparticles were tested against dual-loaded (Pira and Gem) mPEG-b-PLA–LA nanoparticles (Fig. 8B), the latter demonstrated lower IC50 values and greater cytotoxicity in all Pira-to-Gem ratios, indicating a synergistic therapeutic effect in combined Pira and Gem treatment against various cancer types. The IC50 values for single versus dual-loaded Pira–Gem mPEG-b-PLA–LA nanoparticles have been provided in Table 2.
 |
| | Fig. 8 (A) Comparison of the IC50 values for free Pira/Gem and Pira/Gem-loaded mPEG-b-PLA–LA nanoparticles, following cytotoxicity assessment in various breast cancer cell lines using the MTT assay (n = 3); p > 0.05 considered not significant (ns). (B) Dose-dependent cytotoxicity curves for single- and dual drug-loaded nanoparticles with varying Pira![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gem ratios (1 Gem ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3 1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1 1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) after 72 h treatment. (C) Combination index (CI) versus fraction affected (Fa) scatter plots for Pira–Gem co-loaded mPEG-b-PLA–LA nanoparticles (at ratios of 1 3) after 72 h treatment. (C) Combination index (CI) versus fraction affected (Fa) scatter plots for Pira–Gem co-loaded mPEG-b-PLA–LA nanoparticles (at ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3 1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1 1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), analyzed using CompuSyn software, in different breast cancer cell lines. 3), analyzed using CompuSyn software, in different breast cancer cell lines. | |
Table 2 IC50 values of Pira/Gem–mPEG-b-PLA–LA in single- versus dual-loaded nanoparticles
| Nanoparticles |
SUM-149 |
MDA-MB-468 |
MCF-7 |
| Gem |
Pira |
Gem |
Pira |
Gem |
Pira |
| Pira Nps |
— |
0.3188 |
— |
0.2899 |
— |
0.2093 |
| Gem Nps |
1.600 |
— |
0.6840 |
— |
1.173 |
— |
Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps 1) Nps |
0.1571 |
0.0628 |
0.0968 |
0.0387 |
0.0786 |
0.1190 |
Pira–Gem (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps 1) Nps |
0.0228 |
0.0603 |
0.0720 |
0.1899 |
0.0119 |
0.1671 |
Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Nps 3) Nps |
0.0916 |
0.0274 |
0.0881 |
0.0264 |
0.0855 |
0.0724 |
3.10. Combination index evaluation
The combination of pirarubicin and gemcitabine in dual-loaded nanoparticles demonstrated synergistic therapeutic efficacy in MCF-7 cells across all three tested drug ratios (Fig. 8C). However, in the other breast cancer cell lines SUM-149 and MDA-MB-468, synergistic effects were observed only with the Pira and Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mPEG-b-PLA–LA nanoparticle formulation. This finding suggests that while the dual loading of Pira and Gem can enhance the effectiveness of a treatment, the optimal ratio for consistent synergy across various cell types may vary, with the 1
1) mPEG-b-PLA–LA nanoparticle formulation. This finding suggests that while the dual loading of Pira and Gem can enhance the effectiveness of a treatment, the optimal ratio for consistent synergy across various cell types may vary, with the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio being particularly effective across all of the cell lines that were tested (Table 3).
1 ratio being particularly effective across all of the cell lines that were tested (Table 3).
Table 3 Combination index values of Pira and Gem–mPEG-b-PLA–LA dual-loaded nanoparticles
| Nanoparticles |
Combination index (CI values) |
| SUM-149 |
MDA-MB-468 |
MCF-7 |
Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps 1) Nps |
0.7740 |
0.1538 |
0.3434 |
Pira–Gem (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps 1) Nps |
0.5018 |
1.2770 |
0.3242 |
Pira–Gem (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Nps 3) Nps |
1.3764 |
1.3282 |
0.7372 |
4. Conclusion
In conclusion, the developed Pira–Gem co-loaded mPEG-b-PLA–LA nanoparticles were found to be highly stable and non-toxic towards healthy cells. They demonstrated moderate drug loading capacity and superior cellular uptake ability compared to free drug formulations. In vitro drug release studies revealed the pH-responsive release of both Pira and Gem in a controlled and sustained manner. Additionally, the Pira–Gem duo exhibited the highest cell migration inhibitory and apoptotic activity potential on breast cancer cells, indicating their synergistic action. Furthermore, the cytotoxicity studies concluded that the Pira–Gem co-loaded nanoparticles, in every Pira–Gem ratio, possessed greater therapeutic efficacy compared to the single drug-loaded nanoparticles or respective free drugs across multiple breast cancer cell lines. All these findings underscore the significant potential of the Pira–Gem co-loaded mPEG-b-PLA–LA nanoparticles as an advanced therapeutic option for breast cancer. Their synergistic activity, coupled with effective drug release and cellular uptake, warrants further investigation, particularly in preclinical animal studies, to evaluate their efficacy and safety in vivo.
Author contributions
Priya Gupta – conceptualization, methodology, investigation, formal analysis, data curation and writing – original draft. Harshdeep Kaur, Mohammad Anees, Sachchidanand Tiwari and Ankushi Bansal – writing – review & editing. Harpal Singh – supervision, resources, project administration and funds acquisition.
Data availability
The data supporting this article have been included as part of the ESI.†
Conflicts of interest
The are no conflicts to declare.
Acknowledgements
The animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of AIIMS Delhi, with ethical clearance 122/IAEC/2019 and were approved by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA). We acknowledge the Central Research Facility (CRF), IIT Delhi, for TEM and FE-SEM characterizations, and Kusuma School of Biological Sciences (KSBS) for facilitating the FACS analysis. We also extend our gratitude to the Council of Scientific and Industrial Research (CSIR), India, for financial support through the PhD stipend.
References
- M. Arnold, E. Morgan, H. Rumgay, A. Mafra, D. Singh, M. Laversanne, J. Vignat, J. R. Gralow, F. Cardoso, S. Siesling and I. Soerjomataram, Current and future burden of breast cancer: Global statistics for 2020 and 2040, Breast, 2022, 66, 15–23 CrossRef.
- L. A. Korde, M. R. Somerfield, L. A. Carey, J. R. Crews, N. Denduluri, E. S. Hwang, S. A. Khan, S. Loibl, E. A. Morris, A. Perez and M. M. Regan, Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline, J. Clin. Oncol., 2021, 39(13), 1485–1505 CrossRef CAS.
- D. R. Byrd, J. D. Brierley, T. P. Baker, D. C. Sullivan and D. M. Gress, Current and future cancer staging after neoadjuvant treatment for solid tumors, Ca-Cancer J. Clin., 2021, 71(2), 140–148 CrossRef.
- R. Kapoor, A. Saini and D. Sharma, Indispensable role of microbes in anticancer drugs and discovery trends, Appl. Microbiol. Biotechnol., 2022, 106(13), 4885–4906 CrossRef CAS.
- H. Muley, R. Fado, R. Rodriguez-Rodriguez and N. Casals, Drug uptake-based chemoresistance in breast cancer treatment, Biochem. Pharmacol., 2020, 177, 113959 CrossRef CAS.
- M. Anees, S. Tiwari, N. Mehrotra, S. Kharbanda and H. Singh, Development and evaluation of PLA based hybrid block copolymeric nanoparticles for systemic delivery of pirarubicin as an anti-cancer agent, Int. J. Pharm., 2022, 620, 121761 CrossRef CAS PubMed.
- P. Gupta, A. Bansal, H. Kaur, M. Anees, N. Singh and H. Singh, Folic acid-targeted redox responsive polylactic acid-based nanoparticles co-delivering pirarubicin and salinomycin suppress breast cancer tumor growth in vivo, Nanoscale, 2024, 16(43), 20131–20146 RSC.
- M. Anees, P. Gupta, H. Kaur, S. Kharbanda and H. Singh, Concomitant Delivery of Pirarubicin and Salinomycin Synergistically Enhanced the Efficacy of Cancer Therapy and Reduced the Risk of Cancer Relapse, AAPS PharmSciTech, 2024, 25(7), 211 CrossRef CAS PubMed.
- Q. Gao, J. Feng, W. Liu, C. Wen, Y. Wu, Q. Liao, L. Zou, X. Sui, T. Xie, J. Zhang and Y. Hu, Opportunities and challenges for co-delivery nanomedicines based on combination of phytochemicals with chemotherapeutic drugs in cancer treatment, Adv. Drug Delivery Rev., 2022, 188, 114445 CrossRef CAS PubMed.
- A. Maimaitijiang, D. He, D. Li, W. Li, Z. Su, Z. Fan and J. Li, Progress in research of nanotherapeutics for overcoming multidrug resistance in cancer, Int. J. Mol. Sci., 2024, 25(18), 9973 CrossRef CAS.
- T. C. Chou, Drug combination studies and their synergy quantification using the Chou-Talalay method, Cancer Res., 2010, 70(2), 440–446 CrossRef CAS PubMed.
- B. Al-Lazikani, U. Banerji and P. Workman, Combinatorial drug therapy for cancer in the post-genomic era, Nat. Biotechnol., 2012, 30(7), 679–692 CrossRef CAS.
- Y. Zhao, D. Y. Alakhova and A. V. Kabanov, Can nanomedicines kill cancer stem cells?, Adv. Drug Delivery Rev., 2013, 65(13–14), 1763–1783 CrossRef CAS.
- J. Jia, F. Zhu, X. Ma, Z. W. Cao, Y. X. Li and Y. Z. Chen, Mechanisms of drug combinations: interaction and network perspectives, Nat. Rev. Drug Discovery, 2009, 8(2), 111–128 CrossRef CAS.
- Y. Yu, F. Zhang, W. Xiao, Q. Cheng, T. Li, J. Tang, W. Tao and L. Mei, Adaptive design of nanovesicles overcoming immunotherapeutic limitations of chemotherapeutic drugs through poliovirus receptor blockade, ACS Nano, 2024, 18(7), 5915–5929 CAS.
- Y. Zhang, X. Yu, L. Luo, Y. Xu, H. Zhang, Z. Mao, Y. Zhang, C. Yang, L. Wang, P. Zhang and S. Li, Engineered manganese-BODIPY coordinated nanoadjuvants for enhanced NIR-II photo-metalloimmunotherapy, J. Controlled Release, 2024, 376, 1115–1129 CrossRef CAS.
- L. Wang, T. Wang, Y. Zhang, X. Kang, X. K. Ouyang, X. Yu, T. Chen, W. Li and L. Mei, Biomimetic nanosystems harnessing NIR-II photothermal effect and hypoxia-responsive prodrug for self-amplifying and synergistic tumor treatment, Nano Today, 2024, 57, 102395 CrossRef CAS.
- T. E. Yalcin, S. Ilbasmis-Tamer and S. Takka, Antitumor activity of gemcitabine hydrochloride loaded lipid polymer hybrid nanoparticles (LPHNs): In vitro and in vivo, Int. J. Pharm., 2020, 580, 119246 CrossRef CAS PubMed.
- L. Devi, R. Gupta, S. K. Jain, S. Singh and P. Kesharwani, Synthesis, characterization and in vitro assessment of colloidal gold nanoparticles of Gemcitabine with natural polysaccharides for treatment of breast cancer, J. Drug Delivery Sci. Technol., 2020, 56, 101565 CrossRef CAS.
- A. Behl, P. Sarwalia, S. Kumar, C. Behera, M. J. Mintoo, T. K. Datta, P. N. Gupta and A. K. Chhillar, Codelivery of gemcitabine and MUC1 inhibitor using PEG-PCL nanoparticles for breast cancer therapy, Mol. Pharmaceutics, 2022, 19(7), 2429–2440 CrossRef CAS.
- M. Lei, S. Sha, X. Wang, J. Wang, X. Du, H. Miao, H. Zhou, E. Bai, J. Shi and Y. Zhu, Co-delivery of paclitaxel and gemcitabine via a self-assembling nanoparticle for targeted treatment of breast cancer, RSC Adv., 2019, 9(10), 5512–5520 RSC.
- J. Zhang, P. Zhang, Q. Zou, X. Li, J. Fu, Y. Luo, X. Liang and Y. Jin, Co-delivery of gemcitabine and paclitaxel in cRGD-modified long circulating nanoparticles with asymmetric lipid layers for breast cancer treatment, Molecules, 2018, 23(11), 2906 CrossRef.
- S. Tiwari, P. Gupta, M. Anees, H. Kaur, S. Kharbanda and H. Singh, Pluronic modified PLA based hybrid block copolymeric nanoformulation enhanced anti-cancer therapeutic efficacy of Epirubicin, Eur. Polym. J., 2023, 193, 112084 CrossRef CAS.
- M. Anees, N. Mehrotra, S. Tiwari, D. Kumar, S. Kharbanda and H. Singh, Polylactic acid based biodegradable hybrid block copolymeric nanoparticle mediated co-delivery of salinomycin and doxorubicin for cancer therapy, Int. J. Pharm., 2023, 635, 122779 CrossRef CAS.
- C. Zhang, J. Wang, M. Fan, L. Han, B. Xiao, X. Xie, Y. Fu, Y. Zhai, C. Wang, N. Zhang and Z. Xu, Folate functionalized pH-sensitive nanoparticulate system to decrease the toxicity and enhance the anti-cancer activity of pirarubicin for lymphoma therapy via downregulation of H3K18la and H3K9la, J. Drug Delivery Sci. Technol., 2024, 97, 105829 CrossRef CAS.
- L. Palanikumar, S. Al-Hosani, M. Kalmouni, V. P. Nguyen, L. Ali, R. Pasricha, F. N. Barrera and M. Magzoub, pH-responsive high stability polymeric nanoparticles for targeted delivery of anticancer therapeutics, Commun. Biol., 2020, 3(1), 95 CrossRef CAS PubMed.
- A. Nazli, M. Z. Khan, Á. Rácz and S. Béni, Acid-sensitive prodrugs; a promising approach for site-specific and targeted drug release, Eur. J. Med. Chem., 2024, 116699 CrossRef CAS.
- C. Mukherjee, D. Varghese, J. S. Krishna, T. Boominathan, R. Rakeshkumar, S. Dineshkumar, C. B. Rao and A. Sivaramakrishna, Recent advances in biodegradable polymers–properties, applications and future prospects, Eur. Polym. J., 2023, 192, 112068 CrossRef CAS.
- Y. Takakura and Y. Takahashi, Strategies for persistent retention of macromolecules and nanoparticles in the blood circulation, J. Controlled Release, 2022, 350, 486–493 CrossRef CAS.
- M. Hasannia, A. Aliabadi, K. Abnous, S. M. Taghdisi, M. Ramezani and M. Alibolandi, Synthesis of block copolymers used in polymersome fabrication: Application in drug delivery, J. Controlled Release, 2022, 341, 95–117 CrossRef CAS PubMed.
- A. Jordan, K. D. Whymark, J. Sydenham and H. F. Sneddon, A solvent-reagent selection guide for Steglich-type esterification of carboxylic acids, Green Chem., 2021, 23(17), 6405–6413 RSC.
- J. U. Izunobi and C. L. Higginbotham, Polymer molecular weight analysis by 1H NMR spectroscopy, J. Chem. Educ., 2011, 88(8), 1098–1104 CrossRef CAS.
- M. Kus-Liśkiewicz, P. Fickers and I. Ben Tahar, Biocompatibility and cytotoxicity of gold nanoparticles: recent advances in methodologies and regulations, Int. J. Mol. Sci., 2021, 22(20), 10952 CrossRef PubMed.
- K. Kumar, S. G. Rawat, M. Mishra, A. Kumar and R. Chawla, Dual targeting pH responsive chitosan nanoparticles for enhanced active cellular internalization of gemcitabine in non-small cell lung cancer, Int. J. Biol. Macromol., 2023, 249, 126057 CrossRef CAS.
- H. Yousefi, M. Vatanmakanian, M. Mahdiannasser, L. Mashouri, N. V. Alahari, M. R. Monjezi, S. Ilbeigi and S. K. Alahari, Understanding the role of integrins in breast cancer invasion, metastasis, angiogenesis, and drug resistance, Oncogene, 2021, 40(6), 1043–1063 CrossRef CAS PubMed.
- R. Mukhopadhyay, R. Sen, B. Paul, J. Kazi, S. Ganguly and M. C. Debnath, Gemcitabine co-encapsulated with curcumin in folate decorated PLGA nanoparticles; a novel approach to treat breast adenocarcinoma, Pharm. Res., 2020, 37, 1–9 CrossRef.
|
| This journal is © The Royal Society of Chemistry 2025 |
Click here to see how this site uses Cookies. View our privacy policy here.  and
Harpal
Singh
and
Harpal
Singh
 *
*
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resulting in death.1 In addition to surgery or radiation therapy, chemotherapy has been the primary treatment given to patients with solid breast tumors.2,3 Among various categories of chemotherapeutic agents, anthracyclines are the most prominently researched class of drugs for their anti-cancer properties.4 Doxorubicin (Dox), the most well-known anthracycline, is an FDA-approved drug for solid tumors and metastatic breast cancer treatment.5 However, its associated cardiac toxicity and other side effects prompt the ongoing search for less toxic and more efficient analogues of Dox. Interestingly, pirarubicin (THP-Dox) is a semi-synthetic analogue of doxorubicin, with its 4-O position substituted with a tetrahydropyranyl (THP) ring, which has emerged as a potent substitute. According to pre-clinical trials, the latter demonstrated reduced cardiac toxicity while maintaining similar anti-tumor efficacy as that of Dox. Moreover, it showed faster intracellular uptake and competent anti-cancer activity even in Dox-resistant tumor cell lines.6–8
000 resulting in death.1 In addition to surgery or radiation therapy, chemotherapy has been the primary treatment given to patients with solid breast tumors.2,3 Among various categories of chemotherapeutic agents, anthracyclines are the most prominently researched class of drugs for their anti-cancer properties.4 Doxorubicin (Dox), the most well-known anthracycline, is an FDA-approved drug for solid tumors and metastatic breast cancer treatment.5 However, its associated cardiac toxicity and other side effects prompt the ongoing search for less toxic and more efficient analogues of Dox. Interestingly, pirarubicin (THP-Dox) is a semi-synthetic analogue of doxorubicin, with its 4-O position substituted with a tetrahydropyranyl (THP) ring, which has emerged as a potent substitute. According to pre-clinical trials, the latter demonstrated reduced cardiac toxicity while maintaining similar anti-tumor efficacy as that of Dox. Moreover, it showed faster intracellular uptake and competent anti-cancer activity even in Dox-resistant tumor cell lines.6–8

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). The obtained polymer was vacuum dried and stored at −20 °C till further use.
4). The obtained polymer was vacuum dried and stored at −20 °C till further use.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (70
H2O (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) as the mobile phase, 1 mL min−1 as the flow rate, 10 μL as the injection volume, 25 °C as the column temperature and UV detection at λmax = 270 nm. Another calibration curve was plotted using free Gem in the conc. range of 300 μg mL−1 to 1 μg mL−1 mobile phase following the above-mentioned HPLC protocol. In both cases, the unconjugated drug content was quantified using HPLC and the % drug conjugation efficiency was calculated indirectly using the following equation:
30) as the mobile phase, 1 mL min−1 as the flow rate, 10 μL as the injection volume, 25 °C as the column temperature and UV detection at λmax = 270 nm. Another calibration curve was plotted using free Gem in the conc. range of 300 μg mL−1 to 1 μg mL−1 mobile phase following the above-mentioned HPLC protocol. In both cases, the unconjugated drug content was quantified using HPLC and the % drug conjugation efficiency was calculated indirectly using the following equation:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution of 10 mg mL−1 stock solution) were drop-casted on a carbon-coated copper TEM grid, followed by uranyl acetate staining. Additionally, Pira–Gem co-loaded nanoparticles were prepared using the Pira/Gem–mPEG-b-PLA–LA conjugate in three different drug ratios (Pira–Gem as 1
10 dilution of 10 mg mL−1 stock solution) were drop-casted on a carbon-coated copper TEM grid, followed by uranyl acetate staining. Additionally, Pira–Gem co-loaded nanoparticles were prepared using the Pira/Gem–mPEG-b-PLA–LA conjugate in three different drug ratios (Pira–Gem as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), following the afore-mentioned protocol.
3), following the afore-mentioned protocol.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mPEG-b-PLA–LA nanoparticles were performed under two different pH environments, i.e., neutral physiological pH (7.4) and tumor microenvironment pH (5.0). For the study, 30 mg of lyophilized Pira/Gem–mPEG-b-PLA–LA nanoparticles were reconstituted at a concentration of 10 mg mL−1 and equally distributed into 10 mL of each type of buffer. To simulate physiological conditions, the nanoparticles suspension was incubated under dark conditions at 37 °C with gentle stirring at 150 rpm. Thereafter, at pre-determined time intervals, the nanoparticles were filtered and washed using Amicon Centrifuge filters (MWCO 3 kDa, Millipore) at 4000 rpm to separate out the drug released per day. The residual nanoparticles were redispersed in fresh buffer every time, and the collected filtrates were lyophilized and drug content was quantified using the previously described HPLC detection method. The drug release experiments were conducted in triplicate to minimize manual errors. For Pira–Gem co-loaded (1
1) mPEG-b-PLA–LA nanoparticles were performed under two different pH environments, i.e., neutral physiological pH (7.4) and tumor microenvironment pH (5.0). For the study, 30 mg of lyophilized Pira/Gem–mPEG-b-PLA–LA nanoparticles were reconstituted at a concentration of 10 mg mL−1 and equally distributed into 10 mL of each type of buffer. To simulate physiological conditions, the nanoparticles suspension was incubated under dark conditions at 37 °C with gentle stirring at 150 rpm. Thereafter, at pre-determined time intervals, the nanoparticles were filtered and washed using Amicon Centrifuge filters (MWCO 3 kDa, Millipore) at 4000 rpm to separate out the drug released per day. The residual nanoparticles were redispersed in fresh buffer every time, and the collected filtrates were lyophilized and drug content was quantified using the previously described HPLC detection method. The drug release experiments were conducted in triplicate to minimize manual errors. For Pira–Gem co-loaded (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) nanoparticles, the filtrate collected at every time was divided in half and processed separately to quantify the individual drug release.
1) nanoparticles, the filtrate collected at every time was divided in half and processed separately to quantify the individual drug release.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) for adhesion. The cells were observed under a confocal laser scanning microscope (Olympus FV1000) in bright field and fluorescent modes. Additionally, the Pira–mPEG-b-PLA–LA nanoparticles uptake was evaluated by FACS (BD Biosciences, LSRFortessaTM X-20) with the standard 10
1, v/v) for adhesion. The cells were observed under a confocal laser scanning microscope (Olympus FV1000) in bright field and fluorescent modes. Additionally, the Pira–mPEG-b-PLA–LA nanoparticles uptake was evaluated by FACS (BD Biosciences, LSRFortessaTM X-20) with the standard 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events recorded for each sample. Untreated cells were used as a negative control. Software FlowJo (Version 10.10.0) was used to plot and analyze the FACS data.
000 events recorded for each sample. Untreated cells were used as a negative control. Software FlowJo (Version 10.10.0) was used to plot and analyze the FACS data.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) was compared with that of the individually drug-loaded Pira/Gem mPEG-b-PLA–LA nanoparticles to determine the most synergistic combination ratio. GraphPad Prism (Version 9.0) was used to plot the dose-dependent cytotoxicity curves and to calculate the IC50 values.
3) was compared with that of the individually drug-loaded Pira/Gem mPEG-b-PLA–LA nanoparticles to determine the most synergistic combination ratio. GraphPad Prism (Version 9.0) was used to plot the dose-dependent cytotoxicity curves and to calculate the IC50 values.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) were calculated and compared to those of free drug combinations across various breast cancer cell lines; MCF-7, MDA-MB-468 and SUM-149. A drug combination resulting in CI value <1, =1 or >1 indicates synergy, additive effect or antagonism, respectively. Fraction affected (Fa) vs. CI curves were generated using CompuSyn software.
3) were calculated and compared to those of free drug combinations across various breast cancer cell lines; MCF-7, MDA-MB-468 and SUM-149. A drug combination resulting in CI value <1, =1 or >1 indicates synergy, additive effect or antagonism, respectively. Fraction affected (Fa) vs. CI curves were generated using CompuSyn software.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching peak at 1650 cm−1 (Fig. 2B). The –C
O stretching peak at 1650 cm−1 (Fig. 2B). The –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of mPEG-b-PLA–LA was then used to conjugate both pirarubicin or gemcitabine to obtain the Schiff's base polymer drug conjugates. The successful conjugation of both Pira and Gem individually to mPEG-b-PLA–LA was confirmed again using 1H NMR spectroscopy (Fig. 2C and D), where new benzylic proton peaks were observed between δ 7.0–δ 8.0 ppm, along with all the characteristic peaks of the block copolymer, validating the formation of the Pira/Gem–mPEG-b-PLA–LA conjugates.
O group of mPEG-b-PLA–LA was then used to conjugate both pirarubicin or gemcitabine to obtain the Schiff's base polymer drug conjugates. The successful conjugation of both Pira and Gem individually to mPEG-b-PLA–LA was confirmed again using 1H NMR spectroscopy (Fig. 2C and D), where new benzylic proton peaks were observed between δ 7.0–δ 8.0 ppm, along with all the characteristic peaks of the block copolymer, validating the formation of the Pira/Gem–mPEG-b-PLA–LA conjugates.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps
1) Nps![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) co-loaded nanoparticles, the cumulative release of Pira was observed to be ∼21% and ∼28% at pH 7.4 and pH 5.0, respectively. In contrast, the co-loaded nanoparticles released ∼45% and ∼70% of Gem at pH 7.4 and pH 5.0, respectively (Fig. 4C and D). The contrasting physiochemical properties of the drugs, pirarubicin being hydrophobic and gemcitabine being hydrophilic, resulted in repulsive interactions when co-loaded, contributing to the enhanced release of both drugs.
1) co-loaded nanoparticles, the cumulative release of Pira was observed to be ∼21% and ∼28% at pH 7.4 and pH 5.0, respectively. In contrast, the co-loaded nanoparticles released ∼45% and ∼70% of Gem at pH 7.4 and pH 5.0, respectively (Fig. 4C and D). The contrasting physiochemical properties of the drugs, pirarubicin being hydrophobic and gemcitabine being hydrophilic, resulted in repulsive interactions when co-loaded, contributing to the enhanced release of both drugs.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) co-loaded nanoparticles exhibited only 7.46% gap closure under identical conditions (Fig. 6A and B). The substantial reduction in cell migration highlights the synergistic therapeutic potential of the Pira–Gem combination. In control wells without any treatment, the scratch gap was completely closed within 24 hours, underscoring the aggressive migratory behavior of untreated cells. These findings provide compelling evidence of the superior migration inhibitory activity potential of Pira–Gem co-loaded nanoparticles compared to single-drug loaded formulations for combating metastatic breast cancer.
1) co-loaded nanoparticles exhibited only 7.46% gap closure under identical conditions (Fig. 6A and B). The substantial reduction in cell migration highlights the synergistic therapeutic potential of the Pira–Gem combination. In control wells without any treatment, the scratch gap was completely closed within 24 hours, underscoring the aggressive migratory behavior of untreated cells. These findings provide compelling evidence of the superior migration inhibitory activity potential of Pira–Gem co-loaded nanoparticles compared to single-drug loaded formulations for combating metastatic breast cancer.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps
1) Nps![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps
1) Nps![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Nps
3) Nps![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mPEG-b-PLA–LA nanoparticle formulation. This finding suggests that while the dual loading of Pira and Gem can enhance the effectiveness of a treatment, the optimal ratio for consistent synergy across various cell types may vary, with the 1
1) mPEG-b-PLA–LA nanoparticle formulation. This finding suggests that while the dual loading of Pira and Gem can enhance the effectiveness of a treatment, the optimal ratio for consistent synergy across various cell types may vary, with the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio being particularly effective across all of the cell lines that were tested (Table 3).
1 ratio being particularly effective across all of the cell lines that were tested (Table 3).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps
1) Nps![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Nps
1) Nps![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Nps
3) Nps





